Highlight
To illustrate the hypoxia signal mediated by SnRK1.1, phosphoproteomics was used to identify not only the new SnRK1.1 putative targets related to submergence, but also to gain an insight into early hypoxia signalling events.
Key words: Energy starvation, phosphoproteomics, SnRK1, submergence.
Abstract
SNF1 RELATED PROTEIN KINASE 1 (SnRK1) is proposed to be a central integrator of the plant stress and energy starvation signalling pathways. We observed that the Arabidopsis SnRK1.1 dominant negative mutant (SnRK1.1 K48M) had lower tolerance to submergence than the wild type, suggesting that SnRK1.1-dependent phosphorylation of target proteins is important in signalling pathways triggered by submergence. We conducted quantitative phosphoproteomics and found that the phosphorylation levels of 57 proteins increased and the levels of 27 proteins decreased in Col-0 within 0.5–3h of submergence. Among the 57 proteins with increased phosphorylation in Col-0, 38 did not show increased phosphorylation levels in SnRK1.1 K48M under submergence. These proteins are involved mainly in sugar and protein synthesis. In particular, the phosphorylation of MPK6, which is involved in regulating ROS responses under abiotic stresses, was disrupted in the SnRK1.1 K48M mutant. In addition, PTP1, a negative regulator of MPK6 activity that directly dephosphorylates MPK6, was also regulated by SnRK1.1. We also showed that energy conservation was disrupted in SnRK1.1 K48M, mpk6, and PTP1 S7AS8A under submergence. These results reveal insights into the function of SnRK1 and the downstream signalling factors related to submergence.
Introduction
Owing to the limited availability of oxygen in water, plants cannot obtain sufficient energy from the oxidative phosphorylation of mitochondria to maintain their necessary functions, such as growth and defence responses to environmental changes, when submerged. Consequently, in order to avert an energy crisis during submergence plants have to adjust their metabolism. To maintain ATP production, plants switch to anaerobic fermentation. In addition, under hypoxia, plants also accumulate alanine and GABA through the GABA shunt. Alanine and GABA are used to store carbon and nitrogen which are easily lost during oxygen deprivation (Miyashita et al., 2007). To control energy consumption, dynamic polysome loading of mRNAs is reduced by 50% and only specific stress response genes, such as hypoxia core response genes, are selected to the polysome (Mustroph et al., 2009). However, how plants sense an energy crisis and modulate their responses under submergence remains unclear.
Two types of kinases are known to play important roles in oxygen deprivation. The activities of MPK3 and 6 are induced by the ROS released from mitochondria in the early stages of hypoxia and overexpression of MPK6 enhances anoxia tolerance in Arabidopsis (Chang et al., 2012). Furthermore, SNF1 RELATED PROTEIN KINASE 1 (SnRK1) is known to trigger a vast array of transcriptional and metabolic reprogramming in response to declining energy levels (Crozet et al., 2014). SnRK1 is an evolutionarily conserved energy-sensing protein kinase which is known as AMP-DEPENDENT PROTEIN KINASE 1 (AMPK) in mammals and SUCROSE NON-FERMENTING 1 (SNF1) in yeast (Polge and Thomas, 2007). In mammals, AMPK is activated by an increase in the cellular AMP/ATP ratio to enhance glucose uptake. In yeast, SNF1 plays a key role in shifting fermentation to oxidative phosphorylation in order to generate ATP efficiently under glucose starvation (Hardie et al., 2011). In plants, SnRK1 is regulated by AMP (Sugden et al., 1999), Ca2+ signalling (Lu et al., 2007), GEMINIVIRUS RAP INTERACTING KINASE 1/2 (GRIK1/2) (Shen et al., 2009), glucose-6-phosphate (Nunes et al., 2013), and trehalose-6 phosphate (Zhang et al., 2009). In Arabidopsis, two SnRK1s, SnRK1.1 (akin10) and SnRK1.2 (akin11), were proposed to be the central hub of a transcriptional network that responds to abiotic stresses and energy signalling (Baena-Gonzalez et al., 2007). Two amino acids, K48 and T175 of SnRK1.1 and K49 and T176 of SnRK1.2, are essential for kinase activity. The conserved lysine residue (K48) of SnRK1.1 is required for ATP binding and the T175 is located in the activation loop of SnRK1.1 (Baena-Gonzalez et al., 2007). The T175 of SnRK1.1 is phosphorylated by GRIKs (Crozet et al., 2014). Phosphorylated SnRK1.1 has a stronger activity than non-phosphorylated SnRK1.1. SnRK1.1 is responsible for the major part of the activity of SnRK1 in sugar and ABA signalling (Jossier et al., 2009; Rodrigues et al., 2013) and miRNA-mediated energy signalling and development (Tsai and Gazzarrini, 2012; Confraria et al., 2013). Under submergence, SnRK1.1 also regulates ADH1 and PDC1 expression, and the transgenic line that overexpresses inactive SnRK1.1 shows sensitivity to submergence (Cho et al., 2012). In addition, SnRK1.1 forms complexes with β and γ proteins or with β and βγ proteins to phosphorylate the targets (Baena-Gonzalez et al., 2007; Ramon et al., 2013). The well-known SnRK1 targets are SUCROSE PHOSPHATE SYNTHASES (SPS), TREHALOSE PHOSPHATE SYNTHASE (TPS) (Zhang et al., 2009), and FRUCTOSE-2,6 PHOSPHATE BIPHOSPHATASE (F2KP). However, the environmental conditions that trigger SnRK1 to phosphorylate these targets in plants are not clear, and the signalling mechanism of SnRK1.1 is not well elucidated.
Through AMP/ATP measurement and SnRK1 phosphorylation analysis, we found here that energy starvation occurred in the early stage (30min) of submergence in the dark. The transgenic lines, SnRK1.1K48M and SnRK1.1T175A, that overexpress inactive forms of SnRK1.1 were sensitive to submergence and showed disrupted energy homeostasis in the late stage of submergence. We also performed iTRAQ to compare the amounts of phosphorylated peptides in Col-0 and SnRK1.1 dominant negative mutants under submergence. The results suggest that, in the early stage of submergence, SnRK1 plays an essential role in regulating multiple cellular responses, including anaerobic metabolism, PTP1-MPK6 signalling, protein synthesis, and osmotic regulation.
Materials and methods
Plant materials
Transgenic lines expressing SnRK1T175A, SnRK1T175D, and SnRK1K48M in Col-0 were generated by T-DNA transformation containing the 35S promoter driving the ATP binding site mutant [SnRK1K48M] and catalytic site mutants [SnRK1T175D and SnRK1T175A] in the pMDC32 vector. We analysed SnRK1 expression and the submergence phenotype in seedlings of the T2 or T3 generations from at least two independent transgenic lines. The cMYC-SnRK1 K48M overexpression lines were generated by transforming the 35S::cMYC-SnRK1 K48M/pEarleyGate 203 vector into Col-0. The T-DNA insertion lines of mpk6 (SALK-127507) and ptp1 (SALK-118658C) were obtained from the ABRC, Ohio State University. The PTP1S7AS8A overexpression line was generated by transforming the 35S::PTP1 S7AS8A -YFP/pEarleyGate 103 construct into ptp1.
All seeds were sterilized with 0.5% sodium hypochlorite for 10min and washed with sterilized water and then incubated with sterilized water at 4 °C in the dark for 3 d to achieve uniform germination. Seeds were sown on plates with 0.55% phytagel (Sigma-Aldrich) in half-strength Murashige and Skoog (MS) medium (Duchefa Biochemie) containing 0.5% sucrose at pH 5.7. The plates were transferred to a growth chamber at 22 °C with a 16h light (81 μmol m−2 s−1)/8h dark cycle and placed vertically.
