SUMMARY
We have previously demonstrated that lagging-strand synthesis in budding yeast is coupled with chromatin assembly on newly synthesized DNA. Using a strain of S. cerevisiae in which DNA ligase I can be conditionally depleted, we can enrich and purify Okazaki fragments. We delineate a method to extract, end-label and visualize Okazaki fragments using denaturing agarose gel electrophoresis. Furthermore, we describe an ion-exchange chromatographic method for purification of fragments and preparation of strand-specific sequencing libraries. Deep sequencing of Okazaki fragments generates a comprehensive, genomic map of DNA synthesis, starting from a single asynchronous culture. Altogether this approach represents a tractable system to investigate key aspects of DNA replication and chromatin assembly.
Keywords: Okazaki fragments, DNA replication, lagging strand, budding yeast, chromatin
1. INTRODUCTION
Eukaryotic DNA replication has been traditionally studied using synchronization of cells followed by measurements of S-phase dependent incorporation of modified nucleotides, increases in copy number, single-stranded DNA or binding of specific replisome proteins. However, conventional methods lack information about dynamics of individual replication forks and provide little insight into nucleosome organization during replication.
DNA replication at the replisome is inherently asymmetric with the leading strand synthesized in advance of the lagging strand. Okazaki fragment synthesis on the lagging strand necessitates the repeated production of single stranded DNA and polymerization in the opposite direction to fork progression. Each Okazaki fragment is initiated by polymerase α-primase (Pol α-primase), which creates a short, ~35-50nt, mixed RNA-DNA primer(1). After priming, elongation is stimulated by RFC (the clamp loader), which loads PCNA onto the 3’ primer end and exchanges Pol α-primase for the processive polymerase δ (Pol δ)(2). Pol δ extends the 3’ end of the fragment until it meets the 5’ end of the preceding Okazaki fragment. Part of the RNA primer is likely removed by the action of RNase H, yet Pol δ also extends through the 5’ portion of the preceding fragment, displacing a single stranded flap(3). Strand displacement by Pol δ is stimulated by the structure specific Flap Endonuclease 1 (Fen1) nuclease, which cleaves the flap and generates a nick in the duplex DNA, which is ultimately sealed by the replicative ligase Lig1 (Cdc9)(3-5).
Using a doxycycline-repressible CDC9 allele, we have recently shown that unligated Okazaki fragments can be massively enriched (6); we have developed two assays to characterize them: The first is a low-resolution agarose gel based approach that allows us to visualize general properties of the population (6). The second is a single-nucleotide resolution, deep sequencing approach that allows precise characterization of millions of individual DNA molecules (6; 7). Our published data shows that nucleosome assembly occurs very rapidly on nascent DNA and that polymerization by Pol δ is often impeded by nucleosomes. Consequently, Okazaki fragment ends correlate with nucleosome positions and have a periodicity reminiscent of the nucleosome repeat (6). In addition, global analysis of Okazaki fragments purified from asynchronous cultures reveals key properties that underlie genome-wide DNA replication (7).
The analysis of Okazaki fragments offers a powerful approach to study not only DNA replication but also chromatin assembly pathways.
2. MATERIALS
Prepare all solutions using ultrapure water and analytical grade reagents. Prepare and store all reagents at room temperature unless otherwise indicated. Follow all waste disposal regulations.
