Abstract
The relation between gene body methylation and gene function remains elusive. Yet, our understanding of this relationship can contribute significant knowledge on how and why organisms target specific gene bodies for methylation. Here, we studied gene body methylation patterns in two Daphnia species. We observed both highly methylated genes and genes devoid of methylation in a background of low global methylation levels. A small but highly significant number of genes was highly methylated in both species. Remarkably, functional analyses indicate that variation in methylation within and between Daphnia species is primarily targeted to small gene families whereas large gene families tend to lack variation. The degree of sequence similarity could not explain the observed pattern. Furthermore, a significant negative correlation between gene family size and the degree of methylation suggests that gene body methylation may help regulate gene family expansion and functional diversification of gene families leading to phenotypic variation.
Keywords: gene function, DNA methylation, Daphnia
Introduction
While the number of available genomes is readily increasing, the molecular mechanisms that translate the genomic information to organismal stress responses and phenotypic plasticity often remain to be elucidated. This lack of knowledge can partly be attributed to the complexity of gene functions and the molecular mechanisms that are generally the result of interactions at the DNA, RNA, and protein level. However, our improved understanding of epigenetic mechanisms has generated an appreciation for the complexity of functional regulation of the genome (Cubas et al. 1999; Feil and Fraga 2012; Heyn et al. 2013).
At present, gene body methylation, referring to methylation in transcription units, is considered a basal evolutionary pattern in eukaryotes yet the function remains unclear (Suzuki et al. 2007; Feng et al. 2010; Sarda et al. 2012, Zemach et al. 2010). In vertebrates and plants, gene body methylation, as opposed to methylation of upstream promoter regions, is associated with actively transcribed genes (Jones 2012, Zemach et al. 2010). Gene body methylation has also been put forward as a potential mechanism to regulate alternative splicing in several animal genomes (Flores et al. 2012; Jones 2012). In invertebrates, the potential role of gene body methylation is less obvious, studies have demonstrated associations between gene body methylation patterns and higher biological functions including caste specificity in honey bees and ants (Elango et al. 2009; Lyko et al. 2010; Bonasio et al. 2012). Thus far, gene body methylation in invertebrates seems to be targeted to a nonrandom subset of genes (Sarda et al. 2012; Takuno and Gaut 2013), which suggests important functional consequences of DNA methylation. Previous studies in closelyrelated plants (closest common ancestor 40–53 million years) and distantly related invertebrates (closest common ancestor 300 million to 1 billion years) have found that gene body methylation is conserved among orthologous genes and that protein sequence conservation of highly methylated genes is a common feature in invertebrate taxa (Sarda et al. 2012; Takuno and Gaut 2013). Furthermore, these studies also observed significant enrichment of genes with essential functions in the set of conserved highly methylated genes.
Yet, it remains unclear whether conserved gene body methylation across orthologs is driven by gene function or gene sequence (Sarda et al. 2012; Takuno and Gaut 2013). If conservation of methylation is driven by gene function, the question remains as to what extent the functional divergence and methylation of paralogous genes are affected. Answers to these questions are crucial to understand the function of DNA methylation and its ultimate role in gene regulation and genome biology.
In this study, we attempt to answer these questions by focusing on gene body methylation patterns in two closely related invertebrate species, Daphnia pulex and Daphnia magna (common ancestor 10 million years) (Haag et al. 2009). Daphnia, an ubiquitous freshwater crustacean, is primarily known for its cyclic parthenogenetic reproductive mode, and its ecological and environmental relevance (Harris et al. 2012; Miner et al. 2012). Previous genome-wide studies in Daphnia have revealed functional responses of gene regulation to environmental and ecological challenges that are associated with specific gene families and molecular pathways (Latta et al. 2012; De Coninck et al. 2014; Asselman et al. 2015a) have shown that many genes are under selection (McTaggart et al. 2012) while others demonstrated differences in methylation following exposure to environmental stressors (Asselman et al. 2015b; Schield et al. 2015).
Methods
Culture Conditions
The D. magna strain used was an inbred clonal lineage originating from a rock pool near Tvärminne, Finland (Routtu et al. 2014). This isolate has also been used in an ongoing genome sequence project to develop a D. magna reference genome assembly and a high-density linkage map (Routtu et al. 2014). The D. pulex strain used was a clonal lineage sampled from a pond in Oregon (Paland et al. 2005; Shaw et al. 2007). Both strains have been cultured in our present lab (GhenToxLab) for at least 50 generations under standardized culture conditions that allow for optimal growth and reproduction prior to DNA sampling. In brief, D. magna isolates were cultured in ADaM medium (Klüttgen et al. 1994) at a density of ten animals per liter while D. pulex isolates were cultured in no-N no-P COMBO medium at a density of 15 animals per liter (Kilham et al. 1998; Shaw et al. 2007). All animals were cultured under controlled conditions (20 ± 1°C, 16 h:8 h light–dark cycle at a light intensity of 14 µmoles m−2 s−1). Animals were fed daily ad libitum with an algal mixture consisting of Pseudokirchneriella subcapitata and Chlamydomonas reinhardtii in a 3:1 mixture ratio based on cell numbers. Final feeding concentration was 1.5 mg carbon per liter. Medium was renewed completely every 2 days.
Experimental Setup
Neonates of <24 h old were isolated from the TWO cultures and randomly placed in one of three 8-L aquaria representing three biological replicates for each species at a density of ten animals per liter for D. magna and 15 animals per liter for D. pulex. An additional fourth replicate was set up for the D. pulex strain for genome sequencing as no reference sequence was available for the particular isolate used in this study. All experimental parameters and culture conditions were identical to the parameters of the culture maintenance described above. After 14 days, 30 animals that were not carrying eggs or embryos in their brood chamber were selected and removed from each aquarium for DNA extraction. Selecting animals not carrying eggs or embryos excludes confounding effects due to methylation differences associated with differences in developmental stage or the number of eggs or embryos.