Submergence treatments
For phenotypic assays, data were collected from 36 plants per genotype in six independent experiments. Four-week-old potted plants were put into distilled water at a depth of at least 5cm from the water surface in the dark in a growth chamber under a cycle of 9h light (81 μmol m−2 s−1 at 22 °C) and 15h dark (at 18 °C). After the submergence treatment, plants were put back into a growth chamber under a 9h light (81 μmol m−2 s−1 at 22 °C) and 15h dark (at 18 °C) cycle for 4 d of recovery and then counted for damage. The percentage necrotic area of a leaf was used as the damage index. For the submergence treatment of 9-d-old seedlings, plates with plants on the surface of the medium containing half-strength MS salts and 0.5% sucrose at pH 5.7 were placed into half-strength MS liquid medium that was bubbled with 3% oxygen balanced with nitrogen for 0.5h, 1h, and 3h at room temperature in the dark. The MS liquid medium was pre-bubbled with 3% oxygen for 1h before use (the final oxygen concentration of the medium was 0.006%).
ATP and AMP assay
Whole seedlings (fresh weight 80mg) from 9-d-old Col-0, SnRK1 T175A, SnRK1 T175D, SnRK1 K48M, mpk6, ptp1, and PTP1 S7AS8A /ptp1, treated with different times of submergence, were collected and then used to quantify ATP and AMP -with an LC mass spectrometer. The detailed conditions of LC mass spectrometry are provided in the Supplementary data at JXB online.
Sample preparations for phosphoproteomics and proteomics
Sample preparations of phosphoproteomics and proteomics were performed according to Lan et al. (2012), and the workflow is shown in Supplementary Fig. S3 at JXB online. In brief, total protein was extracted from 9-d-old whole seedlings of Col-0 and SnRK1.1 K48M treated with 0h, 0.5h, 1h, and 3h of submergence. The protein samples were reduced and alkylated by using dithiothreitol (DTT) and iodoacetamide (IAA). Subsequently, endoproteinase Lys-C (Wako) and trypsin (Promega) were added to digest the proteins into peptides. Then, the resulting peptide solution was acidified and desalted.
iTRAQ labelling
For phosphoproteomics, samples of Col-0 that underwent different durations of submergence (0h, 0.5h, 1h, and 3h) were labelled 113, 114, 115, or 116, respectively, and samples from SnRK1.1 K48M-5 that also underwent different durations of submergence were labelled 117, 118, 119, or 121, respectively. To reduce the peptide complexity of the eight samples, after labelling each reaction mixture was aliquoted into two parts of equal volume and these aliquots were mixed into three fractions as follows: fraction 1: 113,114,115, and 116; fraction 2: 117,118,119, and 121; fraction 3: 113,114,117, and 118. The profile of each fraction in different biological repeats is listed in Supplementary Table S1 at JXB online.
For total proteome quantitative analysis, the desalted peptide samples from Col-0 with different treatments were labelled 113–116, and samples from SnRK1.1 K48M-5 with different treatments were labelled 117–121, respectively.
Proteomics database search, quantification, and statistical analysis
Protein identification, phosphorylation modification, and quantitative analyses were analysed by the Proteome Discoverer software (ver. 1.3, Thermo Scientific). MS data were searched against the Arabidopsis protein database, TAIR10, downloaded from the Arabidopsis Information Resource Center. The data were normalized with the value of the 113-labelled sample and the median of the log2 ratio in each sample. The median ratio of each sample was derived from the frequency distribution histograms of log2 protein/phosphorylated peptide ratios of each sample.
In the phosphoproteome quantitative analysis, only unique peptides with a pRS score >50 were used for quantification. To integrate fraction 1 and fraction 3 in each batch, the values of fraction 3 were normalized by the average value of 117/113 in fraction 2 to get the ratio of 118/113, 119/113, and 121/113. The peptides detected in two of the three biological experiments were selected for bootstrap statistical analysis (de la Fuente van Bentem et al., 2008; Cleries et al., 2012). The time point data that were not significantly different (P <0.05) are marked as “nosig”. The time point data that were not detected or detected once in three batches are marked as NA (not available).
Recombinant protein expression and in vitro kinase assay
The recombinant proteins of F2KP, PENTA, eIFiso4G1, and PTP1 were purified from E. coli. For the in vitro kinase assay, Immunoprecipitated SnRK1.1 from Col-0 was incubated with recombinant protein (1–0.8 μg) in 20 μl of kinase buffer for 1h at 30 °C. The kinase reactions were performed with or without λ phosphatase (New England Biolabs) for 1h at 30 °C. All reaction products were resolved in a Phos Tag SDS PAGE (Wako) and S-tag immunoblotting. The details of recombinant protein expression and in vitro kinase assay are provided in the Supplementary data.
Metabolomics extraction and quantification
The ground tissue (100mg) from 9-d-old whole seedlings of Col-0, SnRK1.1 K48M-5, and SnRK1.1 K48M-9 treated for different durations of submergence were re-suspended with 1ml 70% methanol with 12 μg ml–1 ribitol, and then sonicated for 30min at 4 °C. Subsequently, samples were centrifuged at the highest speed at 4 °C for 15min. The supernatant of each sample was collected and vacuum dried. The dried samples were then derivatized by bis(trimethylsilyl)-trifluoroacetamide (BSTFA) containing 1% trimethylchlorosilane (TMCS) and analysed using a Pegasus 4D GCxGC-TOFMS system (LECO, St Joseph, MI, USA). The detailed conditions of GCxGC-TOFMS system are provided in the Supplementary data.
Bimolecular fluorescence complementation
For the BIFC assay, the SnRK1.1, PTP1, and MPK6 coding regions were constructed into the destination expression vectors 3130 (pSAT-35S:: cYFP-DEST) and 3136 (pSAT-35S::nYFP-DEST), respectively. The resulting 3130-target and 3136-SnRK1.1 constructs were mixed and transfected into Arabidopsis leaf protoplasts (Wu et al., 2009). The protoplasts were visualized with a Zeiss LSM510 META laser scanning confocal microscope.
Arabidopsis transient transformation
Arabidopsis transient assays were performed using 7-d-old seedlings infiltrated with Agrobacterium C58Ci carrying 35S::PTP1-YFP, 35S::PTP1S7AS8A-YFP, or 35S::PTP1S7DS8D-YFP in vector pEarlygate 103 according to Wu et al. (2014). For examination of the interaction of MPK6 and transiently expressed PTP1, PTP1S7AS8A-YFP, or PTP1S7DS8D-YFP, and different forms of PTP1 were immunoprecipitated by an anti-GFP antibody. The steps of protein immunoprecipitation were the same as those used in the in vitro kinase assays. The pull-down protein mixture was further examined by MPK6 and GFP immunoblotting.
Accession numbers
The two proteomics datasets were deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaino et al., 2014) with the data set identifier PXD001815 for phosphoprotoemics data and PXD002353 for total proteomics data. The primer sequences used in this study, including gene accession numbers, are listed in Supplementary Table S8.
Results
AMP/ATP ratio increases in Arabidopsis cells under submergence
To control cellular metabolism, energy sensing is usually achieved in the cell by monitoring of the AMP/ATP ratio (Hardie, 2011). To uncover the time at which energy starvation occurs under submergence, we quantified the cellular levels of ATP and AMP by LC-MS. The results showed that the AMP/ATP ratio increased within 30min of submergence, indicating the occurrence of energy starvation during submergence. Notably, after 6h of submergence, the AMP/ATP ratio reached a steady-state level (Fig. 1A). These observations suggest that plants have the ability to adjust energy production and energy consumption under submergence.