Genomic DNA prep
YPD (1% yeast extract, 2% peptone: autoclave and add filter-sterilized 40% glucose to a final concentration of 2%)
100mg/ml Doxycycline hydrochloride stock solution in DMSO (if using Dox-repressible CDC9 construct) (Fisher Scientific, Cat #BP2653-5)
Shaking incubator
250ml Erlenmeyer flasks
SCE: 1M sorbitol, 100mM sodium citrate, 60mM EDTA. pH to 7.0, filter-sterilize and store at 4°C
Zymolyase 20T (Sunrise Science Products, N0766391)
Lysis buffer: 50mM Tris-HCl pH8.0, 50mM EDTA, 100mM NaCl, 1.5% sarkosyl
Proteinase K (Fisher Scientific, cat# BP1700100)
Isopropanol
3M Sodium acetate, pH 5.2
Absolute ethanol
STE: 100mM NaCl, 10mM Tris-HCl pH8.0, 1mM EDTA
Riboshredder RNase blend (Epicentre, cat #RS12500) or RNase A (20mg/ml stock solution)
TE: 10mM Tris-HCl pH8.0, 1mM EDTA
(Optional) Spin-X centrifuge tubes with 0.45um cellulose acetate filter (Costar, cat#8162)
Agarose, 0.5x TAE/0.5x TBE running buffer, and GelRed or ethidium bromide (Invitrogen, Cat#15585-011)
Okazaki fragment labeling and detection
Klenow exo- DNA polymerase (NEB, cat#M0212L)
α-[32P]dCTP 6000Ci/mmol (Perkin Elmer Health Sciences, cat#BLU013Z500UC)
(Optional) illustra™ G-50 microspin columns (GE Healthcare Biosciences, cat #27-5330-02)
2-log ladder (NEB cat#N3200L) end-labeled with and γ-[32P]ATP (Perkin Elmer Health Sciences cat#BLU002H250UC) and T4 polynucleotide kinase (NEB cat #M0201L)
Large-format agarose gel running apparatus (20×25cm)
Agarose (Seakem LE or Invitrogen ultra-pure)
10X denaturing agarose running buffer: 500mM NaOH, 10mM EDTA
6X denaturing agarose loading buffer: 20% Ficoll, 300mM NaOH, 6mM EDTA
10mg/ml bromophenol blue stock solution
Large plastic container for capillary transfer
Plastic wrap (e.g. Saran wrap)
400 mM NaOH or 10X SSC for transfer (20X SSC: 3M NaCl, 300mM Sodium citrate dehydrate)
EA Zeta-probe membrane (BioRad 1620159) or Genescreen™ Hybridization transfer membrane (Perkin Elmer, cat# NEF1018001PK)
Hyblot 3A gel paper 20 X 20cm or 6MW Gel Blot Paper 20×20cm 0.83mm thick (Denvile B6003-20 or cat# 6MW-2020)
Paper towels
Generating sequencing libraries
Source 15Q (GE Healthcare Biosciences, cat# 17-0947-20)
Spin-X centrifuge tubes with 0.45um cellulose acetate filter (Costar cat#8162)
NaCl solutions from 300-1000 mM in 100 mM increments, pH to 12.0 with NaOH
3M NaOAc, pH 5.2
Riboshredder RNase blend (Epicentre, cat #RS12500) or RNase A (20mg/ml stock solution)
(Optional) Glycogen (MP Biomedicals cat# 11GLYCO001)
Sequencing adaptor oligonucleotides with polar random overhangs. See note 10 for adaptor sequences
Reagents for native PAGE purification of adaptor oligonucleotides if not ordered as purified duplexes
DNA ligase (Enzymatics cat# L6030-HC-L)
S-300 microspin columns (GE Healthcare Biosciences, cat# 27-5130-01)
Taq DNA polymerase 2x master mix (NEB cat# M0270L)
Agarose, 0.5x TAE/0.5x TBE running buffer and GelRed
Razor blade or surgical knife
Qiaquick PCR purification kit (Qiagen) or equivalent
Qiaquick gel purification kit (Qiagen) or equivalent
AMPure (Beckman, cat # A63880) or equivalent MagNA beads (see (8), for instructions)
PCR reagents for library PCR (KOD or Q5) – see manufacturer’s instructions for the sequencing method being used.
3. METHODS
Carry out all procedures at room temperature unless otherwise specified.
3.1 Preparing Genomic DNA
Grow cells with doxycycline-repressible/degron-tagged CDC9 (or other repressible/temperature-sensitive CDC9 allele – see note 1) to an O.D of 0.4.
At O.D. 0.4 add freshly made doxycycline solution in water to 40ug/ml. Shake at 30° for 2.5h (see note 1).
Warm SCE buffer to room temperature: assume that you will use 5ml SCE per 50ml culture to be processed. All volumes are for 50 ml cultures: see note 2.
After 2.5hr, collect cells by centrifugation at 3000xg for 5 minutes in a 50ml tube containing 1/10 volume of 0.5M EDTA.
