Abstract
Purpose:
To investigate whether microRNA (MIR)-184 mutations make a substantial contribution to keratoconus (KCN) among affected Iranian patients.
Methods:
A total of 47 Iranian KCN patients, diagnosed based on family history, clinical examinations using slit lamp biomicroscopy, refraction and corneal topography were enrolled in this study. The pri-miR-184 encoding gene obtained from the DNAs of all participants was amplified using polymerase chain reaction and subsequently sequenced by the Sanger dideoxynucleotide protocol. The sequences were compared to MIR184 reference sequence in order to identify sequence variations. The potential effects of a single variation observed on RNA structure was predicted.
Results:
Only one sequence variation, +39G >T, was observed within the pri-miR-184 encoding sequence in one proband. The patient's KCN-affected sister harbored the same variation. The variation was not novel and was recently shown to be present at similar frequencies among large cohorts of KCN patients and control individuals.
Conclusion:
Mutations in MIR-184 are not a major cause of keratoconus among Iranian patients. The pri-miR-184 sequence needs to be screened in larger cohorts in order to establish whether mutations in the gene are present at low frequencies among Iranian patients.
Keywords: MIR-184, Keratoconus, Iran
INTRODUCTION
Keratoconus (Online Mendelian Inheritance in Man [OMIM] #148300) is a non-inflammatory progressive corneal thinning disorder and a major indication for corneal transplantations.[1,2] It is the most common corneal dystrophy with an estimated prevalence of 1 in 2,000 in Western populations. The incidence of keratoconus (KCN) varies among ethnicities and is estimated to be 29-229/100,000, depending on the population studied.[3] Its prevalence and incidence are likely to be higher if novel diagnostic tools which allow the diagnosis of mild cases (i.e. computerized corneal topography and in vivo confocal microscopy) are used.[3,4]
Despite intensive investigations, the underlying cellular and molecular mechanisms of KCN have remained poorly understood.[1,5,6] Although KCN usually appears to be sporadic, twin studies and family-based studies have provided convincing evidence for the existence of a genetic component in the etiology of the disease.[7] Inheritance pattern in familial cases is usually autosomal dominant with incomplete penetrance.[8] It has been proposed that genetic backgrounds in some cases may create a genetic predisposition to KCN which manifests upon exposure to precipitating environmental factors. Candidate gene approaches,[9,10,11,12,13,14] genome-wide association studies,[15,16,17] and family-based linkage analyzes have been used to identify potentially causative genes.[10,18,19,20] VSX1 that encodes visual system homeobox 1 and SOD1 that encodes superoxide dismutase 1 were among candidate genes which received considerable attention; however, their potential contribution are now highly controversial.[9,10,11] Linkage studies in two large KCN pedigrees ultimately led to the identification of mutations in DOCK9 encoding dedicator of cytokinesis 9 and microRNA (MIR)-184 encoding miR-184 as causatives of the disease.[10,18,19,20] The affected members of an Irish family with the MIR184 mutation were diagnosed with combined early-onset autosomal dominant polar cataract and clinically severe KCN.[19] The mutation was identified only after deep sequencing of a 5 Mb linked region.[18] This represented only the second report of a mutation in an miRNA coding gene as the cause of a mendelian disease, the other being a mutation in miR-96 which caused a form of hereditary deafness.[21]
microRNA-184 is of particular interest because putative mutations in the gene were subsequently reported in a family affected with endothelial dystrophy, iris hypoplasia, congenital cataract and stromal thinning (EDICT “endothelial dystrophy, iris hypoplasia, congenital cataract and stromal thinning”; OMIM number 614303),[22] and in two families affected with pure KCN.[23] miR-184 is expressed in the central corneal epithelial basal and suprabasal cells, and in the crystalline lens epithelium.[24,25] miR-184 is the most abundant miRNA in both the cornea and the crystalline lens.[25] It was shown in ex vivo studies that miR-184 can competitively inhibit the binding of miR-205 to mRNA of the inositol polyphosphate phosphatase-like 1,[26] and integrin beta 4 genes.[18] Through this mechanism, miR-184 prevents knock-down by miR-205 and rescues the production of encoded proteins.[18] Both proteins are potentially relevant to corneal functions. Recently, miR-184 was shown to regulate the differentiation of human-induced pluripotent stem cells into corneal epithelial-like cells.[27]
Herein, we aimed to establish whether MIR184 mutations make a substantial contribution to KCN disease among affected Iranian patients by direct sequencing of the entire coding sequence of the pri-miRNA in 47 unrelated patients.
METHODS
The present research was performed in accordance with the Declaration of Helsinki and was approved by the Ethics Board of the University of Tehran, Tehran, Iran. All participants or their responsible guardians consented to participate after being informed of the nature of research.