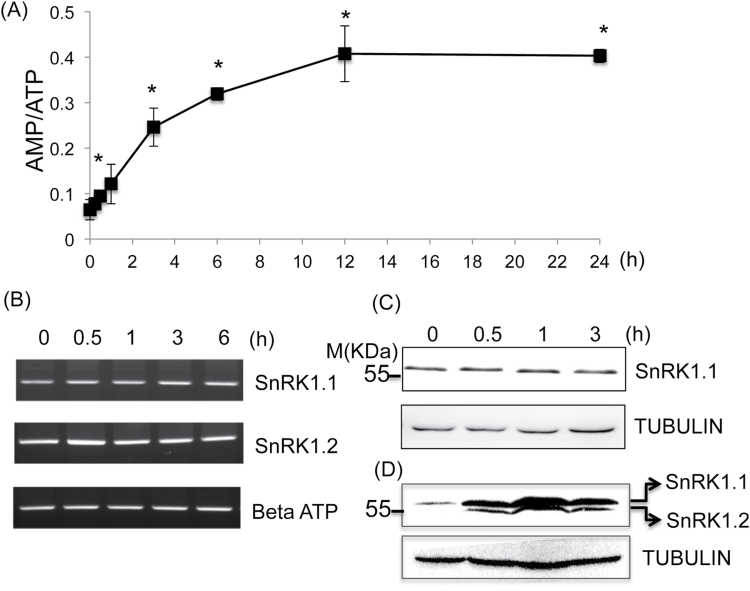
Fig. 1.
Energy starvation occurs in 9-d-old seedlings in the early stage of submergence at room temperature in the dark. (A) The change in the AMP/ATP ratio in Col-0 in the early stage of submergence. (B) The expression of SnRK1.1 and SnRK1.2 in Col-0 under submergence. The expression of βATP was used as an internal control. (C) Protein abundance and (D) phosphorylation of SnRK1 under submergence. According to Baena-Gonzalez et al. (2007) and the molecular weight of SnRK1.1 (61kDa) and SnRK1.2 (58kDa), the upper band represents the phosphorylation of SnRK1.1T175 and the lower band represents the phosphorylation of SnRK1.2T175. TUBULIN was used as an internal control. The data represent four independent biological replicates. Statistical differences between submerged and control samples were determined by Student’s t test. *, P <0.05.
SnRK1 was proposed as a signal hub of energy starvation (Baena-Gonzalez et al., 2007). Therefore, we examined whether SnRK1 is regulated during the early stage of submergence by quantifying the transcripts, protein abundance, and phosphorylation of SnRK1 (Fig. 1B–D; Fig. S1). The transcripts of SnRK1.1 and SnRK1.2 were not changed under submergence (Fig. 1B). Since SnRK1.1 is a major SnRK1 isoform (Jossier et al., 2009), we generated a specific SnRK1.1 antibody to detect the SnRK1.1 protein and used the p-AMPKα (T172) antibody to examine the phosphorylation level of SnRK1 under submergence. The p-AMPKα (T172) antibody recognizes threonine phosphorylation of the two SnRK1 isoforms (T175 of SnRK1.1 and SnRK1.2) (Baena-Gonzalez et al., 2007). The results showed that SnRK1.1 protein abundance was not changed under submergence, but that phosphorylation of SnRK1.1 was induced within 0.5–1h of submergence (Fig. 1C, D). Interestingly, phosphorylation of SnRK1.2 was also induced at the same stage of submergence. These results suggest that SnRK1 is involved in the early signalling response of submergence.
SnRK1.1 is involved in maintaining energy balance under submergence
Prior studies indicated a potential functional redundancy of SnRK1.1 and SnRK1.2 in hypoxia responses (Baena-Gonzalez et al., 2007). Thus transgenic Arabidopsis expressing the inactive forms of SnRK1.1 (SnRK1.1K48M and SnRK1.1T175A) were generated. We selected two SnRK1.1K48M, two SnRK1.1T175A mutants, and one SnRK1.1T175D mutant that have different transcript and protein levels of the SnRK1 transgene, for further analysis (Fig. 2A; Supplementary Fig. S2A). We first examined the submergence tolerance of these transgenic lines by subjecting 4-week-old plants to submergence for 2.5 d (60h) and recovery for 4 d. The percentage necrotic area of a leaf was used as the damage index (Fig. 2B). The results showed that the dominant negative mutants (SnRK1.1K48M and SnRK1.1T175A) were more sensitive to submergence (Fig. 2B, C). To examine whether these physical responses were related to SnRK1.1 abundance and activity, we overexpressed the constitutive active form (SnRK1.1T175D) in Col-0 according to the results of (Avila et al., 2012) showing that SnRK1T175D has stronger activity than SnRK1.1. The transgenic plant SnRK1.1 T175D with higher SnRK1.1 protein and activity had better tolerance toward submergence than Col-0 (Fig. 2C; Supplementary Fig. S2B).
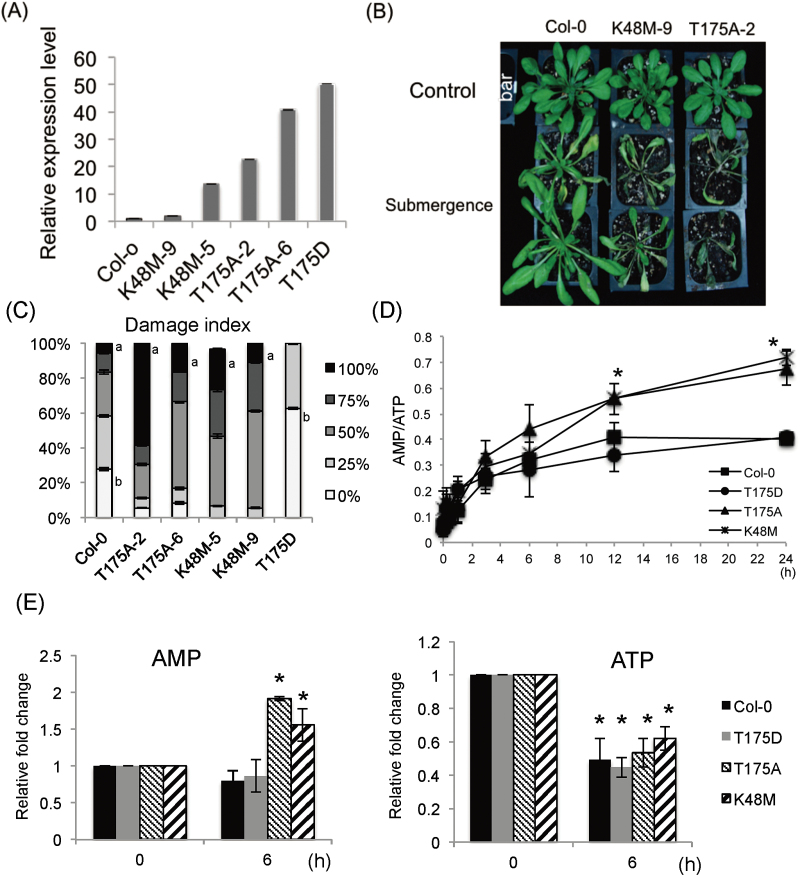
Fig. 2.
