Highlight
The chlorotic phenotype of the paramutated sulfurea tomato plants is due to the epigenetic silencing of the SLTAB2 gene, involved in the translation of photosystem I.
Key words: Auxin, DNA methylation, paramutation, photosynthesis, RdDM, siRNA, SULFUREA, VIGS.
Abstract
The sulfurea (sulf) allele is a silent epigenetic variant of a tomato (Solanum lycopersicum) gene affecting pigment production. It is homozygous lethal but, in a heterozygote sulf/+, the wild-type (wt) allele undergoes silencing so that the plants exhibit chlorotic sectors. This transfer of the silenced state between alleles is termed paramutation and is best characterized in maize. To understand the mechanism of paramutation we mapped SULF to the orthologue SLTAB2 of an Arabidopsis gene that, consistent with the pigment deficiency, is involved in the translation of photosystem I. Paramutation of SLTAB2 is linked to an increase in DNA methylation and the production of small interfering RNAs at its promoter. Virus-induced gene silencing of SLTAB2 phenocopies sulf, consistent with the possibility that siRNAs mediate the paramutation of SULFUREA. Unlike the maize systems, the paramutagenicity of sulf is not, however, associated with repeated sequences at the region of siRNA production or DNA methylation.
Introduction
Paramutation involves the transfer of epigenetic marks from a (paramutagenic) silent gene to the active (paramutable) allele so that it becomes heritably silent and paramutagenic. Several plant species exhibit paramutation and the best characterized examples, the b1 and pl1 loci in maize, have been linked to the process of RNA-directed DNA methylation (RdDM) (Chandler and Stam, 2004; Hollick, 2012) in which paramutagenic small interfering (si)RNAs mediate silencing of the paramutable allele. This simple model does not, however, explain why most siRNA loci are not paramutagenic: there must be other factors.
To shed light on the mechanism of paramutation we are analysing the tomato SULFUREA (SULF) locus. The silent sulf allele has a chlorotic phenotype (Hagemann, 1958) that is associated with reduced auxin (Ehlert et al., 2008). A sulf homozygote is seedling lethal but a viable heterozygous sulf/+ plant has large chlorotic sectors that are due to paramutation of the active allele to a silenced state in early development. This system is like classic maize paramutation because the paramutated state is heritable and paramutagenic (Hagemann, 1969). SULF maps to the pericentromeric heterochromatin of chromosome 2, at approximately 29 cM from the S locus (Solyc02g077390, compound inflorescence) (Hagemann and Snoad, 1971) but the affected gene could not be mapped precisely due to low recombination frequency in this region (The Tomato Genome Consortium, 2012).
A gene orthologue of the Arabidopsis ATAB2 is strongly down-regulated in chlorotic sectors of sulf/+ tomato (Ehlert et al., 2008) but it was previously excluded as the SULF gene because it is still expressed at detectable levels. However, from analysis of transcriptome, methylome, and small RNA populations of wild-type and paramutated tomato leaves we show here that paramutation of SLTAB2 is responsible for the sulf chlorosis and the decrease in auxin levels. SLTAB2 silencing was associated with changes in DNA methylation and siRNA levels at its promoter, a signature of RdDM.
Additional evidence supporting the identification of SLTAB2 as SULF is from virus-induced gene silencing (VIGS) of the SLTAB2 promoter resulting in methylation of the target DNA sequence, silencing of its expression, and a phenocopy of the sulf chlorosis. Together, these results support a causal role of siRNAs and RdDM in paramutation but, unlike the maize examples, the SLTAB2/SULF locus lacked repeated sequences. Mapping of SULF to SLTAB2 and further comparison with maize will help build a general model of paramutation in plants.
Materials and methods
Plant material and growth conditions
Atab2 T-DNA knockouts (GABI-KAT line 354B01) and wild-type Col-0 seedlings were sown on 1/2 strength Murashige–Skoog medium, 1× Nitsch&Nitsch vitamins, 0.8% agar, 1.5% sucrose, pH 6; stratified for 72h at 4 °C in the dark and transferred to short-day conditions (8h light at 23 °C and 50 µmol photons m–2 s−1, 16h dark at 21 °C). Whole seedlings were collected after 7 d of growth. Tomato plants were raised from seeds in compost (Levington M3) and maintained in a growth room at 23 °C with 16/8h light/dark periods with 60% relative humidity, at a light intensity of 150 µmol photons m–2 s−1. Young leaves were collected from 1-month-old plants. Sulf and sulf/+ tissue was collected from sulf/+ plants that had both fully yellow (sulf) and fully green (sulf/+) sectors.
Transcriptome analysis
Total RNA samples were prepared from 100mg of leaf tissue using TRIzol (LifeTechnologies). For qPCR, 5 µg of total RNA was first DNase treated using Turbo DNase (Ambion), following the manufacturer’s guidelines. cDNA was then synthesized using random hexamers and Oligo(dT) and SuperScript III (LifeTechnologies), according to the protocol. qPCR was performed on a Roche LC480 with SYBR in technical triplicates. mRNA abundance was normalized by the geometric mean of two housekeeping genes TIP41 and EXPRESSED (Coker and Davies, 2003). Genotyping of amplified cDNA was performed by digesting 100ng purified SLTAB2 amplicon with BaeI (NEB) for 12h at 25 °C in 1× NEB2.1, 100 µg ml−1 BSA, and 20 µM SAM as per the manufacturer’s instructions, and electrophoresis on a 1.5% agarose gel. Strand-specific RNA-Seq libraries for two wild-types and three pairs of sulf and sulf/+ were made and indexed with the ScriptSeq v2 kit (Epicentre) according to the protocol after RiboZero treatment (Plant leaf, Epicentre), and sequenced as a pool on one lane of HiSeq 2000 100PE. Sequences were trimmed and filtered with Trim Galore! with default parameters and 11–29 million reads per library were concordantly aligned on Heinz genome SL2.50 and ITAG2.4 gene models using TopHat2 v2.0.13 (Kim et al., 2013) (with parameters -r 200 --mate-std-dev 100 -N 3 --read-edit-dist 3 --library-type fr-firststrand --solexa1.3-quals, and version 2.2.4.0 of Bowtie2; alignment rate 66–73%; see Supplementary Table S1 at JXB online). Differential expression analysis was performed on raw counts on annotated mRNAs (ITAG2.4) with DESeq2 v1.8.1 (Love et al., 2014). Genes were considered differentially expressed when the adjusted P-value was <0.05. Hierarchical correlation clustering of the genes differentially expressed between wt and sulf was performed in SeqMonk (v0.32.0). Gene Ontology analysis was performed with the goseq package (v1.20.0, Young et al., 2010) using previously published gene ontology annotation (Koenig et al., 2013), normalizing with mRNA length and running with the following parameters: method=Wallenius, repcnt=2 000, use_genes_without_cat=F. Categories were considered to be over-represented if the associated P-value was <0.05 after Benjamini–Hochberg correction.
