Highlight
Plastid RPS5 affects proteins involved in photosynthesis and translation machinery and mediates cold stress tolerance in Arabidopsis.
Key words: Arabidopsis, cold stress tolerance, photosynthesis, plastid ribosomal protein, proteomics, RPS5.
Abstract
Plastid ribosomal proteins are essential components of protein synthesis machinery and have diverse roles in plant growth and development. Mutations in plastid ribosomal proteins lead to a range of developmental phenotypes in plants. However, how they regulate these processes is not fully understood, and the functions of some individual plastid ribosomal proteins remain unknown. To identify genes responsible for chloroplast development, we isolated and characterized a mutant that exhibited pale yellow inner leaves with a reduced growth rate in Arabidopsis. The mutant (rps5) contained a missense mutation of plastid ribosomal protein S5 (RPS5), which caused a dramatically reduced abundance of chloroplast 16S rRNA and seriously impaired 16S rRNA processing to affect ribosome function and plastid translation. Comparative proteomic analysis revealed that the rps5 mutation suppressed the expression of a large number of core components involved in photosystems I and II as well as many plastid ribosomal proteins. Unexpectedly, a number of proteins associated with cold stress responses were greatly decreased in rps5, and overexpression of the plastid RPS5 improved plant cold stress tolerance. Our results indicate that RPS5 is an important constituent of the plastid 30S subunit and affects proteins involved in photosynthesis and cold stress responses to mediate plant growth and development.
Introduction
Ribosomes are large ribonucleoprotein complexes that are essential for protein synthesis in all living cells. Cytoplasm, plastids, and mitochondria are the three major locations for protein synthesis in plants. Plastid protein synthesis utilizes a bacterial-type 70S ribosome that comprises one small (30S) and one large (50S) ribosomal subunit (Schippers and Mueller-Roeber, 2010; Tiller and Bock, 2014). The chloroplast 30S subunit contains a single 16S rRNA and 24 plastid ribosomal proteins (RPs). The 50S subunit consists of three rRNAs (23S, 5S, and 4.5S) and 33 plastid RPs. While all four chloroplast rRNAs are encoded by the plastid genome, the plastid RPs are encoded by both plastid and nuclear genomes. Of the 24 RPs in the 30S small subunit (RPS), half are nuclear encoded and the remainder are encoded by the plastid genome (Yamaguchi et al., 2000). However, 24 of the plastid RPs in the 50S large subunit (RPL) are encoded by the nuclear genome, whereas only nine are encoded by the plastid genome (Yamaguchi and Subramanian, 2000). Plastid RPs have been shown to play versatile roles in plant growth and development (Fleischmann et al., 2011; Tiller et al., 2012). While some RPs are not essential for ribosome accumulation and translation, the others are required for basal ribosome activity and influence specific processes in plant growth and development (Tiller and Bock, 2014).
A range of phenotypic effects are observed as a consequence of the lack of some individual plastid RPs, as shown with cytosolic RPs (Carroll, 2013; Schippers and Mueller-Roeber, 2010). Seven Arabidopsis plastid RP mutants (prps20 and prpl1, l4, l21, l27, l28, and l35) are embryo lethal, indicating an essential role of these plastid proteins in embryo development (Romani et al., 2012; Yin et al., 2012). The tobacco prps18 knockout mutant shows misshapen leaves and abnormal leaf blades, which reveals the importance of this plastid protein in leaf development (Rogalski et al., 2006). The rps21 mutation in Arabidopsis impairs photosynthesis and chloroplast development, and is hypersensitive to glucose (Morita-Yamamuro et al., 2004). A defect in photosynthesis, pale green leaves, and a drastic reduction in growth rate are observed in the Arabidopsis prpl11 mutant (Pesaresi et al., 2001) and a number of other prp mutants (prps1, prps17, and prpl24) (Romani et al., 2012; Tiller et al., 2012). While some plastid RP gene mutations cause reductions in rRNA levels with impaired ribosome function, some others do not affect plastid rRNA accumulation and the assembly of plastid polysomes (Pesaresi et al., 2001; Romani et al., 2012; Tiller et al., 2012). Clearly, these studies reveal diverse roles of plastid RPs in plant growth development. However, how plastid RPs regulate these processes is not fully understood, and the functions of some individual plastid RPs remain largely unknown.
Chloroplasts, as the most conspicuous plastid type, carry out many essential metabolic processes, including photosynthesis, fatty acid synthesis, amino acid synthesis, and carotenoid metabolism, and play an important role in plant development (Rolland et al., 2012). To identify genes that affect chloroplast development and pigment synthesis, in this study we screened an Arabidopsis ethyl methanesulfonate (EMS)-mutagenized M2 population for leaf color mutants. Here, we report the isolation and functional characterization of a pale yellow inner leaf rps5 mutant in Arabidopsis, which encoded a plastid RP S5 (RPS5). rps5 showed serious impairment of chloroplast 16S rRNA processing and accumulation. A global and quantitative examination of proteomes via comparative proteomic analysis revealed that RPS5 affected the accumulation of a large number of photosystem I and II proteins and plastid RPs. In addition, it affected some proteins associated with cold stress responses, and overexpression of RPS5 resulted in enhanced cold tolerance. Our data reveal that RPS5 is important for plastid ribosome function and is involved in photosynthesis, plant development, and cold stress resistance in Arabidopsis.
Materials and methods
Plant materials and growth conditions
All Arabidopsis thaliana plants used in this study were Columbia ecotype except where otherwise mentioned. Seeds of Arabidopsis were mutagenized by EMS and the M2 population was screened for the yellow leaf phenotype. Arabidopsis plants were grown on soil under controlled chamber conditions with a 16h light/8h dark photoperiod at 25 °C. Arabidopsis T-DNA homozygous lines of Salk_095863 from the ABRC stock center were identified by PCR using gene-specific primer sets (see Supplementary Table S1 at JXB online).
For cold treatment, the surface-sterilized seeds were germinated and grown in Murashige and Skoog agar medium (4.3g l−1 Murashige and Skoog salts and 12g l−1 agar, pH 5.8) plates with 10g l−1 sucrose at 4 °C under light. After 6 weeks of growth, the cold-treated seedlings were photographed and analyzed.
Pigment quantification and seedling weight measurement
Total chlorophyll from 4-week-old rosette leaves (25mg) was extracted with 80% acetone. The absorbance of the supernatants was measured at 645 and 663nm, and chlorophyll content was calculated by simultaneous equations as described by Inskeep and Bloom (1985). Carotenoids from leaf tissue of the same age (50mg) were extracted and analyzed as described previously (Li et al., 2012). The samples were analyzed with three biological replicates.
For cold-treated seedling weight measurement, 15 seedlings from each line were combined and weighed. The analysis was performed with four biological replicates.
Map-based cloning
An F2 mapping population was generated by crossing the rps5 mutant in Columbia background with wild-type Arabidopsis ecotype Landsberg erecta. Genomic DNA from a total of 1258 homozygous rps5 F2 plants was isolated and used for fine mapping. Linkage analysis was conducted using simple sequence length polymorphism markers based on the TAIR database (http://www.arabidopsis.org/) and Arabidopsis Mapping Platform (http://amp.genomics.org.cn/).
Plasmid construction and plant transformation
For molecular complementation of the rps5 mutant, a full-length RPS5 (At2g33800) cDNA was amplified by PCR and cloned into pCAMBIA1300s (Zhou et al., 2011) to generate the overexpression plasmid 35S:RPS5. A native promoter complementation construct of RPS5:RPS5 was generated, which carried 2.6kb genomic DNA including the native promoter, 5ʹ-untranslated region (UTR), coding region, 3ʹ-UTR, and 3ʹ flanking sequences of RPS5. The PCR amplification primer sets used are shown in Supplementary Table S1. All the constructs were confirmed by sequencing.
The plasmids were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation and transformed into Arabidopsis plants by floral dip. Positive transgenic plants were screened on Murashige and Skoog plates containing 30mg l−1 carbenicillin and 50mg l−1 hygromycin (Yuan et al., 2015).
Subcellular localization
Full-length cDNAs of RPS5 and rps5 were cloned into the pGPTVII.GFP vector (Walter et al., 2004) and electroporated into A. tumefaciens strain GV3101. Agrobacterium cells carrying the 35S:RPS5-GFP or 35S:rps5-GFP construct were infiltrated into 4-week-old tobacco leaves for transient expression. After infiltration, leaves of transgenic tobacco expressing RPS5-GFP and rps5-GFP fusion proteins were analyzed by a Leica TCS SP5 laser scanning confocal microscope as described by Zhou et al. (2015). The primers used for the plasmid construction are listed in Supplementary Table S1.
RNA isolation and qRT-PCR
Total RNA from 4-week-old wild-type, rps5, and Salk_095863 rosette leaves was extracted and used for cDNA synthesis with random hexamer primers as described by Zhang et al. (2015). Quantitative real-time RT-PCR (qRT-PCR) was performed using iTaqTM Universal SYBR Green Supermix (Bio-Rad) with gene-specific primer sets (Supplementary Table S1). Thermal cycling conditions consisted of a first step of denaturation at 95 °C for 10min, followed by 40 cycles of denaturation for 15s at 95 °C and annealing/extension for 1min at 60 °C. Relative expression levels were calculated using the △△Ct method (Lyi et al., 2007) and normalized first with a ubiquitin control and then with the expression of wild-type controls. Each sample was quantified in triplicate with three biological replicates.