Dominant negative mutants are more sensitive to submergence than Col-0. (A) The expression of SnRK1.1 in Col-0, SnRK1.1 K48M, SnRK1.1 T175A, and SnRK1.1 T175D quantified by qRT-PCR. TUBULIN mRNA was the internal control. (B) The phenotype of 4-week-old Col-0 and dominant negative mutants (SnRK1.1 K48M, SnRK1.1 T175A, and SnRK1.1 T175D), after submergence for 60h in the dark at 22 °C and recovery for 4 d, bar =1cm. (C) After submergence for 60h and recovery for 4 d, the percentage of necrotic leaf area was used as the damage index. The data represent means ±SE from six independent biological replicates. Data from the lines with the same lowercase letters were significantly different from Col-0, which was determined by Student’s t test (P <0.05). (D) The change in the AMP/ATP ratio in 9-d-old seedlings of Col-0 and SnRK1.1 mutants at different stages of submergence at room temperature in the dark. (E) The relative fold change of ATP and AMP in 9-d-old seedlings of Col-0 and SnRK1.1 mutants under submergence. The data represent means ±SD from four independent biological replicates. Statistical differences between submerged and control samples were determined by Student’s t test. *, P <0.05.
We quantified the ATP and AMP levels in Col-0, SnRK1.1 K48M-5, SnRK1.1 T175A-2, and SnRK1.1 T175D plants. At 0.5–3h of submergence, the AMP/ATP ratios were similar in the mutants and Col-0. After 6h of submergence, the AMP/ATP ratio in Col-0 and SnRK1.1 T175D reached a steady state, but the AMP/ATP ratio in SnRK1.1 K48M-5 and SnRK1.1 T175A-2 continued to increase (Fig. 2D). These results demonstrated that inactive SnRK1.1 disrupted the ability of the cells to maintain the energy balance during the later stages of submergence. Interestingly, under submergence, the ATP level decreased in all four lines (Fig. 2E), suggesting that the energy reduction caused by oxygen deprivation was not impacted by SnRK1.1. However, the AMP accumulated to higher levels in SnRK1.1 K48M-5 and SnRK1.1 T175A-2 than in Col-0 and SnRK1.1 T175D (Fig. 2E), indicating that SnRK1.1 was involved in energy conservation under submergence.
SnRK1.1 is a central phosphorylation hub under submergence
To determine the functions of SnRK1.1 under submergence, we applied iTRAQ to determine the changes in phosphorylation and the abundance of cellular proteins in Col-0 and SnRK1.1 K48M-5. The workflow of this analysis is illustrated in Supplementary Fig. S3. In total proteomics, 5 574 proteins were identified. In phosphoproteomics, the phosphorylation peptides with a pRS score (the possibility of phosphorylation in the peptide) ≥50 that were identified in two out of three repeats were selected. A total of 615 unique phosphopeptides that belong to 485 proteins was identified by phosphoproteomics (Supplementary Tables S1, S2). Three steps were used to generate a list of up- or down-regulated phosphopeptides from these 615 peptides. (i) We normalized all the data with the value of Col-0 in normal conditions. (ii) Because the standard deviations of three biological repeats were E0.3 (Supplementary Table S1), the proteins with an average fold-change in phosphorylation level >1.3 or <0.7 and that passed the bootstrap analysis (P <0.05) were selected as being significantly different under submergence. This generated 57 ‘proteins with up-regulated phosphorylation’ (PUP) and 27 ‘proteins with down-regulated phosphorylation’ (PDP) in Col-0 under submergence (Supplementary Tables S3, S4). These proteins were mainly located in the nucleus and plasma membrane (Fig. 3A) and were involved in carbohydrate metabolism (FDR 2.0E-05), gene expression (FDR 6.20E-05), nitrogen compound metabolism (FDR 5.5E-05), protein metabolism (FDR 1.60E-03), and transport (FDR3.4E-04) (Supplementary Tables S1, S3). We also identified 47 PUP and 60 PDP in SnRK1.1K48M-5 under submergence (Supplementary Table S1). (iii) Since we analysed the phosphorylated proteins profiles of Col-0 and SnRK1.1 K48M-5 by LC MS/MS separately, it meant that the phosphorylation of some proteins was only detected in one of the lines. Therefore, these proteins were excluded from the lists of PDP and PUP. In addition, some phosphorylated proteins in SnRK1.1 K48M-5 had pRS scores lower than 50, but not in Col-0, suggesting that their phosphorylation was disrupted by SnRK1.1 K48M-5. We added these phosphorylated proteins to the PUP lists. Taken together, there were 50 PUP and 18 PDP in Col-0 and 29 PUP and 42 PDP in SnRK1.1 K48M-5 under submergence (Fig. 3B).
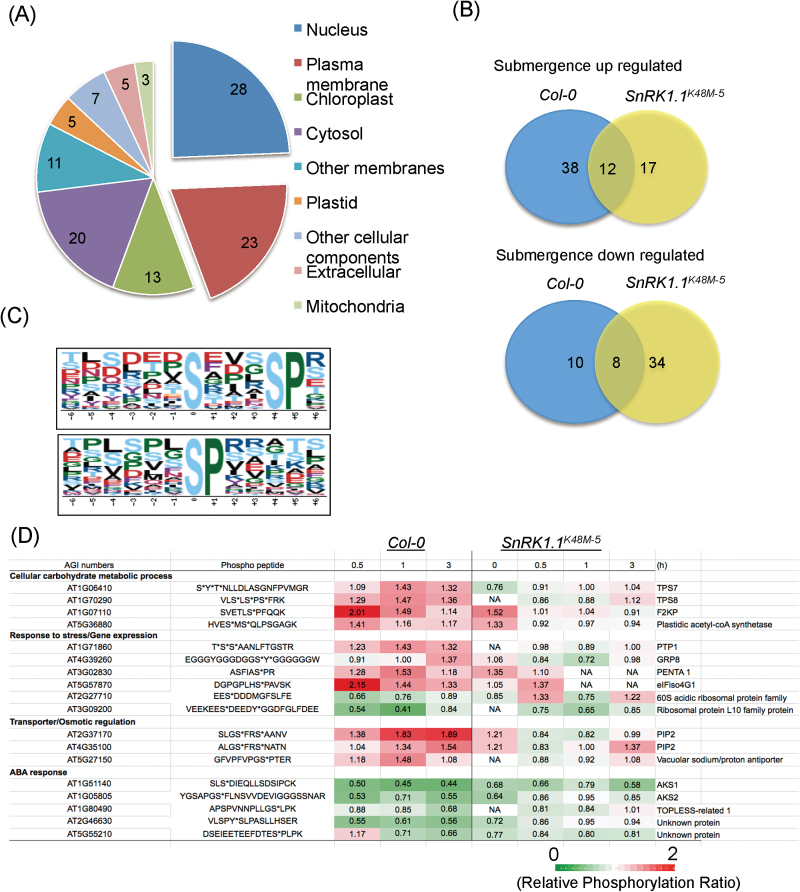
Fig. 3.
Phosphoproteomic profiles of the SnRK1.1 K48M mutant and Col-0 under submergence. The proteins with phosphorylation levels that were significantly different at any time point under submergence (at room temperature in the dark) were selected to for analysis. (A) Gene ontology (GO) catalogues of the submergence-changed phosphorylated proteins in Col-0. (B) Venn diagram illustrating the overlap of up-regulated (upper diagram) and down-regulated (lower diagram) phosphorylated proteins identified in Col-0 and SnRK1 K48M. (C) Sequence logos of the phosphorylation sites seen in submergence up-regulated phosphopeptides. (D) The four gene ontology groups in the SnRK1.1 phosphorylation list, where the statistical differences (P value) of protein phosphorylation in treatment and control of Col-0 was smaller than 0.05, but that in SnRK1 K48M-5 was not;. ‘*’ represents the phosphorylation site. The numbers indicate the average value of the phosphorylation fold change of all the biological repeats. The time point data not detected in the three batches or only detected once in three batches is marked as NA (not available).