DNA Extraction, Library Construction, and Sequencing
Per aquarium, all animals were pooled and DNA was extracted immediately using the MasterPure kit (Epicentre, Madison, WI). Sequencing and library preparation was done at the BGI sequencing facility in Hong Kong. In brief, the extracted DNA was fragmented by sonication to a mean size of ∼300 bp. After blunt ending and 3′-end addition of dA, Illumina methylated adapters (Illumina, San Diego, CA) were added according to the manufacturer’s instructions for all samples. For bisulfite sequencing, the bisulfite conversion (C → U) was carried out using the EZ DNA methylation Gold kit (Zymo Research, Irvine, CA) according to manufacturer’s instructions. During the bisulfite conversion, 5 ng of unmethylated lambda DNA per microgram of DNA sample was added to assess the bisulfite conversion error rate. Ultra-high-throughput pair-end sequencing for all samples was carried out using the Illumina HiSeq-2000 (Illumina) according to the manufacturer’s instructions. Raw sequencing data were processed by the Illumina 1.5 base-calling pipeline, resulting in 90 bp reads. The bisulfite-treated sequence data have been deposited to NCBI GEO under reference GSE60475 while the other sequence data have been deposited to NCBI SRA under reference PRJNA281096.
Quality Assessment, Preprocessing, and Mapping
Overall quality of the reads was evaluated using the FastQC software (Babraham Institute, Cambridge, UK). Reads containing >5% N bases were omitted. The remaining reads were dynamically trimmed to the longest stretch of bases which had a Phred score higher or equal to 30 (i.e., ∼99.9% base-call accuracy) using Trim Galore! 0.3.2 software (Babraham Institute) with standard settings. In addition to removal of poor-quality bases, adaptor sequences were trimmed from the reads. For bisulfite-treated samples, trimmed reads were subsequently transformed into fully bisulfite-converted forward (C -> T conversion) and reverse read (G -> A conversion of the forward strand) versions, before being mapped to similarly converted versions of the genome (also C -> T and G -> A converted) using Bowtie2 v.2.1.0 (Langmead and Salzberg 2012) while setting the scoring function as −score_min L, 0, −0.6. These four mapping processes were run in parallel and only the unique best mapping of each read was withheld. Reads from the nonbisulfite-treated samples did not need conversion and were mapped to the nonconverted version of the genome using the same scoring function. Nonuniquely mapping reads were discarded for further analysis. For bisulfite-treated samples, reads that might have occurred as PCR duplicates were removed using the Bismark deduplicate script (Krueger and Andrews 2011). The D. pulex filtered reference genome assembly with ∼5,000 scaffolds (Dappu1; Colbourne et al. 2011) was obtained from the DOE Joint Genome Institute (JGI) Genome Portal. The D. magna reference genome assembly v2.4, which was based on the exact same isolate, was used for mapping the D. magna data (http://arthropods.eugenes.org/EvidentialGene/daphnia/daphnia_magna/, last accessed April 4, 2016). The above-described procedure was applied to each biological sample separately.
Bisulfite Conversion Error Rate
The conversion error rate (supplementary table S3, Supplementary Material online) was defined as the percentage of reads mapping to the unmethylated lambda phage control DNA and which yielded a methylation call.
Single Nucleotide Polymorphisms and Heterozygosity Sites
The available reference genome for D. pulex was developed using a different isolate than the one used here. Therefore, additional non-bisulfite converted DNA sequencing was done to identify and exclude single nucleotide polymorphisms between the reference genome and the isolate at all cytosine sites. The mapped DNA reads of the nonbisulfite-treated sample were processed with GATK (McKenna et al. 2010) and all single nucleotide polymorphisms at cytosine sites and heterozygous C/T sites identified through GATK were flagged and removed from the bisulfite sequenced data on both the forward and reverse strand.
Methylation Levels
For each read covering a cytosine site the methylation state of that site was inferred using the Bismark 0.9.0 software (Krueger and Andrews 2011) by comparing the uniquely mapped read to the original, nonconverted reference genome. To obtain high reliability and high resolution of the methylation level across all cytosines and not only rely on an average raw coverage of 17× at the CpG level (supplementary tables S1 and S2, Supplementary Material online), only cytosine sites with a minimum coverage of 5× in all three biological replicates were considered for further downstream analyses. After filtering, 99.9% of the gene models have an average coverage of ≥10× (D. pulex) or ≥25× (D. magna) per cytosine. A binomial distribution was used to distinguish true methylated reads from false positives using the calculated bisulfite conversion error rate for each replicate (Lyko et al. 2010; Bonasio et al. 2012). P values were corrected for multiple testing using a Benjamini–Hochberg correction. Similar to Bonasio et al. (2012), true methylated cytosines were assigned a methylation ratio defined by the number of methylated reads at the cytosine site divided by the total number of reads at the cytosine site.
Gene Body Methylation Levels
Gene models were extracted from the 2011 frozen annotation version of the D. pulex reference genome downloaded from the DOE JGI Genome Portal. Given the fragmented state of the D. pulex reference genome, there is a probability that current gene numbers and gene copies within a family are inflated (Denton et al. 2014). We therefore filtered these gene models to a conservative but representative gene list using the following criteria based on suggestions by Denton et al. (2014). All gene models that occur within poorly covered regions or having gapped alignments were removed. In particular, all genes with 50 or more consecutive unidentified bases (labeled as N) were excluded. In addition, only gene models with protein sequences containing both a start and stop codon were retained. Finally, only D. pulex gene models that have a significant hit with a reciprocal blast (cutoff e-value 1e−05) against the available D. magna gene set were retained (http://arthropods.eugenes.org/EvidentialGene/daphnia/daphnia_magna/, last accessed April 4, 2016). These filtering steps resulted in a conserved D. pulex gene set of 14,102 genes and a conserved orthologous D. magna gene set of 8,800 genes generated through the reciprocal blast. Genes within the D. pulex set have been transcriptionally validated through several microarray experiments (Colbourne et al. 2011; Latta et al. 2012; Asselman et al. 2015a) while D. magna gene models have been validated using extensive RNAseq experiments (Orsini et al. submitted for publication). To evaluate potential bias in the conservative gene set we used BUSCO, a software developed by Simão et al. (2015) to provide quantitative measures of gene set completeness. This software uses single copy orthologs from OrthoDB, called benchmarks, to evaluate the completeness of a gene set. We used BUSCO to evaluate how representative the conserved gene sets were compared with the complete nonfiltered gene set as reported by in http://buscos.ezlab.org/arthropoda_table.html (last accessed April 4, 2016). We found 72% of the benchmark single-copy orthologs as defined by BUSCO in the conserved D. magna gene set and 69% in the conserved D. pulex gene set while 94% of the orthologs were present when using all available gene models (30,940 genes). By using a conserved gene set, rather than the full gene set, we reduce the chance of inflating gene copy numbers and gene family size to due errors in sequence assembly (Denton et al. 2014). Cytosine-specific methylation levels for each gene body within the conservative set were obtained by overlapping these gene models through BEDtools 2.17.0 (Quinlan and Hall 2010) with cytosine-specific methylation levels as determined above. The methylation level of a gene was inferred as sum of all methylation rates within the gene divided by the total number of cytosines covering the feature according to Bonasio et al. (2012).