RNA gel blotting
Total RNA (3 μg) from 4-week-old leaves was separated on 1.5% denaturing agarose gels containing 2.2M formaldehyde, transferred to a positively charged nylon membrane (Hybond-N, Amersham), and fixed by UV crosslinking. 16S, 18S, 23S, 5S, and 4.5S rRNA probes were amplified by PCR using gene-specific primers as described by Yu et al. (2008). 32P-labeled cDNA probes were generated using a random primer DNA-labeling kit (Life Technologies) following the manufacturer’s protocol. Filters were pre-hybridized for 1h at 42 °C using NorthernMax prehybridization/hybridization buffer (Life Technologies), hybridized with 32P-labeled DNA probes overnight at 42 °C, and washed as described by Lyi et al. (2005). Storm 860 PhosphorImager (Molecular Dynamics) was used to detect the signals. The intensity of the image bands was quantified using ImageJ (http://www.di.uq.edu.au/sparqimagejblots).
Proteomics analysis
Total proteins from 4-week-old rosette leaves of wild-type and rps5 (Supplementary Figure S1) were extracted using the phenol extraction method (Yang et al., 2007). Isobaric tags for relative and absolute quantification (iTRAQ)-based proteomic analysis including protein purification, digestion, iTRAQ labeling, protein identification, and quantification was performed following methods detailed previously (Yang et al., 2011). The iTRAQ-labeled samples were analyzed on Orbitrap Elite using a two-dimensional (2D) LC-MS/MS approach. In addition, a gel-based proteomic analysis including 2D gel electrophoresis, comparative 2D gel image analysis using SameSpots software (Nonlinear Dynamics), in-gel digestion, and 2DGeLC sample analysis on Synapt HDMS coupled with nan-Acquity was carried out as described (Wang et al., 2013; Yang et al., 2007, 2011). Proteins extracted from RPS5-overexpressing lines were analyzed on Orbitrap Elite using a label-free approach and the relative quantification of PRS5 was determined by extracted ion chromatograms as described by Vaidya et al. (2013). The peak areas of detected precursor ions at each specific m/z corresponding to the targeted peptides were obtained with mass tolerance at 5ppm using Xcalibur 2.2 software.
The functional classification of identified proteins was conducted based on the bincodes of MapMan (http://mapman.gabipd.org/web/guest/mapman). Protein subcellular localizations were predicted by TAIR10 (https://www.arabidopsis.org/) and the PPDB database (http://ppdb.tc.cornell.edu/).
Results
The rps5 mutant shows pale yellow inner leaves and a developmental defect
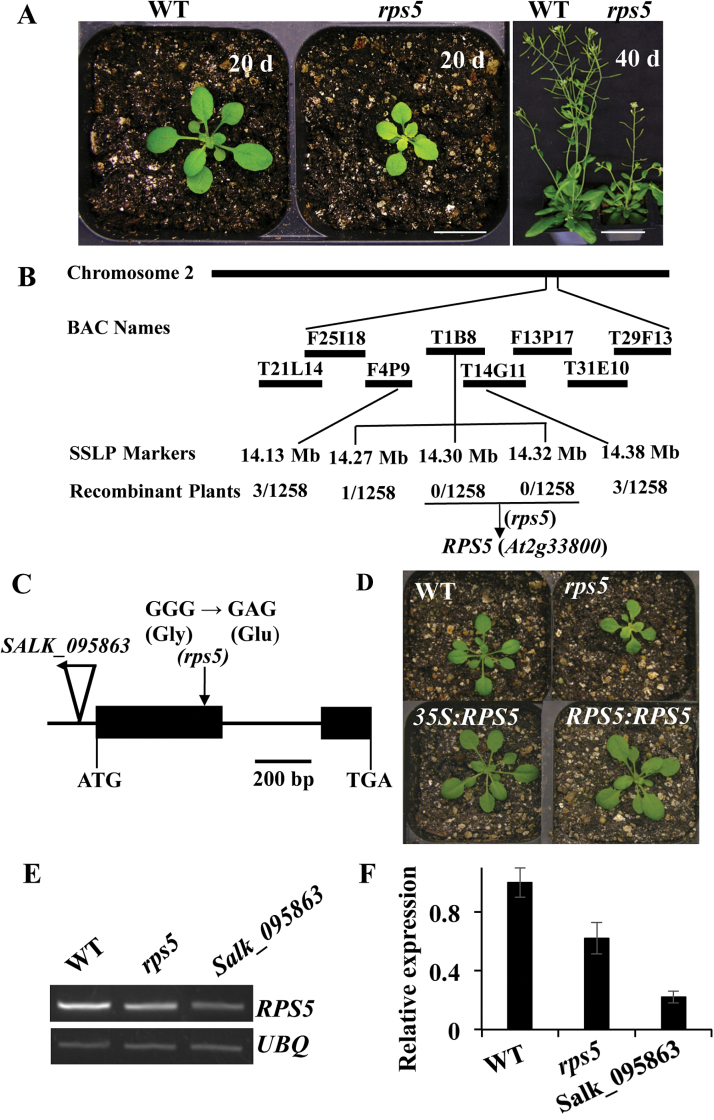
In an effort to identify genes involved in chloroplast development and pigment synthesis, we screened an EMS-mutagenized M2 population for mutants with abnormal leaf color. This screen uncovered a pale yellow inner leaf color mutant, rps5, whose phenotype was controlled by a single nuclear recessive gene. In comparison with wild-type plants, this mutant had pale yellow inner young leaves (Fig. 1A). The yellow color was distinct at the seedling stage and gradually became pale green during plant growth. In addition, a delay of 2 days in first true leaf emergence in the mutant plants was observed.
Fig. 1.
Molecular cloning of rps5. (A) Growth phenotype of wild-type and rps5 mutant plants at 20 and 40 days old, respectively. Scale bars=1cm. (B) Positional cloning of rps5. Homozygous rps5 F2 seedlings from a total of 1258 plants were used for fine mapping. (C) RPS5 gene structure and the locations of the missense mutation for rps5 and T-DNA insertion for Salk_095863. Exons and introns are shown as solid boxes and lines, respectively. The T-DNA insertion is indicated with a triangle and the single nucleotide change in rps5 is indicated with a vertical arrow. (D) Complementation of the rps5 mutant. The rps5 phenotype was complemented with RPS5 cDNA (35S:RPS5) or genomic DNA (RPS5:RPS5). Plants were 4 weeks old. (E, F) Expression of RPS5 in wild-type and rps5 plants detected by semi-quantitative RT-PCR and qRT-PCR, respectively. Data represent means±SD from three biological replicates with three technical repeats. (This figure is available in colour at JXB online.)
It is well established that pale green leaves are associated with impaired photosynthetic performance, which subsequently affects plant growth and development (Romani et al., 2012; Tiller et al., 2012). Consistent with this, the total chlorophyll and carotenoid levels were significantly lower in rps5 than those in the wild-type plants (Supplementary Figure S2). The mature rps5 mutant plants had a reduced growth rate and were much smaller than wild-type plants (Fig. 1A).
The rps5 mutant results from a missense mutation of ribosomal protein S5 (RPS5)
To isolate the gene responsible for rps5, a map-based cloning strategy was employed following the generation of an F2 mapping population by crossing rps5 in Columbia ecotype with wild-type in Landsberg erecta background. A total of 1258 homozygous rps5 mutant plants were selected and used for fine mapping. The rps5 locus was mapped to the long arm of chromosome 2 between BACs T21L14 and T29F13 using simple sequence length polymorphism markers from the TAIR database and Arabidopsis Mapping Platform, and further narrowed to BAC clone T1B8 with two co-segregation markers (Fig. 1B). Thirteen candidate genes between the 14.30Mb and 14.32Mb regions on chromosome 2 from the rps5 mutant were sequenced. A missense mutation was found in At2g33800 due to a G to A base substitution in the first exon in rps5, resulting in an amino acid Gly to Glu change (Fig. 1C). At2g33800 encodes a RP S5 family protein (RPS5). Thus, the mutant was named as rps5.
To determine whether this missense mutation in RPS5 was responsible for the mutant phenotype of rps5, a complementation test was performed in rps5 using the wild-type RPS5 cDNA under control of the cauliflower mosaic virus 35S promoter. Over 20 positive transgenic lines overexpressing RPS5 were obtained. RPS5 restored the rps5 mutant defects and the transgenic lines showed a completely wild-type phenotype (Fig. 1D). To further validate the cloned gene, we also performed a complementation test using the genomic DNA fragment containing the wild-type RPS5 coding region and the associated flanking sequences. The genomic DNA construct was introduced into the rps5 mutant. Again, the positive transgenic lines with endogenous promoter also displayed the wild-type phenotype (Fig. 1D). These results indicated that RPS5 was responsible for the mutant phenotypes.
