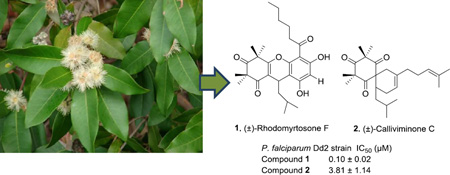
Abstract
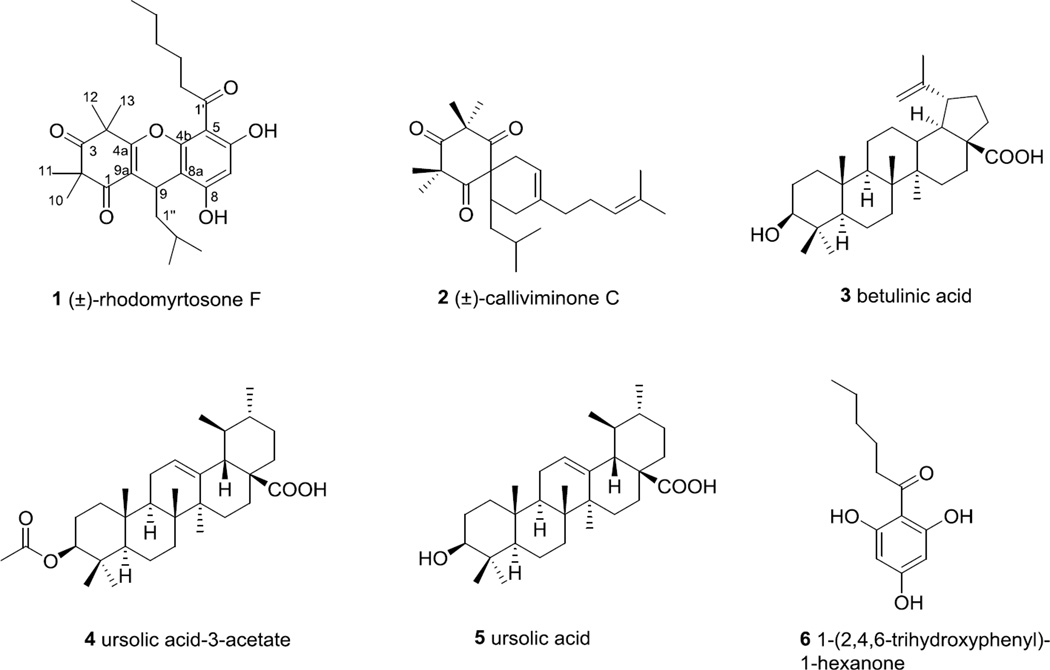
Bioassay guided fractionation of a MeOH extract of the stem bark of Syncarpia glomulifera (Myrtaceae) led to the isolation of the two new phloroglucinol derivatives (±)-rhodomyrtosone F (1) and (±)-calliviminone C (2), the three known triterpenes, betulinic acid (3), ursolic acid-3-acetate (4), and ursolic acid (5), and 1-(2,4,6-trihydroxyphenyl)-1-hexanone (6). Compound 1 exhibited strong antiplasmodial activity, while compounds 2 – 4 were moderately active and 5 and 6 were inactive in this assay. The structures of 1 and 2 were elucidated based on analyses of their mass spectrometric data, 1D and 2D NMR spectra, and comparison with related compounds.
Keywords: Antiplasmodial activity, Phloroglucinol, Syncarpia glomulifera (Myrtaceae
Graphical Abstract
Syncarpia glomulifera Flowers and leaves at Kahakapao Reservoir Haleakala Ranch, Maui - Credit: Forest and Kim Starr - Plants of Hawaii - Image licensed under a Creative Commons Attribution 3.0 License, permitting sharing and adaptation with attribution
1. Introduction
Malaria is a life threatening disease that has afflicted human beings for thousands of years, with references to periodic fevers as far back as 2700 BC in China.1 The disease is caused by protozoan parasites of the Plasmodium genus and transmitted by mosquitoes infected with these parasites.2 More than 200 million people are estimated to be infected with malaria every year, and over 400,000 of them die, with women and children being the most vulnerable.3–4 One of the major families of antimalarial drugs is based on chloroquine, but chloroquine resistant P. falciparum (the deadliest of the five species that infect humans) has been detected since the 1950s in Southeast Asia and South America,5–6 and resistance has developed to nearly all of the other currently available antimalarial drugs, such as sulfadoxine/pyrimethamine, mefloquine, halofantrine, and quinine.7 The most effective current treatments are artemisinin-based combination therapies (ACTs). Unfortunately, resistance against artemisinin was reported on the Thai-Cambodian border in 2009, where it has been used for more than 20 years.8 Because antimalarial resistance can emerge quickly, it is critical to have new drugs with novel mechanisms of action in the antimalarial pipeline.