Identification of Zero and Hyper-Methylated Gene Bodies
To identify gene bodies that are, with a high reliability, zero- or hyper-methylated a strategy of making use of the independent biological replication was applied. Only gene bodies that showed consistently 0 or high methylation levels in all three biological replicates were considered as being either zero- or hyper-methylated in the respective species. Gene bodies were considered zero-methylated if no methylation was detected in all three replicates (i.e., if not a single methylated cytosine was detected in any read in any of the three replicates for all cytosines in that gene body) and hyper-methylated if a methylation level of at least 50% in each of the three biological replicates of the respective species was detected.
Differential Methylation Analysis
To determine which gene bodies were differentially methylated between the two species, the Dispersion Shrinkage for Sequencing data package in R was used (Feng et al. 2014). Prior to differential methylation analysis, all genes with zero methylation in all three replicates in both species were removed from the dataset. These genes were removed to reduce the number of genes to be tested as zero methylated genes in both species can never be statistically differentially methylated. Not removing these would lead to a less stringent multiple testing correction as the number of genes is smaller. Second, data were smoothed using the BSmooth function and statistically differentially methylated gene bodies were identified using the function callDML. In brief, these functions use a beta-binomial distribution to model the sequencing data including information from all biological replicates while dispersion is estimated using a Bayesian hierarchical model. Finally, a Wald-test is conducted to calculate P values and false discovery rates.
Functional Analyses
Annotation from the reference D. pulex genome was used to study functional patterns of gene families, defined as sharing a full annotation definition. Over- and underrepresentation analyses consisted of Fishers-exact tests combined with Benjamini–Hochberg multiple testing corrections by comparing the proportion of a gene family among the differentially methylated genes versus the proportion of that gene family within the conserved gene set. Patterns of methylation variation within and across gene families were evaluated using a bootstrap procedure described in Asselman et al. (2015a). In brief, for every gene family, methylation variation was compared with a distribution of variations in 1,000 artificial gene families with the exact same size constructed by randomly sampling gene bodies from the conserved gene set. Gene families with a variation smaller than the 2.5 percentile were defined as having a variation significantly smaller than expected by chance whereas gene families with a variation significantly larger than the 97.5 percentile were defined as having a variation larger than expected by chance.
CpG Observed/Expected Ratio and Comparison with Other Invertebrate Species
CpG Observed/Expected ratios have been reported to be a good indicator of methylation levels when no methylation data are available (Gladstad et al. 2011; Sarda et al. 2012). Furthermore, the CpG O/E ratio is an indicator of methylation over evolutionary time, and therefore allows to study functional and evolutionary mechanisms of gene body methylation (Gladstad et al. 2011; Sarda et al. 2012). The CpG O/E ratio is defined as the frequency of CpG dinucleotides divided by the product of the frequency of C nucleotides and the frequency of G nucleotides for the genomic region of interest (Sarda et al. 2012). Here, we calculate the CpG O/E ratios for gene bodies.
Gene Expression Data
We downloaded publically available data from GEO using the whole genome nimbleGen array GPL11278, which comprises 12 GEO series, all using D. pulex, and a total of 49 conditions. M values and q values were extracted and used for analysis.
Results
Distribution of Gene Body Methylation Levels in D. magna and D. pulex
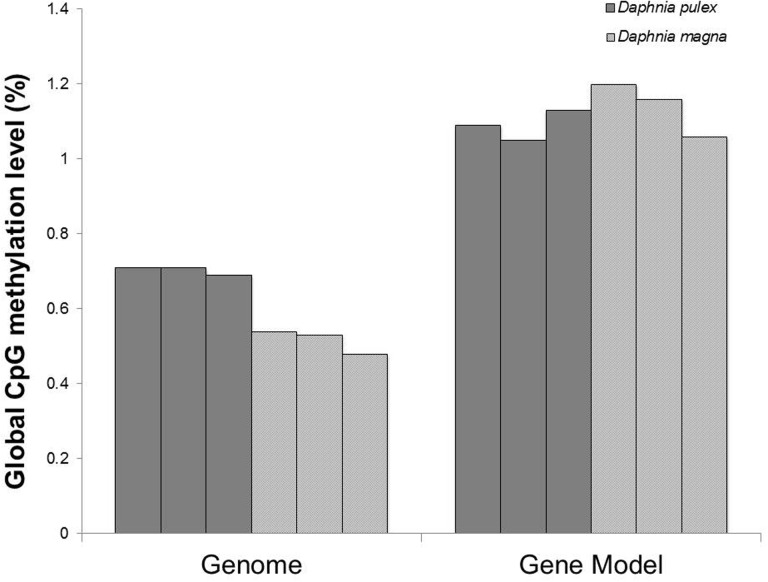
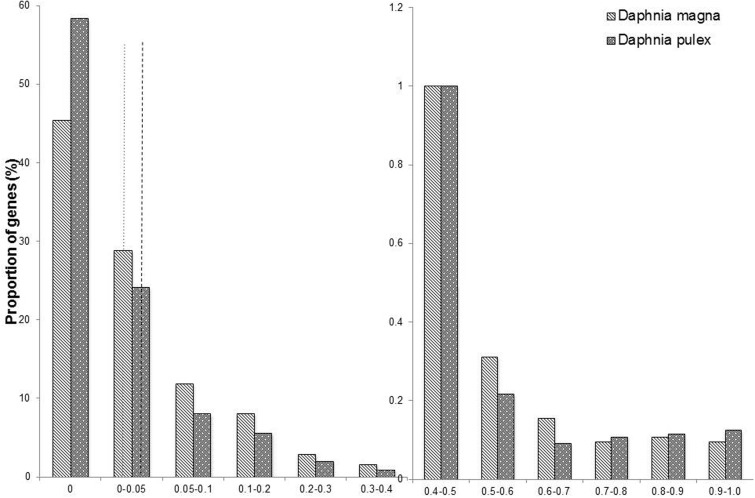
The average global cytosine methylation within CpG context was 0.70% in D. pulex and 0.52% in D. magna while global cytosine methylation was negligible in CHG and CHH, with H being a nucleotide other than G, contexts in both species (fig. 1, supplementary tables S1–S3, Supplementary Material online). Cytosine methylation within CpG contexts in these conserved gene models follows a bimodal distribution in the two species with a high number of cytosines showing no methylation. The distribution of methylation levels of gene bodies was significantly different between the two species (Kruskal–Wallis test: P value <2.2e−16, fig. 2). In particular, we observed significant differences in the distribution of gene bodies with methylation levels lower than 5% (P value <2.2e−16, fig. 2) between D. pulex and D. magna whereas the distributions of gene bodies with a methylation level higher than 5% were comparable across the two species (Pvalue = 0.91, fig. 2). Both species contained a small proportion of highly methylated gene bodies (methylation level >50%, D. magna = 0.63% of all genes, D. pulex = 0.69% of all genes, fig. 2).