While cells are spinning, add 0.7ul 2-mercaptoethanol per ml to the room-temperature SCE buffer. Also weigh out 5mg zymolyase 20T per 50ml. Resuspend each 5mg zymolyase in 100ul SCE.
Wash cell pellet with 1ml SCE, transferring cells to a 2ml eppendorf tube in the process. Collect cells via a short spin in a microcentrifuge.
Wash cell pellet with another 1ml SCE.
Resuspend pellet in 900ul SCE (pipette up and down to resuspend) then add the 100ul (5mg) zymolyase solution. Invert a few times to mix and incubate at room temperature for 2-3 minutes.
Collect spheroplasts and wash twice with 1ml SCE, each time via a short spin in a microcentrifuge.
Resuspend spheroplast pellet in 480ul lysis buffer by gently pipetting up and down: pellet is often clumpy.
Add 20ul 10mg/ml proteinase K stock solution and mix by inverting a few times.
Incubate at 42°C for 2hr. Occasionally take the tubes out and mix gently by flicking the side of the tube. If any visible cell debris are left after 1.5hr, add 5ul more proteinase K stock solution and incubate at 42° for a further hour.
Add 200ul 5M KOAc to each tube. Mix by inverting a few times and cool on ice for 5 min. Spin at 4° for 20min at 16,000xg.
Collect supernatant (~700ul) to a fresh 2ml tube. Add 0.5ml isopropanol, mix by inverting a few times and spin at 4° for 10 min at 16,000g. You should obtain a large pellet and perhaps some precipitate on the side of the tube.
Wash pellet with 0.5ml 70% ethanol. Leave pellets to dry at room temperature for a few minutes. Do not over-dry the pellets as they will become extremely difficult to resuspend.
Add 250ul STE, making sure to wash it down the side of the tube that has precipitate on it. Mix gently by flicking the tube but do not try to actively resuspend the pellet (especially by pipetting up and down), as this will shear the DNA.
Add 1U Riboshredder or 1ul of 20mg/ml RNAse A and mix gently again. Spin for a few seconds at 5,000xg to bring all liquid to the bottom of the tube, and incubate at 37°C for 1 hour. As an alternative, the RNase digestion can be carried out overnight at 4°C.
Add 30ul 3M NaOAc (pH 5.2) and 900ul absolute ethanol and invert a few times to mix. A large white bundle should immediately form unless the starting volume of cells was very small (<10ml). Spin at room temperature for 10min at 5,000xg. Wash pellet twice with 70% ethanol and dry. Again, do not over-dry the pellet
Resuspend in 1ul TE per ml of original culture. Leave DNA to dissolve at room temperature for 30-60 minutes, or for several hours/overnight at 4°C.
Spin tubes at 16,000xg for 5 min. Transfer supernatant to a fresh 1.5ml tube, avoiding the gelatinous pellet as far as possible. For original culture volumes >50ml, pool the DNA from separate preps and remove the gelatinous pellet by filtration (Costar Spin-X tube).
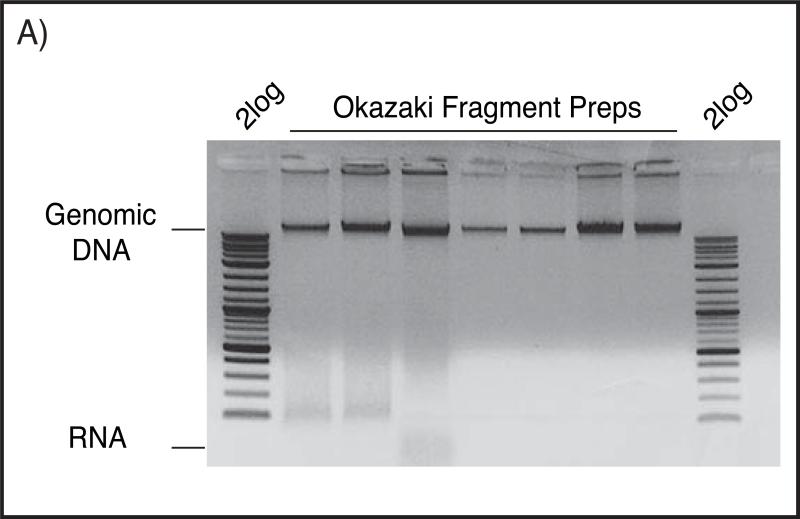
Run a small aliquot on a 0.7% agarose gel to assay concentration/quality. Quantitate using the high-Mw DNA band that migrates at about 10-20kb (Fig. A). Because RNase digestion does not go to completion, spectrophotometric quantitation of DNA is often highly inaccurate.