Venn diagram analysis was then used to distinguish between SnRK1.1-dependent and -independent phosphorylation. The analysis of PUP in Col-0 and SnRK1.1 K48M-5 under submergence (Fig. 3B, upper panel; Supplementary Table S1) showed that 38 only appeared in Col-0, 12 appeared in both Col-0 and SnRK1.1 K48M-5, and 17 appeared only in SnRK1.1K48M-5. These results suggested that 55 PUP were under SnRK1 regulation. Noticeably, the protein levels of 38 ‘Col-0 only PUP’ were not changed significantly in Col-0 and SnRK1.1 K48M-5 under submergence (Fig. 3B; Supplementary Tables S5, S6), suggesting that SnRK1.1 only regulated their phosphorylation. On the other hand, in the analysis of PDP (Fig. 3B, lower panel; Supplementary Table S1), there were 10 PDP in Col-0 only, eight in both Col-0 and SnRK1.1 K48M-5, and 34 in SnRK1.1 K48M-5 only. These results suggested that the phosphorylation of 44 PDP was under SnRK1 regulation. The protein levels of 10 PDP in Col-0 only were not changed significantly in Col-0 and SnRK1.1 K48M-5 under submergence (Fig. 3B; Supplementary Tables S5, S6), revealing that SnRK1.1 was only involved in their phosphorylation reduction. Therefore, under submergence, 99 phosphorylated proteins, including 55 PUP and 44 PDP, were under SnRK1.1 regulation (Fig. 3B). In addition, two significantly enriched phosphorylation motifs were extracted from the lists of up-regulated phosphopeptides in Col-0 under submergence by the motif finding algorithm, Motif-X (Fig. 3C; Supplementary Table S7; Schwartz and Gygi, 2005). The second motif was also identified in the SnRK1.1-dependent phosphorylated proteins list, including TPS8 and F2KP, two well-known SnRK1 substrates.
Identification of putative SnRK1.1 substrates
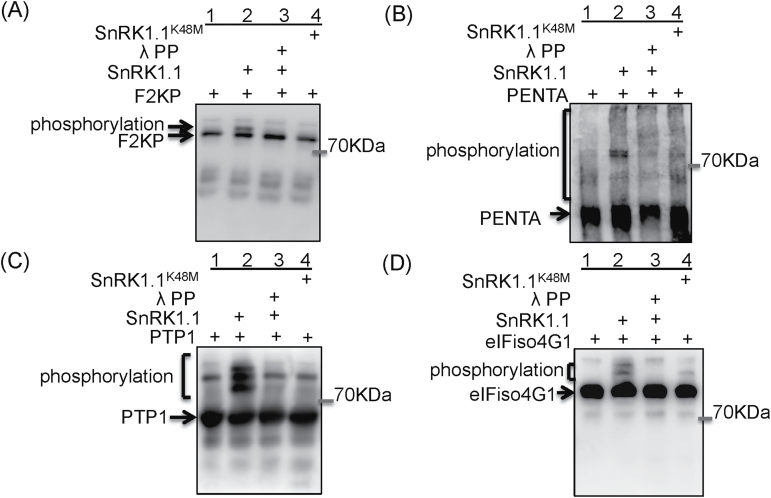
The majority of PUP under submergence was SnRK1.1-dependent, and their protein levels were not changed significantly in Col-0 and SnRK1.1 K48M-5 under submergence (Fig. 3D; Supplementary Tables S5, S6). To test whether they are direct targets of SnRK1.1, we selected four candidates in different functional categories, eIFiso4G1 (a translation initiation factor), PENTA (an RNA stability regulator), PTP1 (PROTEIN TYROSINE PHOSPHATASE 1, a primary target of ROS) (Supplementary Fig. S4A), and F2KP, which is a well-known SnRK1.1 substrate, for the in vitro kinase assay. These proteins were fused with His and S tags and were obtained by E. coli expression systems and affinity-purification. In addition, the active SnRK1.1 and inactive SnRK1.1K48M were immunoprecipitated from Col-0 and cMYC-SnRK1 K48M-1 that had been submerged for 1h. The specificity of the IP products was examined by SnRK1.1 immunoblotting following Phos Tag SDS PAGE (Supplementary Figs S4B, S5). After mixing the four recombinant proteins with SnRK1.1 (IP-SnRK1.1-S) respectively, through S tag immunoblotting following Phos Tag SDS PAGE, the four recombinant proteins (Lane 2, Fig. 4A–D) showed mobility retardation when compared with the lanes in which only the recombinant proteins were loaded (Lane 1, Fig. 4A–D). To validate that the mobility retardation of these proteins was caused by SnRK1.1 phosphorylation, the mixture of IP-SnRK1.1-S and recombinant proteins were treated with λ phosphatase (Lane 3, Fig. 4A–D), or the recombinant proteins were mixed with inactive SnRK1.1K48M (Lane 4, Fig. 4A–D). The mobility retardation of eIFiso4G1, PENTA, and PTP1 was shown only in the mixture of SnRK1, demonstrating that these three proteins were the substrates of SnRK1.1 in vitro.
Fig. 4.
In vitro phosphorylation assays of SnRK1. The recombinant proteins fused with the S tag, (A) F2KP, (B) Penta, (C) PTP1, and (D) eIFiso4G1, were mixed with immunoprecipitated active SnRK1.1 (IP-SnRK1.1-S) or inactive SnRK1.1K48M and the phosphorylation of these recombined proteins was examined with Phos-Tag page and S-tag immunoblotting. Phosphorylated protein mobility is retarded in Phos-Tag PAGE and this retardation can be reduced by the addition of λ phosphatase. Lane 1 of each blot was loaded with recombinant protein only, representing the mobility of non-phosphorylated substrate. In the second and fourth lanes of each blot, the four candidates showed mobile retardation in IP-active SnRK1.1 treatment but not in inactive SnRK1.1K48M treatment. In the third lane of each blot, the retardation of the band shift was reduced with λ phosphatase treatment. At least three independent experiments were performed with similar results.
SnRK1 regulates anaerobic metabolism under submergence
Two known SnRK1 substrates, F2KP and TPS7, had increased phosphorylation in Col-0, but were unchanged in SnRK1.1 K48M-5 under submergence (Supplementary Table S1). F2KP is involved in glycogenesis (Fig. 5A), and its phosphorylation by SnRK1 resulted in a decrease in its activity, leading to reduced energy consumption (Halford and Hey, 2009). TPS7, is a Class II TPS (Ramon et al., 2009) and has been shown to have T6P binding affinity (Harthill et al., 2006), but not clear TPS activity (Ramon et al., 2009). T6P is an essential factor of metabolic signalling and an inhibitor of SnRK1.1 (Zhang et al., 2009). T6P accumulation and the expression of Class II TPS are regulated by increased cellular sucrose (Ramon et al., 2008). Thus, the Class II TPS protein is proposed to be involved in T6P sensing (Harthill et al., 2006; Paul et al., 2008). Interestingly, the reported phosphorylation site, the 5th serine of TPS7 recognized by SnRK1 and 14-3-3 in vitro (Harthill et al., 2006) was also identified in our data (Supplementary Table S5). This phosphorylation site was conserved in TPS5, 6, and 7 and was phosphorylated in response to glycolysis inhibitor (2-deoxyglucose) that stimulates the activity of SnRK1 (Harthill et al., 2006). In our data (Supplementary Table S1), the other Class II TPS, TPS8, also had increased phosphorylation and was also phosphorylated at the consensus sequence of SnRK1 (phiXXXXS/TXXXphi), which is a 10-residue motif with phi being a hydrophobic residue (M, L, V, I, or F), and X representing random amino acids (Halford and Hardie, 1998). These data indicated that, under submergence, SnRK1 might modulate metabolic signalling by phosphorylating these important metabolism factors.
