Abstract
Like many other Bacteroides species, Bacteroides vulgatus strain mpk, a mouse fecal isolate which was shown to promote intestinal homeostasis, utilizes a variety of mobile elements for genome evolution. Based on sequences collected by Pacific Biosciences SMRT sequencing technology, we discuss the challenges of assembling and studying a bacterial genome of high plasticity. Additionally, we conducted comparative genomics comparing this commensal strain with the B. vulgatus type strain ATCC 8482 as well as multiple other Bacteroides and Parabacteroides strains to reveal the most important differences and identify the unique features of B. vulgatus mpk. The genome of B. vulgatus mpk harbors a large and diverse set of mobile element proteins compared with other sequenced Bacteroides strains. We found evidence of a number of different horizontal gene transfer events and a genome landscape that has been extensively altered by different mobilization events. A CRISPR/Cas system could be identified that provides a possible mechanism for preventing the integration of invading external DNA. We propose that the high genome plasticity and the introduced genome instabilities of B. vulgatus mpk arising from the various mobilization events might play an important role not only in its adaptation to the challenging intestinal environment in general, but also in its ability to interact with the gut microbiota.
Keywords: Bacteroides vulgatus, mobile elements, genome plasticity, horizontal gene transfer, CRISPR/Cas
Introduction
The mammalian gut represents a complex and densely populated ecosystem in which two commensal bacterial phyla are predominant: Firmicutes and Bacteroidetes (Ottman et al. 2012; Kamada et al. 2013; Coyne et al. 2014). The latter are represented by at least 25 different species in the human gut (Coyne et al. 2014). In the mouse gut species diversity of Bacteroides might be similar to the human gut as the NCBI-NT (National Center for Biotechnology Information - Nucleotide) database includes similar numbers of Bacteroides species that are associated with the mouse host.
Although some Bacteroides sp. are obligate pathogens (Thomas et al. 2011), others have potential health benefits for their hosts, for example, Bacteroides fragilis, which is able to suppress intestinal inflammatory responses (Mazmanian et al. 2008), or Bacteroides vulgatus mpk, which has been shown to prevent Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient (IL2−/−) mice (Waidmann et al. 2003) by induction of host anti-inflammatory immune responses (Waidmann et al. 2003; Bohn et al. 2006; Muller et al. 2008). The pool of different Bacteroides sp. in the gut ecosystem provides a diverse collection of phenotypic strain variations with various fitness advantages. Besides the abovementioned immunomodulatory functions, Bacteroides species have the potential to contain enterotoxins (e.g., in B. fragilis), different gene clusters to degrade a variety of dietary polysaccharides (Coyne et al. 2014), and many mobile genetic elements (Nguyen and Vedantam 2011).
In competitive environments like the gastrointestinal tract, commensal microbes are constantly forced to adapt and survive upon different challenges like competition for nutrients or phage and pathogen attacks. Bacteria which are able to constantly reshape their genome architecture provide improved adaptation to their microenvironment, thus exhibiting a selective survival advantage (Xu et al. 2007). Recently, some evidence was provided that intestinal Bacteroides has exchanged DNA among each other within the human gut microbiota. Further it was proposed that some of the genes that might have been acquired contribute to bacterial fitness (Coyne et al. 2014). However, it is of high interest to prove whether the DNA exchange in human and mouse gut microbial communities is similar or significantly different because mice are model organisms to study gut microbiota.
Although B. vulgatus is a common constituent in the mouse and human gut microbiota, only few findings on genome composition and horizontal gene transfer (HGT) have been reported (Xu et al. 2007).
Here we report on the high quality genome draft of B. vulgatus strain mpk—further referred to as B. vulgatus mpk—which was isolated from mouse feces. Hitherto, only one complete genome reference was available for the B. vulgatus type strain ATCC 8482, a human isolate (Xu et al. 2007). Besides, several fragmented sequencing projects of other Bacteroides strains are available in the databases. We suspected that the high fragmentation rates of these available short-read assemblies are caused by the large amount of mobile elements, which are frequently found in Bacteroides genomes, and therefore increasing complexity of the assembly (Kingsford et al. 2010). Thus we decided to use long-read sequencing in order to produce a draft genome of B. vulgatus mpk which is suitable for further analysis, including comparative genomics. We achieved this aim (fig. 1) by using Pacific Biosciences (PacBio) SMRT sequencing, error correction using the PBCR pipeline (Koren et al. 2012), and assembly with Celera Assembler (Myers et al. 2000). Here we report on the results of B. vulgatus mpk whole-genome sequencing and provide a detailed analysis of the extraordinary repertoire of mobile genetic elements and how these different mobile structures might contribute to HGT and genome evolution. We show that B. vulgatus mpk harbors in fact a large number of repetitive sequences in the form of mobile elements like conjugative transposons, insertion sequences (ISs), a transposable bacteriophage, a CRISPR/Cas system, and numerous integrases and transposases. Further we provide evidence for different externally and internally driven genome rearrangements. We consider B. vulgatus mpk to be a potent competitor of the mouse microbiota due to its repertoire of mobile genetic elements and the different genome rearrangements we found.