sRNA-Seq
sRNAs were cloned from 10 µg total RNA (from the same tissue as used for RNA-Seq, for two wild-types and two pairs of sulf and sulf/+) using the Illumina TruSeq Small RNA cloning kit and libraries were indexed during the PCR step (12 cycles) according to the manufacturer’s protocol. Gel size-selected, pooled libraries were sequenced on a HiSeq 2000 50SE. Sequences were trimmed and filtered with Trim Galore! (with the adapter parameter -a TGGAATTCTCGGGTGCCAAGG) and 14–20 million reads per library were mapped without mismatches and clustered on Heinz genome SL2.50 using the ShortStack software v2.1.0 (Axtell, 2013; Supplementary Table S2). sRNA counts on the defined loci were analysed with DESeq2 v1.8.1. Uniquely mapping reads on DMR1 and DMR2 were normalized with edgeR’s implementation of TMM size factors, on all sRNAs present in all libraries and with at least 10 total counts (Robinson et al., 2009), and a Poisson regression was applied to the normalized counts (generalized linear model in R, with the genotype variable taking values wt, sulf/+, and sulf).
Methylome analysis
DNA was extracted from 100mg of leaf tissue (from the same sampling as for RNA-Seq and sRNA-Seq, for two wild-types and two sulf) using the Puregene kit (QIAGEN). Bisulfite library preparation was performed with a custom protocol similar to Urich et al. (2015).1.2 µg DNA was sonicated on a Covaris E220 to a target size of 400bp and purified on XP beads (Ampure, ratio 1.8). DNA was end-repaired and A-tailed using T4 DNA polymerase and Klenow Fragment (NEB) and purified again using XP beads (ratio 1.8×). Methylated Illumina Y-shaped adapters for paired-end sequencing were ligated using Quick-Stick Ligase (Bioline). 450ng of purified (ratio 1.8×), adapter-ligated DNA was bisulfite-converted using the EZ DNA Methylation-Gold Kit (Zymo Research) according to the manufacturer’s instructions. DNA was barcoded using 12 cycles of PCR amplification with KAPA HiFi HotStart Uracil+Ready Mix (Kapa Biosystems) with PE1.0 and custom index primers (courtesy of the Sanger Institute). Pooled libraries were sequenced to a depth of about 5× on a HiSeq 2500 125PE. Sequences were trimmed and filtered with Trim Galore! (default parameters), then mapped on Heinz genome SL2.50 using Bismark v0.14.3 (Krueger and Andrews, 2011) (first in paired-end mode with options --score-min L,0,-0.2 -p 4 --reorder --ignore-quals --no-mixed --no-discordant -X 1500 --unmapped --ambiguous, then unmapped read1 was mapped in single-end mode with the same quality parameter -N 1, Supplementary Table S3). Reads were deduplicated with bismark_deduplicate and methylation calls were extracted using Bismark methylation_extractor (with options -r2 2 for paired end reads). Methylated and unmethylated counts for cytosines of both strands were pooled into contiguous 200bp bins and separated by context (CG, CHG, and CHH) with a custom python script. Bins with fewer than 10 counts were excluded from the analysis. Bins are considered differentially methylated if the maximum P-value of the two chi-square tests (wt1 versus sulf 1, wt2 versus sulf 2) is <0.01. Analysis of methylation by McrBC was performed as previously described by Bond and Baulcombe (2015). For Sanger bisulfite sequencing, 450ng of DNA was bisulfite-converted, amplified with primers specific to the region of interest, A-tailed, and cloned into pGEM-T easy (Promega) following protocols similar to the library preparation. Sequences aligned with MUSCLE were then analysed with CyMATE (Hetzl et al., 2007).
VIGS
DMR1a (606bp) and DMR1b (562bp) genomic inserts were cloned into the binary TRV RNA2 vector using the KpnI and XhoI restriction sites of the multiple cloning site as described previously (Liu et al., 2002; Bond and Baulcombe, 2015). Cotyledons of tomato seedlings were agro-infiltrated 10 d after sowing with a 1:1 mixture of Agrobacterium tumefaciens (strain GV3101:pMP90+pSOUP) carrying TRV RNA1 and RNA2 at OD600=1.5. Symptoms of SLTAB2 silencing were visible from 2 weeks post-infection.
Auxin quantification
Endogenous levels of free IAA were detected by LC-MS/MS method as described in Novák et al. (2012). Briefly, 10–20mg fresh tissue of the control and mutant lines were collected, extracted in ice-cold 50mM sodium phosphate buffer (pH 7) and purified by SPE on hydrophilic–lipophilic balance reversed-phase sorbent columns (Oasis HLB, 1 cc/30mg, Waters). To each extract, 5 pmol of 13C6-IAA were added as internal standards to validate the quantification. Purified samples were analysed by the LC-MS/MS system consisting of an ACQUITY UPLC System (Waters, Milford, MA, USA) and Xevo TQ-S (Waters) triple quadrupole mass spectrometer. Quantification was obtained using a multiple reaction monitoring (MRM) mode of selected precursor ions and the appropriate product ion.
