Abstract
Comparative genomics analyses empowered by the wealth of sequenced genomes have revealed numerous instances of horizontal DNA transfers between distantly related species. In eukaryotes, repetitive DNA sequences known as transposable elements (TEs) are especially prone to move across species boundaries. Such horizontal transposon transfers, or HTTs, are relatively common within major eukaryotic kingdoms, including animals, plants, and fungi, while rarely occurring across these kingdoms. Here, we describe the first case of HTT from animals to plants, involving TEs known as Penelope-like elements, or PLEs, a group of retrotransposons closely related to eukaryotic telomerases. Using a combination of in situ hybridization on chromosomes, polymerase chain reaction experiments, and computational analyses we show that the predominant PLE lineage, EN(+)PLEs, is highly diversified in loblolly pine and other conifers, but appears to be absent in other gymnosperms. Phylogenetic analyses of both protein and DNA sequences reveal that conifers EN(+)PLEs, or Dryads, form a monophyletic group clustering within a clade of primarily arthropod elements. Additionally, no EN(+)PLEs were detected in 1,928 genome assemblies from 1,029 nonmetazoan and nonconifer genomes from 14 major eukaryotic lineages. These findings indicate that Dryads emerged following an ancient horizontal transfer of EN(+)PLEs from arthropods to a common ancestor of conifers approximately 340 Ma. This represents one of the oldest known interspecific transmissions of TEs, and the most conspicuous case of DNA transfer between animals and plants.
Keywords: lateral transmission, transposable elements, Dryads, loblolly pine
Introduction
In the absence of mating, the transfer of genes across species barriers is considered rare in eukaryotes. Although such horizontal transfer (HT) events have been reported in several nuclear and organelle genes (Andersson 2005; Keeling and Palmer 2008), the majority of eukaryotic genes indeed show no evidence of HT. Transposable elements (TEs) form a group of nearly ubiquitous repetitive DNA sequences in eukaryotes that, contrary to genes, is HT-prone. A number of independent HTT events have been documented in animals (Kordis and Gubenek 1995; Casola et al. 2007; Schaack et al. 2010; Thomas et al. 2010; Gilbert et al. 2012; Sormacheva et al. 2012; Walsh et al. 2013), angiosperms (Diao et al. 2006; Fortune et al. 2008; El Baidouri et al. 2014), and fungi (Novikova et al. 2009, 2010), and a recent survey estimated that millions of HTTs could have occurred in angiosperms alone (El Baidouri et al. 2014). However, only a few instances of HTTs between eukaryotic kingdoms—hereafter defined following Simpson and Roger (2004)—have been described thus far (Gorinsek et al. 2004; Llorens et al. 2009; Novikova et al. 2010; Parisot et al. 2014).
The intrinsic ability of TEs to self-propagate through transposition has a major impact on the genome landscape in many eukaryotes. For example, TE proliferation is responsible for the large genome size observed in numerous animals, fungi, and plants, including the enormous conifer genomes (De La Torre et al. 2014). Retroelements, one of the two known TE classes (Wicker et al. 2007), are the primary drivers of genome size expansion in eukaryotes. Retroelements transpose through a so-called “copy-and-paste” mechanism initiated by the reverse transcription of the element’s RNA into a cDNA molecule that is then inserted in a novel genomic location (Eickbush and Jamburuthugoda 2008). These key enzymatic reactions are carried out by the reverse transcriptase (RT) and the integrase/endonuclease (EN) domains encoded in the retroelements’ protein. Retroelements are classified according to their structure and sequence conservation in two major groups, long-terminal repeat (LTR) and non-LTR elements, with the former group characterized by the distinctive LTRs flanking the coding region (Eickbush and Jamburuthugoda 2008). These two groups account for the majority of TEs in many eukaryotes (Deininger and Batzer 2002; Martin et al. 2010; Sun et al. 2012; Nystedt et al. 2013; Neale et al. 2014) and have been implicated in numerous HTT events (Kordis and Gubenek 1995; Novikova et al. 2009, 2010; Schaack et al. 2010; Walsh et al. 2013; El Baidouri et al. 2014; Parisot et al. 2014).
Penelope-like elements (PLEs) represent a third group of retroelements originally isolated in the fruit fly Drosophila virilis, wherein they have been associated with a hybrid dysgenesis syndrome (Evgen’ev et al. 1997). Several HTT events of PLEs have been documented in Drosophila (Evgen’ev et al. 2000; Morales-Hojas et al. 2006). Two types of PLEs have been found in eukaryotes. Elements of the first type encode both an RT domain and an EN domain belonging to the GIY-YIG family of ENs, which is unrelated to the EN domain of other retroelements (Arkhipova 2006). We will refer hereafter to this group as EN(+)PLEs following the Gladyshev and Arkhipova nomenclature (2007). EN(+)PLEs are widespread across metazoans, yet have not been detected in other eukaryotes in previous bioinformatics surveys (Arkhipova et al. 2003; Arkhipova 2006; Gladyshev and Arkhipova 2007). The second lineage of PLEs was discovered in a variety of eukaryotes and is represented by elements that encode only the RT domain, named EN(−)PLEs (2007). Phylogenetic analyses indicated that the RT domains encoded by both PLE types are closely related to the same domain of telomerases, the enzymes responsible for the stability of telomeres in eukaryote chromosomes (Arkhipova et al. 2003). Intriguingly, EN(−)PLEs show an insertion preference toward telomeric regions of the host chromosomes (Gladyshev and Arkhipova 2007). It remains debated whether telomerases evolved from a group of EN(−)PLEs or vice versa (Gladyshev and Arkhipova 2007, 2011). Interestingly, the RNA encoded by both types of PLEs have been recently found to contain self-cleaving structures such as the Hammerhead ribozyme (Cervera and De la Pena 2014).
PLEs have been reported in the recently sequenced genome of the loblolly pine tree (Wegrzyn et al. 2013; Neale et al. 2014), but no further evolutionary investigation has been carried out on these elements. Here, we perform an in-depth analysis of conifer genomes to characterize the diversity and phylogenetic relationships of PLEs, and in particular the EN(+)PLE types, which we denominated Dryads.
Our investigation reveals that Dryads occur in most conifer lineages, but are absent in other gymnosperms. Furthermore, Dryads are closely related to a group of EN(+)PLEs that mainly inhabit arthropod genomes. Bioinformatics searches on 1,928 fully sequenced genomes from 14 major eukaryotic lineages showed no occurrence of EN(+)PLEs outside animals and conifers. These results suggest that Dryad elements originated from an EN(+)PLE lineage in arthropods that invaded the genome of a conifers’ ancestor approximately 340 Ma.
Materials and Methods
Specimens and DNA Extraction
Specimen descriptions and their sources are listed in table 1. DNA extraction from needles was performed at the AgriGenomics Laboratory at Texas A&M University using the standard protocol in the DNeasy Plant Mini Kit (Qiagen).
Table 1.
EN(+) PLE Sequences Used in Phylogenetic Analyses Based on DNA Alignments
| Species | Abbreviation (supplementary figure S6, Supplementary Material online) | Common Name | Order |
|---|---|---|---|
| Abies lasiocarpa | Abies | Rocky mountain fir | Pinales |
| Acromyrmex echinatior | Aech | Fungus-growing ant | Hymenoptera |
| Agrilus planipennis | Aplan | Emerald ash borer | Coleoptera |
| Anolis carolinensis | Aca | Green anole | Squamata |
| Anoplophora glabripennis | Agla | Asian long-horned beetle | Coleoptera |
| Blattella germanica | Bger | German cockroach | Blattodea |
| Cephus cinctus | Ceph | Stem sawfly | Hymenoptera |
| Diabrotica undecimpunctata | Diabro | Spotted cucumber beetle | Coleoptera |
| Gerris buenoi | Gbue | Water Strider | Heteroptera |
| Harpegnathos saltator | Harpe | Indian jumping ant | Hymenoptera |
| Juniperus deppeana | Junipe | Alligator juniper | Pinales |
| Ladona fulva | Lful | Scarce Chaser | Odonata |
| Leptinotarsa decemlineata | Ldec | Colorado potato beetle | Coleoptera |
| Loxosceles reclusa | Lrec | Brown recluse spider | Chelicerata |
| Oncopeltus fasciatus | Ofas | Milkweed bug | Hemiptera |
| Onthophagus taurus | Otaur | Bull-headed dung beetle | Coleoptera |
| Petromyzon marinus | Pmar | Marine lamprey | Petromyzontiformes |
| Picea abies | MA | Norway spruce | Pinales |
| Picea sitchensis | Psi | Sitka spruce | Pinales |
| Pinus taeda | Pita | Loblolly pine | Pinales |
| Pinus taeda | Pt | loblolly pine | Pinales |
| Pseudotsuga menziesii | Psme | Douglas fir | Pinales |
| Pseudotsuga menziesii | Pseudo | Douglas fir | Pinales |
| Solenopsis invicta | Sinv | Fire ant | Hymenoptera |
| Taxodium mucronatum | Taxodi | Montezuma cypress | Pinales |
| Thuja occidentalis | Thuja | White cedar | Pinales |
Annotation of Dryads
The 258 Penelope-like families originally annotated in loblolly pine (Neale et al. 2014) were retrieved from the pier-2.0.fa file containing all TE families from this species and deposited on TreeGenes (http://dendrome.ucdavis.edu/ftp/Genome_Data/genome/Repeats/, last accessed April 6, 2016). The fasta header of all these families begins with “>PtRPX” in the pier-2.0.fa file. Annotated Penelope-like families in animals were obtained from Repbase (Jurka et al. 2005) in March 2014 and used for searches with the standalone BLAST+ v2.2.29 (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/, last accessed April 6, 2016) against the 258 putative PLE families from loblolly pine (tBLASTx search, e-value 0.0001). To search for the presence of typical Penelope-like RT and EN domains in Dryads, we first translated the six frames of all Dryad DNA sequences with the six frame translation tool available at the Max-Planck Institute for Developmental Biology website (http://toolkit.tuebingen.mpg.de/sixframe, last accessed April 6, 2016); these protein sequences were then used as queries in searches at the National Center for Biotechnology Information (NCBI) CDD database (http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, last accessed April 6, 2016).
