Abstract
Mutations in TP53, encoding the master tumor suppressor p53, have posed a developmental therapeutic dilemma due to inability to target loss of function. Inhibition of WEE1 or CHK1 kinase, negative regulators of the G2–M checkpoint, selectively sensitizes p53-deficient cells to exogenous DNA damage, abrogating G2 arrest and precipitating mitotic catastrophe.
In this issue of Clinical Cancer Research, Moser and colleagues (1) report an interspecies functional kinomics approach to identify conserved, druggable survival kinases in TP53-mutated head and neck squamous cell carcinoma (HNSCC). High-throughput kinome-wide siRNA viability screens were performed on human HNSCC cell lines bearing TP53 mutations and murine SCC cell lines from DMBA/TPA-induced tumors in mice harboring germline p53 pathway mutations. Kinase targets were selected on the basis of impaired viability following kinase knockdown, interspecies commonality, and ontologic association with HNSCC pathogenesis. Putative survival kinases included signaling proteins within the focal adhesion and integrin, PI3-kinase, SRC, and G2–M cell-cycle regulatory pathways. WEE1, a nonreceptor tyrosine kinase regulating mitotic entry, was implicated as a critical survival kinase for TP53 mutant (TP53mut) HNSCC cells. Treatment with a WEE1 inhibitor caused unscheduled mitotic entry and apoptotic death selectively in TP53mut versus wild-type (TP53wt) cell lines and potentiated cisplatin in a TP53mut orthotopic xenograft—an example of synthetic lethality amenable to clinical translation.
TP53, encoding the tumor suppressor p53, is the most commonly mutated gene in human cancer in general and HNSCC in particular (2). Because of its master role in cell-cycle regulation, triggering cell-cycle arrest, apoptosis, or senescence after DNA damage, p53 is considered to be the guardian of the genome. TP53 mutations can be classified according to functional impact upon DNA binding. Both nonconservative mutations in the DNA-binding domain and stop codons, predicted to disrupt p53’s direct regulation of gene expression, are classified as disruptive, whereas others are considered nondisruptive (3). In a multicenter trial evaluating molecular biomarkers in patients with HNSCC undergoing curative-intent surgery and heterogeneous adjuvant therapy, disruptive TP53 mutations were associated with reduced survival, independent of pathologic nodal stage or primary tumor site (3). Disruptive TP53 mutations were validated as a prognostic biomarker in an institutional cohort of patients treated homogeneously with primary surgery and postoperative radiotherapy (4). In an orthotopic murine model evaluating 48 validated HNSCC cell lines, disruptive TP53 mutations correlated with higher growth rate, cervical nodal metastases, and decreased survival, suggesting a biologic basis for inferior prognosis (5).
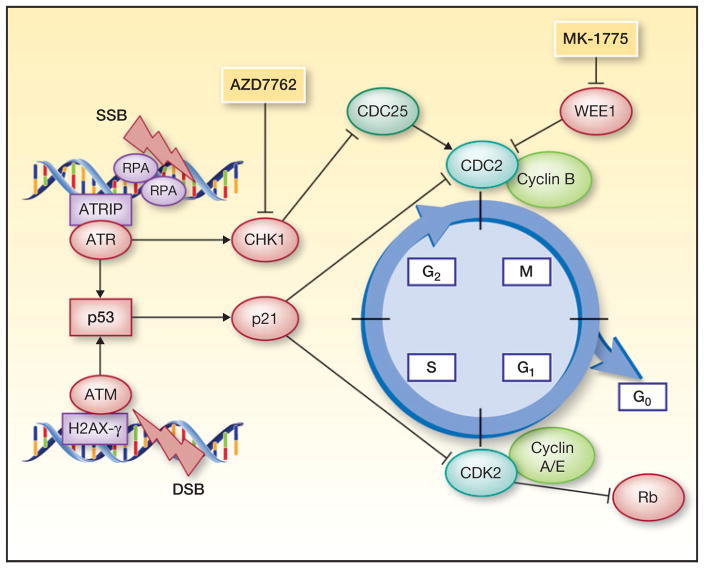
Recognition of TP53 mutation status as a prognostic biomarker in HNSCC creates a classic dilemma in developmental therapeutics: Unlike gain-of-function oncogene mutations, loss-of-function tumor-suppressor mutations cannot be directly targeted. This gives rise to the principle of “synthetic lethality,” a phenomenon in genetics whereby two mutations lead to cell death, whereas neither individual mutation is fatal. Observation of synthetic lethality implies that cells harboring the mutated gene of interest, in this case TP53, are dependent upon the product of the second gene. Cells without functional p53 lack an effective G1 checkpoint and demonstrate excess reliance on the G2 checkpoint to arrest cell-cycle progression and repair DNA damage (Fig. 1). Coordinated progression through the cell cycle is dependent upon the activation state of cyclin-dependent kinases (CDK) in complex with cyclins. At the G2 check-point, mitotic transition occurs when CDK1 (CDC2) is in its active or dephosphorylated state. G2–M arrest can be initiated by two druggable mechanisms: direct phosphorylation of CDC2 by WEE1 kinase, or inactivation of CDC2’s upstream phosphatase, CDC25, by CHK1.