Oligonucleotides
Please refer to Supplementary Table S4.
Accession codes
All sequencing data have been deposited in the Sequence Read Archive under the BioProject SRP066362.
Results
Pericentromeric SLTAB2 is strongly down-regulated in sulfurea
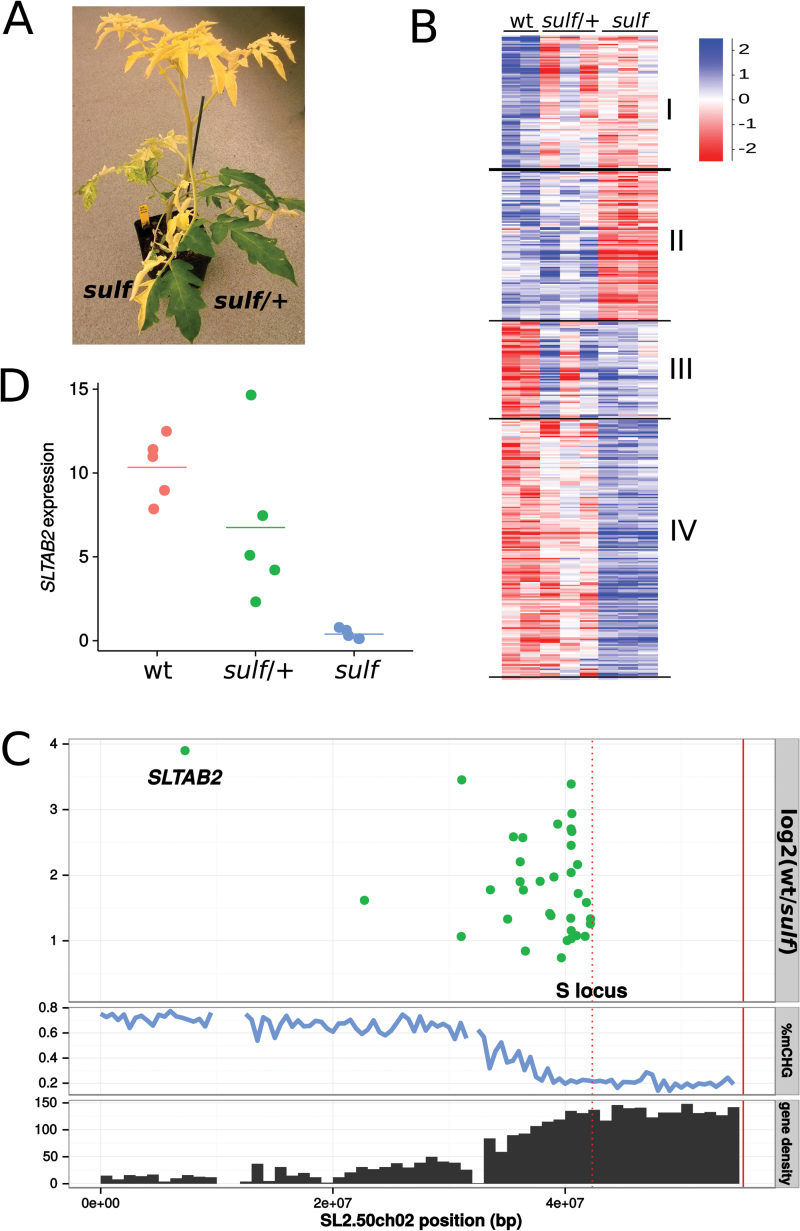
The tomato lines in this study had either unsilenced SULF loci (wild type) or they were the progeny of a cross between the wild type and a plant with sectors with silent sulf. Some of the F1 plants were wild type and fully green or, like the sectored parent, they had green and chlorotic sectors consistent with a heterozygous sulf/+ epigenotype with paramutation. The chlorotic sectors would have had the homozygous sulf/sulf epigenotype (referred to as sulf) and the non-paramutated green sectors would be sulf/+ (Fig. 1A).
Fig. 1.
(A) Fully paramutated yellow sulf tissue and non-paramutated green sulf/+ tissue on a plant grown from a heterozygous seed. (B) Hierarchical clustering of 2 237 differentially expressed genes between wt and sulf. With a correlation coefficient of 0.7, 2 223 genes fall into four broad categories: down-regulated in sulf and sulf/+ (I), down-regulated in sulf only (II), up-regulated in sulf and sulf/+ (III), and up-regulated in sulf only (IV). Values are log2-transformed library-normalized/median-normalized counts. (C) Down-regulated genes in sulf on chromosome 2. Log2 fold-change wt/sulf of significantly down-regulated genes (adjusted P <0.05). Percentage CHG methylation and gene density (genes/Mb) are plotted along chromosome 2 to show the distribution of heterochromatin. The S locus (red dotted line) marks the right-most border for the possible location of SULFUREA (Hagemann and Snoad, 1971). The end of the chromosome is marked by a solid red line. (D) SLTAB2 expression in wt, sulf/+ (green) and sulf (yellow) leaves. Horizontal bar shows the mean of five biological replicates. 26-fold reduction in sulf compared with wt (P-value=0.0002324, two–tailed t test), while the difference between wt and sulf/+ is not significant (P-value=0.1771).
Based on the understanding of paramutation in maize, the SULF locus would be suppressed in sulf (paramutated yellow sectors), partially silent in sulf/+ (non-paramutated green sectors) and fully expressed in the wild type (wt). It would also be located in the pericentromeric heterochromatin of chromosome 2 upstream of the euchromatic S locus (Hagemann and Snoad, 1971). To find loci with these characteristics, we analysed transcripts of wild-type, non-paramutated sulf/+, and paramutated sulf leaves using mRNA-seq. We identified 2 237 differentially expressed genes between sulf and wt (P <0.05) that clustered into four main categories with distinct Gene Ontology enrichments (Fig. 1B; Table 1). Consistent with a decrease in photosystem I and the quantity of pigment in sulf leaves (Ehlert et al., 2008), many photosynthesis-related genes were down-regulated (Table 1, class II). The down-regulation of photosystem I was also detectable in the non-paramutated heterozygous sulf/+ leaves (Table 1, class I). Genes associated with various stress responses were up-regulated in sulf (Table 1, class IV) and were probably a secondary consequence of the sulf phenotype.