Bioinformatics Identification of PLEs in Pinaceae and Other Organisms
The genomes of loblolly pine and Norway spruce were downloaded from the TreeGenes (Wegrzyn et al. 2008) ftp website (http://dendrome.ucdavis.edu/ftp/Genome_Data/genome/pinerefseq/Pita/transcriptome/, last accessed April 6, 2016) and the Congenie website (ftp://plantgenie.org/ConGenIE/, last accessed April 6, 2016), respectively. To identify and retrieve Dryad elements and EN(−)PLEs from these genomes, we first performed tBLASTn (default settings except e-value = 1e-10) searches using the consensus sequence of 19 loblolly pine Dryad families originally annotated by Wegrzyn et al. (2014) that were evolutionary distant according to the phylogeny shown in supplementary figure S1, Supplementary Material online. These specific Dryad families were selected because they showed the least number of disabling substitutions in their coding region (supplementary file S1, Supplementary Material online). The BLAST results were parsed with Perl scripts to retrieve the DNA sequences of multiple copies used in subsequent analyses (supplementary files S2 and S3, Supplementary Material online).
EN(−)PLE copies were obtained from loblolly pine, Norway spruce and white spruce genomes by searching their assemblies with the Selaginella moellendorffii EN(−)PLE protein sequences Sm1_1p, Sm1_2p, Sm2_1p and Sm2_2p using the BLAST server in TreeGenes (http://dendrome.ucdavis.edu/resources/blast/, last accessed April 6, 2016) with default settings except e-value = 1e-10 and no filtering for low complexity regions. The second open-reading frame (ORF) of both S. moellendorffii elements encodes a putative protein containing the RT domain. DNA sequences of the conifer hits with putative complete EN(−)PLE ORFs were retrieved from the genome assemblies using the BedTools suite (Quinlan 2014).
Novel animal EN(+)PLE elements were obtained from tBLASTn searches using Penelope-like elements annotated in Repbase (Jurka et al. 2005) and in loblolly pine against several databases, including NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed April 6, 2016), EMBL ENA Sequence (http://www.ebi.ac.uk/Tools/sss/ncbiblast/nucleotide.html, last accessed April 6, 2016), insect genomes deposited at the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC; https://www.hgsc.bcm.edu/arthropods/i5k-pilot-project-summary, last accessed April 6, 2016), and the Fourmidable ant genomes database (http://www.antgenomes.org/, last accessed April 6, 2016). These BLAST searches were performed using default settings (including a Blosum62 matrix setting) except for the e-value = 1e-10, number of alignments = 100, and filtering for low complexity regions.
To determine whether EN(+)PLEs distantly related to Dryad families occur in conifers, we searched the genome assemblies of loblolly pine (V1.01), Norway spruce (V1.0), and white spruce (V1.0) with 14 protein sequences from distantly related PLE lineages (supplementary file S4, Supplementary Material online) using the BLAST server in TreeGenes with default settings except e-value = 1e-10 and no filtering for low complexity regions. The protein sequences of the 19 loblolly pine Dryad families with intact or nearly intact coding sequences were also blasted (tBLASTn, e-value = 1e-10, no filtering for low complexity regions, 1,000 target sequences). The BLAST score values of the top 50 hits from each PLE and telomerase protein against were compared with the score values from the 1,000 hits of each Dryad family, in each genome separately. Hits longer than 300 amino acids and showing higher BLAST score with non-Dryad PLE sequences were further investigated by building phylogenies including PLE and telomerase protein sequences (see fig. 2 and supplementary fig. S6A–D, Supplementary Material online). All the resulting trees indicated that these divergent elements belonged either to the Dryad lineage or the EN(−)PLE group (supplementary fig. S2, Supplementary Material online).
Fig. 2.—
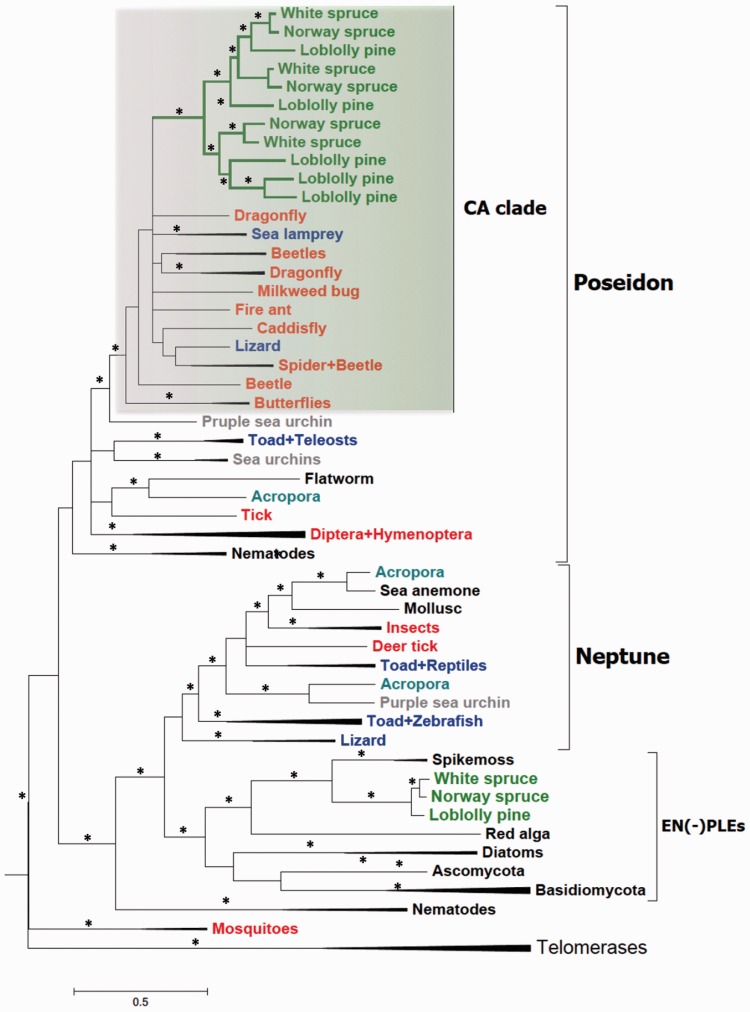
Phylogenetic tree of Dryad, other PLE, and telomerase protein sequences. Bayesian phylogeny of PLE and telomerase RT domains based on the alignment of 266 residues. Major PLE clades are indicated (see also text), including Dryad elements (green branches). Conifer, arthropod and vertebrate PLEs are in green, red and blue, respectively. Other taxa, that is, the coral Acropora and the purple sea urchin, which harbor multiple PLE lineages, are highlighted with specific colors. Asterisks highlight nodes with posterior probabilities ≥0.9. CA, Conifers + Arthropods clade (highlighted by the green box). The expanded version of this tree is shown in supplementary figure S6A, Supplementary Material online.
To assess the distribution of EN(+)PLEs across eukaryotes, tBLASTn searches were performed on both NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed April 6, 2016) and EMBL ENA Sequence (http://www.ebi.ac.uk/Tools/sss/ncbiblast/nucleotide.html, last accessed April 6, 2016) databases using 14 protein sequences (supplementary file S4, Supplementary Material online) and default settings except e-value = 0.001 and no filtering for low complexity regions. Both nr and wgs databases were searched on NCBI. The wgs searches were performed on each eukaryote lineage indicated in figure 5 separately, with prokaryotes (taxid:2), metazoans (taxid:33208), and conifers (taxid:3312) excluded.
Fig. 5.—
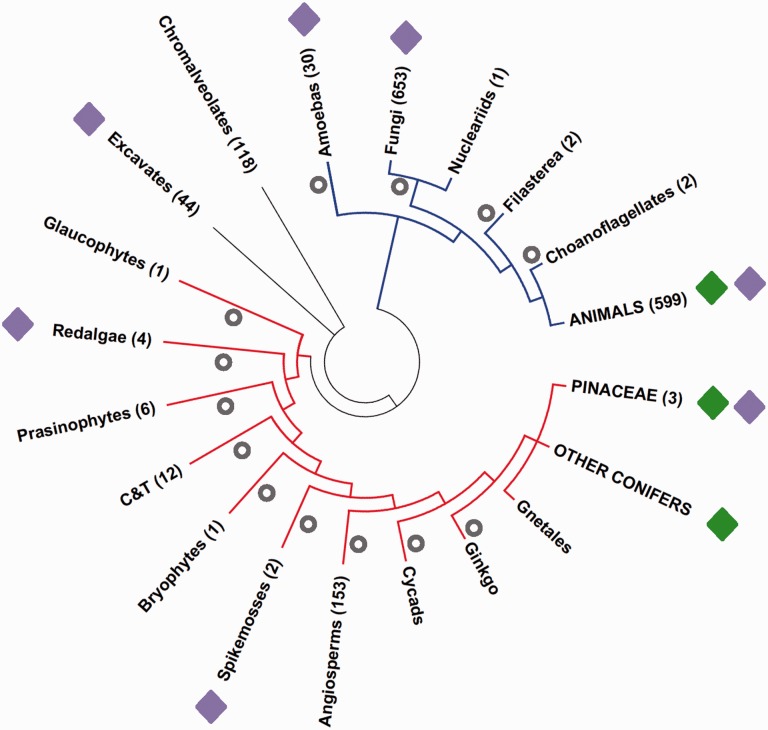
Distribution of PLEs in fully sequenced eukaryote genomes. The number of available genomes for each lineage is shown in parenthesis. Green and purple diamonds indicate lineages with EN(+)PLEs and EN(−)PLEs, respectively. Gray circles show losses of EN(+)PLEs assuming a vertical transmission scenario. Red lineages: Archeaplastids. Blue lineages: Unikonts. C&T groups: Chlorophyceae and Trebouxiophyceae. The tree was based on the eukaryotes phylogeny from the Tree of Life project (http://tolweb.org/Eukaryotes/, last accessed April 6, 2016).