In pursuit of this goal, we obtained a MeOH extract from the stem bark of a Hawaiian specimen of Syncarpia glomulifera (Myrtaceae). The extract was selected for investigation based on its antiplasmodial activity against the Dd2 drug-resistant strain of P. falciparum. Previous studies have reported the isolation of antibacterial and cytotoxic triterpenoids from the bark extract of S. glomulifera from Paluma, North Queensland, Australia.9 However, no work has been reported on the isolation of any antimalarial agent from this species.
We describe herein the isolation, structure elucidation and antiplasmodial activity of two new phloroglucinols, named as (±)-rhodomyrtosone F (1) and (±)-calliviminone C (2), along with four known compounds (Fig. 1).
Figure 1.
Structures of compounds from Syncarpia glomulifera
2. Results and Discussion
Bioassay guided fractionation of an extract of the stem bark of S. glomulifera yielded the four known compounds betulinic acid (3),10 ursolic acid-3-acetate (4),11 ursolic acid (5),11 and 1-(2,4,6-trihydroxyphenyl)-1-hexanone (6),12 and the two new phloroglucinol derivatives 1 and 2. The structures of compounds 3–6 were identified by comparison of their 1H NMR and mass spectrometric data with literature data.10–12 The structures of 1 and 2 were elucidated by interpretation of mass spectrometric and NMR data as described below. Compound 1 showed strong antimalarial activity, while compounds 2–4 showed moderate to weak antimalarial activity against the Dd2 drug-resistant strain of Plasmodium falciparum. Compounds 5 and 6 were inactive at 40 and 80 µM, respectively, in this assay.
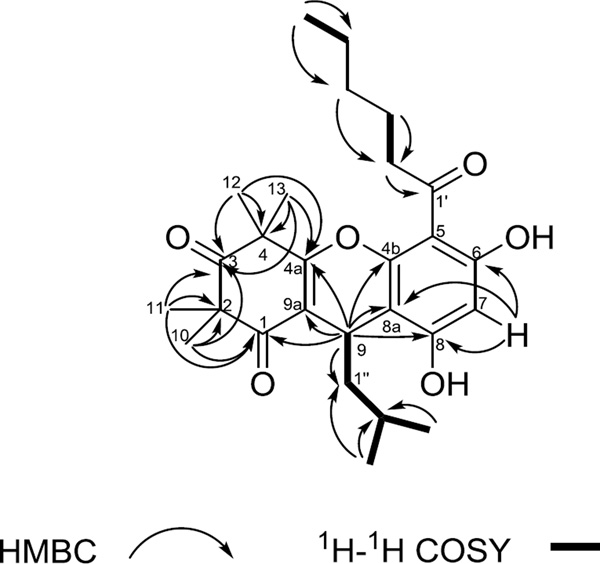
Compound 1 was isolated as a yellowish gum. The protonated molecule peak at m/z 457.2591 [M+H]+ (calcd for C27H37O6 +, 457.2585) suggested a molecular formula of C27H36O6 with 10 degrees of unsaturation. The 1H NMR, 13C NMR and HSQC spectra (Table 1) indicated the presence of 3 carbonyl, 9 quaternary, 3 methine, 5 methylene and 7 methyl carbons in the structure. The cyclohexene-1,3-dione moiety of compound 1 was suggested by HMBC correlations of 10- and 11-Me (δH 1.36 and 1.39, each 3H, s) to C-1, C2 and C-3 (δC 197.6, 56.2 and 212.3), and of 12- and 13-Me (δH 1.43 and 1.54, each 3H, s) to C-3, C-4 (δC 47.3) and C-4a (δC 166.9). In addition H-9 (δH 4.24, d, J = 5.6 Hz) correlated to C-1, C-9a (δC 114.3) and C-4a (δC 166.9). An aromatic ring was assigned to connect to C-9 due to the deshielded chemical shift of H-9 along with HMBC correlations of H-9 to C-4b, C-8a and C-8 (δC 162.6, 106.9 and 155.8). The placement of the aromatic proton H-7 was determined based on its HMBC correlations with C-8a, C-8 and C-6 (δC 158.5). C-4a, C-4b, C-6, C-8 were oxygenated due to their low field signals (δC 155.8~166.9). The identifications and placements of the acyl and isobutyl groups were confirmed by COSY and HMBC correlations (Fig. 2) along with comparison of NMR data with those of related compounds.13–14 An oxygen atom was assigned to connect C-4a and C-4b to give a tricyclic skeleton, based on the 13C NMR signals of C-4a and C-4b, and on the degree of unsaturation.