Table 1.
Enriched gene ontology terms in the 4 hierarchical clusters
| Down-regulated in sulf and sulf/+ (I, 467 genes) | Down-regulated in sulf (II, 524 genes) | Up-regulated in sulf and sulf/+ (III, 339 genes) | Up-regulated in sulf (IV, 893 genes) |
|---|---|---|---|
| Sucrose metabolic process | Photosystem II | Tyramine N-feruloyltransferase activity | Nucleolus |
| Starch metabolic process | Photosynthesis, light harvesting | Plasma membrane part | Ribosome biogenesis |
| Microtubule | Chlorophyll binding | Killing of cells of other organism | Mitochondrion |
| Microtubule-based movement | Protein-chromophore linkage | N-acetyltransferase activity | Translation |
| Endomembrane system | Photosystem I | Unfolded protein binding | |
| Plasma membrane | Chloroplast thylakoid membrane | Ribosome | |
| Microtubule motor activity | Integral component of membrane | Response to heat | |
| Cellulase activity | Light-harvesting complex | RNA binding | |
| Protein-chromophore linkage | Metal ion binding | Arsenate reductase (glutaredoxin) activity | |
| Mannan synthase activity | Membrane | Structural constituent of ribosome | |
| Photosystem I | Plastoglobule | Chloroplast | |
| Metal ion transport | Sterol biosynthetic process | snoRNA binding | |
| Chlorophyll binding | Photosystem II antenna complex | Response to stress | |
| Carbohydrate metabolic process | Water transport | RNA processing | |
| Response to red light | Protein folding | ||
| Response to blue light | Plastid chromosome | ||
| Response to water deprivation | Protein disulfide oxidoreductase activity | ||
| Water channel activity | DNA-directed RNA polymerase activity | ||
| Plasma membrane light-harvesting complex | Binding | ||
| Electron carrier activity | Purine nucleobase metabolic process | ||
| Chlorophyll biosynthetic process | Cytosolic large ribosomal subunit | ||
| Osmosensor activity | Protein refolding | ||
| Non-photochemical quenching | Cell redox homeostasis | ||
| Transferase activity, transferring hexosyl groups | Nucleoid | ||
| ATP-dependent helicase activity | |||
| Pyrimidine nucleobase metabolic process | |||
| Translation elongation factor activity | |||
| Protein processing | |||
| Plastid organization | |||
| Endonuclease activity | |||
| Cobalt ion binding |
Of these differentially expressed genes, 36 were both down-regulated in sulf and located upstream of the S locus on chromosome 2. Among these candidates for SULF, Solyc02g005200 particularly stood out as being the most repressed in sulf (15-fold reduction) and at the predicted map position of SULF (29 cM from S locus, when the centromere-S distance is 30 cM). The other candidates mapped to the euchromatin or the transition zone between heterochromatin and euchromatin (Fig. 1C; Supplementary Table S5). Further qPCR analysis confirmed the strong down-regulation of Solyc02g005200 in sulf (26-fold) and revealed variable levels in sulf/+, compatible with mono-allelic expression (Fig. 1D). Solyc02g005200 is the orthologue of Arabidopsis thaliana ATAB2 that is likely involved in the translation of mRNAs for both photosystems (Barneche et al., 2006) and we refer to it as SLTAB2.
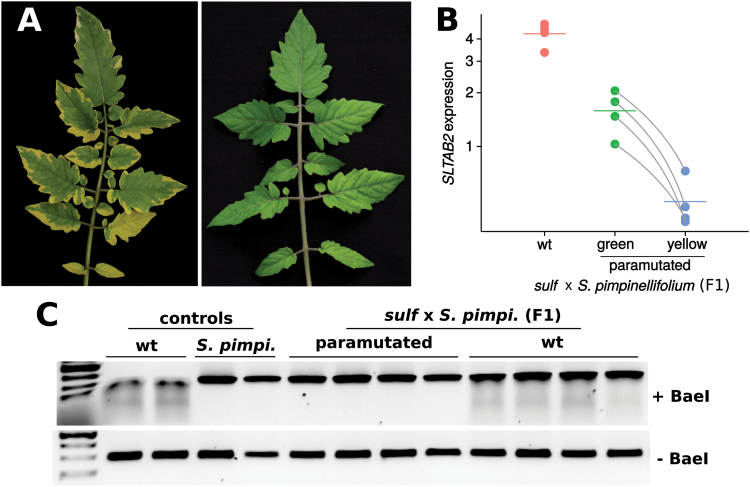
To confirm the heritability of the SLTAB2 silent epiallele, we analysed SLTAB2 in the F1 progeny of chlorotic sulf/+ (S. lycopersicum cv. Lukullus) crossed with S. pimpinellifolium. Out of 22 F1 plants, 8 displayed a paramutated phenotype with yellowing of parts of the leaves (Fig. 2A). The expression of SLTAB2 in these chlorotic plants was reduced by about half in their green sectors compared to wild-type plants, and 9-fold in their yellow sectors (Fig. 2B). Furthermore, in a PCR test that differentiated the polymorphic alleles from the two parents, we only detected expression from S. pimpinellifolium (Fig. 2C). These data are consistent with SLTAB2 being SULFUREA: in the green tissue the S. pimpinellifolium allele would have been expressed (but not the silent allele from the sulf/+ S. lycopersicum parent), and in the chlorotic tissue, it would have been paramutated. In addition, by confirming the heritable silencing of the S. lycopersicum allele in the chlorotic plants, these data confirm that SLTAB2 silencing is not merely a consequence of the sulf phenotype.