We included 47 unrelated patients diagnosed with KCN. The diagnosis made by a cornea subspecialist (NN) and was based on clinical findings including slit lamp biomicroscopy and refraction, and corneal topography [Figure 1]. Out of 47 Iranian KCN patients, 25 had a positive family history of KCN where at least one individual was diagnosed with KCN in their families. In the remaining 22 subjects, the disease was apparently sporadic. Mean age at the time of diagnosis was 21.8 (range 9-41) years and 20 (range 4-32) years in familial and sporadic patients, respectively. Eighteen patients (38%) were female.
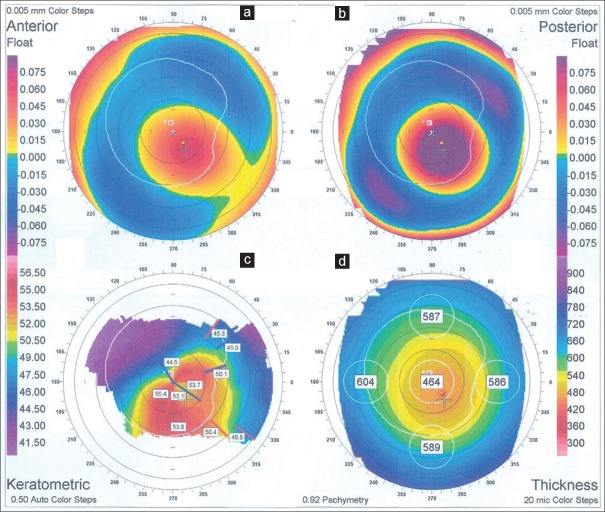
Figure 1.
Corneal tomography image of a patient with miR.184 +39G>T variation confirms the diagnosis of keratoconus; anterior and posterior corneal elevation maps (a and b), corneal curvature map (c), and corneal pachymetry map (d) of the left eye.
DNA was isolated from peripheral blood leukocytes by standard protocols. A 465 bp DNA fragment which includes the entire priRNA encoding sequence of miR-184 (miRBase sequence MI0000481; http://www.mirbase.org/) was amplified from the DNAs by polymerase chain reaction. The sequences of forward and reverse primers used were 5 ' - G A G G C C A G A G C A A A G T A G A A G G - 3 ' and 5 ' - A G A C C C T A A A C C C A G T C G C A - 3 ', respectively. Sanger sequencing of the amplicons was performed and the sequence results were compared to the reference sequence (miRBase MI0000481) using Sequencher version 4.7 software (Ann Arbor, MI, USA).
The effect of the single sequence variation, observed within the pre-miRNA encoding region on secondary structure, was computationally predicted using mFold. (http://unafold.rna.albany.edu/?q = mfold/rna-folding-form) and RNAfold (http://www.rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
RESULTS
Only one variation in the pri-miR-184 encoding gene was observed in one female patient who was a familial case [Figure 2]. The variation was present in heterozygous state. The same variation which was + 39G>T (position 1 = first nucleotide of pri-miRNA) was also observed in her affected sister. The two sisters were diagnosed at the age of 17 and 27 years. Neither of the parents was available for examination, but the sisters reported that their parents were not affected with an ocular disease. The variation was at a site preserved in the pre-miRNA but not preserved in the mature miRNA. The potential deleterious effects of the variation were assessed using the bioinformatics tools mFold and RNAfold. It was predicted that the variation caused an increase in the size of the terminal loop of the pri-miRNA by one nucleotide and removed a bulge in the proximity of the Dicer cutting site [Figure 3]. In terms of free energy, both programs predicted very similar values for the wild type and variant pri-miRNAs (mFold: −35.5 and −35.0 Kcal/mole for wild type and variant molecules, respectively; RNAfold: −35.3 and −35.1 Kcal/mole for wild type and variant molecules, respectively).
Figure 2.
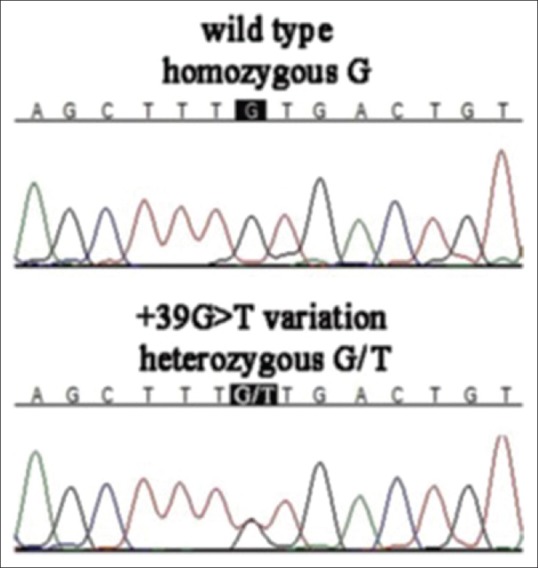
Chromatograms show homozygous microRNA (MIR).184 +39G genotype and MIR184 heterozygous +39G>T variation.