Table 1.
1H and 13C NMR Data (δ, ppm) for compounds 1 and 2 (500 and 125 MHz) in CDCl3
| Position | 1 |
Position | 2 |
||
|---|---|---|---|---|---|
| δH’ (J in Hz)a | δC’ typeb | δH’ (J in Hz)a | δC’ typeb | ||
| 1 | 197.6, C | 1 | 208.3, C | ||
| 2 | 56.2, C | 2 | 56.3, C | ||
| 3 | 212.3, C | 3 | 213.1, C | ||
| 4 | 47.3, C | 4 | 56.9, C | ||
| 4a | 166.9, C | ||||
| 4b | 162.6, C | ||||
| 5 | 107.5, C | 5 | 208.3, C | ||
| 6 | 158.5, C | 6 | 67.7, C | ||
| 7 | 6.06 s | 95.0, CH | 7 | 2.49 brd (17.2), 2.17 m | 29.5, CH2 |
| 8 | 155.8, C | 8 | 5.31 brs | 116.1, CH | |
| 8a | 106.9, C | ||||
| 9 | 4.24, t (5.6) | 25.3, CH | 9 | 136.7, C | |
| 9a | 114.3, C | ||||
| 10 | 1.36 s | 24.3, CH3 | 10 | 2.13 m, 1.98 m | 30.7, CH2 |
| 11 | 1.39 s | 24.7, CH3 | 11 | 2.31 m | 34.1, CH |
| 12 | 1.43 s | 24.7, CH3 | 12 | 1.37 s | 26.5, CH3 |
| 13 | 1.54 s | 24.9, CH3 | 13 | 1.39 s | 26.3, CH3 |
| 14 | 1.33 s | 24.5, CH3 | |||
| 15 | 1.39 s | 25.8, CH3 | |||
| 1′ | 206.9, C | 1′ | 1.93 m | 37.3, CH2 | |
| 2′ | 3.09 t (7.2) | 44.6, CH2 | 2′ | 2.03 m | 26.1, CH2 |
| 3′ | 1.71 m | 24.4, CH2 | 3′ | 5.03 tq (7.0, 1.4) | 124.1,CH |
| 4′ | 1.36 m | 31.8, CH2 | 4′ | 131.7, C | |
| 5′ | 1.36 m | 22.7, CH2 | 5′ | 1.67 d (1.4) | 25.8, CH3 |
| 6′ | 0.91 t (7.1) | 14.2, CH3 | 6′ | 1.59 s | 17.9, CH3 |
| 1″ | 1.43 m | 46.0, CH2 | 1″ | 1.39 m, 0.78 m | 39.2, CH2 |
| 2″ | 1.35 m | 25.3, CH | 2″ | 1.63 m | 25.6, CH |
| 3″ | 0.84 d (6.0) | 23.3, CH3 | 3″ | 0.87 d (7.0) | 24.4, CH3 |
| 4″ | 0.87 d (6.1) | 23.6, CH3 | 4″ | 0.85 d (7.0) | 21.1, CH3 |
Data (δ) measured at 500 MHz; s = singlet, br s = broad singlet, d = doublet, br d = broad doublet, t = triplet, tq= triplet of quartets, m = multiplet. J values are in Hz and are omitted if the signals overlapped as multiplets. The overlapped signals were assigned from HSQC and HMBC spectra without designating multiplicity.