Figure 1.
G2–M cell-cycle arrest. After DNA damage, ATR and ATM initiate cell-cycle arrest following their respective activation at sites of single-strand (SSB) or double-strand breaks (DSB). ATR directly phosphorylates checkpoint kinase-1 (CHK1), whereas ATM activates p53 and CHK2, although there is extensive cross-talk between these pathways. At the G2 checkpoint, G2–M arrest is triggered when CHK1 inhibits the activator of CDC2, CDC25, or when WEE1 directly inactivates CDC2. Proteins contributing to cell-cycle progression are depicted in green, and to cell-cycle arrest in red.
Following exogenous DNA damage by cisplatin or radiation, the cornerstone nonsurgical therapeutics in HNSCC, TP53mut cells evade p53-mediated cellular senescence and initiate G2–M arrest, allowing time for DNA repair (4, 6). Accordingly, the WEE1 and CHK1 kinases are candidates for synthetic lethality. This principle was first demonstrated in TP53mut colorectal carcinoma, in which the preclinical WEE1 kinase inhibitor, PD0166285, potentiated radiation by abrogating G2-–M arrest and forcing premature mitotic entry (7). Of unique interest to HNSCC, in which human papillomavirus (HPV) causes the majority of oropharynx squamous cell carcinoma, this approach was also effective in TP53wt ovarian carcinoma cell lines in which p53 was inactivated by transfection with HPV E6. Similarly, Moser and colleagues observed that HPV+ HNSCC and TP53mut cell lines shared comparable sensitivity to MK-1775, a selective WEE1 inhibitor in clinical development, implying that p53 deficiency arising from either p53 degradation or TP53 mutation may be exploitable by this synthetic lethal strategy. In an isogenic ovarian cancer cell line model, in which the parent was TP53wt and its derivative p53-deficient due to stable p53 mRNA knockdown, MK-1775 selectively sensitized p53-deficient cells to DNA damage from platinum or gemcitabine (8). In analogous experiments, MK-1775 preferentially sensitized TP53mut versus TP53wt lung cancer cell lines and xenografts to radiation by accelerating mitotic entry with unrepaired DNA lesions (9). In HNSCC cell lines, p53-deficient cells circumvented senescence upon cisplatin exposure and underwent mitotic cell death when cisplatin was combined with the CHK1 inhibitor AZD7762 (6). The context specificity of WEE1 or CHK1 inhibition, in which defective p53 is required for sensitization to DNA-damaging therapy, raises hope for a clinically meaningful therapeutic index.
MK-1775 has undergone phase I evaluation in combination with gemcitabine, carboplatin, or cisplatin in 156 biomarker-unselected patients with advanced solid tumors (10). A recommended dose was identified for each combination. The most common adverse events included myelo-suppression, nausea, and fatigue, also class toxicities for chemotherapy. Target engagement, manifested by >50% reduction in phosphorylated CDC2 in serial skin biopsies, was demonstrated across cohorts. AZD7762 was evaluated in combination with gemcitabine in two phase I studies in biomarker-unselected patients; however, development was abandoned due to limited preliminary activity and unexpected cardiac toxicity, potentially related to off-target inhibition of calcium/calmodulin-dependent protein kinases important for cardiac contractility (11). Phase II development of MK-1775 in combination with cisplatin is proceeding in patients with recurrent/metastatic HNSCC amenable to biopsy (NCT01935037). Although patients who are being enrolled are biomarker unselected, this population is naturally enriched for p53-deficient tumors. Rigorous correlative studies will illuminate whether synthetic lethality, as demonstrated in preclinical models, is manifested in patients with TP53mut or HPV-associated cancers.