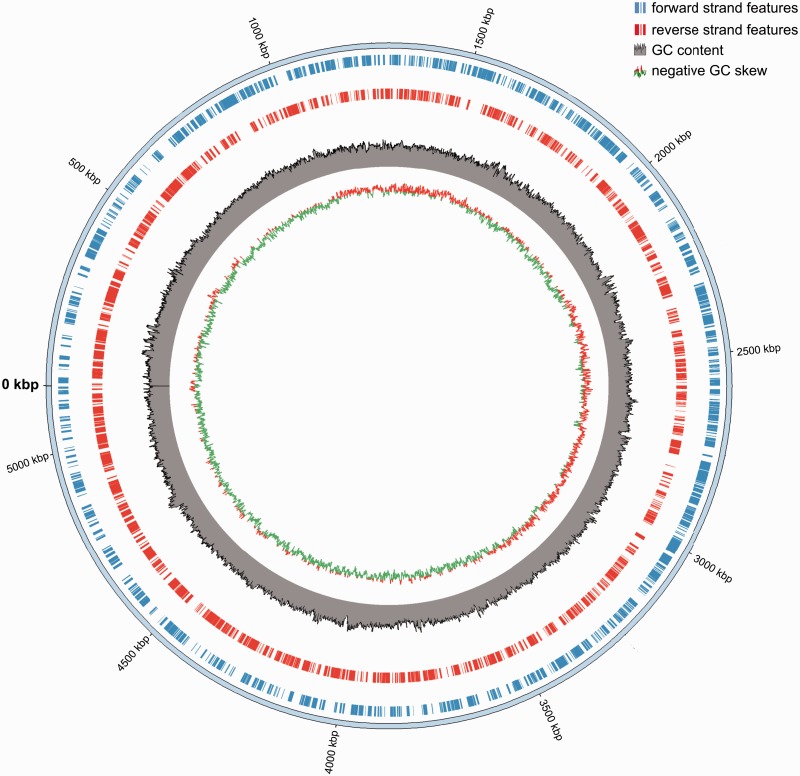
Fig. 1.—
Circular representation of the Bacteroides vulgatus mpk genome. The forward strand features are shown in blue, reverse strand features in red, Guanine-Cytosine (GC) content in gray, and positive and negative GC skew in red-green.
Materials and Methods
Bacteroides vulgatus mpk and ATCC 8482 Growth Conditions
Bacteroides vulgatus mpk was initially isolated from the fecal material of a healthy mouse (Waidmann et al. 2003). Bacteroides vulgatus ATCC 8482 (DSM-1447) type strain, which was isolated originally from human feces, was purchased from DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig). Both strains were grown on brain heart infusion (BHI) broth or agar plates at 37 °C anaerobically in a growth chamber.
Genomic DNA Isolation and PacBio Sequencing
The genomic DNA was extracted from bacteria grown on BHI agar with Genomic Tip 100/G (Qiagen, Hilden, Germany) according to manufacturer’s instructions. DNA concentration was determined fluorometrically in the Tecan Infinite 200 PRO (Tecan, Mainz, Germany) using a Quant-It DNA assay kit (Life Technologies, Darmstadt, Germany). For library preparation at least 300 ng/µl DNA was used. DNA was used to construct a 10-kb insert size library according to PacBio standard protocols. Sequencing of the libraries was performed on five SMRT cells using PacBio RSII with P4-C2 chemistry achieving a genome coverage of 330-fold. PacBio sequencing was performed at Eurofins (Ebersberg, Germany).
Draft Genome Assembly
Quality assessment of the sequences was done using FastQC (Andrews 2010). The reads were self-corrected using the PBCR pipeline (Koren et al. 2012) and downsampled to 25-fold coverage, keeping only the longest corrected reads. Those reads were assembled using Celera Assembler v8.1 (Myers et al. 2000), resulting in 33 contigs. By mapping the full set of corrected reads back to the assembly, 25 small contigs were identified having a 10-fold or less average coverage and those were excluded from further analysis. The remaining 8 contigs were closely checked for misassemblies and manually broken into 15 contigs. Remapping the corrected reads to the 15 curated contigs showed that more than 96% of the data was accounted for. The 15 curated contigs were ordered using the Mauve ContigMover (Rissman et al. 2009) guided by the B. vulgatus type strain ATCC 8482 and then manually curated into a 5.1-Mbp-long draft genome. The Guanine-Cytosine content is 42.2%. The sequence is available on NCBI under the accession CP013020.
Functional Annotation
Automated annotation of the draft genome sequence was done using the RAST server (Aziz et al. 2008), additionally using BaSys (Van Domselaar et al. 2005) and xBase (Chaudhuri et al. 2008), using the published genome of B. vulgatus ATCC 8482 as a reference. The three obtained annotations were merged, selecting the correct annotation by criteria determined by a previous assessment and comparison of the methods. The highest weight in the merging process was given to RAST annotations. The final annotation was curated removing genes following a selection by strong (<150 bp, annotated as hypothetical protein, no protein homology in comparison with selected related strains) and weak criteria (suspicious start codons, overlapping another well-annotated gene, different reading direction compared with all neighboring genes). An annotation was removed if it matched either a total of four criteria or all three of the strong criteria.
Comparative Genomics
Phylogenetic tree construction was based on a multiple sequence alignment of 15 Bacteroides species (supplementary table S1, Supplementary Material online) and Parabacteroides distasonis, using partial 16S ribosomal Ribonucleic acid (rRNA) gene sequences acquired from GenBank. The tree was calculated using maximum likelihood with 100 bootstrap replications. The resulting tree was rerooted using P. distasonis as an outgroup. For comparative analysis, nine related strains from the order Bacteroidales were chosen for protein homology search. Protein sequences for each of those strains were compared using pairwise BLASTP and filtering for matches covering at least 60% of the longer protein sequence. Those potential homologs were filtered for 50%, 90%, and 98% protein sequence identity to increase the specificity of the analysis. Another pairwise BLASTP comparison of the full set of all predicted B. vulgatus mpk protein sequences against itself was done to identify paralogs. By identifying colocalized groups of homologous proteins, synteny regions were detected to identify structural differences and be able to screen the genomes for potential HGT (supplementary table S2, Supplementary Material online). Multiple sequence alignments were done using ClustalΩ (Sievers et al. 2011). Visualization of the results used AnnotationSketch (Steinbiss et al. 2009) and Circos (Krzywinski et al. 2009) and RNA structure prediction used the RNAfold server (Lorenz et al. 2011).