Data (δ) measured at 125 MHz; CH3, CH2, CH, and C multiplicities were determined by HSQC experiments.
Figure 2.
Key HMBC and COSY correlations of compound 1.
Compound 1 had an optical rotation of zero, indicating that it was isolated as a racemate. It was thus assigned the structure (±)-4,9-dihydro-6,8-dihydroxy-2,2,4,4-tetramethyl-5-hexanoyl-9-isobutyl-1H-xanthene-1,3(2H)-dione and named as (±)-rhodomyrtosone F based on its similarity to rhodomyrtone15 and rhodomyrtosones B14 and E.16
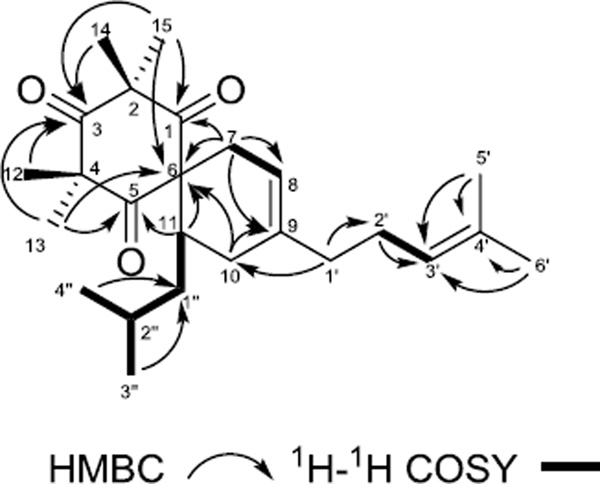
Compound 2 was isolated as light yellow gum. The protonated molecule peak at m/z 387.2896 [M+H]+ (calcd for C25H39O3+, 387.2894) suggested a molecular formula of C25H38O3 with 7 degrees of unsaturation. The 1H NMR, 13C NMR and HSQC spectra of 2 showed 3 carbonyl, 5 quaternary, 4 methine, 5 methylene and 8 methyl carbons in the structure. The structure of compound 2 was elucidated to contain a 1,3,5-cyclohexanetrione moiety based on HMBC correlations of 14- and 15-Me (δH 1.33 and 1.39, each 3H, s) to C-1, C-2 and C-3 (δC 208.3, 56.3 and 213.1), of 12- and 13-Me (δH 1.37 and 1.39, each 3H, s) to C-3, C-4 and C-5 (δC 213.1, 56.9, 208.3), and of 13- and 15-Me to C-6 (δC 67.7). Additional HMBC correlations of H-7a (δH 2.17, 1H, m) to C-6, C-8 and C-9 (δC 67.7, 116.1 and 136.7); H-7b (δH 2.49, 1H, brd, J = 17.2 Hz) to C-1, of H-10a (δH 2.13, 1H, m) to C-6, C-8 and C-9, and of H-11 (δH 2.31, 1H, m) to C-5 and C-6 indicated a spiro-[5,5]-undec-8-ene skeleton. The presence of an isobutyl group was determined by the cross peaks between H-1″ (δH 0.78 and 1.39, each 1H, m) to H-2″ (δH 1.63, 1H, m), and of H-2″ to 3″-Me (δH 0.87, 3H, d, J = 7.0 Hz) and 4″-Me (δH 0.87, d, J = 7.0 Hz) in the 1H-1H COSY spectra. The placement of the isobutyl group was confirmed by a 1H-1H COSY correlation of H-1″a (δH 0.78, 1H, m) to H-11. The presence of the 4-methylpentene unit was established by HMBC data. Key HMBC correlations include H-1′ (δH 1.93, 2H, m) to C-2′ (δC 26.1); H-2′ (δH 2.03, 2H, m) to C-3′ (δC 124.1); 5′-Me (δH 1.67, 3H, d, J = 1.4 Hz), 6′-Me (δH 1.59, 3H, s) to C-3′ and C-4′ (δC 131.7). The 4-methyl-pentene unit was placed at C-9 on the basis of 3J HMBC coupling between H-1' and C-10 (δC 30.7) (see Fig. 3). Compound 2 was a racemate at C-11 based on its zero optical rotation, and its structure was thus elucidated as (±)-2,2,4,4-tetramethyl-11-isobutyl-9-(4-methyl-3-penten-1-yl)- spiro[5.5]undec-8-ene -1,3,5-trione, and named as (±)-calliviminone C, based on its similar structure to (±)-calliviminones A and B.17
Figure 3.