Analyzed sequenced eukaryotic genomes were downloaded from the NCBI website ftp://ftp.ncbi.nlm.nih.gov/genomes/GENOME_REPORTS/eukaryotes.txt, last accessed April 6, 2016 and are listed in the supplementary file S5, Supplementary Material online.
Chromosome Preparation and Fluorescent In Situ Hybridization
Actively growing root tips, about 1.5 cm long, were collected from two pine clones (loblolly pine 20-10-10 and slash pine 8-7) and immediately pretreated in 0.15% colchicines (Sigma, P-9754) for 7.5 h at room temperature in the dark, then fixed in 4:1 (95% ethanol:glacial acetic acid) fixative. The fixed root tips were digested with cell-wall degrading enzyme to prepare pine chromosome spreads (Jewell and Islam-Faridi 1994; Islam-Faridi et al. 2007), with the following enzyme solution formulation specific for pine root tips: 40% (v/v) Cellulase (C2730, Sigma), 20% (v/v) Pectinase (P2611, Sigma), 2% (w/v) Cellulase RS (SERVA Electrophoresis GmbH), 2% (w/v) Macerozyme R10 (Yakult Pharmaceutical, Japan), and 1.5% (w/v) Pectolyase Y23 (Kyowa Chemical, Japan) in 0.01 M citrate buffer (pH 4.8).
Either whole pGmr3 plasmid DNA including 18S–28S Glycine max rDNA insert or PtRPX_125 Dryad family DNA was labeled by nick translation, using either biotin-16-dUTP (Biotin-Nick Translation Mix; Roche, Indianapolis, IN) or digoxigenin-11-dUTP (Dig-Nick Translation Mix; Roche) in accordance with the manufacturer’s instructions. A standard fluorescent in situ hybridization (FISH) technique was used as previously reported (Islam-Faridi et al. 2009; Reddy et al. 2013). FISH preparations were mounted with Vectashield containing DAPI (Vector Laboratories, USA) to prevent photo bleaching of the fluorochromes. Digital images were recorded using an epi-fluorescence microscope (AxioImager M2; Carl Zeiss, Germany) with suitable filter sets (Chroma Technology, USA) and a Cool Cube high performance CCD camera, and processed with ISIS V5.1 (MetaSystem Inc., USA) and Adobe Photoshop CS v8 (Adobe System, USA).
Sequence Alignments, Editing, and Phylogenetic Analyses
Protein sequences of Dryad elements and novel EN(+)PLEs and EN(−)PLEs were obtained by translating their DNA sequences using The Sequence Manipulation Suite (Stothard 2000). Alignments of these protein sequences with full-length PLEs were used to identify possible frameshifts and stop codons and correct them manually. Other EN(+)PLEs and EN(−)PLEs protein sequences were retrieved from their correspondent Repbase entries (Jurka et al. 2005). Repbase entries with frameshifts/stop codons were also reinspected to identify possible errors in the translation. Only proteins with no more than three putative stop codons and frameshifts were used in subsequent alignments and phylogenies. Alignments of protein and DNA sequences were performed with MUSCLE (Edgar 2004), MAFFT (Katoh and Standley 2013), and Clustal Omega (Sievers et al. 2011), without modifying default settings. Protein alignments were edited to remove regions outside the RT domain, or both the RT and GIY-YIG domains, using CLC Sequence Viewer 7 (CLC Bio-Qiagen, Aarhus, Denmark). In addition, alignments with a set of highly conserved protein regions were obtained with Gblocks (Castresana 2000). DNA alignments were edited with SeaView 4 (Gouy et al. 2010).
Protein substitution models were evaluated using ProtTest3 (Darriba et al. 2011). For all protein alignments, LG was the best fitting rate matrix (Le and Gascuel 2008). We built maximum-likelihood (ML) phylogenies using the PhyML software (Guindon et al. 2009) available through the ATGC bioinformatics platform (http://www.atgc-montpellier.fr/phyml/, last accessed April 6, 2016). For each PhyML analysis, 100 bootstrap samplings were performed. The following models were implemented in PhyML for the trees shown in supplementary figure S6, Supplementary Material online: LG + I+G + F (S6B, F, and H); LG + I+G (S6D).
Bayesian trees were built using MrBayes3.2 (Ronquist et al. 2012) available in the Cipres Science Gateway (Miller et al. 2010). Because the Cipres MrBayes version at the time of the analyses did not implement the LG matrix, we set up instead a mixed model, with other parameters estimated according to the models specified above. Phylogenetic trees were visualized and edited with FigTree (http://tree.bio.ed.ac.uk/software/figtree/, last accessed April 6, 2016) and MEGA6.0 (Tamura et al. 2013).
DNA substitution models were evaluated using jModelTest2 (Darriba et al. 2012) implemented in the Cipres Science Gateway (Miller et al. 2010). For phylogenies made with either PhyML or MrBayes, we applied a GTR (general time reversible) + I+F + G model. One hundred bootstrap samplings were performed for PhyML phylogenies. In MrBayes, we run 5,000,000 generations and sampled every 100 trees for each analysis.
Primers Design, PCR Experiments, and PCR Bands Purification
All polymerase chain reaction (PCR) experiments were performed using the Phusion High-Fidelity DNA Polymerase (New England Biolabs). The conserved 353-bp-long region found in 125 Dryads and their closely related animal EN(+)PLEs was used to design the primer pair PLE_353bp_136tx_F1 (ATGGGHTCMCCHYTHTCHCC) and PLE_353bp_136tx_R1 (YTGDBHNGGRWRRTGRTGKG). The following touch-down PCR cycling conditions were used in a total volume of 50 μl: Initial denaturation at 98 °C for 1 min; 6 cycles with 98 °C for 10 s, 72 °C for 30 s, 72 °C for 10 s, with annealing temperature decreasing by 0.5 °C at each cycle; ten cycles with 98 °C for 10 s, 68 °C for 30 s, 72 °C for 10 s, with annealing temperature decreasing by 1 °C at each cycle; 20 cycles at 98 °C for 10 s, 57 °C for 30 s, 72 °C for 10 s; final extension at 72 °C for 5 min. DNA amounts for PCR analyses were normalized across species to 30–40 ng/μl, whenever possible. PCR results were run on 1% agarose gel, using GelRed (Biotium) for staining.
Universal primers for gymnosperms were designed using 28S sequences downloaded from GenBank belonging to 57 species representative of all the major gymnosperm lineages (supplementary table S5, Supplementary Material online). The two primers F2_28S_gymno (CGAACCGGGARSAGCCC) and R1_28S_gymno (GCCTCCRTYCGCTTCCC) amplify a region of approximately 335 bp in the 28S gene of gymnosperms (fig. 5). PCR cycling conditions were the same as described above for the 353-bp region, except for a total PCR volume of 20 μl.
Primers for the Dryad family Pt125, CCL_PLE_Pt125_F1 (CACCCTCAGGGCAATAAGGTG) and CCL_PLE_Pt125_R2 (TGGATGTAAGGCAGGTTAACACCC) were designed on the multialignment of nine PtRPX_125 family copies and used to amplify a region of 1,442 bp. The following touch-down PCR cycling conditions were used in a total volume of 50 μl: Initial denaturation at 98 °C for 1 min; ten cycles with 98 °C for 10 s, 66 °C for 30 s, 72 °C for 45 s, with annealing temperature decreasing by 0.5 °C at each cycle; 25 cycles with 98 °C for 10 s, 61 °C for 30 s, 72 °C for 45 s; final extension at 72 °C for 5 min. PCR results were run on 1% agarose gel, using GelRed (Biotium) for staining.
PCR reactions of the 353-bp-long EN(+)PLE region and the 28S were purified using the QIAquick PCR Purification Kit (Qiagen). PCR reactions of the Pt125 family were eluted from gel and purified using the QIAquick Gel Extraction Kit (Qiagen). DNA sequencing of purified PCR bands was performed using the ABI BigDye Terminator V3.1 reaction kit on an ABI Genetic Analyzer 3130xl.
Estimates of Dryad Copy Numbers
BLAST searches were performed using a cut-off e-value of 10−05. The results were parsed and analyzed using in-house Perl scripts. Hits shorter than 50 bp were removed and overlapping hits were merged to eliminate redundancy. Hits matching multiple Dryad families were assigned to the family with highest BLAST score value.