Fig. 1.—
CpG methylation levels in all three biological replicates for the two species across the entire genome and within the conserved gene models.
Fig. 2.—
Proportion of gene bodies within categories of discrete CpG methylation levels averaged across the three biological replicates for the two species (proportions were calculated relative to the number of conserved gene models within each species). Dotted line indicates in which discrete category the global methylation level in D. magna (0.52%) falls, while the dashed line indicates in which discrete category the global methylation level in D. pulex (0.70%) falls, see also figure 1.
Differential Methylation Between D. magna and D. pulex
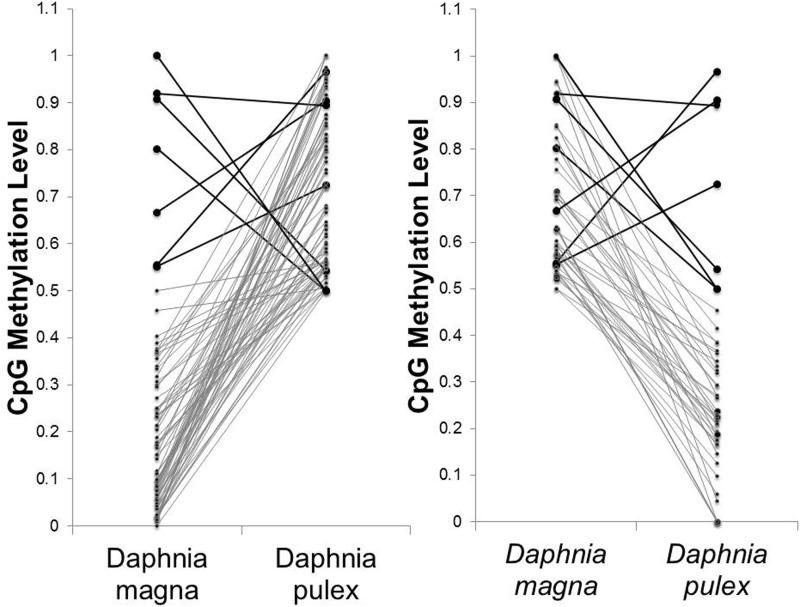
Only seven genes were highly methylated in both species, but this number is higher than expected by chance (fig. 3, P value = 2.38e−08, hypergeometric test). Pairwise comparison of gene models revealed 1,711 gene models that showed significantly different methylation levels between the two species at a false discovery level of 0.01. While the majority of these genes only showed small differences in methylation between the two species, 387 genes had a difference in methylation level of at least 20% and 72 genes showed >50% difference in methylation. The correlation between the difference in methylation levels and sequence identity and the correlation between the difference in methylation levels and difference in CpGs were weak, 0.14 and −0.23, respectively.
Fig. 3.—
Left: Median methylation levels of highly methylated genes in D. pulex (n = 83) and their corresponding methylation levels in D. magna. Right: Median methylation levels of highly methylated genes in D. magna (n = 53) and their corresponding methylation levels in D. pulex. Black bold lines highlight genes that are highly methylated in both species.
Functional Analysis of Gene Body Methylation Patterns in Daphnia
Functional analysis of differentially methylated gene bodies between the two species revealed significant over- and underrepresentation of differentially methylated genes in 55 specific functional categories (table 1). Six gene families lacked genes that were differentially methylated between both species, that is, they contained only genes that in one species demonstrated similar methylation patterns to their orthologous gene in the other species. Twenty-one gene families had only genes that were differentially methylated between both species, including methylases and glutathione-S-tranferases. Gene families without differentially methylated genes were significantly larger than gene families with only differentially methylated genes (P value = 5.6e−08). In particular, family size of gene families without differentially methylated genes varied between 24 and 98 genes with an average of 51 genes per family while family size of gene families with only differentially methylated genes varied between 2 and 65 with an average gene family size of eight genes. We observed a negative correlation between gene family size and the proportion of significantly differentially methylated genes within the gene family (r = −0.82, P < 2.2e−16) for these gene families (supplementary fig. S2, Supplementary Material online).
Table 1.