Key HMBC and COSY correlations of compound 2.
Compounds 1–4 showed strong to weak inhibition of the growth of the drug-resistant Dd2 strain of P. falciparum (Table 2), with compound 1 exhibiting the strongest activity with an IC50 of 0.10 ± 0.02 µM. Acylphloroglucinols with similar structures to compound 1 have been reported to show antibacterial activity,18–19 and examples of phloroglucinol derivatives as antimalarial agents can be found in the literature,20–21 but compound 1 is an order of magnitude more potent than tomentosone A,21 the closest analog with reported antimalarial activity. Compound 1 was also tested for cytotoxicity toward human embryonic kidney cells (HEK) and no inhibition was detected up to 3.125 µM, and only 58% inhibition was observed at the highest dose tested (50 µM). Compound 1 thus appears to be potent and relatively non-toxic antimalarial agent. Compound 2 contains the unusual spiro-[5,5]-undec-8-ene skeleton. The first examples of such carbon Diels-Alder adducts between a phloroglucinol and a terpenoid were (±)-calliviminones A and B, isolated from Callistemon viminalis.17 Compound 2 exhibited moderate antiplasmodial activity (IC50 3.81 ± 1.14 µM), which is the first reported antiplasmodial activity of a phloroglucinol with the spiro-[5,5]-undec-8-ene skeleton. Compound 2 did not show any toxicity to HEK cells up to the highest dose tested (100 µM). Betulinic acid (3) showed moderate inhibitory effect on the Dd2 strain of P. falciparum, and it has previously been reported to inhibit the 3D7 stain of P. falciparum, a chloroquine-sensitive strain.22 Ursolic acid (5) and its 3-acetate (4) have been reported to be antibacterial and antioxidant agents,23 but these compounds were inactive against the Dd2 strain of P. falciparum, as previously observed.24 Compound 6, 1-(2,4,6-trihydroxyphenyl)-1-hexanone, is a possible biosynthetic precursor of 1; it was inactive against P. falciparum at the highest dose tested (90 µM).
Table 2.
Antiplasmodial activity data of compounds 1 – 6
| Compound | P. falciparum Dd2 strain IC50 (µM) |
|---|---|
| (±)-Rhodomyrtosone F (1) | 0.10 ± 0.02 |
| (±)-Calliviminone C (2) | 3.81 ± 1.14 |
| Betulinic acid (3) | 3.44 ± 0.42 |
| Ursolic acid-3-acetate (4) | 12.09 ± 0.08 |
| Ursolic acid (5) | Not active (> 40) |
| 1-(2,4,6-Trihydroxyphenyl)-1-hexanone (6) | Not active (> 80) |
3. Experimental
3.1. General experimental procedures
UV spectra were measured on a Shimadzu UV-1201 spectrophotometer. IR spectra were measured on a MIDAC M-series FTIR spectrometer. NMR spectra were recorded in CDCl3 on a Bruker Avance 500 spectrometer in CDCl3; chemical shifts are given in δ (ppm), and coupling constants are reported in Hz. Mass spectra were obtained on an Agilent 6220 LC-TOF-MS in the positive ion mode. Optical rotations were recorded on a JASCO P-2000 polarimeter. Open column chromatography was performed using Sephadex LH-20 (I.D×L 3×50 cm). Semipreparative HPLC was performed on a Shimadzu LC-10AT instrument with a semipreparative C18 Phenomenex Luna column (5 µm, 250 Χ10 mm). All isolated compounds were evaluated for purity by HPLC (both UV and ELSD detection) and by NMR before bioactivity assay. Purity of each compound was checked on a Phenomenex Luna C18 column (5 µm, 250 ×10 mm) and a Cogent Bidentate C18 column (4 µm, 76 Χ 4.6 mm).