Figure 3.
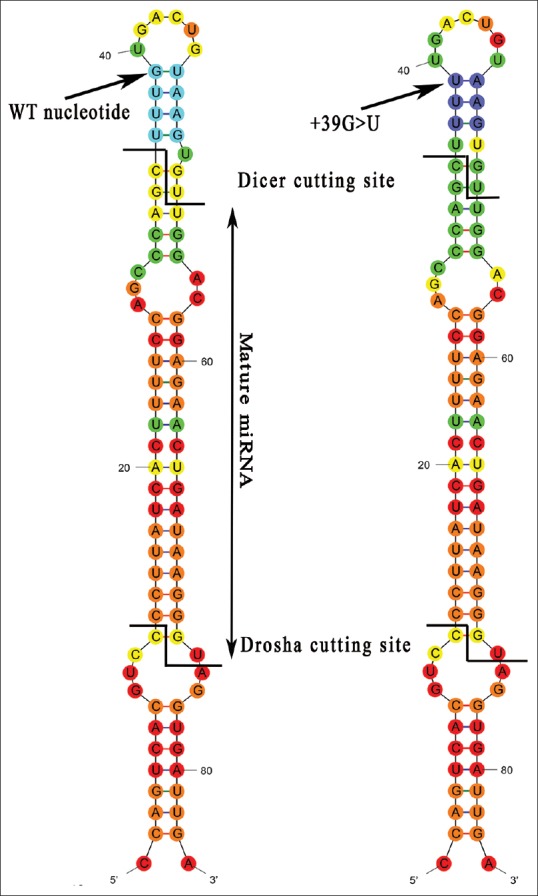
RNA-Fold prediction of the effect of +39G>T variation on pri-miR-184 secondary structure; the variation is shown by arrow. It is predicted that the size of the terminal loop would be increased by one nucleotide to delete a bulge in the proximity of the Dicer cleavage site.
DISCUSSION
The potential deleterious effect of the variation found in the present study was assessed using bioinformatics analysis which did not provide definitive evidence on the functional consequence of the single observed variation. The + 39G > T variation is not novel (rs41280052) and has been reported at a low frequency of 0.006 in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/). Prompted by its position within the pre-miRNA and its low frequency, other researchers during the course of our study determined and compared its frequency in large cohorts including 692 Caucasian KCN patients and 1865 control subjects.[23] The frequencies in the two groups were 1.3% and 1.7%, respectively which are not significantly different. This finding strongly argues against a potential role for the rs41280052 variation in KCN etiology. As such, we surmise that the + 39G > T variation in the two Iranian KCN siblings was not the cause of their ocular condition. Therefore, we did not find a disease-causing mutation in the pri-miR184 encoding gene among 47 Iranian patients studied. This indicates that mutations that affect miR-184 function are not a major cause of keratoconus among Iranian patients. Clearly, the pri-miR-184 sequence needs to be screened in larger cohorts in order to determine whether mutations in the gene are present at low frequencies among Iranian patients. Furthermore, because mutations in the pri-miR-184 have been identified as a causative factor in KCN in some studies, the target genes of this RNA should be considered as potential contributors to the etiology of keratoconus.[18,23]
Financial Support and Sponsorship
Nil.
Conflicts of Interest
There are no conflicts of interest.
Acknowledgments
We acknowledge the Ophthalmic Research Center of Shahid Beheshti University of Medical Sciences for funding this research. We also thank the patients for participating in this study.