Gene Families that Are Significantly over (+) or under (-) Represented for Differentially Methylated Genes, their P Values and the KOG Category (Eukaryotic Orthology Groups as Defined by the Joint Genome Institute)
| Name | P value | FDR <0.01 | FDR >0.01 | Proportion (%) with FDR <0.01 | Over/under represented | KOG category |
|---|---|---|---|---|---|---|
| Trypsin | 7.91E−04 | 0 | 75 | 0 | – | Amino acid transport and metabolism |
| Chitinase | 2.85E−02 | 3 | 59 | 4.84 | – | Cell wall/membrane/envelope biogenesis |
| Collagens (type IV and type XIII) | 7.54 E − 06 | 1 | 97 | 1.02 | – | Extracellular structures |
| Bestrophin | 3.96 E − 02 | 0 | 24 | 0 | – | General function prediction only |
| FOG: 7 transmembrane receptor | 4.61 E − 04 | 1 | 70 | 1.41 | – | General function prediction only |
| Low-density lipoprotein receptors | 2.78 E − 02 | 0 | 29 | 0 | – | Intracellular trafficking, secretion, and vesicular transport |
| Nucleolar GTPase/ATPase p130 | 4.97 E − 03 | 1 | 52 | 1.89 | – | Nuclear structure |
| Cytochrome P450 CYP4/CYP19/CYP26 subfamilies | 3.96 E − 02 | 0 | 24 | 0 | - | Secondary metabolites biosynthesis, transport and catabolism |
| C-type lectin | 3.98 E − 02 | 3 | 56 | 5.08 | – | Signal transduction mechanisms |
| Fibroblast/platelet-derived growth factor receptor | 3.96 E − 02 | 0 | 24 | 0 | – | Signal transduction mechanisms |
| RNA polymerase II. large subunit | 3.99 E − 02 | 2 | 48 | 4 | – | Transcription |
| 1-pyrroline-5-carboxylate dehydrogenase | 2.03 E − 02 | 2 | 0 | 100 | + | Amino acid transport and metabolism |
| Cysteine desulfurase NFS1 | 5.85 E − 05 | 5 | 0 | 100 | + | Amino acid transport and metabolism |
| Delta-1-pyrroline-5-carboxylate dehydrogenase | 2.03 E − 02 | 2 | 0 | 100 | + | Amino acid transport and metabolism |
| Cell cycle-regulated histone H1-binding protein | 2.03 E − 02 | 2 | 0 | 100 | + | Cell cycle control, cell division, chromosome partitioning |
| Cyclin B & related kinase-activating proteins | 2.31 E − 02 | 3 | 2 | 60 | + | Cell cycle control, cell division, chromosome partitioning |
| DNA topoisomerase (ATP-hydrolysing) | 2.89 E − 03 | 3 | 0 | 100 | + | Chromatin structure and dynamics |
| DNA topoisomerase type II | 3.10 E − 04 | 5 | 1 | 83.33 | + | Chromatin structure and dynamics |
| Actin regulatory protein | 2.31 E − 02 | 3 | 2 | 60 | + | Cytoskeleton |
| Actin-binding protein Coronin | 2.31 E − 02 | 3 | 2 | 60 | + | Cytoskeleton |
| Von Willebrand factor & related coagulation proteins | 1.23 E − 03 | 0 | 47 | 0 | – | Defense mechanisms |
| Predicted membrane protein | 1.50 E − 02 | 11 | 26 | 29.73 | + | Function unknown |
| Uncharacterized conserved protein with CXXC motifs | 2.03 E − 02 | 2 | 0 | 100 | + | Function unknown |
| F-box protein containing LRR | 7.40 E − 04 | 8 | 8 | 50 | + | General function prediction only |
| FOG: Zn-finger | 5.40 E − 05 | 22 | 43 | 33.85 | + | General function prediction only |
| HMG box-containing protein | 1.94 E − 02 | 5 | 7 | 41.67 | + | General function prediction only |
| Methylase | 2.03 E − 02 | 2 | 0 | 100 | + | General function prediction only |
| Predicted methyltransferase | 1.85 E − 05 | 8 | 3 | 72.73 | + | General function prediction only |
| Sulfotransferases | 2.03 E − 02 | 2 | 0 | 100 | + | General function prediction only |
| H(+)-transporting two-sector ATPase | 2.03 E − 02 | 2 | 0 | 100 | + | Inorganic ion transport and metabolism |
| P-type ATPase | 1.00 E − 02 | 4 | 3 | 57.14 | + | Inorganic ion transport and metabolism |
| Emp24/gp25L/p24 membrane trafficking proteins | 2.03 E − 02 | 2 | 0 | 100 | + | Intracellular trafficking, secretion, and vesicular transport |
| Karyopherin (importin) alpha | 1.15 E − 07 | 11 | 3 | 78.57 | + | Intracellular trafficking, secretion, and vesicular transport |
| Sphingosine N-acyltransferase | 2.03 E − 02 | 2 | 0 | 100 | + | Lipid transport and metabolism |
| Beta-tubulin folding cofactor D | 1.82 E − 03 | 4 | 1 | 80 | + | Posttranslational modification, protein turnover, chaperones |
| Glutathione transferase | 2.89 E − 03 | 3 | 0 | 100 | + | Posttranslational modification, protein turnover, chaperones |
| Molecular chaperone (HSP90 family) | 9.56 E − 04 | 5 | 2 | 71.43 | + | Posttranslational modification, protein turnover, chaperones |
| Thioredoxin-like protein | 4.12 E − 04 | 4 | 0 | 100 | + | Posttranslational modification, protein turnover, chaperones |
| Ubiquitin-protein ligase | 4.74 E − 04 | 6 | 3 | 66.67 | + | Posttranslational modification, protein turnover, chaperones |
| Nuclear 5-3 exoribonuclease-interacting protein | 2.03 E − 02 | 2 | 0 | 100 | + | Replication, recombination and repair |
| FtsJ-like RNA methyltransferase | 2.03 E − 02 | 2 | 0 | 100 | + | RNA processing and modification |
| Heterogeneous nuclear ribonucleoprotein R | 1.69 E − 07 | 10 | 2 | 83.33 | + | RNA processing and modification |
| Leucine rich repeat proteins | 1.15 E − 06 | 15 | 13 | 53.57 | + | RNA processing and modification |
| Putative N2.N2-dimethylguanosine tRNA methyltransferase | 2.03 E − 02 | 2 | 0 | 100 | + | RNA processing and modification |
| TPR repeat-containing protein | 1.03 E − 02 | 3 | 1 | 75 | + | RNA processing and modification |
| Dehydrogenases (related to short-chain alcohol dehydrogenases) | 4.47 E − 03 | 5 | 4 | 55.56 | + | Secondary metabolites biosynthesis, transport and catabolism |
| Ca2+/calmodulin-dependent protein phosphatase | 2.03 E − 02 | 2 | 0 | 100 | + | Signal transduction mechanisms |
| Failed axon connections (fax) proteins | 2.89 E − 03 | 3 | 0 | 100 | + | Signal transduction mechanisms |
| Predicted GTPase-activating protein | 2.85 E − 02 | 4 | 5 | 44.44 | + | Signal transduction mechanisms |
| Tyrosine kinases | 2.31 E − 02 | 3 | 2 | 60 | + | Signal transduction mechanisms |
| RNA polymerase II transcription initiation factor TFIIH | 2.03 E − 02 | 2 | 0 | 100 | + | Transcription |
| Site-specific DNA-methyltransferase | 2.03 E − 02 | 2 | 0 | 100 | + | Transcription |
| Ubiquitin/60s ribosomal protein L40 | 2.03 E − 02 | 2 | 0 | 100 | + | Translation, ribosomal structure and biogenesis |
Genes are defined as differentially expressed at a false discovery rate (fdr) smaller than 0.01.