Identification of Recently Active Dryads and Transcribed Copies
To identify Dryad sequences that potentially inserted recently in the loblolly pine genome we first performed a tBLASTn search against the genome assembly with the protein sequence of 19 Dryad families that show intact or almost intact coding regions in their consensus sequence, setting an e-value threshold of 10−05. We considered a relatively intact coding region and a high similarity to the protein sequence of the consensus of the corresponding family as valid proxy for a recent transposition activity. We were able to identify 250 elements from 12 Dryad families that encode a bona fide full-length PLE protein (supplementary table S1, Supplementary Material online). The divergence from the consensus sequence in these families ranges between 1.9% and 9.7% (table 2).
Table 2.
Dryad Families with Copies Showing Intact Coding Region
| Family Id | Number of Copies | Sequence Conservation |
|---|---|---|
| PtRPX_11 | 20 | 91.98 |
| PtRPX_11_C | 11 | 91.85 |
| PtRPX_47 | 8 | 94.97 |
| PtRPX_64 | 27 | 90.27 |
| PtRPX_4 | 5 | 96.65 |
| PtRPX_42 | 1 | 98.12 |
| PtRPX_59 | 46 | 92.89 |
| PtRPX_62 | 3 | 95.20 |
| PtRPX_61 | 3 | 94.28 |
| PtRPX_46 | 7 | 95.99 |
Putative Dryad transcripts were searched for in expressed sequence tag (EST) databases (http://dendrome.ucdavis.edu/treegenes/transcriptome/transcr_summary.php, last accessed April 6, 2016) and in transcriptome sequences (http://dendrome.ucdavis.edu/ftp/Genome_Data/genome/pinerefseq/Pita/transcriptome/; last access 06/04/2016) of loblolly pine retrieved from TreeGenes. BLASTn searches using the consensus sequence of the 175 Dryad families as queries against ESTs and transcriptome sequences were performed, applying an e-value threshold of 10−10. EST and transcriptome sequences matching at least one Dryad family were inspected for the presence of RT and EN domains in their encoded protein sequences with the NCBI CDD database (http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, last accessed April 6, 2016). Proteins containing both EN(+)PLE domains were considered derived from bona fide Dryad transcripts.
Results
Dryads Form a Diverse Group of PLEs in Loblolly Pine Tree and Other Conifers
A total of 258 families of PLEs were annotated in the initial analysis of the loblolly pine genome (Neale et al. 2014). To better characterize these families, we performed BLAST searches against TE protein sequences deposited in Repbase (Jurka et al. 2005). This analysis revealed that 175 of 258 families have best similarity hits with known EN(+)PLEs and encode at least part of the EN(+)PLE protein (supplementary table S1, Supplementary Material online). These 175 families, or Dryads, have been used in all subsequent analyses. To further confirm the presence of Dryad elements in the loblolly genome, we performed FISH experiments using probes from one of the Dryad families that highlighted the interspersed organization of these retroelements (fig. 1A). Dryads appear as interspersed signals across all 12 pairs of loblolly pine chromosomes, similarly to other previously characterized retroelements (Morse et al. 2009).
Fig. 1.—
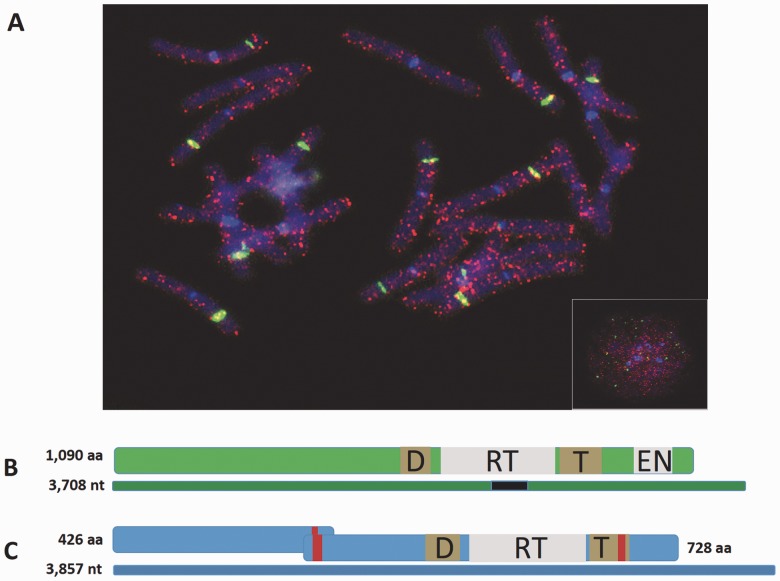
Chromosomal localization of a Dryad family and structure of PLEs in loblolly pine. (A) In situ hybridization showing the localization of PtRPX_125 Dryad copies (red signals) in metaphasic chromosomes of loblolly pine. Inset: Interphase nucleus. Green signals: 18S-28S rDNA; Blue signal: DAPI. (B) Structure of the DNA sequence and the putative protein of the Dryad family PtRPX_46. (C) Structure of the DNA sequence and the putative protein of a full-length EN(−)PLE. Consensus DNA sequences are represented by thin rectangles (green: PtRPX_46; blue: EN(−)PLE). Thick rectangles indicate putative encoded proteins. Gray boxes: RT and EN domains; brown boxes: conserved DKG (D) and Thumb (T) domains; red bars: nuclear localization signals. The black rectangle in the PtRPX_46 consensus sequence indicates the position of the 353-bp-long conserved region.
The phylogeny of 64 representative Dryad families is resolved into two clades with high statistical support (supplementary fig. S1, Supplementary Material online). These Dryad families share between 54% and 97% sequence identity. The high degree of sequence and phylogenetic divergence among Dryads likely reflects an ancient colonization and subsequent diversification of these families in pine trees. In line with this observation, Dryad sequences distantly related to the loblolly pine families were detected in the Norway spruce (Picea abies) genome assembly, and in the Douglas-fir (Pseudotsuga menziesii) transcriptome, pointing to a high diversity of Dryads across conifers (see also below).
The annotation of PLEs is particularly challenging because of the lack of both unambiguous signatures of their insertion, such as target site duplications, and sequence features equivalent to terminal repeats. In fact, the LTRs originally characterized in some PLEs have been later shown to represent artifacts (pseudo-LTRs) due to tandem insertions of two PLE copies, with the upstream copy usually missing most of the 5′-region (Arkhipova 2006; Gladyshev and Arkhipova 2007). Accordingly, we observed some annotation errors in loblolly pine Dryads, which also underscore the complexity of TE annotation in the very large conifer genomes. We built improved consensus sequences for a few Dryad families to better determine the structure and sequence organization of these elements in loblolly pine. Although most full-length Dryad sequences encode a putative protein approximately 650 amino acids long, a few atypical Dryad families possess coding regions extending to the 5′-end and encode an N-terminal region with a nuclear localization signal (fig. 1B). The C-terminus of predicted full-length Dryad proteins contain both the RT domain and the PLE-specific GIY-YIG EN domain (fig. 1B and supplementary fig. S3, Supplementary Material online) (Arkhipova 2006). Conserved amino acid motifs found in RT domains of retroelements and telomerases were also present in Dryad proteins (supplementary fig. S3, Supplementary Material online).
EN(−)PLE Copies Are Also Present in Conifer Genomes
Together with Dryad elements, we identified two full-length and approximately 40 truncated EN(−)PLE copies distributed in the loblolly pine genome assembly. Similarly to the EN(−)PLEs found in the spikemoss S. moellendorffii (Gladyshev and Arkhipova 2007), the most complete loblolly pine EN(−)PLE sequence contains two ORFs (fig. 1C). The 5′-end ORF1 encodes a protein with no similarity to functionally characterized protein domains, whereas the 3′-end ORF2 encodes a protein with a typical PLE RT domain, but no EN domain. Most of these elements are arranged in short tandem arrays and contain one or several telomeric repeats (TTTAGGG)n (supplementary fig. S4, Supplementary Material online), similarly to what was observed in PLEs from other organisms (Gladyshev and Arkhipova 2007). In the Norway spruce genome, we identified 12 full-length and 256 truncated EN(−)PLE elements. The two predicted proteins encoded by Norway spruce EN(−)PLE full-length copies share approximately 50% and approximately 77% identity with the ORF1-protein and ORF2-protein from loblolly pine EN(−)PLEs, respectively (supplementary fig. S5, Supplementary Material online). Telomeric repeats were found in 79/268 Norway spruce EN(−)PLEs. Furthermore, novel EN(−)PLE sequences were identified in database surveys in red algae and Ascomycota, where no PLEs have been previously reported (supplementary table S2, Supplementary Material online).
Dryads Form a Monophyletic Clade with a Group of EN(+)PLEs Present in Arthropods and Vertebrates
To determine the evolutionary relationships between Dryad elements and other PLEs, we built both Bayesian and ML phylogenies based on the amino acid alignments of the RT domain from a total of 97 elements including loblolly pine and Norway spruce Dryad families, animal EN(+)PLEs, EN(−)PLEs and telomerase proteins (supplementary file S6, Supplementary Material online). In Bayesian trees, the two PLE types are separated from telomerases and group together with a high statistical support (fig. 2; supplementary fig. S6A and C, Supplementary Material online). Importantly, Dryad proteins consistently form a monophyletic lineage embedded within a major animal EN(+)PLE group named Poseidon (fig. 2; supplementary fig. S6A and C, Supplementary Material online).