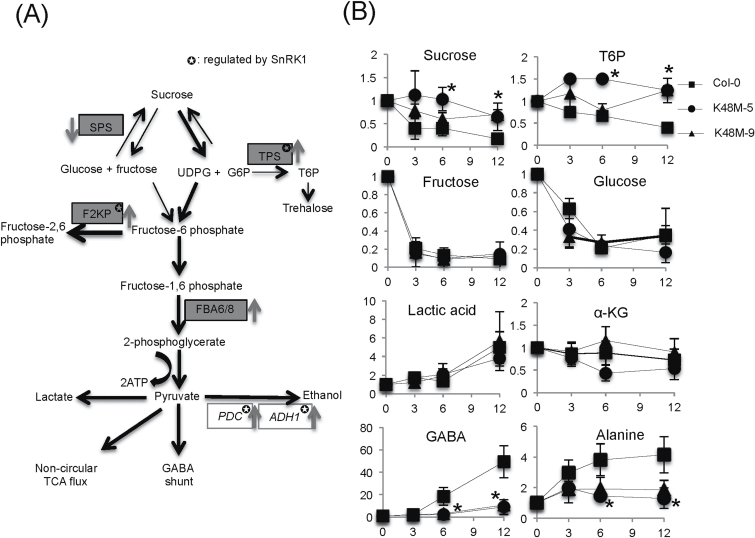
Fig. 5.
Effects of SnRK1.1 on the phosphorylation of glycolytic enzymes during submergence. (A) Schematic model of carbohydrate metabolism under submergence in Arabidopsis. Grey boxes indicate the gene under phosphorylation and white boxes represent the gene under transcriptional regulation. The arrows near the boxes show up- or down-regulation in Col-0 under submergence. Bold arrows indicate the promoting reaction under submergence. (B) The profile of soluble sugars, T6P, alanine, GABA, lactic acid, and α-KG in Col-0 and SnRK1.1 K48M under submergence. The data represent means ±SD from three independent biological replicates. Statistical differences between Col-0 and mutant samples were determined by Student’s t test. *, P <0.05.
Next, we quantified soluble sucrose and T6P in seedlings of Col-0, SnRK1.1 K48M-5, and SnRK1.1 K48M-9. Under submergence, plants initiate fermentation to regenerate NAD+. In addition to the fermentation products ethanol and lactic acid, plants also convert sucrose into alanine and alpha-ketoglutaric acid (α-KG). Alanine is accumulated, but α-KG is also used in the GABA shunt and non-circle TCA (Miyashita and Good, 2008).Therefore, we also quantified lactic acid, α-KG, alanine, and GABA in Col-0 and the two SnRK1.1 K48M lines to examine the effect on glycolysis. Our data showed that sucrose and T6P were reduced in Col-0 in the early stage of submergence; however, there was a smaller decrease in sucrose and T6P in SnRK1.1 K48M-9 and no decrease in the stronger dominant negative mutant SnRK1.1 K48M-5 at the early stage (0–6h) of submergence (Fig. 5B). Glucose and fructose showed the same reduction patterns in Col-0 and SnRK1.1 K48M under submergence (Fig. 5B), and the accumulation of lactic acid in Col-0 and SnRK1.1 K48M was not significantly different, indicating that glucose and fructose were still being converted into lactic acid in SnRK1.1 K48M. Notably, the content of α-KG was not significantly changed in Col-0 and SnRK1.1 K48M under submergence, but alanine and GABA were accumulated in Col-0, but were less accumulated in SnRK1.1 K48M-5 and SnRK1.1 K48M-9 (Fig. 5B). Accordingly, SnRK1.1 modulated sucrose and T6P degradation and altered GABA and alanine accumulation under submergence.
SnRK1 communicates with MPK signalling under submergence
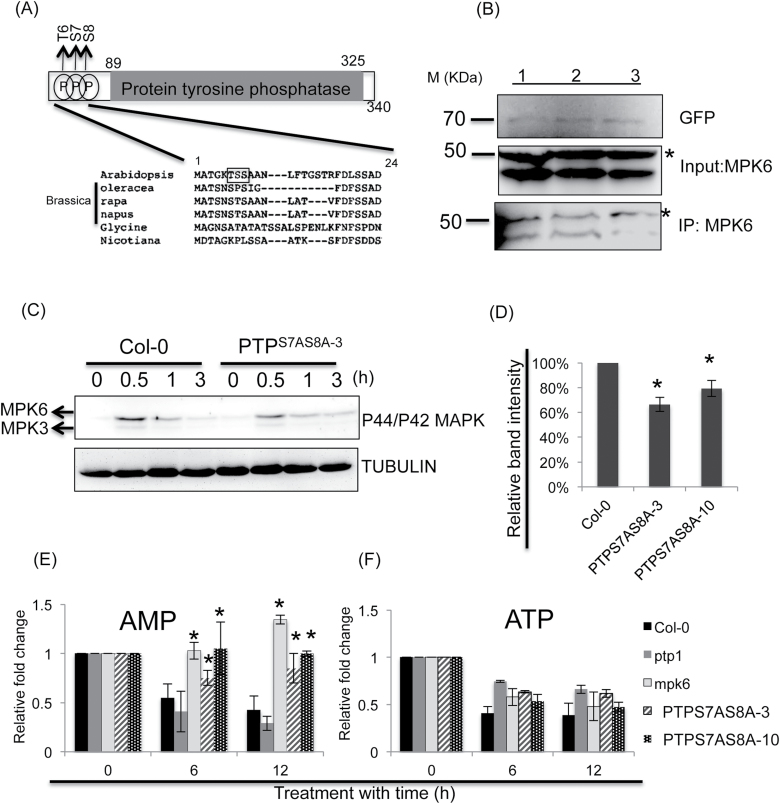
The other prominent group of putative SnRK1.1 substrates comprised proteins related to the stress response (Supplementary Table S5), including PROTEIN TYROSINE PHOSPHATASE 1 (PTP1). PTP1 is a primary target of H2O2 (Gupta and Luan, 2003) and a negative regulator of MPK3/6 activities (Bartels et al., 2009). Recent studies showed that MPK3 and 6 are the central signalling components involved in different abiotic stresses (Taj et al., 2010; Pucciariello et al., 2012a). In the early stages of oxygen deprivation, MPK3 and 6 are activated by the interruption of the oxidative phosphorylation pathway in the mitochondria (Chang et al., 2012). The level of phosphorylated PTP1 in SnRK1.1 K48M-5 is significantly lower than in Col-0 (Supplementary Table S5), and PTP1 was directly phosphorylated by SnRK1.1 in vitro (Fig. 4C; Supplementary Fig. S4C). Through MS-MS analysis, the phosphorylation sites of PTP1 by the in vitro kinase assay were identified at the 6th threonine (PTPT6) and 7th and 8th serine (PTPS7 and PTPS8) (Fig. 6A; Supplementary Table S1), which were also identified in our phosphoproteomics study (Supplementary Table S1). We compared the Arabidopsis PTP1 sequence with orthologues from other plants (Fig. 6A). Only the 7th amino acid of PTP1 in Brassicaceae and soybean (Glycine max) were serine or threonine, which can be phosphorylated. Interestingly, the phosphorylation possibility of PTPS8 is the highest in the in vitro kinase assay and two of three proteomic biological repeats. We therefore focused on the examination of phosphorylation of PTP1S7 and PTPS8.