3.2. Plant material
Stem bark from S. glomulifera (Sm.) Nied was collected by Gary J. Ray in Hawaii in June 1998 near Pupukea Road, Pupukea (21.38.39 N, 158.00.54 W). A specimen is on deposit at the New York Botanical Garden under the accession number GR01020c (ID 89558).
3.3. Extraction and isolation
Dried, powdered plant material was exhaustively extracted with MeOH at room temperature to give an extract designated 0040244-10F; a total of 3 g of this extract was made available to Virginia Tech and 1.2 g of the material was used in the bioassay guided fractionation. Extract 0040244-10F (1200 mg, IC50 around 1.25 µg/mL, tested in antimalarial assay) was suspended in aqueous MeOH (MeOH- H2O, 9:1, 100 mL) and extracted with hexane (5×100 mL portions) to give an active hexane-soluble fraction (288.6 mg, IC50 < 1.25 µg/mL). The aqueous fraction was then diluted to MeOH-H2O, 6:4 (150 mL) and further extracted with CH2Cl2 (5×100 mL portions) to yield the CH2Cl2-soluble fraction (84.4 mg, IC50 approximately 2.5 µg/mL) and the aqueous fraction (824.7 mg, IC50 > 10 µg/mL). The active hexane fraction was then subjected to a size exclusion open column chromatography on Sephadex LH-20 (I.D×L 3×50 cm) eluted with CH2Cl2: MeOH (1:1). Three fractions (F1-1: 122.7 mg, F1-2: 65.5mg, F1-3: 99.0 mg) were collected based on TLC analyses. The active fraction F1-3 (IC50 < 1.25 µg/mL) was further separated by HPLC on a semipreparative C18 column (Phenomenex Luna column, 5 µm, 250 Χ10 mm) eluted with a solvent gradient from CH3OH:H2O, 75:25 to 85:15 from 0 to 10 min, to 95:5 from 10 to 20 min, ending with 100% CH3OH from 20 to 40 min at a flow rate of 2.5 mL/min. This process gave compounds 3 (4.8 mg, tR 23.5 min), 4 (4.2 mg, tR 31 min), and semi-pure compound 1 (5.1 mg, tR 26 min) and 2 (2.0 mg, tR 27.5 min). The collected semi-pure compound 1 was then further purified by HPLC on the same column eluted with a solvent gradient from CH3CN:H2O, 85:15 to 90:10 from 0 to 15 min, to 100:0 from 15 to 25 min, ending with 100% CH3CN from 25 to 40 min at a flow rate of 2.5 mL/min. This process gave compounds 1 (3.6 mg, tR 28 min) and 5 (1.3 mg, tR 26 min). Compound 2 was also purified by HPLC on the semipreparative C18 column with aqueous acetonitrile as above. This process gave purified compound 2 (1.4 mg, tR 36 min).
The moderately active dichloromethane fraction was also subjected to the size exclusion open column using the method described above, to yield three sub-fractions (F2-1: 35.6 mg, F2-2: 21.5 mg, F2-3: 21.6 mg). Fraction F2-3 (IC50 around 2.5 µg/mL) was separated by HPLC on the Luna C18 column, eluted with a solvent gradient from CH3OH:H2O, 60:40 to 70:30 from 0 to 10 min, to 90:10 from 10 to 30 min, ending with 100% CH3OH from 30 to 40 min at a flow rate of 2.5 mL/min. This process yielded compound 6 (2.1 mg, tR 24.5 min) and a small amounts of compounds 1 (0.2 mg, tR 33.0 min) and 4 (0.4mg, tR 41.5 min).
3.4. Antimalarial bioassay
The effect of each fraction and pure compounds on in vitro parasite growth of Dd2 strain was measured in a 72 h growth assay in the presence of inhibitor as described previously with minor modifications.25–26 Ring stage parasite cultures (100 µL per well, 1% hematocrit and 1% parasitaemia) were grown for 72 h in the presence of increasing concentrations of the inhibitor in a humidified chamber at 37 °C and low oxygen conditions (5.06% CO2, 4.99% O2, and 89.95% N2). After 72 h in culture, parasite viability was determined by DNA quantitation using SYBR Green I as described previously.26 The half-maximum inhibitory concentration (IC50) values were calculated with KaleidaGraph software using a nonlinear regression curve fitting. IC50 values are the average of three independent determinations with each determination in duplicate, and are expressed ± S.E.M. Artemisinin was used as the positive control with an IC50 of 6.2 ±1.2 nM.