Antibiotic Sensitivity Tests
Bacterial suspensions of B. vulgatus mpk and ATCC 8482 at the same optical density (McFarland 1) were spread on fresh BHI agar plates and pads containing different antibiotics were put on the plates. Antibiotic susceptibility tests were performed according to the EUCAST criteria. After anaerobic incubation for 2 days at 37 °C, susceptibility or resistance toward the respective antibiotic was evaluated.
Results and Discussion
General Characteristics and Phylogenetic Placement of Bacteroides vulgatus mpk
The genome of B. vulgatus mpk is 5,165,891 bp in size and does not contain any extrachromosomal structures (fig. 1). Like other members of the Bacteroides genus, it encodes a high number of starch utilization systems (sus) for sugar degradation, for example, one SusR-regulated cluster and several SusC- and SusD-like clusters to degrade different sugars (fucose, rhamnose, galactose) (supplementary table S3, Supplementary Material online) (Xu et al. 2003; Martens et al. 2009). The genome contains nine different loci coding for capsular polysaccharides (e.g., glycosyltransferases; supplementary table S4, Supplementary Material online), which are common traits for Bacteroides sp. and are the most polymorphic genomic regions (Xu et al. 2007). Furthermore B. vulgatus mpk harbors an abundance of structures to mobilize DNA-like IS elements, conjugative transposons, a transposable bacteriophage, various transposases and integrases, and other unassigned mobile element proteins (table 1). Additionally, a CRISPR/Cas system was found. Mobile DNA structures make up 8% of the 4,233 protein-coding genes. Bacteroides vulgatus mpk harbors more mobile element genes than B. vulgatus ATCC 8482, which encodes 16 integrases, 89 transposases, 1–3 conjugative transposons, 22 IS elements, and 19 mobilization proteins (table 1).
Table 1.
Genome Content of Bacteroides vulgatus mpk
| Feature | |
|---|---|
| Genome length | 5,165,891 bp |
| GC content | 42.24% |
| Plasmids | None |
| Protein-coding genes | 4,233 |
| tRNAs | 79 |
| rRNAs | 21 genes in 7 operons |
| IS elements | IS21-like 16 |
| IS4-like 14 | |
| Conjugative transposons | 1 (+ 2 truncated) |
| Complete Mu phages | 1 |
| Transposases (including putative) | 92 |
| Integrases | 33 |
| Mobilizable proteins | 3 pairs BmgA/B (and 11 orphan proteins) |
| Mobile element proteins | 17 |
| CRISPR/Cas systems | 1 type I-C/Dvulg |
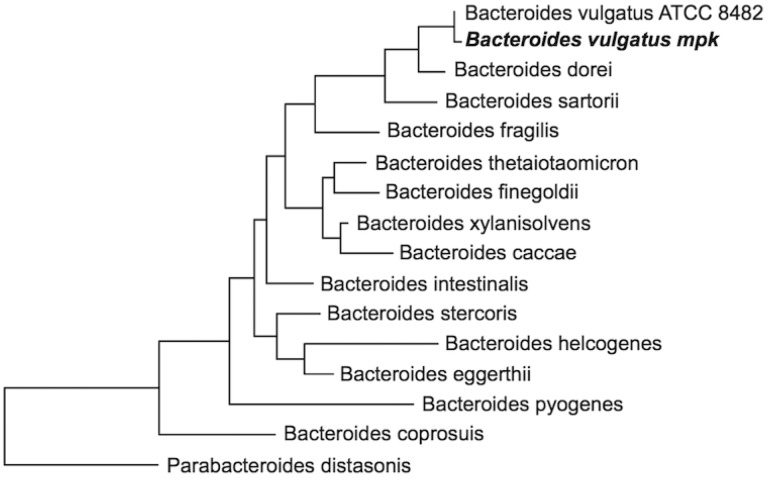
For a phylogenetic placement, we compared B. vulgatus mpk with different Bacteroides isolates from human and porcine fecal samples as well as from a dog oral cavity isolate. As a distant relative, P. distasonis (formerly Bacteroides distasonis) was used for a phylogenetic placement to emphasize the different classification of the Bacteroides and Parabacteroides genus (fig. 2). Bacteroides vulgatus mpk is very closely related to B. vulgatus ATCC 8482 and Bacteroides dorei on the basis of 16s rRNA.
Fig. 2.—
Phylogenetic tree of different Bacteroides strains. Phylogenetic placement of Bacteroides vulgatus mpk, 14 other Bacteroides species, and Parabacteroides distasonis, based on alignment of 1,488 aligned characters of 16S rRNA and calculated using maximum likelihood. Rooting was performed according to known taxonomy.
An analysis of gene synteny between B. vulgatus and close B. dorei strains (HS1_L_1_B_010 and HS1_L_3_B_079) indicates that B. vulgatus mpk shares multiple regions with B. dorei strains including the CRISPR/Cas system, which are missing in B. vulgatus ATCC 8482 (supplementary table S2, Supplementary Material online). This leads to the assumption that the relation between B. vulgatus mpk and B. dorei might be even closer than to B. vulgatus ATCC 8482 due to the evolutionary diversification of these species.
External- and Internal-Driven Genome Rearrangements
For Bacteroides species it was shown that conjugative transposons play an important role for HGT and hence for genome plasticity (Salyers et al. 1995; Coyne et al. 2014). However, they are also characterized by a high internal mobility of transposable elements as discussed below. Comparative genomics by protein homology showed that most of the sequences specific for B. vulgatus mpk compared with B. vulgatus ATCC 8482 are comprised of mobile elements. This also holds true for the comparison of B. vulgatus with multiple other Bacteroides species.