Fig. 2.
SLTAB2 paramutation in the cross between a heterozygous sulf/+ and S. pimpinellifolium. (A) Phenotypes of a paramutated (left) and wild-type leaf (right) in the F1. (B) SLTAB2 expression in the F1. Paramutated plants showed reduced expression in the green sectors compared with wt plants (P=7.8e-4, two-tailed t test), and even further reduction in the chlorotic sectors (P=6.6e-3, paired t test between green and yellow samples). Data are plotted on a log2 scale, the mean is represented by a horizontal bar. Paired data points for paramutated plants (green and yellow sectors) are joined by grey lines. (C) Only the S. pimpinellifolium SLTAB2 allele is expressed in paramutated F1 plants. The S. lycopersicum allele is sensitive to digestion by the BaeI restriction enzyme, resulting in a smear. Two SNPs in the S. pimpinellifolium SLTAB2 allele make it resistant to BaeI treatment. F1s expressing the S. lycopersicum allele show a smear, whereas F1s in which the S. lycopersicum allele is silent do not.
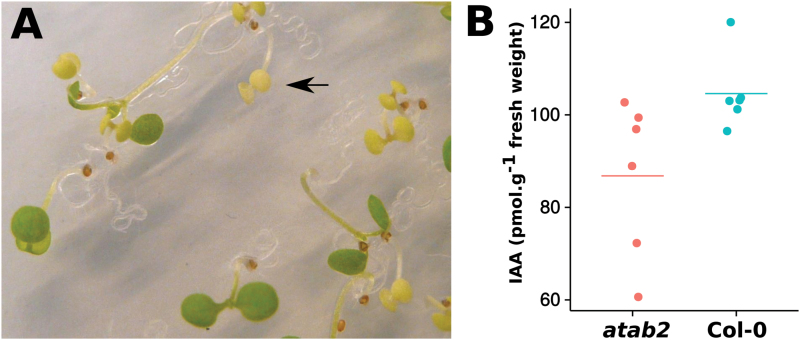
Furthermore, consistent with equivalence of SULF and SLTAB2, the Arabidopsis T-DNA knockout of ATAB2 is seedling-lethal in heterotrophic conditions and deficient in green pigment (Barneche et al., 2006; Fig. 3A). This mutant also has less auxin than the wild type (Fig. 3B). These three phenotypes all resemble sulf.
Fig. 3.
Arabidopsis atab2 mutants resemble tomato sulfurea. (A) Segregating atab2 mutation in seedlings. Homozygous atab2 mutants (arrow) are chlorotic and seedling-lethal on heterotrophic medium. (B) Decreased auxin in atab2 (P-value=0.02497, Kruskal-Wallis rank sum test).
Paramutation is associated with changes in SLTAB2 promoter DNA methylation
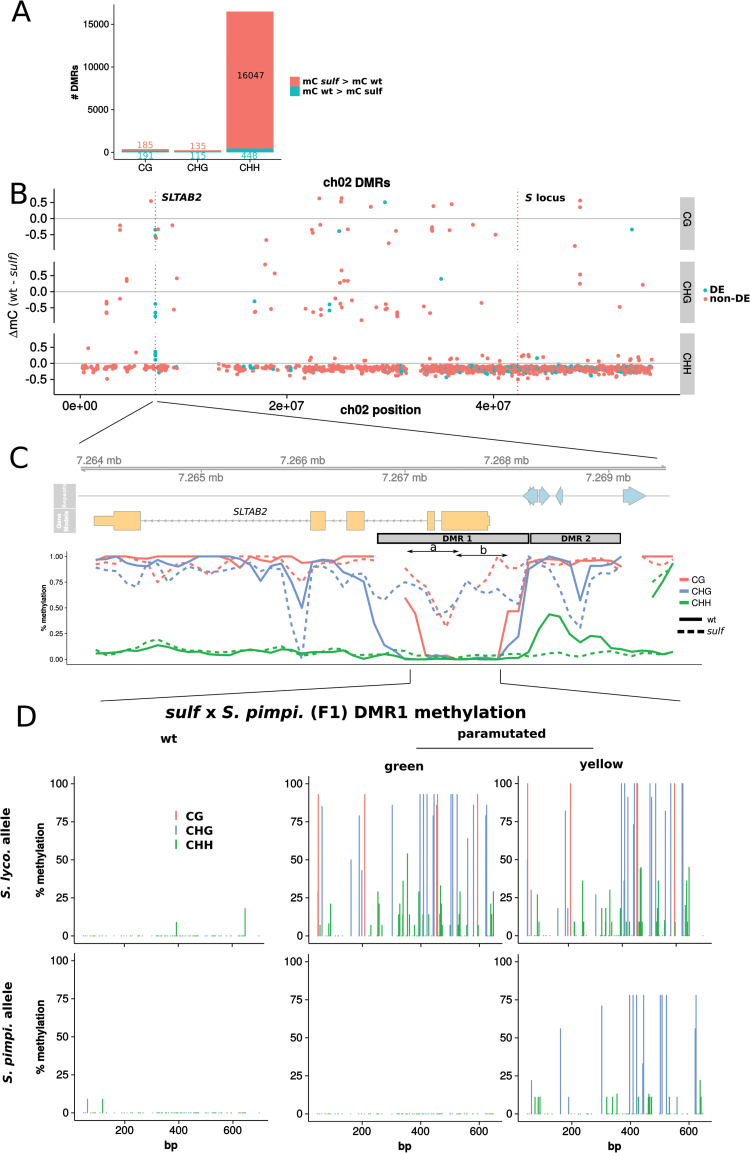
We predicted, based on the analysis of the maize b1 gene (Stam et al., 2002), that the DNA of the paramutagenic sulf would be hypermethylated. After a genome-wide analysis of differentially methylated regions (DMRs) between wt and sulf, we looked for candidate loci in the appropriate region of chromosome 2 and adjacent to genes that were differentially expressed. Genome-wide there were thousands of such DMRs in the CHH context and hundreds in the CG and CHG contexts, with the CHH DMRs being predominantly hypermethylated in sulf whereas CG and CHG DMRs were evenly split between hyper- and hypo-DMRs (Fig. 4A). On chromosome 2 there were several differentially expressed genes with adjacent CHH DMRs but only the SLTAB2 locus had strong DMRs in all cytosine contexts (Fig. 4B).