Fig. 6.
SnRK1.1 regulated MPK6 signalling by phosphorylating PTP1 under submergence. (A) Comparison of three phosphorylation sites (Thr6, Ser7, and Ser8) of Arabidopsis PTP1 with orthologues in other Brassica species, Glycine max and Nicotiana tomentosiformis by ClustalW. The black frame in the amino acid alignment indicates the phosphorylation sites of Arabidopsis PTP1. (B) The interaction of PTP1 and MPK6 was disrupted by PTP1S7DS8D. Upper panel: after immune precipitation, the abundance of (1) PTP1–GFP, (2) PTPS7AS8A–GFP, and (3) PTP1–S7DS8D–GFP conjugated in the beads of each transient samples. Middle panel: the abundance of MPK6 in the three PTP1 transient expression samples. Lower panel: the pull-down MPK6 were detected in PTP1–GFP and PTPS7AS8A–GFP transient expression samples, but not in PTP1–S7DS8D–GFP. *: Non-specific band. At least three biological repeats were performed and showed similar patterns. (C) Phosphorylation of MPK3/6 in Col-0, and PTP1 S7AS8A-3 under submergence. (D) The quantification of band intensity of (C) Western blot by GelEval software (FrogDance). The band intensity of 0.5h submergence in Col-0 and PTP1 S7AS8A lines was quantified, and TUBULIN was used as an internal control. The AMP (E) and ATP (F) profiles in Col-0, ptp1, mpk6, and two PTP1 S7AS8A lines under submergence. Statistical differences between Col-0 and mutant samples were determined by Student’s t test. *, P <0.05.
Since MPK6 could interact with PTP1 (Gupta and Luan, 2003), we examined whether the phosphorylation of PTP1 interrupted its interaction with MPK6. We transiently expressed three forms of GFP-tagged constructs into the ptp1 seedlings: wild-type PTP1–GFP; PTP1S7AS8A–GFP, in which two serines were replaced by alanines and could not be phosphorylated; and PTP1S7DS8D–GFP, which mimics phosphorylated PTP1,. Cellular extracts, with equal amounts of endogenous MPK6, were prepared from these seedlings and used for immunoprecipation of PTP1–GFP, PTP1S7AS8A–GFP, and PTP1S7DS8D–GFP. With the same amounts of endogenous MPK6 input (Fig. 6B, middle panel) and immnuoprecipitated PTP1–GFP, PTP1S7AS8A–GFP, and PTP1S7DS8D–GFP (Fig. 6B, upper panel), MPK6 could be pulled down by PTP1–GFP and PTP1S7AS8A–GFP (Fig. 6B, lanes 2 and 3, lower panel), but not by PTP1S7DS8D –GFP (Fig. 6B, lane 3, lower panel). This indicated that the interaction of PTP1 and MPK6 was disrupted by phosphorylation of the 7th serine and 8th serine of PTP and the interaction of PTP1–GFP with MPK6 was not caused by the GFP fusion tag. These results suggested that, through phosphorylating PTP1, SnRK1 disrupted the interaction of PTP1 and MPK6 to enhance the MPK6 signalling.
To examine this hypothesis, we transformed PTP1 S7AS8A into ptp1 to generate transgenic lines and two transgenic lines were selected that showed expression levels of PTP1 close to those of Col-0 (Supplementary Fig. S6A). We compared the MPK3/6 phosphorylation in Col-0, ptp1, and the two PTP1 S7AS8A lines. MPK6 was phosphorylated in Col-0 after 30min of submergence, but the phosphorylation of MPK3 was not significant (Fig. 6C). The increases in phosphorylation levels of MPK6 in ptp1 and Col-0 were not significantly different (Supplementary Fig. S6B). However, the increases in MPK6 phosphorylation in the two PTP1 S7AS8A transgenic lines were lower than in Col-0. (Fig. 6C, D), indicating that non-phosphorylated PTP1 S7AS8A could prevent MPK6 from being phosphorylated during submergence. To validate the impact of MPK6 phosphorylation in PTP1 S7AS8A under submergence, we also compared the cellular levels of AMP and ATP in the two PTP1 S7AS8A lines, mpk6, ptp1, and Col-0 (Fig. 6E). The ATP was reduced in all of these five lines, demonstrating that energy reduction caused by oxygen deprivation was not impacted by MPK6 and PTP1. Interestingly, AMP was reduced in Col-0 and ptp1 after 6h submergence, but AMP was accumulated more in mpk6 and was not significantly reduced in the two PTP1 S7AS8A lines. These results suggested that MPK6 was involved in energy conservation under submergence and PTP1 S7AS8A interfered with MPK6 signalling by preventing MPK6 phosphorylation.
SnRK1.1 interacts with MPK6 and PTP1
We performed bimolecular fluorescence complementation (BiFC) assays to examine the interaction of SnRK1.1, PTP1, and MPK6. The C-terminal region of Agobacterium tumefaciens Vird2 containing the bipartite nuclear localization signal (NLS) fused with mCherry was used as a nuclear marker and a transient effect control (Fig. 7, lane 3) (Wu et al., 2009). To examine the specificity in BiFC, the pairs of SnRK1.1-nYFP and vector 3130 (pSAT-35S::cYFP-DEST), MPK6-nYFP and PIP2-cYFP, MPK6-cYFP and PIP2-nYFP, PTP1-cYFP and PIP2-nYFP were co-transfected into protoplasts, respectively, as a negative control (Fig. 7, panels 1, 2, 6, 7). On the other hand, three pairs, MPK6-nYFP and PTP1-cYFP, SnRK1.1-nYFP and PTP1-cYFP, and SnRK1.1-nYFP and MPK6-cYFP (Fig. 7, panels 3, 4, 5) were transfected into protoplasts, respectively. The results demonstrated that SnRK1.1 mainly interacts with PTP1 or MPK6 in the cytoplasm (Fig. 7, lane 4, panels 3, 5), but PTP1 interacted with MPK6 in the cytoplasm and nucleus (Fig. 7, lane 4, panel 4). We also examined the interaction of MPK6 with PTP1S7AS8A or PTP1S7DS8D. However, we could not distinguish the interaction of these proteins, due to the high background signal of PTP1S7AS8A and PTP1S7DS8D with the negative control (Supplementary Fig. S7). Taken together, these results suggested that SnRK1.1 directly interacted with PTP1 to phosphorylate PTP1.
Fig. 7.
Protein–protein interactions between PTP1, SnRK1.1, and MPK6. Examination of the interactions between PTP1 and SnRK1.1, PTP1 and MPK6, and MPK6 and SnRK1.1 in protoplasts by BiFC. SnRK1.1-nYFP+cYFP, MPK6-nYFP+PIP2-cYFP, MPK6-cYFP+PIP2-nYFP, and PTP1-cYFP+PIP2-nYFP were the negative controls. The images have different interference contrasts: (lane 1) YFP fluorescence by BiFC, (lane 2) chloroplast auto fluorescence (Chl), (lane 3) mCherry fluorescence as a nuclear marker and transient effect control, and (lane 4) merged image.