Horizontally Transferred Genome Evolution
The most prominent mobile element of B. vulgatus mpk is the complete conjugative transposon. By protein homology analysis, the B. vulgatus mpk complete conjugative transposon region was revealed to have the closest relation to conjugative transposons found in Bacteroides xylanisolvens XB1A as well as in B. dorei isolate HS1_L_1_B_010 and B. fragilis YCH46. It has high similarity to the Bacteroides conjugative transposon type CTn341 (Bacic et al. 2005), which was originally isolated from a clinical human B. vulgatus isolate.
The B. vulgatus type strain ATCC 8482 also includes such conjugative transposon proteins, although it has a conjugative transposon which lacks the important TraP and TraD homologs and another conjugative transposon which additionally lacks TraB and TraE (supplementary table S5, Supplementary Material online). In comparison with the B. fragilis CTn341 sequence (AY515263.1), the conjugative transposon region contains an insertion of 11 proteins between the RteA and BmhA region and lacks a protein with significant homology to TetQ (supplementary table S6, Supplementary Material online). This protein is also absent in B. vulgatus ATCC 8482, but can be found in both of the compared B. dorei genomes. The inserted sequence includes transcriptional regulators as well as one glyxoxalase family protein (BvMPK_0111) and a β-lactamase gene BvMPK_0116. The insertion is absent from any of the compared Bacteroides species, with only BvMPK_102 to BvMPK_106 having low homology to proteins in B. dorei, B. xylanisolvens, and B. fragilis YCH46. The inserted proteins between BvMPK_0101 and BvMPK_0117 have no paralogs with significant similarity in the B. vulgatus mpk genome. The analysis of different antibiotic resistance genes suggests that both B. vulgatus mpk and ATCC 8482 encode the same genes for β-lactamases except BvMPK_0116, as its homolog is absent in ATCC 8482 (table 2). Testing of different antibiotics revealed that both strains exhibit similar resistance mechanisms against certain antibiotics like different aminoglycoside antibiotics (amikacin, gentamicin, or tobramycin) and some penicillins (penicillin, ampicillin, oxacillin) (supplementary table S7, Supplementary Material online). Only B. vulgatus mpk is resistant against different second-, third-, and fourth-generation cephalosporins. This indicates an additional resistance mechanism in B. vulgatus mpk mediated by BvMPK_0116 which might have been acquired by HGT.
Table 2.
Genes Coding for β-Lactamases in Bacteroides vulgatus mpk and ATCC 8482
| Annotation | PFAM Domain | Bacteroides vulgatus mpk | Bacteroides vulgatus ATCC 8482 |
|---|---|---|---|
| AmpG protein, beta-lactamase induction signal transducer | PF07690.11 (MFS_1) | BvMPK_0055 | BVU_0613 |
| Beta-lactamase | PF13354.1 (Beta-lactamase2) | BvMPK_0116 | Not found |
| Metal-dependent hydrolases of the beta-lactamase superfamily I | PF12706.2 (Lactamase_B_2) | BvMPK_1767 | BVU_2240 |
| Putative metallo-beta-lactamase | PF00753.22 (Lactamase_B) | BvMPK_2668 | BVU_3020 |
| Metallo-beta-lactamase family protein | PF00753.22 (Lactamase_B) | BvMPK_2756 | BVU_3071 |
| TPR repeat-containing protein | PF08238.7 (Sel1) | BvMPK_2840 | BVU_3175 |
| Beta-lactamase | PF13354.1 (Beta-lactamase2) | BvMPK_2895 | BVU_3257 |
We also found evidence of a potential HGT of a genomic island with more than 40 kb in B. vulgatus mpk between BvMPK_2233 and BvMPK_2278. The region may have been transferred among B. vulgatus mpk, B. dorei isolate HS1_L_3_B_079, and Bacterioides thetaiotaomicron VPI-5482, because those strains are the only significant hits in the NCBI-NT database filtered for all bacteria and a query coverage over 55%. As no other B. vulgatus or B. dorei strains include more than 53% percent of this region, the transfer might have occurred in a common ancestor of B. vulgatus mpk, B. dorei, or B. thetaiotaomicron and then got partially lost in all other subsequent strains. The island harbors different mobile element proteins and the encoded genes have a mosaic-like distribution. This random gene arrangement is a quite common characteristic for such genomic islands acquiring genes from possibly different donors (Darmon and Leach 2014).
It is surprising how similar—even on nucleotide level—that particular genomic island is in both mouse B. vulgatus mpk and human B. thetaiotaomicron. This could point toward a possible transmission route of Bacteroides sp. from humans to mice or vice versa, for example, in animal facilities.
The chromosome of B. vulgatus mpk contains a region encoding a transposable bacteriophage (Mu phage) absent in all other sequenced Bacteroides strains. Their integration into the genome occurs randomly, that is why they might influence downstream genes and operons and are able to inactivate genes. Mu phages are also able to drive further genomic rearrangements and promote mobility of other phages or stimulate recombination events between other transposable elements (Darmon and Leach 2014). Besides the mutative character of such phages, it is also reported that integration can occur simultaneously with the uptake of, for example, toxins which are prevalent in gut viral metagenomic data sets (Minot et al. 2011).
Most HGT events are suggested to be neutral or detrimental and are therefore lost rapidly. Such events just remain within a population if it brings an advantage for the bacteria (Darmon and Leach 2014). We therefore propose that the integration of the additional β-lactamase gene and the transfer of the genomic island should provide a fitness benefit for B. vulgatus mpk.