Animal EN(+)PLEs from several arthropods and two vertebrates, the lizard Anolis carolinensis and the lamprey Petromyzon marinus, cluster together with Dryads in a highly supported group that we have named Conifers + Arthropods, or CA, clade (fig. 2; supplementary fig. S6A and C, Supplementary Material online). The two other animal EN(+)PLE groups, Neptune and Nematis, also appear to be monophyletic in these phylogenies, although the former group tends to have relatively low posterior probabilities. On the contrary, EN(−)PLE sequences appear paraphyletic in one Bayesian tree. Nevertheless, conifer EN(−)PLEs group with elements from the spikemoss S. moellendorffii and the red algae Chondrus crispus, which is indicative of a vertical, rather than horizontal, transmission modality in plants and red algae. This scenario implies that EN(−)PLEs have been lost in angiosperms and possibly other green plant lineages. In line with previous observations (Arkhipova 2006; Gladyshev and Arkhipova 2007), we noticed that the relationships among major animal lineages and between them and EN(−)PLEs remain poorly resolved, although all our trees show that PLEs are monophyletic with respect to telomerases.
ML trees share all the key topology features described in Bayesian trees, albeit bootstrap values tend to be relatively low for the CA clade (supplementary fig. S6B and D, Supplementary Material online). However, the inspection of all the trees generated in the bootstrap analyses revealed that this depends either on rearrangements in the topology of branches within the CA clade or on the inclusion within this clade of a closely related sequence from the sea urchin Strongylocentrotus purpuratus. CA clade sequences also share a deletion of approximately ten amino acids in the RT domain that is absent in other animal EN(+)PLEs (supplementary fig. S3, Supplementary Material online).
To further analyze the phylogenetic relationships within EN(+)PLE sequences, we generated protein alignments that include both RT and EN domains using 71 EN(+)PLE and Dryad sequences (supplementary file S7, Supplementary Material online). Bayesian and ML trees built on these data confirmed the clustering of Dryads and several animal EN(+)PLEs in the CA clade, and supported the monophyly of both Poseidon and Neptune groups (supplementary fig. S6E–H, Supplementary Material online). Similarly to the trees based only on the RT domain, the overall topology within the CA clade is unresolved, and no single animal sequence is consistently partnering with the group of Dryads (supplementary fig. S6E–H, Supplementary Material online).
The protein-based phylogenies highlighted several animal phyla harboring multiple EN(+)PLE lineages; for example, the lizard A. carolinensis and the toad Xenopus tropicalis host both Poseidon and Neptune elements (fig. 2 and supplementary fig. S6, Supplementary Material online). To assess whether loblolly pine and Norway spruce genomes may also maintain non-Dryad EN(+)PLEs, we performed tBLASTn searches against the assemblies of these two conifers with ten EN(+)PLE protein sequences belonging to distantly related EN(+)PLE lineages. All retrieved BLAST hits belonged to Dryad families, indicating that no other EN(+)PLE lineages are present in these two conifer genomes (supplementary fig. S2, Supplementary Material online).
DNA-Based Phylogenies Support a Single Origin of Dryads
To further investigate the origin of Dryads and their relationships with arthropods’ EN(+)PLEs, we built new phylogenies using DNA sequences. We reasoned that contrary to protein alignments, DNA alignments would not be affected by the phylogenetic noise introduced when translating TE consensus sequences that typically harbor frameshifts and other disabling substitutions. We also searched for novel animal EN(+)PLEs in an attempt to improve the resolution of several nodes within the CA clade and to identify potential sister EN(+)PLE lineages of Dryads. For this purpose, we surveyed several databases of draft genome sequences, including the i5k data set of insect and other arthropod genomes and the ant genomics database (see Materials and Methods), which enabled us to retrieve previously uncharacterized EN(+)PLEs from several taxa (table 1).
Despite the overall low DNA identity between EN(+)PLEs in the CA clade, we identified a 353bp-long sequence that encodes part of the RT domain and is conserved across this clade (fig. 1B). Alignments of these DNA segments that include multiple PLE copies from each species were used to build new phylogenetic trees of the CA clade. In both Bayesian and ML phylogenies based on 141 DNA sequences, Dryads formed a separate group from animal elements (fig. 3 and supplementary fig. S7, Supplementary Material online), similarly to what observed in protein-based phylogenies.
Fig. 3.—
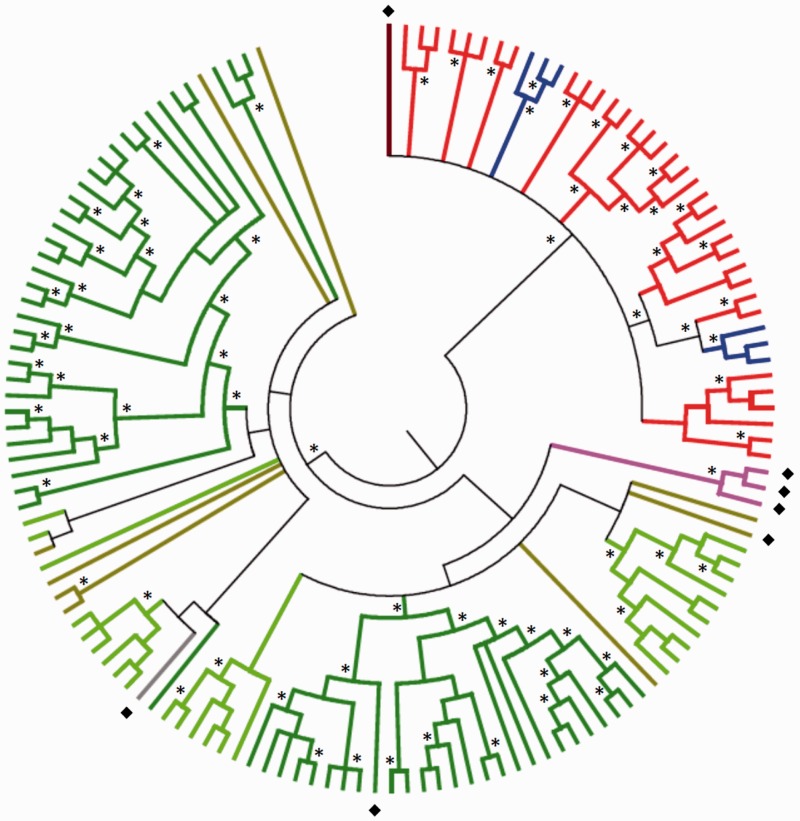
Phylogenetic tree of Dryads and CA clade PLE sequences. Bayesian phylogeny of the conserved EN(+)PLE DNA region in the CA clade. Dark green: loblolly pine. Light green: Norway spruce. Brown: Douglas fir. Gray: Abies lasiocarpa. Pink: Cupressaceae. Light red: arthropods. Light blue: vertebrates. Dark red: Diabrotica undecimpunctata (beetle). Diamonds indicate sequences obtained through PCR amplification. Asterisks highlight nodes with posterior probabilities ≥0.9.
In general, trees based on either DNA or protein alignments failed to resolve the phylogenetic relationships between EN(+)PLEs from different species in the CA clade, possibly because of a complex history of reticulated evolution in this clade.
Distribution of Dryads in Gymnosperms
In order to establish the approximate timing of conifer invasion by EN(+)PLEs, we first screened existing sequence databases to find Dryad elements in conifers other than loblolly pine and Norway spruce. Dryad copies were identified in transcriptomic data obtained from the TreeGenes database (https://dendrome.ucdavis.edu/treegenes/, last accessed April 6, 2016) of Douglas fir and few other Pinaceae (supplementary table S3, Supplementary Material online). Phylogenetic analyses showed that these novel elements group within the Dryad clade, with Douglas-fir sequences dispersed in multiple lineages (fig. 3 and supplementary fig. S7, Supplementary Material online), supporting both Dryads monophyly and the ancient colonization of conifer genomes by these retroelements.