3.5. In vitro cytotoxicity against HEK293 cells
Compounds were evaluated for their cytotoxicity against normal cell line HEK293 (Human Embryonic Kidney). Briefly, 10,000 HEK cells per well were plated in a clear-bottom 96 well plate. After allowing the cells to adhere, the media was replaced with 100 µL of media containing varying amounts of the test compound and incubated for 24 hours. Later, 10 µL of resazurin sodium salt (Sigma) at 0.125 mg/mL was added to each well and incubated for 2 h. Cell viability was determined by measuring the fluorescence at 585 nm after excitation at 540 nm.
3.6. (±)-5-Hexanoyl-6,8-dihydroxy-9-isobutyl-2,2,4,4-tetramethyl-4,9-dihydro-1H-xanthene-1,3(2H)-dione, (±)-rhodomyrtosone F (1)
Yellowish gum; [α]D23 0 (c = 0.22, MeOH); UV (MeOH) λmax (log ε) 223 (3.31), 261 (3.01), 292 (2.51), 333 (1.40) nm; IR (film) νmax 2935, 2872, 1721, 1629, 1430, 1389, 1108 and 1091 cm−1; 1H and 13C NMR data, see Table 1; positive ion HRESIMS m/z 457.2591 [M+H]+ (calculated for C27H37O6+, 457.2585); 474.2831 [M+NH4]+ (calcd for C27H40NO6+, 474.2851).
3.7. (±)-11-Isobutyl-2,2,4,4-tetramethyl-9-(4-methylpent-3-en-1-yl)- spiro[5.5]undec-8-ene-1,3, 5-trione, (±)-calliviminone C (2)
Yellowish gum; [α]D23 0 (c = 0.15, MeOH); UV (MeOH) λmax (log ε) 214 (3.53), 297 (2.67); IR (film) νmax 2357, 2335, 2146 and 1699 cm−1; 1H and 13C NMR data, see Table 1; positive ion HRESIMS m/z 387.2896 [M+H]+ (calcd for C25H39O3+, 387.2894).
Supplementary Material
Acknowledgments
This project was supported by the National Center for Complementary and Integrative Health (NCCIH) under awards 1 R01 AT008088-01. It was also supported by the National Science Foundation under Grant No. CHE-0619382 for purchase of the Bruker Avance 500 NMR spectrometer and Grant No. CHE- 0722638 for the purchase of the Agilent 6220 mass spectrometer. Funding for this work was also provided in part by the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. We thank Mr. B. Bebout for obtaining the mass spectra, and we gratefully acknowledge Gary J. Ray of the New York Botanical Garden for the provision of plant material.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data associated with this article, consisting of HPLC chromatograms and NMR spectra, can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.2016.xx.yy.
References and notes
- 1.Neghina R, Neghina AM, Marincu I, Iacobiciu I. Am. J. Med. Sci. 2010;340:492. doi: 10.1097/MAJ.0b013e3181e7fe6c. [DOI] [PubMed] [Google Scholar]
- 2.Wright AD, König GM, Angerhofer CK, Greenidge P, Linden A, Desqueyroux-Faúndez R. J. Nat. Prod. 1996;59:710. doi: 10.1021/np9602325. [DOI] [PubMed] [Google Scholar]
- 3. [accessed February 18, 2016];10 facts on malaria. World Health Organization. http://www.who.int/features/factfiles/malaria/en/
- 4.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. Nature. 2005;434:214. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alker AP, Lim P, Sem R, Shah NK, Yi P, Bouth DM, Tsuyuoka R, Maguire JD, Fandeur T, Ariey F. Am. J. Trop. Med. Hyg. 2007;76:641. [PubMed] [Google Scholar]
- 6.Mehlotra RK, Mattera G, Bockarie MJ, Maguire JD, Baird JK, Sharma YD, Alifrangis M, Dorsey G, Rosenthal PJ, Fryauff DJ. Antimicrob. Agents Chemother. 2008;52:2212. doi: 10.1128/AAC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. [accessed October 30, 2015];Drug Resistance in the Malaria-Endemic World. http://www.cdc.gov/malaria/malaria_worldwide/reduction/drug_resistance.html.