Fig. 4.
DMRs between wild-type and sulfurea. (A) Hypo- and hyper-DMRs between wild-type and sulfurea. DMRs in the CHH context are the most abundant (1% of tested bins), and heavily biased for hypermethylation in sulf. (B) Methylation difference (wt–sulf) of chromosome 2 DMRs. Negative values indicate hypermethylation in sulfurea, positive values hypomethylation. DMRs whose downstream gene is differentially expressed (DE) between wild-type and sulfurea are coloured in blue, while DMRs whose downstream gene is not differentially expressed (non-DE) are coloured red. (C) Methylation over SLTAB2 promoter, plotted in 200bp sliding windows (step of 100bp). Repeats are plotted as blue arrows. (D) Wild-type sulf×S. pimpinellifolium F1s bear no methylation on either the S. lycopersicum or S. pimpinellifolium alleles at DMR1 (region 7 267 172 to 7 267 898 encompassing the S. pimpinellifolium SNPs, 11 clones per allele). In the green sectors of paramutated F1s only the S. lycopersicum allele is methylated (14 clones, 7 clones for S. pimpinellifolium allele), whereas in the yellow sectors methylation is found on both alleles (11 and 9 clones).
Closer inspection revealed that there are two adjacent DMRs in the immediate promoter of SLTAB2, DMR1 and DMR2 (Fig. 4C; Table 2). DMR2 overlaps annotated repeats directly upstream of SLTAB2 and was hypomethylated in sulf in the CHG and CHH contexts (78% and 4% methylation, respectively, compared with 93% and 27% in the wild type). DMR1, by contrast, overlaps the transcriptional start site of SLTAB2 from 300bp upstream, encompassing the first two exons and introns, and it was hypermethylated in sulf in all contexts (73% mCG, 62% mCHG, and 4% mCHH compared with 24%, 4%, and 0.7%, respectively, in wt). This hypermethylation of the transcriptional start site of SLTAB2 is consistent with a decrease in transcription in sulf and it further strengthens the case that SLTAB2 is SULF.
Table 2.
SLTAB2 DMR methylation
Percentage methylation from two replicates. P-values from a logistic regression analysis on raw methylated and unmethylated cytosine counts.
| DMR | Context | wt | sulf | P-value |
|---|---|---|---|---|
| DMR1 | CG | 21.4; 27.3 | 71.4; 76.8 | <2×10–16 |
| CHG | 2.3; 5.8 | 61.9; 62.4 | <2×10–16 | |
| CHH | 0.5; 0.8 | 3.8; 4.8 | 1.12×10–7 | |
| DMR2 | CG | 95.6; 96.4 | 96.6; 93.7 | 0.557 |
| CHG | 94.4; 90.9 | 78.0; 78.4 | 7.34×10–4 | |
| CHH | 25.6; 29.3 | 5.5; 3.6 | <2×10–16 |
Additional evidence for the relevance of DMR1 methylation for paramutation is our finding that the silencing of the S. pimpinellifolium allele in the chlorotic F1 (sulf/+×S. pimpinellifolium) coincided with a hypermethylation of this region (Fig. 4D). The silent S. lycopersicum allele was methylated in all parts of the paramutated plants, whereas the S. pimpinellifolium allele was unmethylated in green sectors and methylated in yellow sectors.
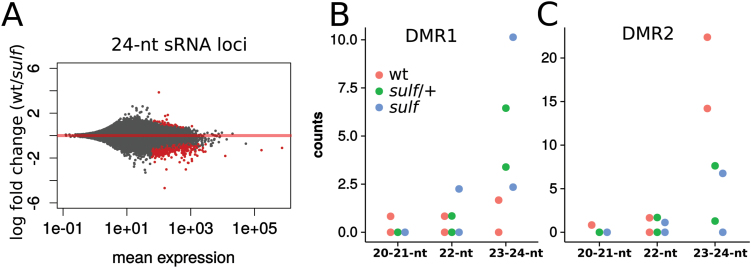
We also predicted, based on the maize paramutation examples, that sulf would correlate with 24-nt siRNAs. At a genome-wide scale, there was a distinct increase in 23–24-nt siRNAs in sulf compared with the wild type (Fig. 5A), in line with the pattern of CHH hypermethylation. At DMR1 of SLTAB2, the 23–24-nt siRNAs were more abundant in paramutated sulf (Fig. 5B) than in the wild type whereas at DMR2 the 23–24-nt siRNAs were less abundant than in the wild type (Fig. 5C).
Fig. 5.
sRNAs in sulfurea. (A) MA plot of 24-nt sRNA loci in wt and sulf. Of the differential sRNA loci between wt and sulf (in red, adjusted P <0.05), 498 had more abundant sRNAs in sulf, while only 48 had more abundant sRNAs in the wild type. (B) sRNA counts on DMR1 in the wild type, heterozygous sulf/+, and homozygous paramutated sulf leaves. 23–24-nt sRNAs are rare but more abundant in sulf (P=0.0146, Poisson regression). (C) sRNA counts on DMR2. 23–24-nt siRNAs are reduced in sulf (P=5.48e − 05, Poisson regression). Counts in (B) and (C) are for normalized, uniquely mapping reads.