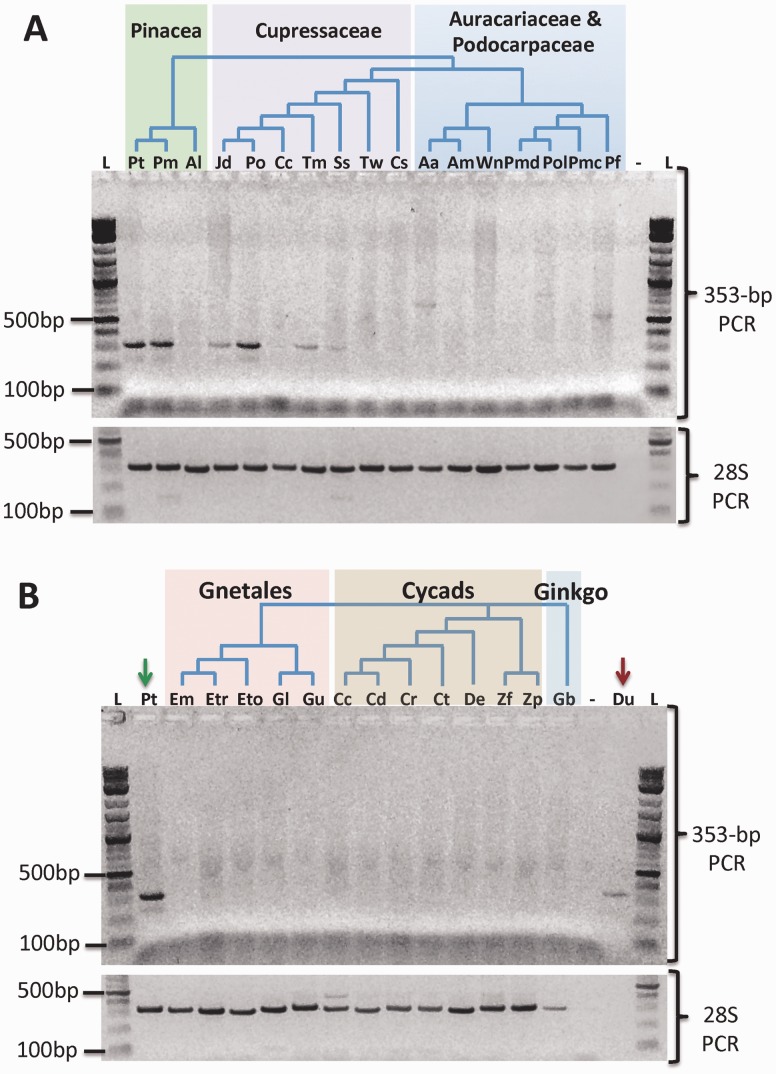
Second, we developed a PCR assay based on primers designed on the conserved 353 bp in elements of the CA clade (see Materials and Methods). The PCR results across a panel of more than 30 gymnosperm species confirmed Dryads presence in Pinaceae and extended their taxonomic distribution to non-Pinaceae conifers (supplementary table S4, Supplementary Material online, and fig. 4). PCR amplicons from several non-Pinaceae species were purified and directly sequenced to generate a Dryad consensus sequence from each analyzed species. The same purification and sequencing procedure was tested on the amplicon obtained from loblolly pine genomic DNA. The sequenced amplicons share a minimum of 68% identity with Dryad elements retrieved from either loblolly pine or Norway spruce assemblies. Moreover, all conifers sequenced amplicons group within Dryad elements retrieved from genome assemblies in phylogenetic trees (fig. 3 and supplementary fig. S7, Supplementary Material online). In these phylogenies, loblolly pine and Douglas-fir amplicon sequences cluster with elements from the same species (fig. 3 and supplementary fig. S7, Supplementary Material online). Taken together, these results suggest that the direct sequencing of Dryad PCR amplicons produced bona fide Dryad consensus sequences from conifer species.
Fig. 4.—
Dryads’ distribution across gymnosperms. PCR amplifications of the conserved Dryads 353-bp fragment. (A) 353-bp PCR (upper panel) and 28S PCR (lower panel) in conifers. The three major conifer groups are highlighted by different colors. (B) 353-bp PCR (upper panel) and 28S PCR (lower panel) in nonconifer gymnosperms, with the three nonconifer groups highlighted by different colors. The green arrow shows the loblolly pine (Pt: Pinus taeda L.) lane, and the red arrow points to the beetle (Du: Diabrotica undecimpunctata) lane. L: 1-kb ladder. Minus symbol: PCR negative control. Species name abbreviations as in supplementary table S4, Supplementary Material online.
Dryad-specific amplicons were not detected in PCRs from six species belonging to the conifer’s families Araucariaceae and Podocarpaceae (fig. 4). Similarly, Dryad bands were not retrieved from DNA samples of the 13 nonconifer gymnosperm species, including seven cycads, five gnetales and the only extant member of the Ginkgoales order, Ginkgo biloba (fig. 4). PCR amplifications using 28S universal gymnosperm primers indicated that all the samples tested contained DNA (fig. 4), and sequencing of 28S bands from several nonconifer species confirmed that the isolated DNA samples corresponded to the expected species or genera. Furthermore, we successfully amplified an approximately 350-bp band from two chrysomelid beetles, and verified through sequencing and subsequent phylogeny reconstruction that the band generated in one of these two PCR reactions represents a bona fide EN(+)PLE consensus that groups with other arthropods’ sequences in the CA clade (fig. 3 and supplementary fig. S7, Supplementary Material online).
EN(+)PLEs Are Absent in Nonmetazoan and Nonconifer Genomes
To determine whether EN(+)PLEs exist in other eukaryotic taxa besides conifers and animals, we performed extensive sequence searches using BLAST on both NCBI and EMBL databases (http://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed April 6, 2016; http://www.ebi.ac.uk/Tools/sss/ncbiblast/, last accessed April 6, 2016). Beside animal and conifer genomes, we retrieved hundreds of putative EN(+)PLE fragments and a few full-length EN(+)PLEs from a variety of taxa (supplementary tables S6–S8 and figs. S8–S11, Supplementary Material online). After careful examination, both through computational analyses and in one case through a PCR test, we conclude that DNA contamination with animal or conifer DNA is the most likely source of these putative EN(+)PLEs (see supplementary material, Supplementary Material online, for details on these sequences’ analysis). This is not unexpected given the occurrence of DNA contamination in many draft genome sequences (Longo et al. 2011; Schmieder and Edwards 2011; Merchant et al. 2014; Orosz 2015).
Although most putative EN(+)PLEs were distantly related from Dryads, several sequences from the genome of the rust fungus Melampsora pinitorqua formed a phylogenetic cluster with the conifer elements (supplementary fig. S10, Supplementary Material online). Given the relevance of these potential Dryad-like elements to our study, we present the analysis of the M. pinitorqua genome in the following paragraphs. We first sought to find out if Dryad-like sequences occurred in other Melampsora genomes. BLAST searches against the M. larici-populina (Duplessis et al. 2011) and the M. lini (Nemri et al. 2014) genome assemblies revealed no significant match with EN(+)PLEs. Subsequent BLAST searches of the three Melampsora genomes with consensus sequences of 61,561 TE families (43,988 from Repbase and 17,573 from the loblolly pine TE annotation) showed an abundance of genome matches with loblolly pine TEs in M. pinitorqua (3,704 matches) compared with M. larici-populina (8 matches) and M. lini (19 matches), whereas matches with Repbase TEs were comparable among the three Melampsora species (519, 563, and 522, respectively). We also searched for homologs of 5,020 loblolly pine high-quality gene models in the three Melampsora assemblies. The highest proportion of matches was again found in M. pinitorqua (75 genes) compared with both M. larici-populina (54 genes) and M. lini (57 genes). Thus, the M. pinitorqua genome appears to include a much higher proportion of pine-like sequences than other Melampsora genomes. This was further supported by k-mer spectrum analyses of M. pinitorqua contigs. We found that contigs with matches to loblolly pine TEs showed the same k-mer distribution of randomly chosen loblolly pine scaffolds, whereas the remaining M. pinitorqua contigs exhibited a different k-mer spectrum (supplementary fig. S11, Supplementary Material online). BLASTn searches showed that only approximately 5% of the 474 pine-like M. pinitorqua contigs shared sequence similarity with the two other Melampsora species genomes, compared with approximately 18% (87/474) randomly chosen M. pinitorqua contigs (supplementary table S9, Supplementary Material online).
We also screened the M. pinitorqua genome with sequences from a Roche 454 genomic DNA library of Pinus sylvestris, a common M. pinitorqua host species (Vialle et al. 2013). Despite the small sample size (270,898 reads), several P. sylvestris 454 sequences showed high similarity with M. pinitorqua contigs. Melampsora pinitorqua contigs matching both P. sylvestris reads and loblolly pine genes/TEs had a much higher identity with the former. These finding can be explained by either a massive horizontal DNA transfer from P. sylvestris or a closely related pine tree to M. pinitorqua, or contamination of the M. pinitorqua genome assembly with pine tree DNA. Because rust fungi are obligate biotrophs that are tightly connected to the host cells through their hyphal tips (Szabo and Bushnell 2001), we argue that in all likelihood the samples used to generate the M. pinitorqua genome assembly were contaminated with pine tree DNA. High levels of DNA contamination in the M. pinitorqua genome were also supported by BLAST searches against the human genome, showing more M. pinitorqua contigs with high similarity to human DNA than in the other Melampsora species (supplementary material and table S10, Supplementary Material online). This suggests that DNA from multiple sources has been incorporated in the M. pinitorqua genome assembly.
Taken together, the analyses of genome sequences retrieved from GenBank and the Glaucophyte genome of Cyanophora paradoxa not deposited in GenBank indicated no evidence of EN(+)PLE copies in a total of 1,928 assemblies from 1,029 nonmetazoan fully sequenced eukaryotic genomes, with the only exception of the three conifer genomes (fig. 5 and supplementary file S5, Supplementary Material online).
Similarly, no EN(+)PLEs were identified in nonmetazoan and nonconifer transcriptomic databases. If EN(+)PLEs were vertically transmitted since the separation of conifers and animals, their current distribution across eukaryotes could only be explained assuming a minimum of 11 independent losses along eukaryotic lineages (fig. 5). Taking into account the PCR data about Dryads distribution across gymnosperms, a total of 13 independent losses would be required to explain the distribution on EN(+)PLEs according to the vertical transmission hypothesis (fig. 5). Molecular analyses have repeatedly associated Gnetophyta to conifers, as a group closely related to Pinaceae (Bowe et al. 2000; Chaw et al. 2000) or non-Pinaceae conifers (Lu et al. 2014). Accordingly, Gnetophyta’s position in figure 5 is shown as uncertain.
Dryads Copy Number and Activity in Loblolly Pine
To obtain a reliable estimate of the minimum copy number of Dryad elements in loblolly pine, we performed a BLASTn search on the 415 genomic scaffolds longer than 1 Mb using the coding region of the 175 loblolly pine Dryad families as queries. We identified 2,394 Dryad copies accounting for 1,347,879 bp in approximately 760 Mb of genomic DNA. Extrapolated to an approximate diploid genome size of 46 Gb, this corresponds to about 145,000 Dryad copies occupying more than 80 Mb of nuclear DNA, or approximately 0.2% of the genome. The range of copies per families varies between 60 and 7,120 (supplementary table S1, Supplementary Material online).