To determine whether the point mutation in rps5 affected the transcript level of RPS5, its expression in 4-week-old plants was analyzed by semi-quantitative RT-PCR (Fig. 1E) and qRT-PCR (Fig. 1F). A Salk_095863 line that carries a T-DNA insertion in the 5ʹ-UTR of RPS5 (Fig. 1C) was also included. The Salk_095863 mutant showed growth and developmental defects, with smaller plants than rps5 (Supplementary Figure S3), showing allele variation. Interestingly, the expression analyses revealed that the transcript level of RPS5 in rps5 was decreased in comparison with wild-type plants, although rps5 was a missense mutation. Missense mutations have also been reported to affect gene expression in other studies (Takano et al., 2010; Wang et al., 2015). As expected, the Salk_095863 line exhibited low transcript levels.
RPS5 structure and subcellular localization
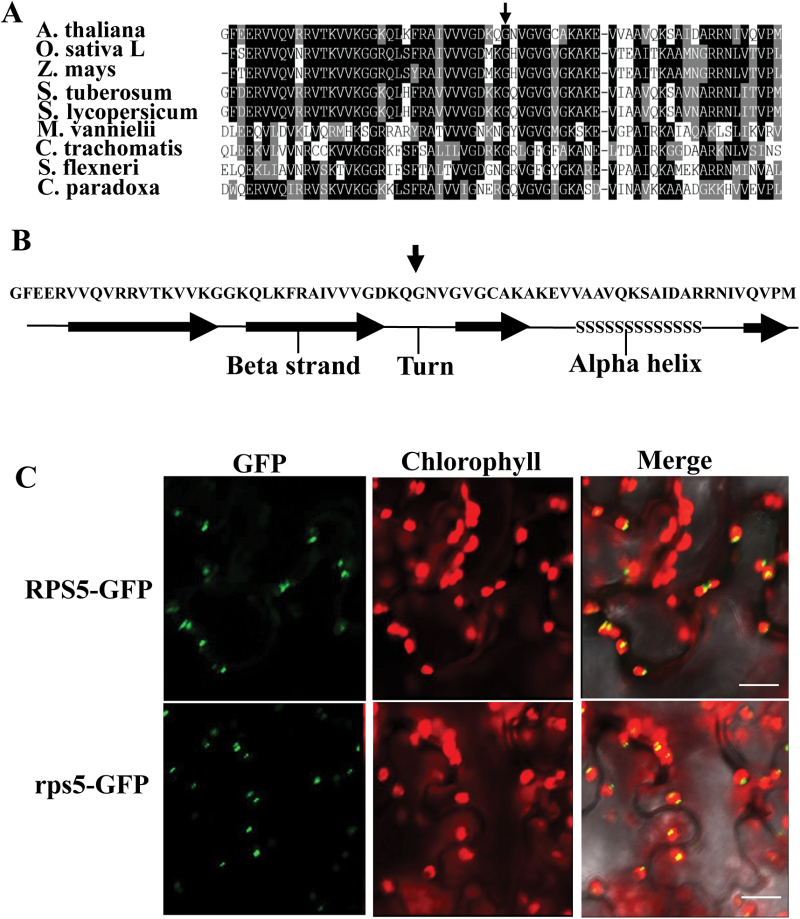
RPS5 is a component of the chloroplast ribosome 30S small subunit (Yamaguchi et al., 2000). The molecular mass of RPS5 mature protein was 32.6 kD. A sequence similarity search showed that only one RPS5 gene orthologous to the Escherichia coli S5 protein gene existed in the Arabidopsis genome (Supplementary Table S2). Interestingly, the N-terminal region of RPS5 proteins was highly conserved among plants and bacterial species (Fig. 2A). The secondary structure for the N-terminal sequence and a three-dimensional 3D structure model for RPS5 protein were constructed using the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index; Kelley and Sternberg, 2009). The predicted secondary structure revealed that the amino acid substitution of RPS5 from rps5 was located in the turn element between two beta strand sheets (Fig. 2B). We also predicted the 3D structures of RPS5 from wild-type and mutant plants using the Phyre2 server. The same 3D structures were observed (Supplementary Figure S4). One explanation for this result is that the software prediction was based on published data and the mutant site might not have been analyzed.
Fig. 2.
Partial sequence alignment, secondary structure prediction, and subcellular localization of RPS5. (A) N-terminal domain sequence alignment of RPS5 from Arabidopsis (A. thaliana, AEC08888), rice (Oryza sativa, NP_001050474), maize (Zea mays, NP_001150762), potato (Solanum tuberosum, XP_006347020), tomato (Solanum lycopersicum, XP_004232896), Methanococcus vannielii (P14036), Chlamydia trachomatis (P0A4C8), Shigella flexneri (P0A7W6), and Cyanophora paradoxa (P23402). The vertical arrow indicates the mutation site. (B) Predicted secondary structure of the RPS5 N-terminal domain. The vertical arrow indicates the Glu substitution site in the mutant protein. (C) Wild-type RPS5 (top) and mutant rps5 (bottom) are localized in chloroplasts of 4-week-old tobacco leaves transiently expressing GFP fusion proteins. Bars=25 µm. (This figure is available in colour at JXB online.)
RPS5 protein was predicted to be localized in chloroplasts (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2007). To validate this, we investigated the subcellular location of RPS5 by transiently expressing the RPS5-GFP construct in tobacco leaves. The fusion protein signal was monitored by confocal microscopy. Merging the chlorophyll autofluorescence and green fluorescence showed that the signals were clearly observed in chloroplasts (Fig. 2C), consistent with the previous indication of a chloroplast thylakoid membrane location in a chloroplast subproteome analysis (Friso et al., 2004).
Plastid localization can be altered by single amino acid changes (Shumskaya et al., 2012). We investigated whether the single amino acid substitution of RPS5 could affect the localization of RPS5. The rps5-GFP construct was infiltrated into tobacco leaves and expression of the fusion protein was examined. The signal pattern of rps5-GFP fusion protein was identical to that of RPS5-GFP (Fig. 2C). These results showed that the RPS5 mutation did not alter the subcellular location of the protein.
RPS5 mutation significantly reduces the efficiency of 16S rRNA processing
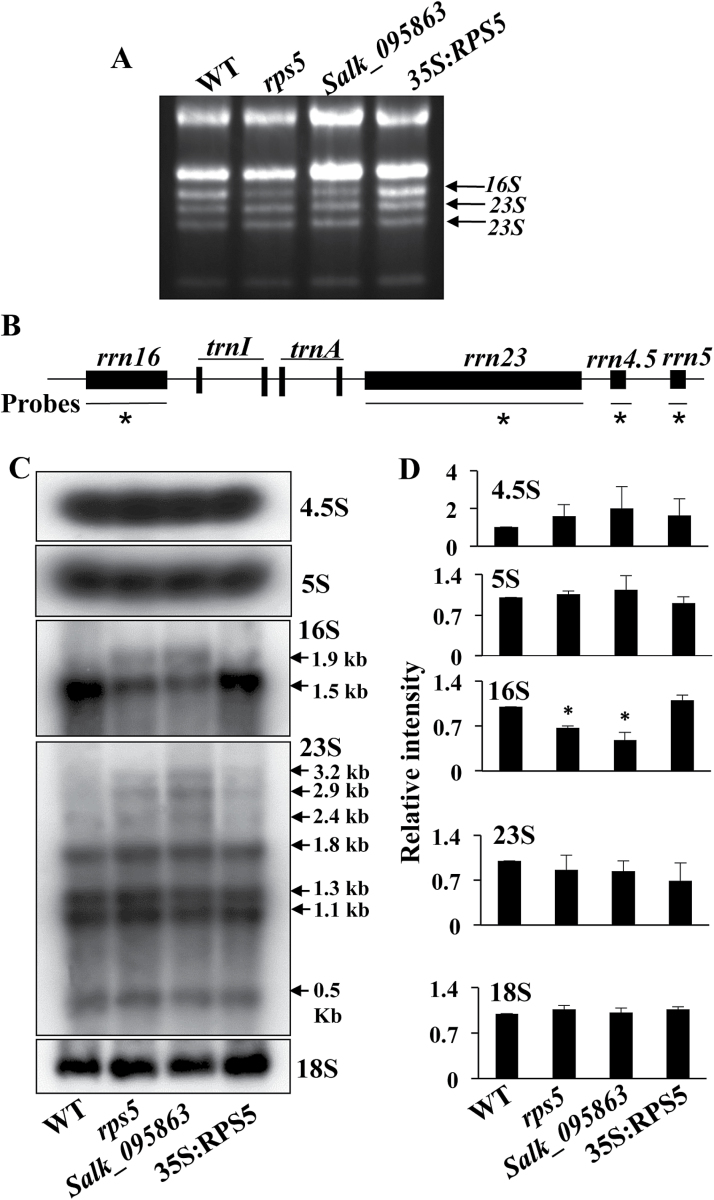
To investigate whether mutation in the plastid RPS5 would impair chloroplast 16S rRNA processing, we initially examined the pattern of total RNA by running denaturing agarose gels and staining them with ethidium bromide. As shown in Fig. 3A, chloroplast 16S rRNA was decreased in the rps5 and Salk_095863 mutants relative to wild-type plants. The reduction could be rescued by overexpressing RPS5 in the rps5 mutant plants. These results indicated that RPS5 had an effect on 16S rRNA processing.