CRISPR/Cas Systems Provide a Potential to Prevent Integration of Invading External Mobile Elements
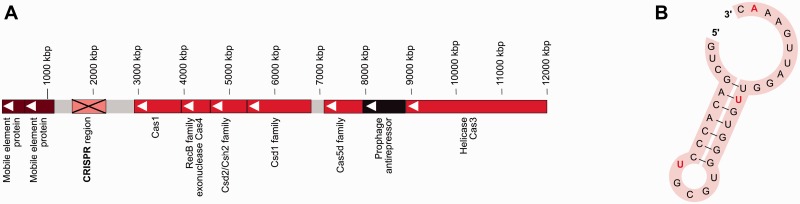
We have identified a complete type I-C CRISPR/Cas system (fig. 3A), comparable with the one identified in Bacillus halodurans C-125 (Nam et al. 2012). The same system is also found in B. dorei and in some of the incomplete B. vulgatus assemblies, but not in B. vulgatus ATCC 8482.
Fig. 3.—
The CRISPR/Cas system of Bacteroides vulgatus mpk. (A) The Cas system is a complete type I-C/DVULG system as postulated in Bacillus halodurans C-125. The Cas2 annotation was manually added, as it was missed by the automated annotation pipelines. The prerequisite parts of the CRISPR/Cas type I-C system are shown in the operon: The helicase Cas3, Cas5d which is involved in interacting with pre-crRNA, Csd1 family protein, Csd2/Csh2 familiy protein, Cas4 belonging to the family of RecB exonuclases, and Cas1. The operon is followed downstream by the CRISPR region, and two mobile element proteins.(B) The CRISPR repeat secondary structure was predicted from the repeat consensus RNA sequence obtained from the CRISPR region. It shows the conserved hairpin region which is necessary for recognition of the pre-crRNA by Cas5d to enable cleaving to obtain functional crRNA. Red bold letters indicate the nucleotide changes compared with Bacillus halodurans.
CRISPR/Cas systems are found in about 45% of the sequenced bacterial genomes (Nam et al. 2012). Due to their high prevalence they might be distributed frequently through HGT (Rath et al. 2015). The occurrence of the CRISPR/Cas system in B. vulgatus mpk might also be the result of a horizontal transfer event as a mobile element pair is located directly upstream of the CRISPR region (fig. 3A). The CRISPR/Cas system is a sophisticated adaptive immunity tool of both bacteria and archaea (Barrangou and Marraffini 2014; van der Oost et al. 2014). It offers specific immunization against invading mobile genetic elements and is able to integrate nucleic acid fragments into the CRISPR region (Jore et al. 2012). In the original automated annotation of the B. vulgatus mpk genes, the CRISPR/Cas seemed to be incomplete, as there were only 13 CRISPR repeats and it lacked the Cas2 protein. These components are essential to match any known functional type of CRISPR/Cas system. As the B. vulgatus mpk CRISPR/Cas system resembled the system type I-C (Nam et al. 2012), we manually checked the presence of the Cas2 gene between the repeat region and the Cas1 gene. By sequence comparison, we found a protein which is an exact match to a Bacteroides Cas2 (WP_005852931.1) and manually included this annotation.
The I-C/DVULG CRISPR-Cas system is special in having a Cas5d protein, which is an endoribonuclease important for processing pre-crRNA (pre-CRISPR RNA) into its mature form (Nam et al. 2012). This is guided by the stem-loop-containing secondary structure formed by the repeat regions, with Cas5d cleaving at the 3′ end of the hairpin (Nam et al. 2012). The CRISPR region of B. vulgatus mpk is made up of thirteen 32 nt repeats and 12 spacer sequences of 33–34 nt. The predicted secondary structure for B. vulgatus mpk CRISPR repeats is a 7-bp stem-loop structure, which has also been described for Bacillus halodurans (Nam et al. 2012) (fig. 3B). In comparison with the Bacillus halodurans CRISPR-repeat sequence, Bacillus vulgatus mpk CRISPR region has two changes inside the stem structure: The G4 is changed to A, and the U8 is changed to C (Nam et al. 2012). In the loop region, the U11 is changed to G and A13 changed to G in the presented system. It was shown that the deletion of the last two nucleotides at the 5′ end at the stem loop had little effect on the functionality of Cas5d (Nam et al. 2012). The last nucleotide of the stem loop is changed in B. vulgatus from a U32 to C, while all other bases of the 5′ tail remain conserved. These observations point toward a substrate specificity of Cas5d for the pre-crRNA structure in B. vulgatus mpk and provides theoretical evidence for general CRISPR/Cas system functionality. The system therefore might help B. vulgatus mpk to prevent invading mobile elements to integrate their DNA into the genome.
The specific function of the CRISPR/Cas type I-C is not well studied. The frequent integration of external DNA in B. vulgatus mpk shows that the strain is challenged by external sequences and thus could profit from a mechanism to reduce the challenge to retain stability. However, we can only refer to current literature which did not identify certain function of the CRISPR/Cas type I-C. Most studies about CRISPR/Cas systems are dealing with the system’s potential to protect from integration of invading genetic material. Additionally, these systems are reported to be involved in other processes than immunity-like regulation of virulence and DNA repair (Rath et al. 2015) as the system could even influence genome evolution by targeting the host chromosome and creating mutants with broad genome rearrangements (Rath et al. 2015). It remains unclear when B. vulgatus mpk acquired the CRISPR/Cas system in its evolutionary process and whether the high prevalence of HGT events can be influenced or regulated by the CRISPR/Cas system.