VIGS of SLTAB2 phenocopies sulf
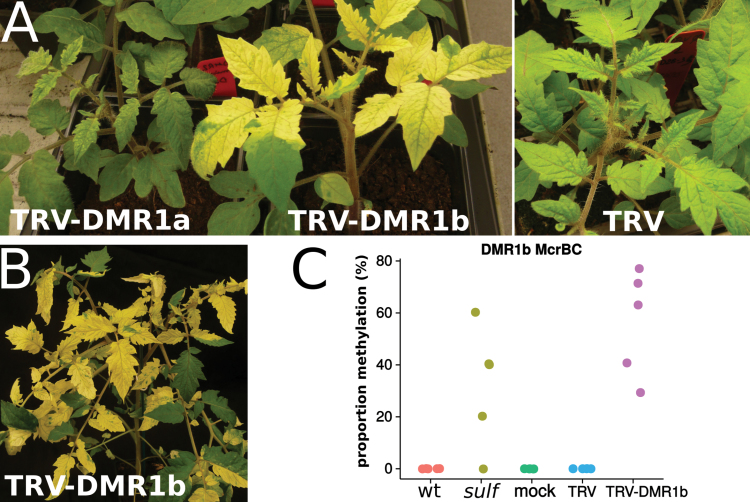
To test the involvement of SLTAB2 in sulf paramutation further, we used virus-induced gene silencing (VIGS). VIGS, when targeted to transcribed sequences, leads to knock-down of mRNA levels by post-transcriptional gene silencing but, when targeted at DNA sequences [e.g. the FWA promoter in A. thaliana (Bond and Baulcombe, 2015)], it can initiate heritable DNA methylation and transcriptional gene silencing. We cloned two segments (a and b, Fig. 4C) of DMR1 into tobacco rattle virus (TRV) RNA2 and inoculated them with TRV RNA1 to wild-type tomato. While infection with TRV-DMR1a caused only mild variegation of the leaves, infection with TRV-DMR1b caused almost all plants to develop large sulf-like chlorotic sectors (Fig. 6A). The similarity of this VIGS phenotype to leaves of sulf is further evidence that SLTAB2 and SULF are equivalent.
Fig. 6.
VIGS of SLTAB2 DMR1. (A) At 3 weeks post-infection, plants infected with TRV-DMR1b displayed large chlorotic sectors. Between 13 and 16 plants were infected for each condition. (B) These sulfurea-like sectors remained in later leaves at 2 months after infection. (C) Methylation of DMR1 is detectable in chlorotic sectors. The proportion of methylation is calculated from the ratio of amplicons in the McrBC-digested versus undigested samples (as determined by qPCR). If all alleles are highly methylated (100% methylation), they will be digested by McrBC and no amplification will occur during qPCR.
DNA methylation analysis of chlorotic sectors by McrBC suggested that there is an epigenetic component to the silencing of SLTAB2 by TRV-DMR1b: the targeted DNA was as strongly methylated as in sulf samples (Fig. 6C). The involvement of epigenetics is further supported by the lasting VIGS phenotype several months post-inoculation (Fig. 6B) during which time the level of the virus vector decreased. From these data we conclude that the DMR1b region of SLTAB2 has the predicted characteristics of the paramutagenic component of sulf because it is susceptible to epigenetic modification.
Discussion
In this paper we present several lines of evidence that SLTAB2 is SULF: it maps closer to SULF than any other genes with the predicted pattern of mRNA accumulation in sulf/+ and sulf tissue (Fig. 1B); it encodes a protein required for photosystem I production that explains the chlorotic phenotype; the orthologous atab2 mutation has the same chlorosis and auxin-deficient phenotype as sulf (Fig. 3); and there is a definite DNA methylation mark at the silent SLTAB2 allele in sulf that is inherited from sulf/+ both in selfed and outcrossed progeny (Fig. 2). This epigenetic mark is transferred to the previously active allele in F1 progeny (Fig. 4D) and is associated with 24-nt siRNAs (Fig. 5). The final evidence for the equivalence of SLTAB2 and SULF is from the finding that VIGS targeted to the SLTAB2 DMR can recapitulate both the physiological and epigenetic features of sulf (Fig. 6).
We envision that paramutation occurs when methylation of DMR1 DNA by paramutagenic siRNAs starts a positive feedback loop in which Pol IV is recruited to the silent locus by SHH1 (Law et al., 2013). The recruited Pol IV (Blevins et al., 2015; Zhai et al., 2015) would transcribe siRNAs that would mediate maintenance of the silent state of SLTAB2 and its transfer to paramutable alleles.
SLTAB2 had been ruled out previously as SULF because it is expressed at detectable levels in sulf and it was thought that, unlike tomato, the mutation of the Arabidopsis orthologue could be rescued on sucrose (Ehlert et al., 2008). An alternative candidate for SULF was implicated in the tryptophan-independent pathway of auxin biosynthesis. With our new data, however, we show that the previous exclusion of SLTAB2 was not valid because targeted suppression by VIGS or mutation of this gene produces an accurate phenocopy of the sulf phenotype including, in the atab2 mutant, an auxin defect and seedling lethality.
A likely explanation for the sulf phenotype based on silencing of SLTAB2 invokes the failure to translate the psaB mRNA as described for the orthologous mutations ATAB2 in Arabidopsis and TAB2 in Chlamydomonas (Dauvillée et al., 2003; Barneche et al., 2006). The PsaB protein is the reaction centre protein of photosystem I and, in its absence, the thylakoid membranes would fail to form, pigments would not accumulate at the normal levels, and the leaves would be chlorotic. An auxin defect of sulf is a likely consequence of the PsaB defect, as observed in the atab2 mutant (Fig. 3). In addition, the genome-wide hypermethylation in the CHH context in sulf is reminiscent of transient hypermethylation in response to stress, already described in phosphate-starved rice (Secco et al., 2015) and virus-infected Arabidopsis (Bond and Baulcombe, 2015).
Several cases of paramutation have been tied to the hypermethylation of regulatory tandem repeats (Stam et al., 2002; Chandler and Stam, 2004). By contrast, the sulfurea paramutation is correlated with increased DNA methylation at the transcriptional start site (DMR1) where there is no repeated DNA (Fig. 4). There are annotated LTR fragments at DMR2 that is directly adjacent to DMR1 but, unlike the classic systems, this region of the paramutagenic sulf allele showed a reduction in sRNAs and hypomethylation. Hypomethylation of DMR2 may be a consequence of a different chromatin state of the silenced allele and, although its repeats may not be directly involved in the silencing of SLTAB2, they and the largely heterochromatic region in which SLTAB2 is embedded may contribute to its paramutability, poising it for silencing.