Figure (A).
Okazaki fragment preparations resolved on 0.7% native agarose gel in 0.5X TBE buffer, stained with Ethidium bromide. Large genomic DNA can be seen above the corresponding 10kb band of 2log ladder, while contaminating RNA typically runs below 100bp as a smear.
Purified DNA can be kept at 4°C for several days (See note 3) or frozen for long-term storage. Avoid repeated freeze-thaw cycles.
3.2 Okazaki fragment detection
Prepare a 1.3% denaturing agarose gel in advance (we use large-format 25×20cm gels): Agarose should be melted in water and allowed to cool to ~60°C before buffer is added.
To 1.5 ml tubes add (in order or as a master-mix to genomic DNA)
Water for a final volume of 30ul
2-4ul Genomic DNA prep
3ul NEBuffer2
3ul α-[32P]dCTP
1U Klenow exo-
Incubate at 37°C for 20 minutes with appropriate shielding.
After 20 minutes, add 1ul 500mM EDTA – see note 4 and (optional) remove free label using an Illustra microspin G50 column. It is not necessary to remove free α-[32P]dCTP, but not doing so will result in the generation of large volumes of liquid and solid radioactive waste in downstream steps.
Add 1ul bromophenol blue stock solution and 6x loading dye for a final concentration of 1x. Mix and leave at room temperature for a few minutes to allow DNA to denature.
Separate labeled material on the denaturing agarose gel. We run large format gels for ~400Vhr (80V for 5h) at room temperature, using 5’ end-labeled (32P) 2-log ladder as a size marker.
Transfer labeled DNA to a neutral nitrocellulose membrane as for a Southern blot. We transfer by capillary action, using 400mM NaOH or 10X SSC as a transfer buffer but most transfer methods are adequate. Alternatively, gels can be dried onto filter paper using a gel drier/vacuum pump apparatus.
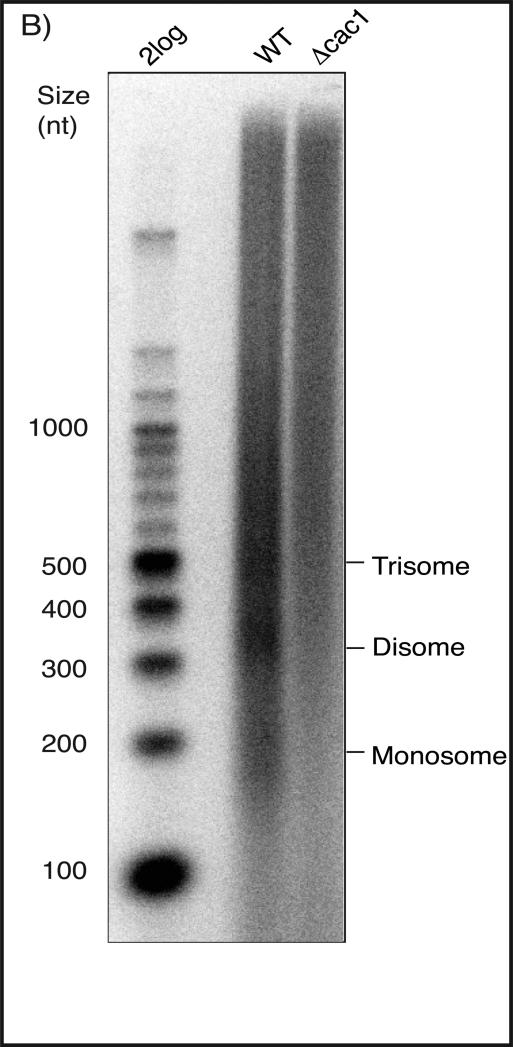
Wrap membrane in saran wrap and expose to film or a phosphor storage screen. See Fig. B for a representative gel image.
Figure (B).