Using BLAST similarity searches against the loblolly genome assembly we were able to identify 250 elements from 12 Dryad families that encode a bona fide full-length PLE protein (supplementary table S1, Supplementary Material online). The divergence from the consensus sequence in these families ranges between 1.9% and 9.7% (table 2). A total of 131 Dryad copies from ten families encode a putative protein with no stop codons, no frameshifts, and at least 90% as long as the consensus protein. The protein sequence conservation for these elements ranged between 90% and 98%. We also identified several transcripts and nine ESTs matching Dryad elements in loblolly pine. Two of these transcripts encoded for proteins containing both RT and GIY-YIG domains. These findings suggest that some Dryad families may currently be active in the loblolly genome, although experimental evidence will be required to confirm the potential activity of Dryad elements.
Discussion
The horizontal transfer of TEs, or HTT, is a widespread phenomenon in plants, animals, fungi, and protists (Diao et al. 2006; Novikova et al. 2009, 2010; Thomas et al. 2010; Walsh et al. 2013; El Baidouri et al. 2014; Parisot et al. 2014). Most documented HTT events occurred in the past few million years. The paucity of known ancient HTTs is likely due to the limited taxonomic distribution of available genome sequences, the decay of TE sequences over short evolutionary periods in many eukaryotes, or a combination of both aspects. The same factors probably determine the deficiency of reported transkingdom HTTs. The few known transkingdom HTT events involve the Tcn1 family of gypsy-like retroelements that has been independently transferred from fungi to spikemosses and to bryophytes (Novikova et al. 2010), a related gypsy-like lineage transmitted from fungi to vertebrates (Gorinsek et al. 2004; Llorens et al. 2009), and the invasion of microsporidians, a group of intracellular parasites, with multiple metazoan TEs (Parisot et al. 2014).
We present in this study a novel transkingdom HTT event involving PLEs of the EN(+) type that we suggest were transferred from arthropods to a common ancestor of modern conifers, which separated from other gymnosperms approximately 340 Ma (Leslie et al. 2012). This represents the first documented HTT from animals to plants. Several lines of evidence support the ancient origin of conifer EN(+)PLEs, or Dryads, through HT, as opposed to a vertical transmission scenario. First, all Dryad elements retrieved from the loblolly pine and Norway spruce genomes, as well as other conifer sequences, form a monophyletic group. Most animal taxa host two or more distantly related EN(+)PLE lineages, highlighting both the ancestry of PLEs among metazoans and the deep evolutionary history of these lineages. The fact that Dryads cluster together in a single group, despite their high copy number and family diversification, indicates a more recent evolutionary history than animal EN(+)PLEs. This is also in agreement with our discovery that Dryads appear to be absent in nonconifer gymnosperms (fig. 4).
Second, in all phylogenies generated in this study, Dryads are embedded within the Poseidon lineage, and cluster with many arthropod and two vertebrate EN(+)PLEs in the CA clade, as expected in the HTT scenario (fig. 2 and supplementary fig. S6, Supplementary Material online). In the alternative hypothesis of vertical transmission, EN(+)PLEs would have been present in the common ancestor of conifers and animals, and Dryads should form a sister lineage of all animal EN(+)PLEs in phylogenetic trees of these retroelements.
Third, no EN(+)PLEs have been identified in 1,925 genomes from 1,026 eukaryotes that do not include animal or conifer assemblies. As explained in detail in the supplementary material, Supplementary Material online, extensive computational and experimental analyses demonstrated that putative EN(+)PLEs retrieved in some of these genomes originated from contamination of these assemblies with insect or conifer DNA. Indeed, the vast majority of these putative EN(+)PLEs are found in very short contigs (often only one contig per species), are absent in closely related species, and/or occur in genome assemblies that harbor other sequences derived from DNA contamination, including widespread contamination with human DNA in some cases (supplementary material, Supplementary Material online). For instance, we observed hundreds pine-like contigs, including Dryad-like sequences, in the rust fungus Melampsora pinitorqua, which infest pine trees and other conifers. Given the obligate biotroph lifestyle of this parasite, either a lateral transfer of DNA from a host species or DNA contamination of the genome assembly with pine sequences is plausible explanation for the occurrence of pine-like sequences in M. pinitorqua. However, our analyses indicate that pine DNA has been incorporated in the sequenced sample, together with some human DNA (supplementary material and fig. S11, Supplementary Material online). These findings are in line with previous observations suggesting widespread DNA contamination in both prokaryote and eukaryote genomes (Longo et al. 2011; Schmieder and Edwards 2011; Merchant et al. 2014; Orosz 2015).
Given the very limited taxonomic distribution of EN(+)PLEs and the phylogenetic relationships between Dryads and other PLEs, a vertical transmission of Dryads from a common ancestor of animals and plants could only be explained by a minimum of 13 independent losses during eukaryotes evolution, rather than the single event required by the HT hypothesis (fig. 5). A scenario of vertical transmission and repeated loss in eukaryotes is instead compatible with the distribution of EN(−)PLEs, which occur in 7/16 major eukaryotic groups with available genome sequences (fig. 5).
A high sequence similarity between TEs found in distantly related species is often used as an independent evidence supporting HTT (Schaack et al. 2010). This criterion is particularly useful in HTTs that occurred in the past few million years, wherein TEs found in donor and recipient species tend to share a higher sequence similarity than the vast majority of orthologous genes. Such criterion is obviously of little use in ancient HTT events, and could not be applied in our analysis of Dryads and animal EN(+)PLEs, which share less than 70% sequence identity even in the conserved region found in the CA clade elements.
Both protein and DNA phylogenies support a CA clade formed by Dryads and EN(+)PLEs found in arthropods and vertebrates (figs. 2 and 3; supplementary figs. S6 and S7, Supplementary Material online). Given that the two vertebrate EN(+)PLEs in this clade are paraphyletic and likely originated from HTT events involving arthropod donor species, the most plausible source of Dryads is an unknown arthropod group. A variety of phytophagous insects are known to feed on tissues of modern conifers, including both female and male cones, and presumably species of many insect orders have been exploiting conifers since their origin (Turgeon 1994). This proximity might have facilitated the transfer of EN(+)PLEs to conifers, directly or through bacteria, fungi, and other vectors harbored in these insects. A broader taxonomic sampling of arthropod genomes may eventually lead to the discovery of a sister EN(+)PLEs lineage of Dryads. However, the host of a possible Dryad sister lineage would not necessarily belong to the taxon that transferred EN(+)PLEs to conifers, given that EN(+)PLEs have likely experienced many HTT and loss events in arthropods since the origin of Dryads.
It could be argued that the arthropod-to-conifer scenario of Dryads origin is somewhat favored by the skewed taxonomic sampling of metazoan-sequenced genomes. Indeed, arthropods represent approximately 37% of sequenced genomes in the NCBI wgs database (219/599 entries as of March 2016). Nevertheless, we think that this is unlikely for two reasons. First, a high number of genomes are also available for vertebrates (265) and nematodes (57), but none of these genomes harbor PLEs closely related to Dryads except two vertebrates that appear to have received these elements through HT from arthropods (see Results section). This is especially remarkable given the prominent ecological interactions between nematodes and conifers and the availability of at least one genome from a nematode pest of pine trees (Kikuchi et al. 2011); this species harbors no EN(+)PLE with high sequence similarity with Dryads (data not shown). Second, in spite of the taxonomic bias in sequenced genomes across metazoans, multiple EN(+)PLE lineages have been described in most sequenced animal phyla, often within the same species (supplementary table S2 and fig. S6, Supplementary Material online). Nevertheless, only some arthropods and the two newly invaded vertebrate species harbor PLEs belonging to the CA clade.
The exceptional transfer of EN(+)PLEs to conifers might either constitute a rare accident or the consequence of a more tolerant genomic environment in these gymnosperms toward TEs. Interestingly, a conserved defense mechanism against TE activity appears to be less effective in conifers than other plants (Dolgosheina et al. 2008; Nystedt et al. 2013). Such deficiency could facilitate the survival of horizontally transferred TEs in these gymnosperms. Indeed, our study indicates that Dryad elements have diversified into a variety of families in conifer genomes and reached a high copy number in loblolly pine and potentially other species (supplementary table S1 and fig. S1, Supplementary Material online). Some Dryad families appear to be still active (table 2), suggesting that these Penelope-like retroelements have survived in conifers for more than 300 Myr. Thus, the invasion and amplification of Dryads have had a significant long-term impact on the evolution of conifer genomes. Preliminary phylogenetic analyses involving all the approximately 17,000 annotated loblolly pine TE families revealed several other potential HTT events (data not shown). Further investigations will be necessary to determine whether Dryads and other horizontally transferred TEs have played an important role in the genome expansion observed in conifers.
Although novel genome sequences and broader TE surveys may facilitate the discovery of further transkingdom HTTs, the paucity of such events in the literature could underlie some intrinsic limitations of TE sequences to proliferate in the genome of species distantly related from their current hosts. TEs employed as functional genomic tools in a broad array of species may provide some experimental evidence in support or against this hypothesis. Some of these studies, which have been mostly carried out with DNA transposons obtained from vertebrates, insects, nematodes, and a few angiosperms, show that TEs can effectively mobilize in genomes of species evolutionarily distant from their native hosts (Osborne and Baker 1995). In a few cases, transkingdom transposition has been achieved (Gueiros-Filho and Beverley 1997; Emelyanov et al. 2006). Although these results suggest that TEs may be capable of transposition in most species following HTT, their chances of survival might be especially low in the long-term after jumping across eukaryotic kingdoms. Future systematic surveys of the distribution of TE groups and the analysis of their phylogenetic relationships in eukaryotes will be needed to determine whether HTT events are indeed extremely rare, have been largely overlooked, or require genomic data from a broader collection of taxa in order to be discovered.