Internal Genome Evolution
Besides studying the synteny of homologous proteins between B. vulgatus mpk and the other selected strains to identify potential HGT events, we also investigated paralogy in B. vulgatus mpk to see if mobile elements were transferred within the genome. With that paralogy analysis we could identify two different types of IS elements occurring multiple times in the genome: IS4-like and IS21-like elements (supplementary table S8, Supplementary Material online). The latter type is composed of two open reading frames (ORFs). Those sequences are able to copy themselves and jump randomly into different genomic regions (De Palmenaer et al. 2008). Integrated IS elements can lead to insertion or deletion of genes, it can alternate gene expression, or cause DNA inversions (Darmon and Leach 2014). IS4-like elements encoded by just a single ORF are found in both B. vulgatus mpk and ATCC 8482 with about the same number of paralogs. There are 14 copies in B. vulgatus mpk and 18 in B. vulgatus ATCC 8482. Three of the IS21-like elements (BvMPK_1852/53, BvMPK_1710/11, BvMPK_1044/45) are paralogs having almost identical IS21-like ATP-binding protein and transposon pairs. BvMPK_1044/45 might be a result of a duplication and transition event. Upstream of the ATP-binding protein we found two proteins (TraG and TraH), which are also part of the conjugative transposon upstream of the BvMPK_1852/53 pair. So it might be possible that TraG and TraH were copied along with BvMPK_1044/45. In B. vulgatus ATCC 8482, two homologous pairs of these IS21-like elements can be found (BVU_0971/72, BVU_3472/73). The other 13 IS21-like gene pairs (BvMPK_0302/03, BvMPK_0856/57, BvMPK_1364/65, BvMPK_1880/81, BvMPK_2015/16, BvMPK_2101/02, BvMPK_2109/10, BvMPK_2229/30, BvMPK_2704/05, BvMPK_2762/63, BvMPK_3201/02, BvMPK_3251/52, BvMPK_4277/78) are paralogs with a different ATP-binding protein and transposon. These paralogous gene pairs share very high sequence similarity. Unlike the previous group of IS21-like elements, it is not possible to identify copies that might have been carried with the IS element proteins as they are always located with different genes. In B. vulgatus ATCC 8482, this pair is only found once (BVU_3194/95). Interestingly, it is reported that IS elements occur more frequently in genomes under low evolutionary pressure and are major drivers for reductive evolution (Vigil-Stenman et al. 2015). IS element clusters could be correlated with areas of recombination and gene losses, for example, in Shigella flexneri (Vigil-Stenman et al. 2015).
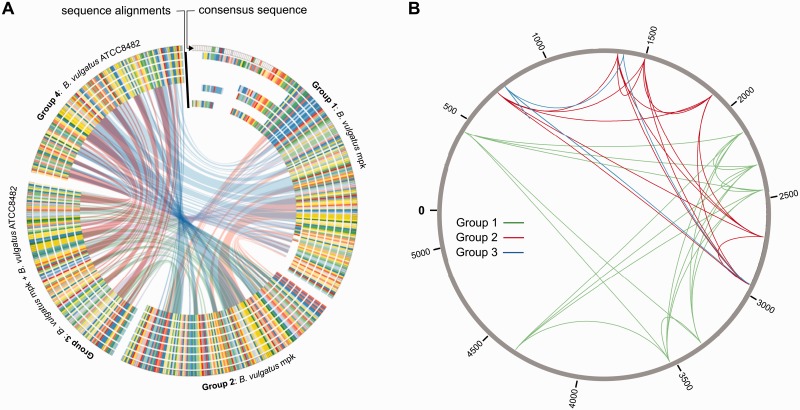
We further studied other paralogous proteins in the genome and found groups of hypothetical proteins that are all direct neighbors of a transposase and share different levels of protein homology. We checked B. vulgatus ATCC 8482 for similar proteins to determine whether they are present with a comparable frequency. We only found six incidences of a comparable hypothetical protein/transposase group in B. vulgatus ATCC 8482, while in B. vulgatus mpk we found 18 of such pairs. By aligning all 24 hypothetical proteins found in B. vulgatus mpk and B. vulgatus ATCC 8482 using ClustalΩ, we found that they can be divided into 4 groups. Subsequently, we generated separate multiple sequence alignments for the four groups (fig. 4A).
Fig. 4.—
Analysis of grouped protein paralogs found in Bacteroides vulgatus mpk and B. vulgatus ATCC 8482. (A) Alignments of hypothetical protein groups found multiple times in the genomes of B. vulgatus mpk and B. vulgatus ATCC 8482. The similarity of protein used for alignments is 90% at minimum. Group 1 includes seven hypothetical proteins from B. vulgatus mpk. Group 2 includes seven closely related hypothetical proteins from B. vulgatus mpk. Group 3 includes four proteins from B. vulgatus mpk as well as two proteins from B. vulgatus ATCC 8482. Group 4 is made up by four proteins found only in B. vulgatus ATCC 8482. The outer circle represents the consensus sequence of each group, and the inner circle shows the multiple sequence alignments of the proteins. Regions of exact matches between the consensus sequences are drawn as links between the groups. (B) Possible movements of the transposons adjacent to group 1 (green), group 2 (red), and group 3 (blue) proteins across the B. vulgatus mpk genome.