1.3% denaturing agarose gel used to resolve Okazaki fragments from wild-type and cac1 mutant straints. Fragments were end-labeled using Klenow enzyme and α-[32P]dCTP. Labeled samples were loaded with 6X denaturing loading buffer and bromophenol blue and resolved for 5h at 80V. The chromatin repeat is easily visualized in the wild-type sample, but is missing in the CAF-1 deficient strain, as expected.
3.3 Purifying fragments and making sequencing libraries
Adaptor duplexes must be purified away from single-stranded adaptors before use. Preannealed, purified duplexes may be purchased directly from oligo--synthesis companies (e.g IDT), or individual oligonucleotides annealed and the resulting duplex purified from a native polyacrylamide gel. We purify adaptor duplexes from 12% native gels, and also gel-purify library PCR primers. See note 10 for adaptor oligonucleotide sequences.
To prepare Okazaki fragment libraries from asynchronous S. cerevisiae cultures, DNA from [100ml x final OD 0.5] cells represents an ample amount of starting material. We have routinely made high-quality libraries from 50ml starting culture. Regardless of starting volume, purify away from the gelatinous pellet using a spin-X filter before starting the purification.
In a 1.5ml tube, spin down ~500ul (wet volume) of Source 15Q at ~8000xg
Remove supernatant (20% ethanol) and resuspend resin in 500ul of 2M NaCl, 50mM NaOH (pH 12). Spin to ~8000xg
Remove supernatant and resuspend resin in 500ul of 50mM NaOH (pH 12). Repeat three times to equilibrate. In the last wash, transfer to a spin-x column
Pool DNA from multiple preps (if needed) and ethanol precipitate to a final volume of 50-100ul.
Denature 50-100ul DNA in TE at 95° for 5min. Let cool for a minute or so, then add 2 volumes of 50mM NaOH (pH 12). Add to freshly-spun Source 15Q
Incubate DNA and resin with rotation or agitation for 10 min at room temperature.
Spin out the flow-through (short spin, ~6 sec is sufficient); transfer it to a fresh 2ml eppendorf tube, and do sequential elutions with 400ul (50mM NaOH + NaCl, pH12): for each elution, pipette up and down to resuspend the resin, leave for 30 seconds, then spin out.
See supplementary Fig. S4 (6) for a representative elution profile at pH 12. We recommend optimizing this step (see notes 5 and 6)
To the fractions of interest (typically, 700, 800 and 900 mM), add 100ul 3M NaOAc pH5.2, 10 mg glycogen (optional), and ethanol to 2ml. Cool briefly at −80°C and spin at full speed in a microfuge at 4°C to precipitate DNA. We do not wash the pellet at this stage. Dry pellet well before proceeding
Resuspend each pellet (note that pellets may be invisible) in 100ul TE. Pipette for ~30 seconds to ensure complete resuspension. Pool fractions.
Treat the pooled fractions with 1U riboshredder or RNAse A for 30 minutes at 37°C to degrade residual RNA. We find that a fairly large amount of RNA survives the purification up to this point.
Ethanol precipitate with 20 ul, 3M NaOAc and 1ml Ethanol (cool briefly at −80°C before spinning). Wash pellet well with 70% ethanol, resuspend in 40ul TE.
Pass prep through G50 column (Illustra microspin) to remove salts and RNA degradation products.
Quantify DNA yield by spectrophotometry. DNA yield is typically low (~1-5 ng/ul):
Take half (20ul) of the purified Okazaki fragments, heat to 95 for 3 minutes, cool on ice and set up a 40ul T4 ligase reaction containing 3ul (1800 units) ligase and 2ug of each purified adaptor pair. It is critical that adaptors are in large excess. Ligation at the 5’ end is quite efficient, but the 3’ adaptor is poorly ligated. See note 7 on ligase choice.
Ligate at 16° overnight.
Pass the ligation reaction through an S300 microspin column to remove buffer and most of the unligated adaptors.
Perform second-strand synthesis on the S300 flowthrough by adding an equal volume of 2x Taq master mix and incubating at 72°C for 10 minutes.