Fig. 3.
Chloroplast rRNA accumulation and processing in wild-type (WT), mutant lines (rps5 and Salk_095863), and a complementation RPS5-overexpressing line (35S:RPS5). (A) rRNA accumulation pattern in an ethidium bromide-stained gel. The mutant lines had reduced levels of 16S rRNA, identified based on Kleinknecht et al. (2014) and Yu et al. (2008). (B) Schematic diagram of the chloroplast rrn operon in Arabidopsis. The probes used for northern blot analysis are marked by black lines under the operon with asterisks. (C) Northern blot analysis of chloroplast 4.5S, 5S, 16S, 23S, and cytoplasmic18S rRNAs. Total RNA (3 µg) from 4-week-old leaves was used for northern blot experiments. Equal loading controls for northern blot analysis are shown in Supplementary Figure S5. (D) Quantification of the mature bands in C using ImageJ (http://www.di.uq.edu.au/sparqimagejblots). Asterisks in 16S indicate significant differences (p<0.05).
Plastid rRNAs include 23S, 16S, 5S, and 4.5S, which form part of the chloroplast rrn operon (Fig. 3B) (Kleinknecht et al., 2014; Yu et al., 2008). These rRNAs are transcribed to yield large RNA precursors that are finally processed to mature rRNAs by a series of processing steps (Harris et al., 1994). To see whether RPS5 mutation affected chloroplast rRNA processing, the plastid rRNA accumulation patterns were analyzed in detail by RNA gel blots using gene-specific probes (Fig. 3B). No significant differences in the abundance of 4.5S and 5S sRNA were observed between rps5 or Salk_095863 and wild-type plants (Fig. 3C, D). In agreement with the results from the ethidium bromide-stained denaturing agarose gels (Fig. 3A), the quantity of mature 16S rRNA in the rps5 and Salk_095863 mutants was dramatically reduced to 65% and 50%, respectively, in comparison with the wild-type control (Fig. 3D), whereas the 16S rRNA 1.9kb precursors were increased in the mutant plants (Fig. 3C). The 16S rRNA processing deficiency in rps5 was rescued by overexpression of RPS5 (Fig. 3C, D). In addition, a slight increase of the 3.2, 2.9, and 2.4kb precursor rRNAs was observed in the rps5 mutants relative to wild-type plants using a 23S rRNA gene-specific probe (Fig. 3C). Cytosolic 18S rRNA was also examined. As expected, similar 18S rRNA levels were observed in these mutant and wild-type plants (Fig. 3C, D), indicating that the RPS5 mutations did not affect cytosolic rRNA accumulation. The loading controls of RNA gel blots are shown in Supplementary Figure S5. Taken together, these data revealed that RPS5 mutations mainly caused the impairment of chloroplast 16S rRNA processing.
Comparative proteomics analysis reveals the role of RPS5 in affecting photosystem I and II core components and plastid RPs
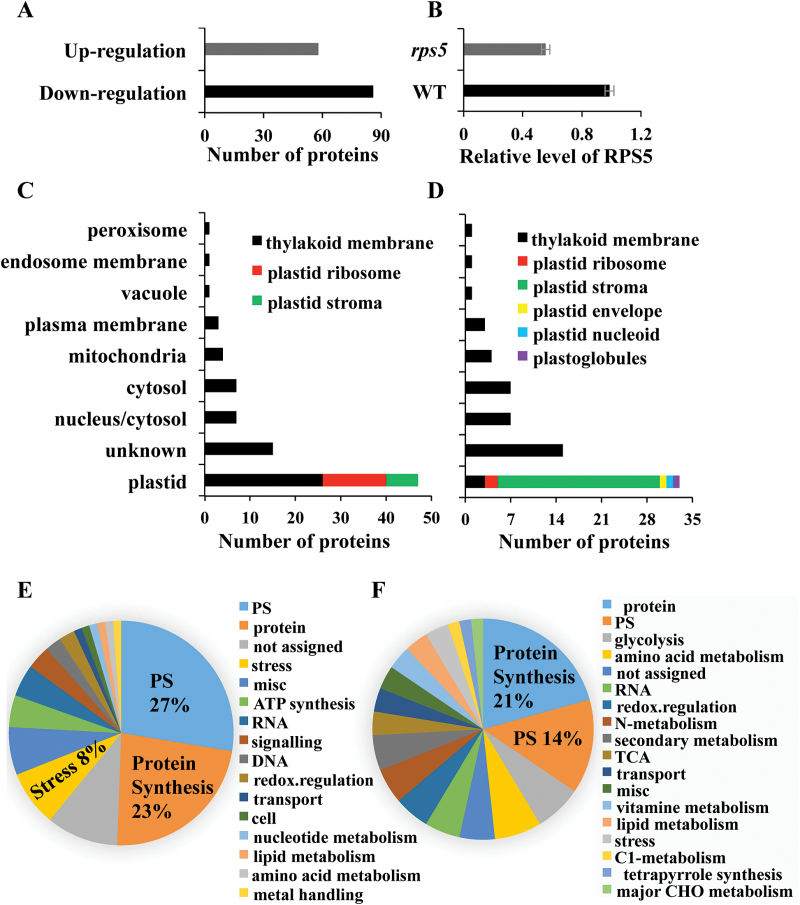
While the effects of RP defects on a number of plastid and nuclear proteins have been examined previously via immunological analysis (Romani et al., 2012; Tiller et al., 2012), there is limited information for their global effects on protein synthesis. Impaired chloroplast 16S rRNA processing can affect chloroplast translation (Kleinknecht et al., 2014). To obtain a global and quantitative view of proteins altered in rps5, a comparative proteomic analysis of total proteins from 4-week-old wild-type and rps5 leaves was carried out using an iTRAQ-based method. A total of 1007 proteins were identified, and 294 proteins were differentially expressed (P<0.05) (Supplementary Table S3). Among the differentially expressed proteins, 58 proteins were up-regulated with at least 1.33-fold change and 86 proteins were down-regulated with 0.75- or less fold change in all three biological replicates of rps5 (Fig. 4A; Supplementary Table S4). Consistent with the reduced RPS5 expression (Fig. 1E, F), the RPS5 protein level was also detected to have ~43% reduction in rps5 compared with wild-type plants (Fig. 4B).
Fig. 4.
Comparative proteomic analysis of the differentially expressed proteins in the rps5 proteome. (A) Numbers of up- and down-regulated proteins showing significant differences between the wild-type and rps5 proteome. (B) Relative level of RPS5 protein in wild-type and rps5 plants. Values are means±SD from three biological replicates. (C, D) Subcellular localization of (C) down-regulated and (D) up-regulated proteins in the rps5 proteome. (E, F) Functional classification of (E) down-regulated and (F) up-regulated proteins in the rps5 proteome. (This figure is available in colour at JXB online.)
The majority of the differentially regulated proteins, in either the down-regulated (Fig. 4C) or up-regulated (Fig. 4D) groups, were localized to plastid. Notably, among the down-regulated plastid proteins, over half (26 of 47) were thylakoid membrane proteins (Fig. 4C). In contrast, a majority (25 of 33) of up-regulated plastid proteins were stroma proteins (Fig. 4D). These differentially expressed proteins were categorized into functional groups using MapMan (http://mapman.gabipd.org/web/guest/mapman). Proteins associated with photosynthesis and protein synthesis represented the most abundant groups (Fig. 4E, F; Supplementary Tables S5 and S6), revealing that the defect in RPS5 exerted profound effects on chloroplast-localized proteins involved in photosynthesis and protein synthesis.