- 8.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. New Engl. J. Med. 2009;361:455. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setzer WN, Setzer MC, Bates RB, Jackes BR. Planta Med. 2000;66:176. doi: 10.1055/s-2000-11129. [DOI] [PubMed] [Google Scholar]
- 10.Ayatollahi AM, Ghanadian M, Afsharypour S, Abdella OM, Mirzai M, Askari G. Iran. J. Pharm. Res. 2011;10:287. [PMC free article] [PubMed] [Google Scholar]
- 11.Gnoatto SC, Dassonville-Klimpt A, Da Nascimento S, Galéra P, Boumediene K, Gosmann G, Sonnet P, Moslemi S. Eur. J. Med. Chem. 2008;43:1865. doi: 10.1016/j.ejmech.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Frišcic T, Drab DM, MacGillivray LR. Org. Lett. 2004;6:4647. doi: 10.1021/ol0484052. [DOI] [PubMed] [Google Scholar]
- 13.Appendino G, Bianchi F, Minassi A, Sterner O, Ballero M, Gibbons S. J. Nat. Prod. 2002;65:334. doi: 10.1021/np010441b. [DOI] [PubMed] [Google Scholar]
- 14.Hiranrat A, Mahabusarakam W. Tetrahedron. 2008;64:11193. [Google Scholar]
- 15.Dachriyamas, Salni, Sargent MD, Skelton BW, Soediro I, Sutisna M, White AH, Yulinah E. Aust. J. Chem. 2002;55:229. [Google Scholar]
- 16.Wang C, Yang J, Zhao P, Zhou Q, Mei Z, Yang G, Yang X, Feng Y. Bioorg. Med. Chem. Lett. 2014;24:3096. doi: 10.1016/j.bmcl.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Luo J, Zhang Y, Zhu M, Wang X, Luo J, Yang M, Yu B, Yao H, Dai Y. Tetrahedron Lett. 2015;56:229. [Google Scholar]
- 18.Rattanaburi S, Mahabusarakam W, Phongpaichit S, Carroll AR. Tetrahedron. 2013;69:6070. doi: 10.1080/14786419.2012.666750. [DOI] [PubMed] [Google Scholar]
- 19.Saising J, Götz F, Dube L, Ziebandt AK, Voravuthikunchai SP. Ann. Microbiol. 2015;65:659. [Google Scholar]
- 20.Harinantenaina L, Bowman JD, Brodie PJ, Slebodnick C, Callmander MW, Rakotobe E, Randrianaivo R, Rasamison VE, Gorka A, Roepe PD, Cassera MB, Kingston DGI. J. Nat. Prod. 2013;76:388. doi: 10.1021/np300750q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiranrat A, Mahabusarakam W, Carroll AR, Duffy S, Avery VM. J. Org. Chem. 2011;77:680. doi: 10.1021/jo201602y. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler HL, Franzyk H, Sairafianpour M, Tabatabai M, Tehrani MD, Bagherzadeh K, Hägerstrand H, Stærk D, Jaroszewski JW. Bioorg. Med. Chem. 2004;12:119. doi: 10.1016/j.bmc.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 23.do Nascimento PG, Lemos TL, Bizerra A, Arriaga Â, Ferreira DA, Santiago GM, Braz-Filho R, Costa JGM. Molecules. 2014;19:1317. doi: 10.3390/molecules19011317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Innocente AM, Silva GN, Cruz LN, Moraes MS, Nakabashi M, Sonnet P, Gosmann G, Garcia CR, Gnoatto SC. Molecules. 2012;17:12003. doi: 10.3390/molecules171012003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett TN, Paguio M, Gligorijevic B, Seudieu C, Kosar AD, Davidson E, Roepe PD. Antimicrob. Agents Chemother. 2004;48:1807. doi: 10.1128/AAC.48.5.1807-1810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. Antimicrob. Agents Chemother. 2004;48:1803. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.