Further analysis of methylation patterns within gene families for each species separately revealed gene families with highly consistent methylation levels across their genes as well as gene families with highly varying methylation levels (supplementary tables S4 and S5, Supplementary Material online). All gene families with less differentially methylated genes than expected (11 in total) also showed highly consistent methylation levels with little variation between the genes within each gene family. In addition, eight overrepresented gene families showed highly varying methylation levels between the genes within the gene family (table 1). We further studied this subset of 19 gene families and observed negative correlations between gene family size and the mean methylation level (rDmagna = −0.3, rDpulex = −0.32) and between gene family size and the standard deviation of the methylation levels within the gene families (rDmagna = −0.1, rDpulex = −0.26) (supplementary figs. S3 and S4, Supplementary Material online). Only the correlation between gene family size and the standard deviation of the methylation levels for D. magna gene families was not significant. We further observed a significant positive correlation between gene family size and mean CpG O/E ratios for both species (rDmagna = 0.43, rDpulex = 0.53) (supplementary fig. S5, Supplementary Material online).
We compared the gene expression of genes within these 19 gene families, over- and underrepresented for differentially methylated genes, by using all publically available D. pulex whole genome microarray data. Only a small proportion of the genes across all gene families (7%) were not differentially expressed in any of the 49 conditions. Although in the majority of the overrepresented gene families all genes were differentially expressed (q value <0.05) in at least one condition, no significant differences between the under and overrepresented gene families were observed (table 2, P value = 0.07). Overall, for the underrepresented gene families, more conditions did have at least one differentially expressed gene (q value <0.05) than for the overrepresented gene families, even when correcting for gene family size (table 2, P value = 0.003). Yet, no significant differences between genes of over- and underrepresented gene families were observed for the average number of conditions in which a gene was differentially expressed (P value = 0.22).
Table 2.
Summary table of the results of the gene expression analysis across 49 conditions organized per gene family for D. pulex
| Gene family | Proportion of genes with no DE | Family size | No. conditions with at least 1 DE gene | Average no. of conditions in which a gene is DE within gene family |
|---|---|---|---|---|
| HMG-Box | 0.06 | 17 | 25 | 5.06 |
| GTPase | 0 | 8 | 20 | 5.13 |
| Cyclin B & related kinase-activating proteins | 0 | 6 | 18 | 6.33 |
| Putative N2.N2-dimethylguanosine tRNA methyltransferase | 0.50 | 2 | 8 | 5 |
| TPR repeat-containing protein | 0 | 6 | 14 | 3.83 |
| Failed axon connections (fax) proteins | 0 | 3 | 11 | 4.67 |
| Tyrosine kinases | 0 | 5 | 8 | 3.6 |
| RNA polymerase II transcription initiation factor TFIIH | 0 | 1 | 2 | 2 |
| Chitinase | 0.04 | 67 | 46 | 5.60 |
| Trypsin | 0.05 | 84 | 46 | 7.32 |
| Collagens (type IV and type XIII). and related proteins | 0.08 | 108 | 40 | 5.14 |
| Bestrophin | 0 | 24 | 25 | 4.46 |
| FOG: 7 transmembrane receptor | 0.15 | 73 | 33 | 4.27 |
| Low-density lipoprotein receptors | 0.03 | 30 | 33 | 7.57 |
| Nucleolar GTPase/ATPase p130 | 0.09 | 54 | 32 | 3.74 |
| Cytochrome P450 CYP4/CYP19/CYP26 subfamilies | 0 | 29 | 35 | 6.34 |
| C-type Lectin | 0.14 | 74 | 43 | 5.46 |
| Fibroblast/platelet-derived growth factor receptor | 0.08 | 24 | 31 | 4.21 |
| RNA polymerase II. Large subunit | 0.04 | 65 | 32 | 4.55 |
A gene is considered as differentially expressed in the array (DE) if it has a q value smaller than 0.05. Gene families above the black line are overrepresented for differentially methylated genes, gene families below the black line are underrepresented for differentially methylated genes (see also table 1).
Discussion
The epigenetic modifications caused by changes in DNA methylation drive essential biological processes including cell development and differentiation through molecular mechanisms such as gene regulation. Yet, we have only limited understanding of the relationship between gene function, gene family size, and DNA methylation. Here, we report DNA methylation patterns in two closely related invertebrate species. Our results are in line with methylation levels reported in other invertebrates including the closely related species Daphnia ambigua and global methylation levels (0.49–0.52%) measured through liquid chromatography coupled with mass spectrometry for two D. magna strains including the isolate used here (Lyko et al. 2010;Xiang et al. 2010; Bonasio et al. 2012; Asselman et al. 2015b; Schield et al. 2015). These results demonstrate that underlying the genome wide levels of methylation there is a complex pattern of mosaic gene body methylation. This pattern is characteristic for invertebrate species in which a few gene bodies are highly methylated in a CpG context while a large group of gene bodies completely lacks methylation. Here, we specifically observed the absence of any methylation in zero methylated gene bodies in both Daphnia species. This concordance across species strongly suggests that zero methylation in these gene bodies is most likely consistent across individuals and across tissues. Thus, mechanisms of gene regulation using DNA methylation are likely targeted to gene bodies having varying methylation levels under control conditions as zero methylated genes lack any methylation. By using a whole body assay, rather than a tissue-specific approach, we are able to better assess general patterns and mechanisms and are not limited to tissue-specific regulation. On the other hand, this approach is limiting in that it can obscure some functional pathways that may be confounded by variation among tissue types.