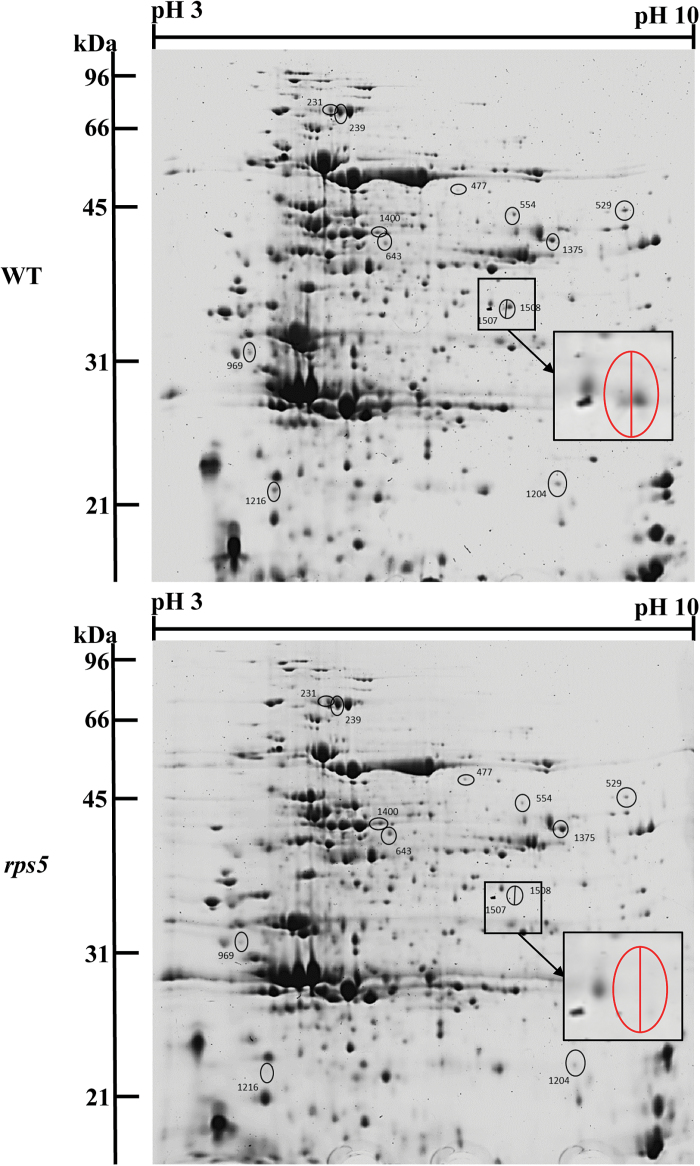
In addition, to provide another approach examining the differentially expressed dominant proteins and validating iTRAQ data, a 2D gel-based quantitative proteomic analysis was conducted using SameSpots image analysis software. Thirteen spots with marked differences between wild-type and rps5 were selected to identify the differentially expressed proteins (Fig. 5). Consistent with the iTRAQ results, seven proteins, including RPS5, photosystem II subunit R (PsbR), ATPase subunit b (PDE334), and Rubisco small chain 1A (RBCS1A), were decreased in rps5, whereas the other seven proteins, including fructose-bisphosphate aldolase 5 (FBA5) and transketolase (ATTKL1), were increased in the mutant plants compared with wild-type plants (Table 1). The cross-validation of these differentially expressed proteins by 2D gel-based proteomic analysis corroborated the iTRAQ results and further confirmed that the amount of RPS5 protein was greatly reduced in rps5 plants relative to wild-type plants (spot 1507/1508 in Fig. 5; Table 1).
Fig. 5.
2D gel-based proteomic analysis of wild-type (top) and rps5 (bottom) plants. The differentially expressed proteins in wild-type and mutant plants are labelled by black circles and the enlarged 1507/1508 spots are indicated in red circles. Spots 1507/1508 indicate RPS5 protein. (This figure is available in colour at JXB online.)
Table 1.
Cross-validation of proteins identified by iTRAQ- and 2D gel-based proteomic analyses
| Gene locus | Protein description | emPAI ratio* | SC_unNor ratio* | Spots | iTRAQ ratio* |
|---|---|---|---|---|---|
| AT1G79040.1 | Photosystem II subunit R | 0.23 | 0.5 | 529 | 0.55 |
| AT2G33800.1 | Ribosomal protein S5 | 0.05 | 0.03 | 1508/1507 | 0.56 |
| AT1G64510.1 | Ribosomal protein S6 | 0.57 | 0.8 | 1216 | 0.6 |
| AT4G32260.1 | ATPase subunit b | 0.55 | 0.7 | 1216 | 0.61 |
| AT3G52150.1 | Plastid-specific ribosomal protein 2 | 0.63 | 0.7 | 1204 | 0.66 |
| AT1G67090.1 | Ribulose bisphosphate carboxylase small chain 1A | 0.41 | 0.7 | 969 | 0.68 |
| AT5G07020.1 | Proline-rich protein family | 0.14 | 0.3 | 969 | 0.73 |
| AT1G11860.1 | Glycine cleavage T-protein | 2.5 | 1.6 | 1375 | 1.33 |
| AT5G26000.1 | Myrosinase 1 | 4.2 | 4 | 239/231 | 1.38 |
| AT2G39730.1 | Rubisco activase | 2.2 | 1.4 | 1400 | 1.48 |
| AT1G23310.1 | Glutamate:glyoxylate aminotransferase | 2.2 | 1.8 | 477 | 1.54 |
| AT5G43940.1 | Alcohol dehydrogenase class-3 | 1.34 | 2.5 | 554 | 1.4 |
| AT4G26530.1 | Fructose-bisphosphate aldolase 5 | 2.6 | 1.9 | 231 | 1.66 |
| AT3G60750.1 | Transketolase | 2.7 | 1.6 | 231 | 1.82 |
emPAI ratio, protein ratio calculated by Exponentially Modified Protein Abundance Index; SC_unNor ratio, protein ratio calculated by Spectrum Counting_unNormalization. * The protein ratio is rps5:wild-type.
Down-regulated proteins in rps5
Among the 86 down-regulated proteins (Supplementary Table S4), numerous proteins (28%) were involved in photosynthesis (Fig. 4E; Supplementary Table S5). Many of these photosynthetic proteins were localized in thylakoid membrane, the site of the light-dependent reactions of photosynthesis. They included six photosystem I polypeptide subunits (PSAD-1, PSAD-2, PSAE-1, PSAN, PSAL, and PSAF), 11 photosystem II polypeptide subunits (PSBTn-1, PSBTn-2, PSBR, psbE, psbH, PSBP-1, PSBP-2, PSBO-1, PSBO-2, PSBQ-1, and PSBQ-2), and three ATP synthases (ATPG, atpA, and atpB) (Table 2). Four (psbE, psbH, atpA, and atpB) were chloroplast-encoded proteins. Proteins that function in the Calvin cycle, such as CP12 domain-containing protein and Rubisco small subunit-4 (RBCS-4), were also dramatically reduced in the rps5 mutant. These results indicate that RPS5 exerts a strong effect on the photosynthetic proteins.
Table 2.
Down-regulated proteins involved in photosynthesis and the chloroplast 30S subunit
| Accession | Protein description | Ratio* | Accession | Protein description | Ratio* |
|---|---|---|---|---|---|
| Proteins involved in photosynthesis | Ribosomal proteins in chloroplast 30S subunit | ||||
| ATCG00710.1 | Photosystem II reaction center protein H | 0.36 | AT4G34620.1 | Ribosomal protein S16B | 0.53 |
| AT2G30790.1 | Photosystem II subunit P-2 | 0.47 | AT2G33800.1 | Ribosomal protein S5 | 0.56 |
| AT3G21055.1 | Photosystem II subunit T | 0.51 | ATCG00900.1 | Ribosomal protein S7B | 0.57 |
| AT1G06680.1 | Photosystem II subunit P-1 | 0.55 | ATCG01120.1 | Ribosomal protein S15 | 0.58 |
| AT1G79040.1 | Photosystem II subunit R | 0.55 | AT1G79850.1 | Ribosomal protein S17 | 0.58 |
| AT1G51400.1 | Photosystem II 5 kD protein | 0.59 | AT1G64510.1 | Ribosomal protein S6 | 0.6 |
| AT3G50820.1 | Photosystem II subunit O-2 | 0.6 | ATCG00770.1 | Ribosomal protein S8 | 0.64 |
| ATCG00580.1 | Photosystem II reaction center protein E | 0.6 | ATCG00650.1 | Ribosomal protein S18 | 0.65 |
| AT1G76450.1 | Photosystem II reaction center PsbP family protein | 0.61 | AT3G15190.1 | Ribosomal protein S20 | 0.66 |
| AT5G66570.1 | PS II oxygen-evolving complex 1 | 0.61 | AT1G74970.1 | Ribosomal protein S9 | 0.67 |
| AT4G05180.1 | Photosystem II subunit Q-2 | 0.64 | AT5G30510.1 | Ribosomal protein S1 | 0.69 |
| AT4G21280.1 | Photosystem II subunit QA | 0.65 | ATCG00380.1 | Ribosomal protein S4 | 0.71 |
| AT1G03130.1 | Photosystem I subunit D-2 | 0.47 | AT5G14320.1 | Ribosomal protein S13 | 0.71 |
| AT4G28750.1 | Photosystem I reaction centre subunit IV | 0.48 | |||
| AT4G02770.1 | Photosystem I subunit D-1 | 0.55 | |||
| AT5G64040.1 | Photosystem I reaction center subunit N | 0.61 | |||
| AT4G12800.1 | Photosystem I subunit l | 0.64 | |||
| AT1G31330.1 | Photosystem I subunit F | 0.74 | |||
| AT1G03130.1 | Photosystem I subunit D-2 | 0.47 | |||
| AT4G28750.1 | Photosystem I reaction centre subunit IV | 0.48 | |||
| AT4G02770.1 | Photosystem I subunit D-1 | 0.55 | |||
| AT5G64040.1 | Photosystem I reaction center subunit N | 0.61 | |||
| AT4G32260.1 | Pigment defective 334 | 0.61 | |||
| ATCG00120.1 | ATP synthase subunit alpha | 0.68 | |||
| ATCG00480.1 | ATP synthase subunit beta | 0.73 | |||
| AT2G47400.1 | CP12 | 0.66 | |||
| AT1G67090.1 | Rubisco small subunit-4 | 0.68 | |||
* The protein ratio is rps5:wild-type from iTRAQ analysis.