Clean up second-strand synthesis reaction using a PCR purification kit (Qiagen) or using AMPure or MagNA beads (8) as follows:
Add beads (1.8x volume of PCR product ) to PCR product in a 1.5ml tube. Pipette 10 times to mix. Incubate on the bench for 5 minutes. Place tube on a magnetic rack for 2 minutes, allowing beads to separate from solution. Keeping the tube on the rack, discard supernatant, and wash beads with 200ul 70% ethanol twice. Remove tube from the rack, and add 40-50ul 1X TE to resuspend beads. Pipette to mix, and incubate on the bench for 2-3minutes. Place tube on magnetic rack, and collect eluent in a fresh 1.5ml tube once beads have fully separated.
(Optional) Run the purified DNA on a 2% agarose gel and purify the 200-600 bp region using a Qiaquick kit according to the manufacturer's instructions and elute in 50 ul. See note 8.
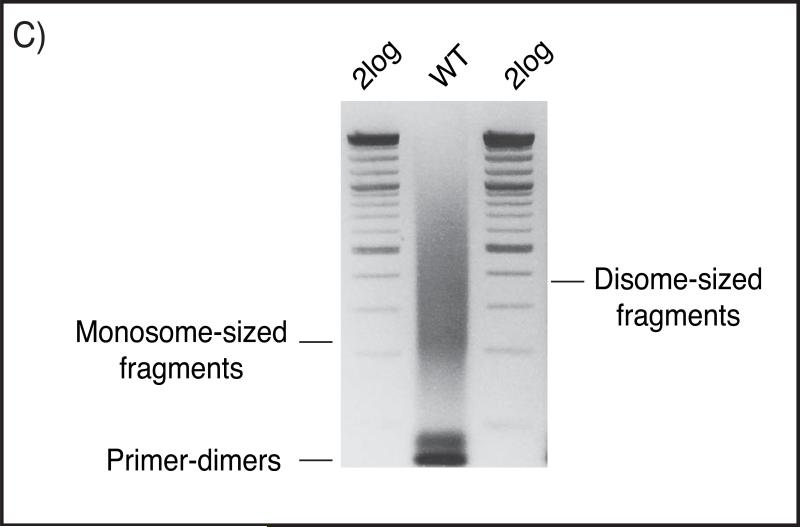
Use your purified material as the template for a test library PCR per the instructions for the sequencing methodology being used. For Illumina libraries, amplify with standard Truseq library primers, adding barcodes at the 3’ end if desired. For Ion Torrent PGM libraries, amplify with standard primers. We run 16 cycles using 1ul and 10ul of purified material as a starting point. 16 cycles on 1ul starting material should show visible product on a gel (Fig C), and the reaction with 10ul purified fragments normally shows significant over-amplification in the form of products significantly larger than 600bp. See note 9.
Figure (C).
2.3% native agarose gel in 0.5X TBE buffer, showing a representative library prep from wild-type Okazaki fragments. The gel is stained with Ethidium Bromide. Monosome- and disome-sized fragments with ligated sequencing-adaptors can be seen along with excess primer-dimer pairs.
Run a preparative library PCR reaction, adding barcodes if desired. We normally find that 10-12 cycles with 10ul purified fragments is optimal.
Precipitate library PCR product, wash with 70% ethanol, resuspend in 25ul and run on a 2.5% agarose gel. Excise DNA of the desired size range and purify using a Qiaquick kit. Alternatively, purify PCR product using AMPure or MagNA beads.
Adaptor oligonucleotides for paired-end Illumina Truseq
5’ ad top strand: ACACTCTTTCCCTACACGACGCTCTTCCGATCT
5’ ad bottom strand: NNNNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT
3’ ad top strand: /5Phos/AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC
3’ ad bottom strand: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNN
Adaptor oligonucleotides Ion Torrent PGM sequencing
5’ ad top strand: CCATCTCATCCCTGCGTGTCTCCGACTCAG
5’ ad bottom strand: NNNNNNCTGAGTCGGAGACACGCAGGGATGAGATGG
3’ ad top strand: /5Phos/ATCACCGACTGCCCATAGAGAGG
3’ ad bottom strand: CCTCTCTATGGGCAGTCGGTGATNNNNNN
ACKNOWLEDGEMENT
This work was supported by NIH grant GM102253 to IW.