We focused on a conserved set of gene models in the two species that are a good representation of the genome based on benchmarking of universal single-copy orthologs through a BUSCO analysis (Simão et al. 2015). As commented by other authors (Denton et al. 2014), the draft genome of Daphnia may contain an inflated number of gene models. We therefore only used a limited gene set with high evidence that allows straightforward comparisons with high confidence between the two species as described in the “Methods” section. While using a reduced gene set may bias our findings, the bias introduced here by using a conserved set is limited as this study focuses on gene body methylation patterns within and between gene families. First, the majority of the gene models (60%) that were excluded did not have any annotation information and could therefore not be assigned to any gene family. Second, 10% of the excluded gene models were single-copy genes. As both single-copy genes and genes without annotation information cannot be used for this analysis focusing on gene families by using annotation information, 70% of the genes filtered out would also be excluded when using the full set. Third, while larger gene families can be more susceptible to misassembly and therefore genes within larger gene families would have a higher chance of being excluded, this was not the case within this study. Indeed, gene family size within the conserved gene set had a correlation coefficient of 0.97 with its gene family size in the full gene set. As the conclusions within this article primarily relate to gene family size, this is the most important indicator and clearly highlights that the findings using conservative filtered set are representative of the full genome set.
Differences in methylation levels between the two species may be a consequence of sequence divergence and thus potential differences in the number of CpGs. For example, one species may contain additional unmethylated CpGs not present in the other species and therefore have a lower methylation level as the methylation level is determined by the number of methylated CpGs divided by the total number of CpGs. Here, we observed weak correlations between methylation differences and sequence divergence, which suggests that sequence divergence is not the major contributor and other factors are likely driving methylation differences between the two species.
Functional analysis of differentially methylated genes highlighted gene families that were over and underrepresented with these genes. Furthermore, underrepresented gene families tend to be significantly larger then overrepresented gene families as we observed a significant correlation between gene family size and the proportion of differentially methylated genes. We further studied distribution of methylation levels within underrepresented gene families as well as overrepresented gene families and observed significant negative correlations between the mean methylation level and gene family size in both species. In D. pulex, we also observed a significant negative correlation between the standard deviation and gene family size. While previous studies have studied gene families and have observed that gene body methylation was strongly conserved among orthologous, these results further suggest a relationship between DNA methylation and gene family size (Takuno and Gaut 2013). Indeed the results suggest that large gene families are more likely to lack methylation and this lack of methylation can be conserved within and between Daphnia species. In contrast, smaller gene families are more likely to express varying methylation levels within and between Daphnia species.
To further understand the functional and evolutionary mechanisms underlying these results, we studied the relationship with CpG O/E ratio. CpG O/E ratio is an indicator of methylation over evolutionary time. Basically, methylated cytosines are subjected to deamination converting methyl-cytosines into thymines resulting in a lower number of CpG islands in region of high methylation than expected (Goulondre et al. 1978). Therefore, genes with a low CpG O/E ratio have less CpG dinucleotides than expected which is likely the result of the known hyper-mutability of methylated cytosines whereas genes with a CpG O/E ratio close to 1 are predicted to be sparsely methylated (Schorderet and Gartler 1992). Here, we observed a significant positive correlation between gene family size and the mean CpG O/E ratio of the gene family for both species. This result suggests that smaller gene families are likely to have become methylated over evolutionary time while larger gene families have been less susceptible to methylation and deamination pressure. The question remains as to why these differences between large and small gene families occur and are conserved between the two Daphnia species. A recent study by Roberts and Gavery (2011) suggests that the sparsely methylated gene bodies specifically allow for increased transcriptional opportunities and thus increased phenotypic plasticity. They postulate that the absence of methylation facilitates random variation that contributes to phenotypic plasticity whereas methylation would therefore limit the transcriptional variation in genes with essential biological functions and protect them for inherent genome wide plasticity (Roberts and Gavery 2011). This implies that methylated genes are more constrained in divergence through duplication. This suggests that when gene regulation or gene function involved methylation it imposes an additional selective constraint on the gene.
Here, we observed that gene families associated with RNA processing and modifications, including post-translational modifications, were overrepresented in differentially methylated genes. In contrast, among the gene families underrepresented in differentially methylated genes are trypsins, collagens, chitinases, and cytochrome P450, which are often noted as differentially expressed in gene expression studies with Daphnia species (Poynton et al. 2008; Jeyasingh et al. 2011; Asselman et al. 2015a; Latta et al. 2012; Yampolsky et al. 2014; Chowdhury et al. 2015).
To further explore the relationship between differential methylation and differential regulation in response to environmental stimuli we studied gene expression patterns within these gene families in publically available D. pulex gene expression data. We restricted our analysis to studies using the same high-density 12-plex NimbleGen array on whole body organisms (Colbourne et al. 2011). From these datasets we were able to analyze gene expression profiles across 49 conditions. Overall, we observed that for small gene families, there was a higher number of conditions in which none of the genes from that gene family were differentially expressed than for larger gene families, even when adjusting for gene family size. Yet, we observed no difference between genes in large and genes in small gene families for the average number of conditions or arrays in which a gene was differentially expressed, suggesting no relation between gene family size and the number of times a gene is differentially expressed. Therefore, these gene expression results do not fully corroborate previous findings that genes with low CpG O/E and high methylation levels tend to be ubiquitously expressed and most likely contribute to housekeeping functions (Gavery and Roberts 2010; Bonasio et al. 2012; Lyko et al. 2010).
Nevertheless, these results do support the assertion of Gavery and Roberts (2010) that the lack of methylation may allow for phenotypic variation while methylation may protect genes from inherent genome-wide plasticity. Here, larger gene families, known to be involved in stress–response based on gene expression studies with Daphnia as discussed above, are sparsely methylated. The low to nonexistent methylation within these gene families, their family size and their involvement in stress response suggests that they contribute to phenotypic variation through mutation, gene family expansion, and alternate regulation of paralogous genes (Colbourne et al. 2011; Asselman et al. 2015a). In contrast, smaller gene families are more likely to be methylated and consequently less likely to contribute to phenotypic variation. Overall, these results suggest that gene body methylation may help regulate gene family expansion and functional diversification of gene families leading to phenotypic variation.