In addition, a high proportion (23%) of the down-regulated proteins were those involved in protein synthesis (Fig. 4E, Supplementary Table S5). Eighteen RPs were down-regulated in the rps5 mutant in comparison to wild-type plants. Most of these (13 of 18) were involved in the chloroplast 30S subunit (Table 2), which might result from RPS5 being a component of the chloroplast 30S subunit. Low plastid RP levels could affect ribosome function and plastid translation, which are essential for plant development and photosynthesis (Horiguchi et al., 2012; Romani et al., 2012; Tiller and Bock, 2014).
Up-regulated proteins in rps5
A total of 58 proteins were up-regulated in rps5, 33 of which were localized to chloroplast (Supplementary Table S4; Fig. 4D). Among these plastid proteins, 25 were in the plastid stroma and mainly involved in protein synthesis, photosynthesis, and nitrogen metabolism (Fig. 4F; Supplementary Table S6). In addition, proteins involved in nitrogen metabolism, secondary metabolism, one-carbon metabolism, vitamin metabolism, tricarboxylic acid cycle, and glycolysis were found only among the up-regulated proteins (Supplementary Table S6).
Transcript levels of photosynthesis-related genes are not affected in rps5
Comparative proteomic analysis indicated that many proteins involved in photosynthesis were altered in rps5. To assess whether the expression of the photosynthesis-related genes was affected in the rps5 plants, the transcript levels of some nuclear- and plastid-encoded genes involved in photosynthesis were tested by qRT-PCR. The genes examined included those involved in photosystem I (psaA, psaB, psaC, PSAD, PSAE, and PSAF) and photosystem II (psbA, psbB, psbC, psbD, and psbE), as well as Rubisco large and small subunits (RbcL and RBCS). As shown in Supplementary Figure S6, no dramatic difference in expression of all the tested genes was observed in rps5 or Salk_095863 compared with wild-type plants. These data, along with the proteomics studies, indicate that the transcript profile of photosynthesis-related genes was not disturbed in the mutant plants and that RPS5 affected photosynthetic proteins at the translational level.
Overexpression of RPS5 enhances cold tolerance
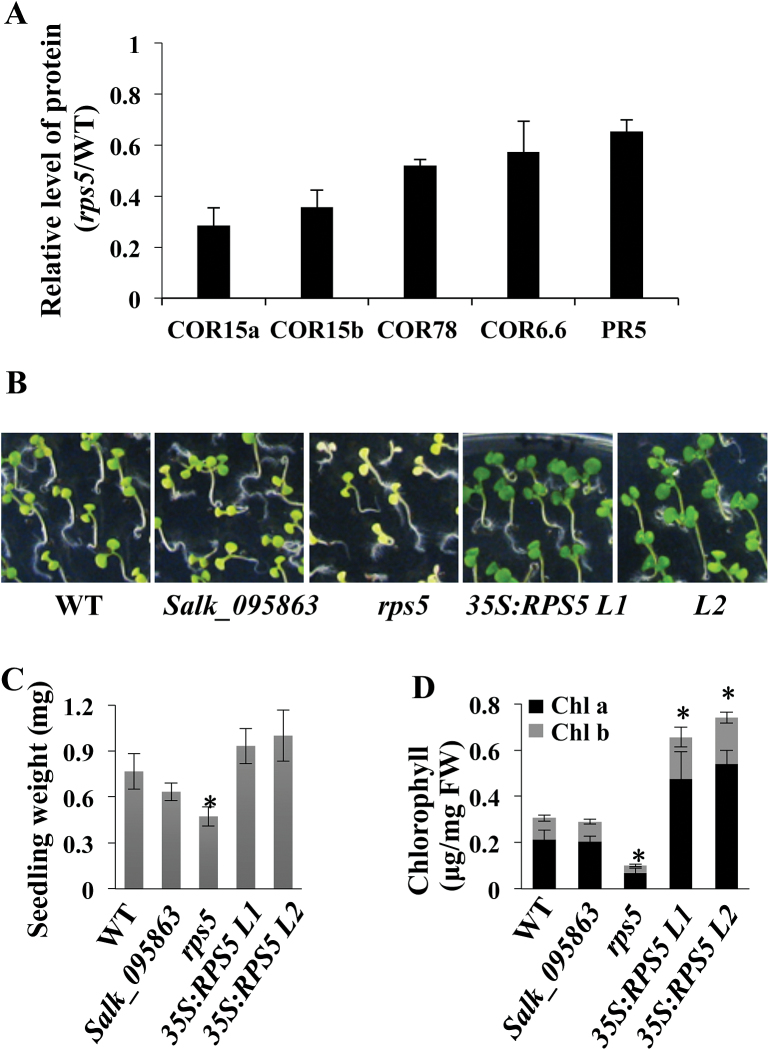
Apart from those proteins associated with photosynthesis and protein synthesis, interestingly, a number of proteins involved in stress responses were found in the comparative proteomic analysis to be reduced in the rps5 mutant (Supplementary Table S4). These included four cold-regulated proteins (COR15a, COR15b, COR78, and COR6.6) and a pathogenesis-related protein (PR5) (Fig. 6A). In addition, two dehydrin family proteins (ERD10 and ERD14) and five glutathione transferases (GST9, GSTF2, GSTF10, GSTU19, and GSTU20) were reduced in the rps5 mutant (Supplementary Table S4). The down-regulation of these stress-related proteins in rps5 suggested a potential role of RPS5 in stress responses.
Fig. 6.
Role of RPS5 in cold stress tolerance. (A) Relative levels of proteins associated with the cold stress response in rps5 plants. Values represent means±SD from three biological replicates. (B) Phenotypes of wild-type (WT), Salk_095863, rps5, and two RPS5 overexpression lines grown on Murashige and Skoog medium at 4 °C for 6 weeks. (C) Average weight per seedling grown at 4 °C for 6 weeks. (D) Chlorophyll content of seedlings grown at 4 °C for 6 weeks. Values are means±SD from four biological replicates. (This figure is available in colour at JXB online.)
To provide evidence in support of this, we compared plant growth of wild-type, Salk_095863, rps5, and two RPS5-overexpressing lines under cold treatment at 4 °C. The two RPS5-overexpressing lines contained 1.53- and 1.78-fold increases in the RPS5 protein levels, respectively, as measured via relative protein quantification by calculating the identified peptide peak areas of RPS5. When these plants were germinated and grown at 4 °C for 6 weeks, the seedlings of the two RPS5-overexpressing lines grew much better and had a dark green color in comparison with those of wild-type plants (Fig. 6B). In contrast, the seedlings of rps5 showed a more yellow and less green color than the seedlings of wild-type control plants (Fig. 6B). The Salk_095863 line had a weak phenotype in comparison with the rps5 mutant (Fig. 6B). The difference in cold tolerance phenotypes between Salk_095863 and rps5 might be due to the different accumulation of functional RPS5 protein in Salk_095863 and the formation of malfunctioning RPS5 protein in rps5 due to the missense mutation. Noticeably, the rps5 mutant appeared to be more severely affected in the cold but less affected under normal growth conditions than the Salk_095863 line, suggesting that the malfunctioning RPS5 protein could function partially under normal growth conditions but was non-functional in cold conditions.
The seedling weight after 6 weeks of growth at 4 °C was measured. The rps5 mutant exhibited significantly lower seedling weight than the wild-type control; in contrast, the two overexpressing lines showed enhanced seedling weight (Fig. 6C). Moreover, the chlorophyll content was also significantly decreased in the rps5 mutant and increased in the two overexpressing lines (Fig. 6D). These data support the potential role of RPS5 in cold stress tolerance.
Discussion
Although a growing number of recent studies support the critical roles of RPs in plant growth and development (Byrne, 2009; Horiguchi et al., 2012; Schippers and Mueller-Roeber, 2010; Tiller and Bock, 2014), it is not fully understood how they affect these processes. In addition, the functions of some individual RPs remain largely unknown. In this study we identified a mutant rps5 with a missense mutation, which resulted in a Gly to Glu substitution in the RPS5 protein and led to a reduced level of RPS5 expression and possible malfunction of the RPS5 protein. The defect in RPS5 enabled us to analyze the roles of this protein. RPS5 is a component of the 30S subunit in chloroplasts and is highly conserved in all species (Ramakrishnan and White, 1992). RPS5 is suggested to be required for translation (Ramakrishnan and White, 1992). In plants, a very preliminary analysis of the rps5 Salk line was briefly described in a large-scale reverse genetic screen (Bryant et al., 2011). In this study, we show that RPS5 is important for plastid ribosome function and is involved in photosynthesis, plant development, and cold stress resistance in Arabidopsis.
RPS5 is expressed at all stages of plant growth and especially highly in the rosette leaves (Supplementary Figure S7). Our data revealed that the rps5 mutation reduced the accumulation of chloroplast 16S rRNA and impaired the processing of 16S rRNA. Our quantitative proteomic analysis showed that the mutation exerted a strong effect on the levels of many proteins involved in photosynthesis and protein synthesis. Such alterations and effects likely contributed to the mutant phenotypes of yellow leaves and overall reduced growth in rps5 plants. The proteomic analysis also suggested that RPS5 likely contributed to abiotic stress resistance, which was supported by the cold tolerance test.