The group 1 proteins were originally annotated as 1-acyl-sn-glycerol-3-phosphate acyltransferase. This annotation was assigned by RAST based on FIGfam similarity. There is another protein (BvMPK_1744) with the annotation of 1-acyl-sn-glycerol-3-phosphate acyltransferase, which is not accompanied by a transposase, but located in a lipoprotein-rich locus. As the other proteins show no significant similarity to this protein, we compared the proteins of group 1 with the NCBI-NR database to verify the annotation. All significant BLASTP hits to group 1 proteins are annotated as hypothetical proteins, with the only exception of a protein from B. vulgatus dnLKV7 that is in fact annotated as 1-acylglycerol-3-phosphate O-acyltransferase. As the proteins from group 1 do not match either of the two FIGfams associated with this function (FIG005243 and FIG135282) and the dnLKV7 annotation was done automatically and is uncurated, we classified this as a misannotation and changed the functional annotation to “hypothetical protein.” We conclude that BvMPK_1744 is the only correctly annotated 1-acyl-sn-glycerol-3-phosphate acyltransferase protein in B. vulgatus mpk.
We also visualized the distribution of transposons colocated with the paralogous proteins to follow-up their integration across the B. vulgatus mpk genome (fig. 4B). The transposons seem to avoid to integrate into certain regions and seem to prefer locations to jump in. Interestingly, the transposons colocating with group 2 and group 3 proteins are randomly located in a certain part of the genome and seem to avoid the other part. We propose that the abovementioned internal transitions influence B. vulgatus mpk’s genetic repertoire. But we cannot conclude if they caused any positive or negative mutations. The dissemination of mobile elements is expected to produce high mutation rates for large deletions with little counteractive selection, and a massive genome reduction might be the consequence. Accordingly, most changes can rather be put down to genetic drift than adaptation (Moran and Plague 2004).
Area of Mobilization Hotspots in the Genome
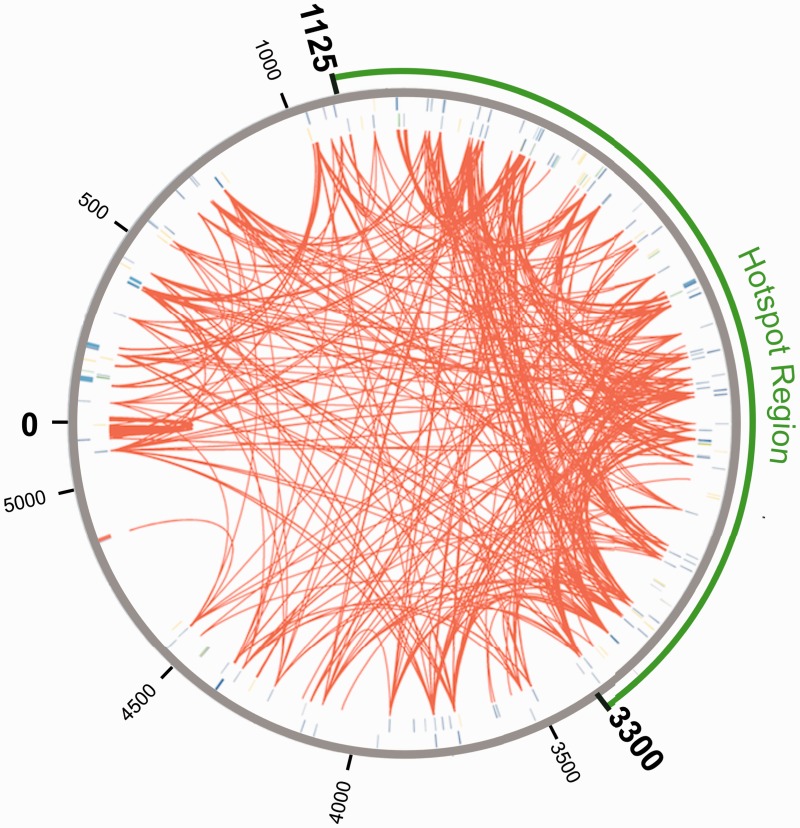
Having identified so many mobile elements, paralogous proteins, and genome reformations among the B. vulgatus mpk genome, it was prudent to determine whether mobile elements are equally distributed over the genome or if they are concentrated at particular regions in the genome like recently proposed for Staphylococcus aureus (Everitt et al. 2014). Correlation of protein paralogy and the location of mobile elements in the genome (fig. 5) reveal that there is in fact an area with a concentration of mobile elements. We identified a region with highly frequent transition sites and a high density of mobile elements. The region is located between 1,125 and 3,300 kbp on the chromosome (fig. 5). It was demonstrated in Bacteroides that integration of different transposase proteins was located at the 3′ end of a Leu-tRNA or Ser-tRNA (Shoemaker et al. 1996; Wang et al. 2000). Therefore we screened the genome for tRNA sequences to identify tRNAs associated with mobile elements (supplementary table S9, Supplementary Material online). Out of 79 tRNA sequences, 17 had an associated integrase or transposase. Eleven of those tRNA-mobile element pairs were located in the 1,125–3,300 kbp region. With this we suggest a connection between the increased occurrence of tRNA-mobile element pairs and the colocalization of transition events. Thus we were able to identify a potential hotspot region for genome transitions and mobile elements. Such regions are areas with high genome instability and are reported to function as a driver for bacterial evolution and adaptation (Darmon and Leach 2014).
Fig. 5.—
A circular view of the Bacteroides vulgatus mpk genome with protein paralogy highlights and display of mobile elements. Red lines indicate paralogy based on at least 90% protein identity. Different mobile elements are shown as lines on the forward (inner circle) and reverse (outer circle) strand: Conjugative transposon proteins, CRISPR/Cas-related proteins, integrases, mobilization proteins, transposases, and other mobile elements. The 1,125–3,300 kbp region with highest density of protein paralogy, occurrence of mobile elements, and the incidence of tRNAs associated with mobile elements was highlighted within a green hotspot area.