Footnotes
We typically transform cells using pPW66R_DOX_Degron plasmid (6) which integrates and replaces the CDC9 promoter with Tet operator promoter sequence and inserts both Ubiquitin and DHFR (9) coding sequence at the N-terminus of the Cdc9 ORF. The integrated sequence also contains a selectable marker and the transcriptional activator – Tet/VP16 – driven by the strong CMV promoter. We and others (10) have labeled fragments enriched via a range of ligase repression/depletion methods, nuclear depletion of Cdc9 using the Anchor-away technique (11) or a temperature-sensitive cdc9-1 allele (4). The Donaldson lab used an auxininducible degron construct to deplete Cdc9 and achieved similar results (10). Although the optimal duration for ligase repression may vary between systems, we find 120-150 minutes to be a good starting point for asynchronous cultures. In our system, addition of doxycycline alone is sufficient for the detection of Okazaki fragments; removal of checkpoint by RAD9 deletion, overexpression of UBR1 and temperature shift to 37° each give a greater signal but are not required.
Volumes are for 50ml cultures: preparation and labeling can be performed successfully (although less reproducibly) with cultures as small as 5ml. For anything below 50ml, we scale reagent volumes down to 40% throughout the spheroplasting, lysis and precipitation steps. Cultures larger than 50ml are split into multiple tubes and processed as separate 50ml cultures. For sequencing we prefer to use 200ml cultures (although 50ml should be adequate).
We routinely store DNA at 4°C for weeks with little to no discernible reduction in quality.
The EDTA is strictly necessary to chelate Mg2+ ions: these form an insoluble Mg(OH)2 precipitate upon addition of the alkaline loading buffer if not chelated, leading to a streaky signal throughout the lane.
In our hands, DNA of the desired length elutes from the Source 15Q resin at 800-900 mM NaCl at pH12. We recommend optimizing this step using DNA spiked an Okazaki fragment prep labeled as in 3.2.
Okazaki fragment purification using the Source 15Q resin can alternatively be carried out at neutral pH: under neutral conditions the desired DNA is eluted at significantly lower salt concentrations than at pH12. Again, we recommend spiking in a radiolabeled prep to optimize binding and elution.
We use T4 DNA ligase from Enzymatics for library preparations. Although we have successfully prepared libraries using the NEB enzyme, average fragment length was unusually short consistent with a small amount of endonuclease contamination or sub-optimal ligation of long fragments.
Typically use of AMPure or MagNA is sufficient to clean up the reaction and remove the adapter dimers. If after PCR there is a dominant band at ~ 70-100nt then gel purification may be necessary.
If the gel slice weighs more than 500mg, use 2 columns. The volume might be large, necessitating multiple loading of the same column. Prior to solubilizing the gel slice, crush it using a blunted p1000 tip. We carry out solubilization at room temperature. We also incorporate an extra wash with buffer QG, and an extra wash with buffer PE due to the large amounts of agarose in the purification.
Oligonucleotides for deep sequencing are listed below:
References
- 1.Nethanel T, Kaufmann G. Two DNA polymerases may be required for synthesis of the lagging DNA strand of simian virus 40. J Virol. 1990;64:5912–5918. doi: 10.1128/jvi.64.12.5912-5918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 3.Ayyagari R, Gomes XV, Gordenin DA, et al. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 4.Johnston LH, Nasmyth KA. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 5.Nasmyth KA. Temperature-sensitive lethal mutants in the structural gene for DNA ligase in the yeast Schizosaccharomyces pombe. Cell. 1977;12:1109–1120. doi: 10.1016/0092-8674(77)90173-8. [DOI] [PubMed] [Google Scholar]
- 6.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuffee SR, Smith DJ, Whitehouse I. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol Cell. 2013;50:123–135. doi: 10.1016/j.molcel.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez-Diaz A, Kanemaki M, Marchesi V, et al. Rapid depletion of budding yeast proteins by fusion to a heat-inducible degron. Sci STKE. 2004;2004:PL8. doi: 10.1126/stke.2232004pl8. [DOI] [PubMed] [Google Scholar]
- 10.Kubota T, Nishimura K, Kanemaki MT, et al. The Elg1 Replication Factor C-like Complex Functions in PCNA Unloading during DNA Replication. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]