Supplementary Material
Supplementary material, files S1–S7, figures S1–S11, and tables S1–S10 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to the following institutions and individuals for providing specimens for DNA samples: The Lovett Pinetum Charitable Foundation, the Missouri Botanical Garden, St. Louis MO, the Texas A&M University S.M. Tracy Herbarium, Michael Arnold, James Carcano, Sofia Casola, Amanda Hulse-Kemp, Jeremy Johnson, Matthew Kent, Dale Kruse, Jesse Meik, Michelle Lawing, Carol Loopstra, Jacob Owen, David Stelly, and James Solomon. They thank Jeffrey Puryear, Mengmeng Lu, and Fei Wang for technical assistance. They are especially grateful to Michelle Lawing, Cedric Feschotte, Ellen Pritham, Tomasz Koralewski, and two anonymous reviewers for comments on the manuscript. This material is based upon work that was supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number TEX0-1-9599, the Texas A&M AgriLife Research, and the Texas A&M Forest Service.
Literature Cited
- Andersson JO. 2005. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 62:1182–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova IR. 2006. Distribution and phylogeny of Penelope-like elements in eukaryotes. Syst Biol. 55:875–885. [DOI] [PubMed] [Google Scholar]
- Arkhipova IR, Pyatkov KI, Meselson M, Evgen’ev MB. 2003. Retroelements containing introns in diverse invertebrate taxa. Nat Genet. 33:123–124. [DOI] [PubMed] [Google Scholar]
- Bowe LM, Coat G, dePamphilis CW. 2000. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales’ closest relatives are conifers. Proc Natl Acad Sci U S A. 97:4092–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Lawing AM, Betran E, Feschotte C. 2007. PIF-like transposons are common in drosophila and have been repeatedly domesticated to generate new host genes. Mol Biol Evol. 24:1872–1888. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Cervera A, De la Pena M. 2014. Eukaryotic penelope-like retroelements encode hammerhead ribozyme motifs. Mol Biol Evol. 31:2941–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, Parkinson CL, Cheng Y, Vincent TM, Palmer JD. 2000. Seed plant phylogeny inferred from all three plant genomes: monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc Natl Acad Sci U S A. 97:4086–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, et al. 2014. Insights into conifer giga-genomes. Plant Physiol. 166:1724–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. 2002. Mammalian retroelements. Genome Res. 12:1455–1465. [DOI] [PubMed] [Google Scholar]
- Diao X, Freeling M, Lisch D. 2006. Horizontal transfer of a plant transposon. PLoS Biol. 4:e5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgosheina EV, et al. 2008. Conifers have a unique small RNA silencing signature. RNA 14:1508–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis S, et al. 2011. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A. 108:9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Jamburuthugoda VK. 2008. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M, et al. 2014. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 24:831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A, Gao Y, Naqvi NI, Parinov S. 2006. Trans-kingdom transposition of the maize dissociation element. Genetics 174:1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgen’ev M, Zelentsova H, Mnjoian L, Poluectova H, Kidwell MG. 2000. Invasion of Drosophila virilis by the Penelope transposable element. Chromosoma 109:350–357. [DOI] [PubMed] [Google Scholar]
- Evgen’ev MB, et al. 1997. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci U S A. 94:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune PM, Roulin A, Panaud O. 2008. Horizontal transfer of transposable elements in plants. Commun Integr Biol. 1:74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Hernandez SS, Flores-Benabib J, Smith EN, Feschotte C. 2012. Rampant horizontal transfer of SPIN transposons in squamate reptiles. Mol Biol Evol. 29:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev EA, Arkhipova IR. 2007. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A. 104:9352–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev EA, Arkhipova IR. 2011. A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci U S A. 108:20311–20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorinsek B, Gubensek F, Kordis D. 2004. Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol. 21:781–798. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Beverley SM. 1997. Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276:1716–1719. [DOI] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 537:113–137. [DOI] [PubMed] [Google Scholar]
- Islam-Faridi MN, Nelson CD, DiFazio SP, Gunter LE, Tuskan GA. 2009. Cytogenetic analysis of Populus trichocarpa–ribosomal DNA, telomere repeat sequence, and marker-selected BACs. Cytogenet Genome Res. 125:74–80. [DOI] [PubMed] [Google Scholar]
- Islam-Faridi MN, Nelson CD, Kubisiak TL. 2007. Reference karyotype and cytomolecular map for loblolly pine (Pinus taeda L.). Genome 50:241–251. [DOI] [PubMed] [Google Scholar]
- Jewell DC, Islam-Faridi MN. 1994. A technique for somatic chromosome preparation and C-banding of maize In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; p. 484–493. [Google Scholar]
- Jurka J, et al. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 9:605–618. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, et al. 2011. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7:e1002219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordis D, Gubenek F. 1995. Horizontal SINE transfer between vertebrate classes. Nat Genet. 10:131–132. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Leslie AB, et al. 2012. Hemisphere-scale differences in conifer evolutionary dynamics. Proc Natl Acad Sci U S A. 109:16217–16221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens C, Munoz-Pomer A, Bernad L, Botella H, Moya A. 2009. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol Direct. 4:41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo MS, O’Neill MJ, O’Neill RJ. 2011. Abundant human DNA contamination identified in non-primate genome databases. PLoS One 6:e16410.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ran JH, Guo DM, Yang ZY, Wang XQ. 2014. Phylogeny and divergence times of gymnosperms inferred from single-copy nuclear genes. PLoS One 9:e107679.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, et al. 2010. Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464:1033–1038. [DOI] [PubMed] [Google Scholar]
- Merchant S, Wood DE, Salzberg SL. 2014. Unexpected cross-species contamination in genome sequencing projects. PeerJ 2:e675.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA; p. 1–8. [Google Scholar]
- Morales-Hojas R, Vieira CP, Vieira J. 2006. The evolutionary history of the transposable element Penelope in the Drosophila virilis group of species. J Mol Evol. 63:262–273. [DOI] [PubMed] [Google Scholar]
- Morse AM, et al. 2009. Evolution of genome size and complexity in Pinus. PLoS One 4:e4332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, et al. 2014. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15:R59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemri A, et al. 2014. The genome sequence and effector complement of the flax rust pathogen Melampsora lini. Front Plant Sci. 5:98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova O, Fet V, Blinov A. 2009. Non-LTR retrotransposons in fungi. Funct Integr Genomics. 9:27–42. [DOI] [PubMed] [Google Scholar]
- Novikova O, Smyshlyaev G, Blinov A. 2010. Evolutionary genomics revealed interkingdom distribution of Tcn1-like chromodomain-containing Gypsy LTR retrotransposons among fungi and plants. BMC Genomics 11:231.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. [DOI] [PubMed] [Google Scholar]
- Orosz F. 2015. Two recently sequenced vertebrate genomes are contaminated with apicomplexan species of the Sarcocystidae family. Int J Parasitol. 45:871–878. [DOI] [PubMed] [Google Scholar]
- Osborne BI, Baker B. 1995. Movers and shakers: maize transposons as tools for analyzing other plant genomes. Curr Opin Cell Biol. 7:406–413. [DOI] [PubMed] [Google Scholar]
- Parisot N, et al. 2014. Microsporidian genomes harbor a diverse array of transposable elements that demonstrate an ancestry of horizontal exchange with metazoans. Genome Biol Evol. 6:2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 47:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy UK, et al. 2013. Cytomolecular characterization of rDNA distribution in various Citrullus species using fluorescent in situ hybridization. Genet Resur Crop Evol. 60:2091–2100. [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. 2010. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 25:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One 6:e17288.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AG, Roger AJ. 2004. The real “kingdoms” of eukaryotes. Curr Biol. 14:R693–R696. [DOI] [PubMed] [Google Scholar]
- Sormacheva I, et al. 2012. Vertical evolution and horizontal transfer of CR1 non-LTR retrotransposons and Tc1/mariner DNA transposons in Lepidoptera species. Mol Biol Evol. 29:3685–3702. [DOI] [PubMed] [Google Scholar]
- Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. [DOI] [PubMed] [Google Scholar]
- Sun C, et al. 2012. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol Evol. 4:168–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo LJ, Bushnell WR. 2001. Hidden robbers: the role of fungal haustoria in parasitism of plants. Proc Natl Acad Sci U S A. 98:7654–7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Schaack S, Pritham EJ. 2010. Pervasive horizontal transfer of rolling-circle transposons among animals. Genome Biol Evol. 2:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon JJ. 1994. Insect fauna of coniferous seed cones: diversity, host plant interactions, and management. Annu Rev Entomol. 39:179–212. [Google Scholar]
- Vialle A, Feau N, Frey P, Bernier L, Hamelin RC. 2013. Phylogenetic species recognition reveals host-specific lineages among poplar rust fungi. Mol Phylogenet Evol. 66:628–644. [DOI] [PubMed] [Google Scholar]
- Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL. 2013. Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci U S A. 110:1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn JL, et al. 2013. Insights into the loblolly pine genome: characterization of BAC and fosmid sequences. PLoS One 8:e72439.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn JL, Lee JM, Tearse BR, Neale DB. 2008. TreeGenes: a forest tree genome database. Int J Plant Genomics. 2008:412875.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8:973–982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.