Realizing the translational promise of synthetic lethality by WEE1 inhibition in p53-deficient cells will be daunting, however, due to the incredible heterogeneity in TP53 mutations and their associated functional phenotypes. Although TP53 mutations have been viewed conventionally as loss-of-function events, complete absence of p53 by TP53 deletion or truncation is clearly not phenotypically identical to the presence of functionally altered p53 protein. Depending upon mutational context, mutant p53 demonstrates aberrant DNA and protein binding, which deregulates gene expression, ultimately conferring gain-of-function oncogenic activity (12). Although categorization of TP53 mutations as disruptive versus nondisruptive in HNSCC is a step forward in recognizing the heterogeneity of mutations and their functional consequences, it remains a relatively crude prognostic biomarker. Moser and colleagues do not distinguish response differences between cell lines with disruptive versus nondisruptive mutations. Considering that 73% (204 of 279) of HNSCC in the The Cancer Genome Atlas dataset contains 243 TP53 mutations spanning the entire gene (www.cbioportal.org), defining associations with drug sensitivity will be challenging. The authors’ data hint at this complexity within human HNSCC; Fig. 4D in their article shows a wide variation in sensitivity to WEE1 inhibition even within TP53mut cell lines (1). A still greater challenge is recognizing that mutant p53 function also depends upon tumor type and genetic context. Identical mutations may generate different functional consequences depending upon coexisting genetic alterations within the tumor.
Although exciting preclinical work remains in the categorization of the myriad TP53 mutations and their exploitable weaknesses, the report by Moser and colleagues is consistent with a growing body of evidence justifying WEE1 as a rational, synthetic lethal target in p53-deficient cells. Clinical trials investigating this lead therapeutic strategy are under way, and will require rigorous correlative science to hone histologic and genetic selection for patients who will benefit.
Acknowledgments
Grant Support
J.E. Bauman is supported by the UPCI SPORE in head and neck cancer (P50CA097190).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: J.E. Bauman, C.H. Chung
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J.E. Bauman, C.H. Chung
Writing, review, and/or revision of the manuscript: J.E. Bauman, C.H. Chung
References
- 1.Moser R, Xu C, Kao M, Annis J, Lerma LA, Schaupp CM, et al. Functional kinomics identifies candidate therapeutic targets in head and neck cancer. Clin Cancer Res. 2014;20:4274–88. doi: 10.1158/1078-0432.CCR-13-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–61. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skinner HD, Sandulache VC, Ow TJ, Meyn RE, Yordy JS, Beadle BM, et al. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res. 2012;18:290–300. doi: 10.1158/1078-0432.CCR-11-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sano D, Xie TX, Ow TJ, Zhao M, Pickering CR, Zhou G, et al. Disruptive TP53 mutation is associated with aggressive disease characteristics in an orthotopic murine model of oral tongue cancer. Clin Cancer Res. 2011;17:6658–70. doi: 10.1158/1078-0432.CCR-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadhikar MA, Sciuto MR, Alves MV, Pickering CR, Osman AA, Neskey DM, et al. Chk1/2 inhibition overcomes the cisplatin resistance of head and neck cancer cells secondary to the loss of functional p53. Mol Cancer Ther. 2013;12:1860–73. doi: 10.1158/1535-7163.MCT-13-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Li J, Booher RN, Kraker A, Lawrence T, Leopold WR, et al. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61:8211–7. [PubMed] [Google Scholar]
- 8.Hirai H, Iwasawa Y, Okada M, Arai T, Nishibata T, Kobayashi M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 9.Bridges KA, Hirai H, Buser CA, Brooks C, Liu H, Buchholz TA, et al. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clin Cancer Res. 2011;17:5638–48. doi: 10.1158/1078-0432.CCR-11-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schellens J, Shapiro G, Pavlick A, Tibes R, Leijen S, Tolaney S, et al. Update on a phase I pharmacologic and pharmacodynamic study of MK-1775, a Wee1 tyrosine kinase inhibitor, in monotherapy and combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J Clin Oncol. 2011;29(suppl):abstr 3068. [Google Scholar]
- 11.Sausville E, Lorusso P, Carducci M, Carter J, Quinn MF, Malburg L, et al. Phase I dose-escalation study of AZD7762, a checkpoint kinase inhibitor, in combination with gemcitabine in US patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;73:539–49. doi: 10.1007/s00280-014-2380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]