Conclusion
In the background of low global methylation levels, gene body methylation in Daphnia species shows a mosaic pattern of both highly methylated genes and genes devoid of any methylation. While general methylation patterns were similar across the two Daphnia species, a significant subset of differentially methylated genes could be detected. Differences in methylation between the two species could not be explained by differences in sequence similarity. Furthermore, functional analysis of methylation levels across gene families highlighted a significant negative correlation between gene family size and methylation. Gene families showing highly variable methylation levels were on average smaller whereas gene families showing highly consistent methylation levels were larger. In addition, we observed a significant positive correlation between gene family size and CpG O/E ratio. These results suggest that methylation may constrain gene family expansion and played a significant role in the functional diversification of gene families contributing to phenotypic variation.
Supplementary Material
Supplementary figures S1–S5 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Jolien Depecker for performing the DNA extractions Jana Asselman is a Francqui Foundation Fellow of the Belgian American Educational Foundation. Funding was received from the Research Foundation Flanders (FWO Project G.0614.11), from BELSPO (AquaStress project: BELSPO IAP Project P7/31). This research contributes to and benefits from the Daphnia Genomics Consortium.
Literature Cited
- Asselman J, et al. 2015a. Conserved transcriptional responses to cyanobacterial stressors are mediated by alternate regulation of paralogous genes in Daphnia. Mol Ecol. 24:1844–1855. [DOI] [PubMed] [Google Scholar]
- Asselman J, et al. 2015b. Global cytosine methylation in Daphnia magna depends on genotype, environment and their interaction. Environ Toxicol Chem. 34:1056–1061. [DOI] [PubMed] [Google Scholar]
- Bonasio R, et al. 2012. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 22:1755–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, et al. 2011. The ecoresponsive genome of Daphnia pulex. Science 331:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury PR, et al. 2015. Differential transcriptomic responses of ancient and modern Daphnia genotypes to phosphorus supply. Mol Ecol 24:123–135. [DOI] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. 1999. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401:157–161. [DOI] [PubMed] [Google Scholar]
- De Coninck DIM, et al. 2014. Genome-wide transcription profiles reveal genotype-dependent responses of biological pathways and gene-families in Daphnia exposed to single and mixed stressors. Environ Sci Technol. 48:3513–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton JF, et al. 2014. Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput Biol. 10:e1003998.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Hunt BG, Goodisman MAD, Yi S. 2009. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A. 106:11206–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. 2012. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 13:97–109. [DOI] [PubMed] [Google Scholar]
- Feng H, Conneely K, Wu H. 2014. A bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acid Res. 42:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 107:8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores K, et al. 2012. Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genomics 13:480.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavery MR, Roberts SB. 2010. DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas). BMC Genomics 11:483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladstad KM, hunt BG, Yi SV, Goodisman MAD. 2011. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol. 20:553–565. [DOI] [PubMed] [Google Scholar]
- Goulondre C, Miller JH, Farabaugh PJ, Gilbert W. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274:775–780. [DOI] [PubMed] [Google Scholar]
- Haag CR, McTaggart SJ, Didier A, Little TJ, Charlesworh D. 2009. Nucleotide polymorphism and within-gene recombination in Daphnia magna and D. pulex, two cyclical parthenongens. Genetics 182:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KDM, Bartlett NJ, Lloyd VK. 2012. Daphnia as an emerging epigenetic model organism. Genet Res Int. 12: article ID 147892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, et al. 2013. DNA methylation contributes to natural human variation. Genome Res. 23:1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyasigngh PD, et al. 2011. How do consumers deal with stoichiometric constratins? Lessons from functional genomics using Daphnia pulex. Mol Ecol 20:2341–2352. [DOI] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 13:484–492. [DOI] [PubMed] [Google Scholar]
- Kilham SS, Kreeger DA, Lynn SG, Goulden CE, Herrera L. 1998. COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia 377:147–159. [Google Scholar]
- Klüttgen B, Dülmer U, Engels M, Ratte HT. 1994. ADaM, an artificial freshwater for the culture of zooplankton. Water Res. 28:743–746. [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta LC, Weider LJ, Colbourne JK, Pfrender ME. 2012. The evolution of salinity tolerance in Daphnia: a functional genomics approach. Ecol Lett. 15:794–802. [DOI] [PubMed] [Google Scholar]
- Lyko F, et al. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8:e1000506.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner B, De Meester L, Pfrender ME, Lampert W, Hairston NG., Jr 2012. Linking genes to communities and ecosystems: Daphnia as an ecogenomic model. Prod R Soc B. 279:1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart SJ, Obbard DJ, Conlon C, Little TJ. 2012. Immune genes undergo more adaptive evolution than non-immune system genes in Daphnia pulex. BMC Evol Biol. 12:63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paland S, Colbourne JK, Lynch M. 2005. Evolutionary history of contagious asexuality in Daphnia pulex. Evolution 59:800–813. [PubMed] [Google Scholar]
- Poynton HC, et al. . 2008. Gene expression profiling in Daphnia magna, Part II: Validation of a copper specific gene expression signature with effluent from two copper mines in California. Environ Sci Technol. 42:6257–6263. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SB, Gavery MR. 2011. Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates? Front Physiol. 2:116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routtu J, et al. 2014. An SNP-based second-generation genetic map of Daphnia magna and its application to QTL analysis of phenotypic traits. BMC Genomics 15:1033.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV. 2012. The evolution of invertebrate gene methylation. Mol Biol Evol. 29:1907–1916. [DOI] [PubMed] [Google Scholar]
- Schield DR, et al. 2015. EpiRADseq: scalable analysis of genomewide patterns of methylation using next-generation sequencing. Methods Ecol Evol. 7:60–69. [Google Scholar]
- Schorderet DF, Gartler SM. 1992. Analysis of CpG suppression in methylated and nonmethylated species. Proc Natl Acad Sci U S A. 89:957–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JR, et al. 2007. Gene response profiles for Daphnia pulex exposed to the environmental stressor cadmium reveals novel crustacean metallothioneins. BMC Genomics 8:477.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Kerr ARW, De Sousa D, Bird A. 2007. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 17:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. 2013. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci U S A. 110:1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H, et al. 2010. Single base–resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol. 28:516–520. [DOI] [PubMed] [Google Scholar]
- Yampolsky, et al. 2014. Functional genomics of acclimation and adaptation in response to thermal stress in Daphnia. BMC Genomics. 15:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.