The opportunity to study paramutation via SLTAB/SULF in tomato has several advantages over the various maize systems that have been most informative until now. First we have a VIGS system so that establishment of the epigenetic mark can be tracked directly in tomato mutants that are defective for components of the RNA silencing pathways. We will also be able to use VIGS on sulf/+ plants to test the role of various tomato genes in the establishment and maintenance of paramutagenicity and paramutability.
A second benefit of the sulf system is the possibility of studying the transfer of the epigenetic mark in vegetative tissue. With the well-studied maize paramutation systems this transfer is likely to occur early in embryo development and is not readily accessible to molecular analysis, whereas, in tomato, it will be taking place in or close to vegetative meristems. It will still not be easy to access the cells in which the allelic transfer is taking place but we may be able to use the DMR1-specific siRNAs as markers of the transfer process. These RNAs are rare in total plant extracts (Fig. 5) but they may be more abundant at the primary sites of paramutation. Having identified the SULF gene, we should also be able to complement the physiological consequences by providing a transgene without the target DNA of paramutation so that we can grow plants with a sulf/sulf epigenotype.
These various experimental tools will allow us to explore the differences of the tomato and maize paramutation systems. For example, the apparent target of paramutation in sulf has no tandem repeats and is close to the transcriptional start whereas, in maize, at the B locus, they are essential and separated from the transcribed region by 100kb. Answers to these and other questions will allow us to explore the frequency of paramutation-like events in plant breeding and evolution. Several recent findings indicate that such events are not restricted to the few well-characterized examples of paramutation in maize and other species (Regulski et al., 2013; Greaves et al., 2014). They may be frequent and have an effect on transgressive and heterotic phenotypes.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Summary of RNA-Seq alignments.
Table S2. Summary of sRNA-Seq alignments.
Table S3. Summary of whole-genome bisulfite sequencing alignments.
Table S4. List of primers.
Table S5. Summary of down-regulated genes in the genetically defined region of SULFUREA.
Acknowledgements
We thank Mel Steer and Jarmila Greplová for technical assistance, Donna Bond and Ottoline Leyser for advice and discussions, Fredy Barneche for sharing atab2 seeds, and Adrian Valli, Jurek Paszkowski, and Krys Kelly for critical reading of the manuscript. IPK Gatersleben kindly provided sulfurea and Lukullus seeds. This work was supported by the European Research Council Advanced Investigator Grant ERC-2013-AdG 340642, the Frank Smart Studentship, and the Ministry of Education, Youth and Sports of the Czech Republic (the National Program for Sustainability I Nr. LO1204). DCB is the Royal Society Edward Penley Abraham Research Professor.
References
- Axtell MJ. 2013. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19, 740–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneche F, Winter V, Crèvecoeur M, Rochaix J-D. 2006. ATAB2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. The EMBO Journal 25, 5907–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, Rusch D, Tang H, Pikaard CS. 2015. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis . eLife 4, e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D, Baulcombe DC. 2015. Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 112, 917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL, Stam M. 2004. Chromatin conversations: mechanisms and implications of paramutation. Nature Reviews Genetics 5, 532–544. [DOI] [PubMed] [Google Scholar]
- Coker JS, Davies E. 2003. Selection of candidate housekeeping controls in tomato plants using EST data. BioTechniques 35, 740–746. [DOI] [PubMed] [Google Scholar]
- Dauvillée D, Stampacchia O, Girard-Bascou J, Rochaix JD. 2003. Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. The EMBO Journal 22, 6378–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert B, Schöttler MA, Tischendorf G, Ludwig-Müller J, Bock R. 2008. The paramutated SULFUREA locus of tomato is involved in auxin biosynthesis. Journal of Experimental Botany 59, 3635–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES. 2014. Inheritance of Trans Chromosomal Methylation patterns from Arabidopsis F1 hybrids. Proceedings of the National Academy of Sciences, USA 111, 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann R. 1958. Somatische Konversion bei Lycopersicon esculentum Mill. Zeitschrift für Vererbungslehre 89, 587–613. [PubMed] [Google Scholar]
- Hagemann R. 1969. Somatic conversion (paramutation) at the sulfurea locus of Lycopersicon esculentum Mill.: IV. The genotypic determination of the frequency of conversion. Theoretical and Applied Genetics 39, 295–305. [DOI] [PubMed] [Google Scholar]
- Hagemann R, Snoad B. 1971. Paramutation (somatic conversion) at the Sulfurea locus of Lycopersicon esculentum: V. The localisation of Sulf . Heredity 27, 409–418. [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. 2007. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. The Plant Journal 51, 526–536. [DOI] [PubMed] [Google Scholar]
- Hollick JB. 2012. Paramutation: a trans-homolog interaction affecting heritable gene regulation. Current Opinion in Plant Biology 15, 536–543. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D, Jiménez-Gómez JM, Kimura S, et al. 2013. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proceedings of the National Academy of Sciences, USA 110, E2655–E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AMS, Strahl BD, Patel DJ, Jacobsen SE. 2013. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. 2002. Virus-induced gene silencing in tomato. The Plant Journal 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K. 2012. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. The Plant Journal 72, 523–536. [DOI] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MT, et al. 2013. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Research 23, 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2009. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Schultz MD, Chiarenza S, Ecker JR, Whelan J, Lister R. 2015. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4, e09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, Belele C, Ramakrishna W, Dorweiler JE, Bennetzen JL, Chandler VL. 2002. The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics 162, 917–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tomato Genome Consortium 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich M, Nery JR, Lister R, Schmitz RJ, Ecker JR. 2015. MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nature Protocols 10, 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, et al. 2015. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.