Conclusions
Here we provide the second draft genome sequence for B. vulgatus mpk generated by PacBio SMRT technology. SMRT sequencing of bacterial genomes for assembly has been proposed as a new standard (Roberts et al. 2013). We and others (Faino et al. 2015; Rhoads and Au 2015) propose it to be a successful technique to sequence genomes with a high content of repetitive sequences. Analysis of the B. vulgatus mpk genome revealed a huge variety of mobile elements and different internally and externally driven genome rearrangements have contributed to shape the genome. We found high copy numbers of certain transposase-hypothetical protein pairs in both B. vulgatus mpk and B. vulgatus ATCC 8482 and propose that these pairs might have evolved simultaneously into four different groups by mostly retaining the full transposase and transposed protein sequence. The finding that these transposase-protein pairs occur in both B. vulgatus mpk and B. vulgatus ATCC 8482 and that they were spread over both genomes might suggest that both species could use that strategy to shape their genomes and are at the same time retaining their genome stability.
With this study we suggest that mouse B. vulgatus mpk and the human isolate ATCC 8482 might be able to constantly adapt to their environment and benefit from their mobilome, the extensive genome evolution, and the introduced genome instability.
The data presented here might help to improve the understanding of Bacteroides sp. genome diversification on a molecular level especially because Bacteroidetes represents a highly abundant phylum in the mammalian gut microbiome. The genome databases include only few complete genomes of these abundant members of the gut microbiota and contain only many fragmented genome projects without being very well annotated or studied. The genome data we generated, the comparison with Bacteroides genomes isolated from humans, and the insights derived from human gut microbiota studies are so far the only way to generate hypotheses for potential HGT between the mouse commensal strain B. vulgatus mpk and other members of the Bacteroides genus.
Supplementary Material
Supplementary tables S1–S9 and figure S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the DFG (DFG FR 2087/6-1, DFG FR 2087/8-1, CRC685, SPP1656), the DFG research training group 1708, the Bundesministerium für Bildung und Forschung, and the German Center for Infection Research.
Literature Cited
- Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data. [Internet]. [cited 2015 May 6]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacic M, et al. 2005. Genetic and structural analysis of the Bacteroides conjugative transposon CTn341. J Bacteriol. 187:2858–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. 2014. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 54:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn E, et al. 2006. Host gene expression in the colon of gnotobiotic interleukin-2-deficient mice colonized with commensal colitogenic or noncolitogenic bacterial strains: common patterns and bacteria strain specific signatures. Inflamm Bowel Dis. 12:853–862. [DOI] [PubMed] [Google Scholar]
- Chaudhuri RR, et al. 2008. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 36:D543–D546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE. 2014. Evidence of extensive DNA transfer between bacteroidales species within the human gut. MBio 5:e01305–e01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon E, Leach DR. 2014. Bacterial genome instability. Microbiol Mol Biol Rev. 78:1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palmenaer D, Siguier P, Mahillon J. 2008. IS4 family goes genomic. BMC Evol Biol. 8:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt RG, et al. 2014. Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nat Commun. 5:3956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino L, et al. 2015. Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome. MBio 6: e00936–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Brouns SJ, van der Oost J. 2012. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harb Perspect Biol. 4: a003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 14:685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford C, Schatz MC, Pop M. 2010. Assembly complexity of prokaryotic genomes using short reads. BMC Bioinformatics 11:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, et al. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 30:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, et al. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 284:24673–24677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625. [DOI] [PubMed] [Google Scholar]
- Minot S, et al. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21:1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 14:627–633. [DOI] [PubMed] [Google Scholar]
- Muller M, et al. 2008. Intestinal colonization of IL-2 deficient mice with non-colitogenic B. vulgatus prevents DC maturation and T-cell polarization. PLoS One 3:e2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, et al. 2000. A whole-genome assembly of Drosophila. Science 287:2196–2204. [DOI] [PubMed] [Google Scholar]
- Nam KH, et al. 2012. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure 20:1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Vedantam G. 2011. Mobile genetic elements in the genus Bacteroides, and their mechanism(s) of dissemination. Mob Genet Elements 1:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottman N, Smidt H, de Vos WM, Belzer C. 2012. The function of our microbiota: who is out there and what do they do? Front Cell Infect Microbiol. 2:104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath D, Amlinger L, Rath A, Lundgren M. 2015. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 117:119–128. [DOI] [PubMed] [Google Scholar]
- Rhoads A, Au KF. 2015. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman AI, et al. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Carneiro MO, Schatz MC. 2013. The advantages of SMRT sequencing. Genome Biol. 14:405.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Shoemaker NB, Li LY. 1995. In the driver's seat: the Bacteroides conjugative transposons and the elements they mobilize. J Bacteriol. 177:5727–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker NB, Wang GR, Salyers AA. 1996. The Bacteroides mobilizable insertion element, NBU1, integrates into the 3′ end of a Leu-tRNA gene and has an integrase that is a member of the lambda integrase family. J Bacteriol. 178:3594–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S, Gremme G, Scharfer C, Mader M, Kurtz S. 2009. AnnotationSketch: a genome annotation drawing library. Bioinformatics 25:533–534. [DOI] [PubMed] [Google Scholar]
- Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. 2011. Environmental and gut bacteroidetes: the food connection. Front Microbiol. 2:93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 12:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Domselaar GH, et al. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33:W455–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil-Stenman T, Larsson J, Nylander JA, Bergman B. 2015. Local hopping mobile DNA implicated in pseudogene formation and reductive evolution in an obligate cyanobacteria-plant symbiosis. BMC Genomics 16:193.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidmann M, et al. 2003. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 125:162–177. [DOI] [PubMed] [Google Scholar]
- Wang J, Shoemaker NB, Wang GR, Salyers AA. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol. 182:3559–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, et al. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. [DOI] [PubMed] [Google Scholar]
- Xu J, et al. 2007. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5:e156.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.