RPS5 is required for chloroplast 16S rRNA processing
Ribosome assembly affects plastid translation, which is essential for plant development and photosynthesis (Schippers and Mueller-Roeber, 2010; Tiller and Bock, 2014). The ribosome contains rRNAs, RPs, and accessory factors, and all ribosomal functions rely on rRNAs. Crystal structure analysis of the ribosome indicates the existence of complicated interactions among the ribosomal components (Yusupov et al., 2001). rRNAs are highly abundant in cells and are easily observed on denaturing agarose gels. Changes in their abundance are often used as an indicator for factors affecting ribosomes (Tiller et al., 2012; Walter et al., 2010). Two plastid RPs (RPS17 and PSRP4) are suggested to be essential for the stability of the 30S ribosomal subunit, as the ratio of 16S rRNA:18S rRNA is greatly decreased and the ratio of 23S rRNA:18S rRNA remains unchanged in the rps17 and psrp4 mutants (Tiller et al., 2012). In addition, a recent study shows that mutation in RBF1, a ribosome-binding factor, leads to defects in chloroplast 16S rRNA processing and reduced accumulation of the 30S ribosomal subunit in Arabidopsis (Fristedt et al., 2014). We found an up to 50% reduction in 16S rRNA in rps5 mutants (Fig. 3D), implying that assembly of the chloroplast 30S subunit was affected by RPS5 (Tiller and Bock, 2014).
Although bacterial RPS5 belongs to the tertiary binding protein in the 30S ribosome assembly map (Shajani et al., 2011), the crystal structure of bacterial RPS5 reveals that its many sites interact with 16S rRNA (Ramakrishnan and White, 1992; Wimberly et al., 2000). This observation provides a basis for the assumption that mutation in RPS5 could cause a defect in the 16S rRNA processing. Indeed, our RNA gel blotting data demonstrated the accumulation of 16S precursor and a much smaller quantity of the mature chloroplast 16S rRNA in rps5 and Salk_095863 mutants (Fig. 3C, D). The reduced processing of 16S rRNA in these mutants could result from the reduced level of RPS5 mRNA, like the Salk T-DNA line. It could be also due to the Gly to Glu mutation in the turn element between two beta strand sheets of RPS5 protein, which might also affect the interaction between RPS5 and 16S rRNA and the normal function of RPS5.
RPs have been shown to exert distinctive effects on rRNA processing in chloroplasts, and not all plastid RPs alter rRNA processing. The abundance of 23S and 16S rRNA precursors is greatly increased in the rps17 and psrp4 mutants, and the accumulation of 4.5S and 5S rRNAs is greatly reduced in psrp3 (Tiller et al., 2012). However, the abundance of chloroplast rRNA and the amount of rRNA processing are not changed in the psrp2 and psrp6 mutants (Tiller et al., 2012). RPs may have different active sites with which to interact with different rRNAs at specific loci, resulting in plastid RPs having different effects on rRNA processing and abundance (Ramakrishnan, 2002).
Global suppression of specific groups of protein synthesis is responsible for the rps5 growth phenotypes
Comparative quantitative proteomics is a powerful tool to discover the proteins and pathways that are associated with or affected by specific factors and processes. As the effects of RPs on protein synthesis at global level have been subjected to limited studies, we performed comparative proteomic analysis and identified the differentially expressed proteins between wild-type and rps5 plants. Proteomic data by both iTRAQ and 2D gel-based analyses revealed that RPS5 was decreased by approximately 50% in rps5 (Fig. 4B; Supplementary Table S4). RPS5 was found to exert a profound effect on a group of both plastid genome-encoded and also nucleic genome-encoded plastid proteins. Since rps5 causes a defect in chloroplast development, this defect might in turn affect distinctive sets of target nuclear gene expression and then protein levels via retrograde plastid-to-nucleus signaling (Chi et al., 2013) to affect the expression of nucleic genome-encoded proteins.
A majority of the differentially regulated proteins were located in plastids. One large group of these down-regulated plastid proteins was associated with photosynthesis (Fig. 4E; Supplementary Table S5). Many proteins involved in photosystems I and II polypeptide subunits were reduced in rps5 (Table 2). The reduction of these core components of photosynthetic proteins explained the yellow leaf and growth defect phenotypes observed in rps5. Indeed, a decrease in the accumulation of several photosynthetic proteins (i.e. psaA, PSAD, PSAF, psbB, Lhca2, and/or rbcL) has been reported to be the cause of defects in photosynthetic performance due to reduced translational capacity (Romani et al., 2012; Tiller et al., 2012).
Another large group of down-regulated plastid proteins in rps5 was plastid RPs, including plastid RP S13, S17, and S20 (Table 2), some of which are known to be associated with growth and developmental defects. PRPS13 and PRPS20 are required for embryo development in Arabidopsis (Bryant et al., 2011; Romani et al., 2012). Similarly, a defect in rice PRPS20 results in albino and seedling-lethal phenotypes (Gong et al., 2013). Mutation in PRPS17 leads to extended leaf longevity and reduced leaf growth rate (Woo et al., 2002). The combination effect of decreased levels of multiple plastid RPs likely caused the phenotypic behavior of rps5.
In addition, most (13 of 18) of those down-regulated RPs were components of the chloroplast 30S subunit. These proteins are known to interact with 16S rRNA (Stern et al., 1989; Wimberly et al., 2000). Down-regulation of a large number of plastid RPs most likely seriously impaired the assembly of the chloroplast ribosome, subsequently reducing plastid translation and affecting overall plant development.
RPS5 expression affects cold stress tolerance
Interestingly, comparative proteomics also identified that some proteins that participate in abiotic stress responses were dramatically decreased in rps5 (Fig. 6A; Supplementary Table S4). These proteins are encoded by nuclear genes and their expression was down-regulated in rps5, which likely resulted from retrograde plastid-to-nucleus signaling due to the defect in chloroplasts in rps5 (Chi et al., 2013). Four cold-regulated proteins (COR6.6, COR15A, COR15B, and COR78) (Thomashow, 1998) were included among the down-regulated proteins; the suppression of their expression in rps5 suggested that RPS5 might play a role in cold stress responses.
Most of the RPs are associated with development, and only a few studies have reported the association of RPs with stress responses. Chloroplast-localized RPS1 protein has been found to be associated with the heat stress response, and knock-down of RPS1 inhibits the expression of HsfA2-dependent heat-stress responses in Arabidopsis (Yu et al., 2012). Tobacco RPL33, a non-essential plastid-encoded RP, has been shown to play a critical role in plant recovery from chilling stress (Rogalski et al., 2008). Here, we found that rps5 showed reduced plant cold tolerance, with significantly decreased seedling weight and chlorophyll content, whereas RPS5-overexpressing plants exhibited enhanced plant cold tolerance, with increased seedling weight and chlorophyll content, suggesting a potential role of RPS5 in stress tolerance. Noticeably, although rps5 with the missense mutation exhibited higher RPS5 expression than Salk_095863, rps5 exhibited significantly less cold tolerance than Salk_095863. This result indicates that the Gly to Glu substitution of RPS5 protein in rps5 produced a malfunctioning protein, contributing to the cold sensitivity phenotype, whereas the low accumulation of functional RPS5 protein in Salk_095863 enabled the plants to better adapt to cold than rps5 plants.
Together, our results show that mutation in RPS5 causes impaired processing of 16S rRNA, the key component of the 30S ribosomal subunit. Mutation of RPS5 greatly affected the expression of core components of photosystem I and photosystem II, and also greatly influenced the level of plastid RPs in the 30S ribosomal subunit. Because both plastid rRNAs and RPs are important components of the chloroplast ribosome, chloroplast 30S subunit assembly was likely affected, resulting in the mutant phenotypes of pigment deficiency and development defect. In addition, our results suggested a role of RPS5 in cold stress tolerance, possibly via a reduced plastid translational capacity.
Supplementary data
Figure S1. The phenotype of 4-week-old wild-type and rps5 mutant plants, and gel evaluation of proteins used for proteomic analysis.
Figure S2. Pigment content of wild-type and the rps5 mutant.
Figure S3. The growth phenotype of wild-type, rps5, and Salk_095863 mutants.
Figure S4. RPS5 3D structure model.
Figure S5. RNA gel blotting loading controls.
Figure S6. Expression of photosynthesis-related genes in wild-type, rps5, and Salk_095863.
Figure S7. Gene expression pattern of RPS5 based on Arabidopsis e-FP Browser.
Table S1. Primers used in this study.
Table S2. Blast analysis of RPS5 in the Arabidopsis genome.
Table S3. List of 294 proteins showing significant difference (p<0.05) detected by iTRAQ proteomic analysis.
Table S4. Proteins significantly down- and up-regulated in rps5 compared with wild-type by iTRAQ proteomic analysis.
Table S5. Detailed information of functional classification of the down-regulated proteins in rps5 proteome.
Table S6. Detailed information of functional classification of up-regulated proteins in rps5 proteome.
Acknowledgements
We thank Dr. Xiangjun Zhou for his generous help and technical advice, and Mamta Srivastava for help with the subcellular localization analysis. JZ acknowledges the support of the China Scholarship Council (CSC).