REFERENCES
- 1.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 2.Eye Bank Association of America. Eye Banking Statistical Report. 2008 [Google Scholar]
- 3.Kennedy RH, Bourne WM, Dyer JA. A 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol. 1986;101:267–273. doi: 10.1016/0002-9394(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 4.Grupcheva CN, Malik TY, Craig JP, Sherwin T, McGhee CN. Microstructural assessment of rare corneal dystrophies using real-time in vivo confocal microscopy. Clin Experiment Ophthalmol. 2001;29:281–285. doi: 10.1046/j.1442-9071.2001.00434.x. [DOI] [PubMed] [Google Scholar]
- 5.Bisceglia L, De Bonis P, Pizzicoli C, Fischetti L, Laborante A, Di Perna M, et al. Linkage analysis in keratoconus: Replication of locus 5q21.2 and identification of other suggestive Loci. Invest Ophthalmol Vis Sci. 2009;50:1081–1086. doi: 10.1167/iovs.08-2382. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Rabinowitz YS, Rotter JI, Yang H. Genetic epidemiological study of keratoconus: Evidence for major gene determination. Am J Med Genet. 2000;93:403–409. [PubMed] [Google Scholar]
- 7.Burdon KP, Vincent AL. Insights into keratoconus from a genetic perspective. Clin Exp Optom. 2013;96:146–154. doi: 10.1111/cxo.12024. [DOI] [PubMed] [Google Scholar]
- 8.Nowak DM, Gajecka M. The genetics of keratoconus. Middle East Afr J Ophthalmol. 2011;18:2–6. doi: 10.4103/0974-9233.75876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aldave AJ, Yellore VS, Salem AK, Yoo GL, Rayner SA, Yang H, et al. No VSX1 gene mutations associated with keratoconus. Invest Ophthalmol Vis Sci. 2006;47:2820–2822. doi: 10.1167/iovs.05-1530. [DOI] [PubMed] [Google Scholar]
- 10.Gajecka M, Radhakrishna U, Winters D, Nath SK, Rydzanicz M, Ratnamala U, et al. Localization of a gene for keratoconus to a 5.6-Mb interval on 13q32. Invest Ophthalmol Vis Sci. 2009;50:1531–1539. doi: 10.1167/iovs.08-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udar N, Atilano SR, Brown DJ, Holguin B, Small K, Nesburn AB, et al. SOD1: A candidate gene for keratoconus. Invest Ophthalmol Vis Sci. 2006;47:3345–3351. doi: 10.1167/iovs.05-1500. [DOI] [PubMed] [Google Scholar]
- 12.Rabinowitz YS, Maumenee IH, Lundergan MK, Puffenberger E, Zhu D, Antonarakis S, et al. Molecular genetic analysis in autosomal dominant keratoconus. Cornea. 1992;11:302–308. doi: 10.1097/00003226-199207000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Fullerton J, Paprocki P, Foote S, Mackey DA, Williamson R, Forrest S. Identity-by-descent approach to gene localisation in eight individuals affected by keratoconus from north-west Tasmania, Australia. Hum Genet. 2002;110:462–470. doi: 10.1007/s00439-002-0705-7. [DOI] [PubMed] [Google Scholar]
- 14.De Bonis P, Laborante A, Pizzicoli C, Stallone R, Barbano R, Longo C, et al. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol Vis. 2011;17:2482–2494. [PMC free article] [PubMed] [Google Scholar]
- 15.Burdon KP, Macgregor S, Bykhovskaya Y, Javadiyan S, Li X, Laurie KJ, et al. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest Ophthalmol Vis Sci. 2011;52:8514–8519. doi: 10.1167/iovs.11-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Bykhovskaya Y, Haritunians T, Siscovick D, Aldave A, Szczotka-Flynn L, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2012;21:421–429. doi: 10.1093/hmg/ddr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bykhovskaya Y, Li X, Epifantseva I, Haritunians T, Siscovick D, Aldave A, et al. Variation in the lysyl oxidase (LOX) gene is associated with keratoconus in family-based and case-control studies. Invest Ophthalmol Vis Sci. 2012;53:4152–4157. doi: 10.1167/iovs.11-9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89:628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes AE, Dash DP, Jackson AJ, Frazer DG, Silvestri G. Familial keratoconus with cataract: Linkage to the long arm of chromosome 15 and exclusion of candidate genes. Invest Ophthalmol Vis Sci. 2003;44:5063–5066. doi: 10.1167/iovs.03-0399. [DOI] [PubMed] [Google Scholar]
- 20.Czugala M, Karolak JA, Nowak DM, Polakowski P, Pitarque J, Molinari A, et al. Novel mutation and three other sequence variants segregating with phenotype at keratoconus 13q32 susceptibility locus. Eur J Hum Genet. 2012;20:389–397. doi: 10.1038/ejhg.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 22.Iliff BW, Riazuddin SA, Gottsch JD. A single-base substitution in the seed region of miR-184 causes EDICT syndrome. Invest Ophthalmol Vis Sci. 2012;53:348–353. doi: 10.1167/iovs.11-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechner J, Bae HA, Guduric-Fuchs J, Rice A, Govindarajan G, Siddiqui S, et al. Mutational analysis of MIR184 in sporadic keratoconus and myopia. Invest Ophthalmol Vis Sci. 2013;54:5266–5272. doi: 10.1167/iovs.13-12035. [DOI] [PubMed] [Google Scholar]
- 24.Karali M, Peluso I, Gennarino VA, Bilio M, Verde R, Lago G, et al. miRNeye: A microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 26.Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci U S A. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shalom-Feuerstein R, Serror L, De La Forest Divonne S, Petit I, Aberdam E, Camargo L, et al. Pluripotent stem cell model reveals essential roles for miR-450b-5p and miR-184 in embryonic corneal lineage specification. Stem Cells. 2012;30:898–909. doi: 10.1002/stem.1068. [DOI] [PubMed] [Google Scholar]