Glossary
List of abbreviations:
- 2D
two-dimensional
- 3D
three-dimensional
- EMS
ethyl methanesulfonate;
- iTRAQ
isobaric tags for relative and absolute quantification
- qRT-PCR
quantitative real-time RT-PCR
- RP
ribosomal protein;
- UTR
untranslated region.
References
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. 2011. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiology 155, 1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME. 2009. A role for the ribosome in development. Trends in Plant Science 14, 512–519. [DOI] [PubMed] [Google Scholar]
- Carroll AJ. 2013. The Arabidopsis cytosolic ribosomal proteome: from form to function. Frontiers in Plant Science 4, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L. 2013. Intracellular signaling from plastid to nucleus. Annual Review of Plant Biology 64, 559–582. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schottler MA, Bock R. 2011. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. The Plant Cell 23, 3137–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. 2004. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. The Plant Cell 16, 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Scharff LB, Clarke CA, Wang Q, Lin CT, Merchant SS, Bock R. 2014. RBF1, a plant homolog of the bacterial ribosome-binding factor RbfA, acts in processing of the chloroplast 16S ribosomal RNA. Plant physiology , 164, 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Jiang Q, Xu J, Zhang J, Teng S, Lin D, Dong Y. 2013. Disruption of the rice plastid ribosomal protein S20 leads to chloroplast developmental defects and seedling lethality. G3: Genes|Genomes|Genetics 10, 1769–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH, Boynton JE, Gillham NW. 1994. Chloroplast ribosomes and protein synthesis. Microbiological Reviews 58, 700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Van Lijsebettens M, Candela H, Micol JL, Tsukaya H. 2012. Ribosomes and translation in plant developmental control. Plant Science 191, 24–34. [DOI] [PubMed] [Google Scholar]
- Inskeep WP, Bloom PR. 1985. Extinction coefficients of chlorophyll a and b in N, N-dimethylformamide and 80% acetone. Plant Physiology 77, 483–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Kleinknecht L, Wang F, Stube R, Philippar K, Nickelsen J, Bohne AV. 2014. RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. The Plant Cell 26, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yang Y, Xu Q, Owsiany K, Welsch R, Chitchumroonchokchai C, Lu S, Van Eck J, Deng X-X, Failla M. 2012. The Or gene enhances carotenoid accumulation and stability during post-harvest storage of potato tubers. Molecular plant 5, 339–352. [DOI] [PubMed] [Google Scholar]
- Lyi SM, Heller LI, Rutzke M, Welch RM, Kochian LV, Li L. 2005. Molecular and biochemical characterization of the selenocysteine Se-methyltransferase gene and Se-methylselenocysteine synthesis in broccoli. Plant Physiology 138, 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyi SM, Zhou X, Kochian LV, Li L. 2007. Biochemical and molecular characterization of the homocysteine S-methyltransferase from broccoli (Brassica oleracea var. italica). Phytochemistry 68, 1112–1119. [DOI] [PubMed] [Google Scholar]
- Morita-Yamamuro C, Tsutsui T, Tanaka A, Yamaguchi J. 2004. Knock-out of the plastid ribosomal protein S21 causes impaired photosynthesis and sugar-response during germination and seedling development in Arabidopsis thaliana. Plant and Cell Physiology 45, 781–788. [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D. 2001. Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. The Plant Journal 27, 179–189. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. 2002. Ribosome structure and the mechanism of translation. Cell 108, 557–572. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, White SW. 1992. The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA. Nature 358, 768–771. [DOI] [PubMed] [Google Scholar]
- Rogalski M, Ruf S, Bock R. 2006. Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Research 34, 4537–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Schottler MA, Thiele W, Schulze WX, Bock R. 2008. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. The Plant Cell 20, 2221–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland N, Curien G, Finazzi G, Kuntz M, Marechal E, Matringe M, Ravanel S, Seigneurin-Berny D. 2012. The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annual Review of Genetics 46, 233–264. [DOI] [PubMed] [Google Scholar]
- Romani I, Tadini L, Rossi F, Masiero S, Pribil M, Jahns P, Kater M, Leister D, Pesaresi P. 2012. Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. The Plant Journal 72, 922–934. [DOI] [PubMed] [Google Scholar]
- Schippers JH, Mueller-Roeber B. 2010. Ribosomal composition and control of leaf development. Plant Science 179, 307–315. [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR. 2011. Assembly of bacterial ribosomes. Annual Review of Biochemistry 80, 501–526. [DOI] [PubMed] [Google Scholar]
- Shumskaya M, Bradbury LM, Monaco RR, Wurtzel ET. 2012. Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. The Plant Cell 24, 3725–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S, Powers T, Changchien L-M, Noller HF. 1989. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science 244, 783–790. [DOI] [PubMed] [Google Scholar]
- Takano S, Niihama M, Smith HM, Tasaka M, Aida M. 2010. gorgon, a novel missense mutation in the SHOOT MERISTEMLESS gene, impairs shoot meristem homeostasis in Arabidopsis. Plant and Cell Physiology 51, 621–634. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. 1998. Role of cold-responsive genes in plant freezing tolerance. Plant Physiology 118, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R. 2014. The translational apparatus of plastids and its role in plant development. Molecular plant 7, 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Weingartner M, Thiele W, Maximova E, Schottler MA, Bock R. 2012. The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. The Plant Journal 69, 302–316. [DOI] [PubMed] [Google Scholar]
- Vaidya AT, Top D, Manahan CC, Tokuda JM, Zhang S, Pollack L, Young MW, Crane BR. 2013. Flavin reduction activates Drosophila cryptochrome. Proceedings of the National Academy of Sciences of the United States of America 110, 20455–20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438 [DOI] [PubMed] [Google Scholar]
- Walter M, Piepenburg K, Schöttler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R. 2010. Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. The Plant Journal 64, 851–863. [DOI] [PubMed] [Google Scholar]
- Wang S, Yang X, Xu M, Lin X, Lin T, Qi J, Shao G, Tian N, Yang Q, Zhang Z, Huang S. 2015. A rare SNP identified a TCP transcription factor essential for tendril development in cucumber. Molecular Plant 8, 1795–1808. [DOI] [PubMed] [Google Scholar]
- Wang YQ, Yang Y, Fei Z, Yuan H, Fish T, Thannhauser TW, Mazourek M, Kochian LV, Wang X, Li L. 2013. Proteomic analysis of chromoplasts from six crop species reveals insights into chromoplast function and development. Journal of experimental botany 64, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Woo HR, Goh CH, Park JH, de la Serve BT, Kim JH, Park YI, Nam HG. 2002. Extended leaf longevity in the ore4-1 mutant of Arabidopsis with a reduced expression of a plastid ribosomal protein gene. The Plant Journal 31, 331–340. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR. 2000. The plastid ribosomal proteins identification of all the proteins in the 50S subunit of an organelle ribosome (chloroplast). Journal of Biological Chemistry 275, 28466–28482. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, von Knoblauch K, Subramanian AR. 2000. The plastid ribosomal proteins identification of all the proteins in the 30S subunit of an organelle ribosome (chloroplast). Journal of Biological Chemistry 275, 28455–28465. [DOI] [PubMed] [Google Scholar]
- Yang Y, Qiang X, Owsiany K, Zhang S, Thannhauser TW, Li L. 2011. Evaluation of different multidimensional LC–MS/MS pipelines for isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of potato tubers in response to cold storage. Journal of Proteome Research 10, 4647–4660. [DOI] [PubMed] [Google Scholar]
- Yang Y, Thannhauser TW, Li L, Zhang S. 2007. Development of an integrated approach for evaluation of 2-D gel image analysis: Impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis 28, 2080–2094. [DOI] [PubMed] [Google Scholar]
- Yin T, Pan G, Liu H, Wu J, Li Y, Zhao Z, Fu T, Zhou Y. 2012. The chloroplast ribosomal protein L21 gene is essential for plastid development and embryogenesis in Arabidopsis. Planta 235, 907–921. [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S. 2008. Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. The Plant Cell 20, 1786–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HD, Yang X-F, Chen S-T, Wang Y-T, Li J-K, Shen Q, Liu X-L, Guo F-Q. 2012. Downregulation of chloroplast RPS1 negatively modulates nuclear heat-responsive expression of HsfA2 and its target genes in Arabidopsis. PLoS Genetics 8, e1002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Owsiany K, Sheeja TE, et al. 2015. A single amino acid substitution of the ORANGE protein causes carotenoid accumulation in Arabidopsis. Plant Physiology 169, 421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate J, Noller HF. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896. [DOI] [PubMed] [Google Scholar]
- Zhang JX, Yuan H, Fei ZJ, Pogson BJ, Zhang LG, Li L. 2015. Molecular characterization and transcriptome analysis of orange head Chinese cabbage (Brassica rapa L. ssp. pekinensis). Planta 241, 1381–1394. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sun TH, Wang N, Ling HQ, Lu S, Li L. 2011. The cauliflower Orange gene enhances petiole elongation by suppressing expression of eukaryotic release factor 1. New Phytologist 190, 89–100. [DOI] [PubMed] [Google Scholar]
- Zhou X, Welsch R, Yang Y, Álvarez D, Riediger M, Yuan H, Fish T, Liu J, Thannhauser TW, Li L. 2015. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proceedings of the National Academy of Sciences of the United States of America 112, 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.