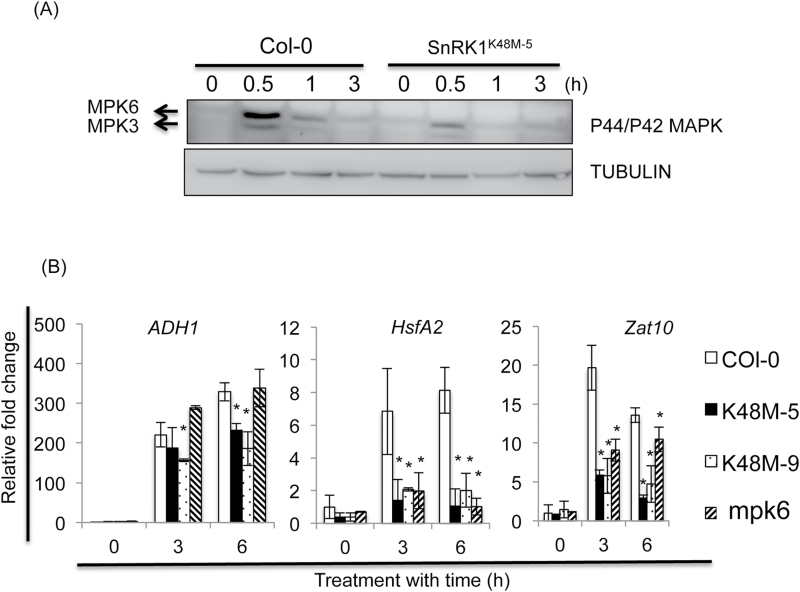
SnRK1.1 regulates MPK6 signalling
To determine whether SnRK1 regulates MPK6 signalling, we compared the phosphorylation of MPK6 in Col-0 and SnRK1.1 K48M-5 (Fig. 8A). Notably, the level of phosphorylated MPK6 was lower in SnRK1 K48M than in Col-0 under submergence (Fig. 8A). The MPK6 protein abundance was not affected in SnRK1 K48M (Supplementary Tables S2, S6). To validate that MPK6 signalling is affected in SnRK1 K48M mutants, we compared the expression of predicted downstream targets of MPK6, HsfA2 and ZAT10 (Pucciariello et al., 2012a), in Col-0, SnRK1.1 K48M-5, and mpk6 under submergence. Both HsfA2 and ZAT10 were induced in Col-0 after 3h and 6h of submergence (Fig. 8B). However, the expression of HsfA2 and ZAT10 were significantly lower in SnRK1.1 K48M and mpk6 than in Col-0 under submergence. In addition, ADH1 had a lower induction level in the two SnRK1.1 K48M mutants than in Col-0, but had a similar level in mpk6 as in Col-0 under submergence (Fig. 8B). Taken together, the results shown in Figs 6–8 indicate that SnRK1.1 enhanced MPK6 signalling by disrupting the interaction of PTP1 and MPK6 under submergence.
Fig. 8.
SnRK1.1 regulated the MPK6 signalling pathway under submergence. (A) Phosphorylation of MPK3 and MPK6 in Col-0 and SnRK1.1 K48M-5 under submergence. The data represent three independent biological replicates. TUBULIN was used as an internal control. (B) Expression of MPK signalling marker genes in Col-0, SnRK1.1 K48M, and mpk6. Transcript levels were detected by qRT-PCR by specific primers. TUBULIN mRNA was the internal control. The data represent means ±SD from four independent biological replicates. Statistical differences between Col-0 and the mutants were determined by Student’s t test. *, P <0.05.
Discussion
SnRK1 is a master regulator of anaerobic metabolism under submergence
SnRK1 has a central role in sugar signalling pathways (Jossier et al., 2009). Two important sugar metabolic enzymes, TPS and F2KP, are phosphorylated by SnRK1 in vitro (Halford and Hey, 2009). We have reported here that these two enzymes are phosphorylated via SnRK1.1 in Arabidopsis under submergence. We have shown that the activity of SnRK1.1 was triggered under submergence and the inactive form of SnRK1.1 disrupted the endogenous SnRK1.1 to phosphorylate its targets. By measuring the cellular levels of various metabolites in Col-0 and SnRK1.1 K48M, we have shown that SnRK1 enhances sucrose and T6P degradation, and alanine and GABA accumulation under submergence (Fig. 5B). The reduction of T6P, a negative regulator of SnRK1 activity (Delatte et al., 2011), suggested that SnRK1.1 might repress T6P synthesis to maintain its activity by phosphorylating TPS7 and 8. However, these TPSs did not have a clear TPS function under normal conditions (Ramon et al., 2009). It will be interesting to know whether they play a role in T6P metabolism under submergence. In addition, one of the SnRK1 targets, F2KP, is a bifunctional enzyme that can generate fructose-6-phosphate or fructose-2,6-bisphosphate. Fructose-2,6-bisphosphate is accumulated under hypoxia and has been proposed to be a signalling molecule for regulating glycolysis (Mertens et al., 1990; Nielsen et al., 2004). Collectively, our results suggest that SnRK1 might phosphorylate F2KP to switch its activity to produce fructose-2,6-bisphophate and then enhance the glycolysis pathway to adapt to energy starvation.
SnRK1 modulates MPK signalling under submergence
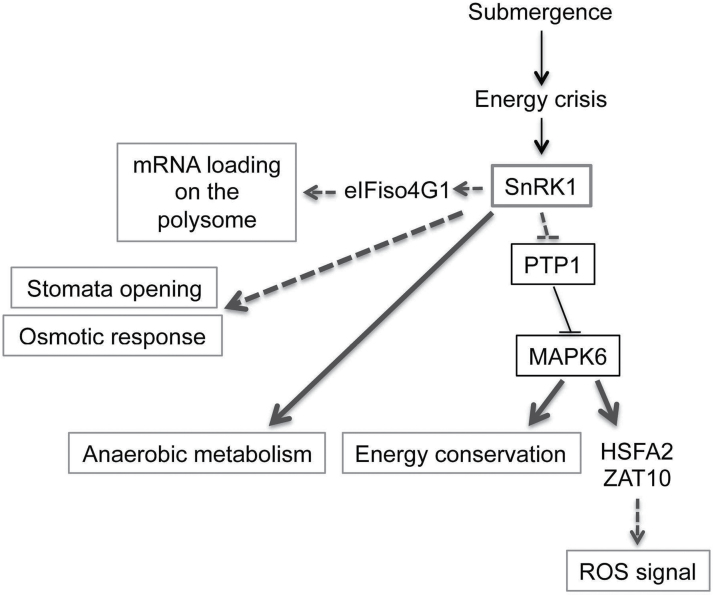
MPK6 signalling is a general stress signal whose phosphorylation can be triggered by the ROS and MKK cascades (Taj et al., 2010). Interestingly, ROS is an important signal that can enhance ethylene signalling and ADH1 expression during oxygen deprivation (Pucciariello et al., 2012a, b; Yang, 2014), and MPK6 is the predominant MPK regulated by ROS released from the disruption of the electron transportation chain in the mitochondria during oxygen deprivation and re-oxygenation. We also showed that the induction levels of two ROS responsive marker genes, HsfA2 and ZAT10 (Pucciariello et al., 2012a, b ), were reduced in mpk6 under submergence. Transgenic lines that overexpress MPK6 are more tolerant than Col-0 under submergence (Chang et al., 2012). These results suggested that the ROS response mediated by MPK6 signalling might contribute to improve submergence tolerance. Interestingly, under submergence, MPK6 phosphorylation was reduced in the SnRK1.1 mutant and PTP1 S7AS8A by the phospho p44/p42 MAPK antibody. This antibody recognizes the (TXY) motif of MPK6 phosphorylation, revealing that the phosphorylation of MPK6T221 and MPK6Y223 was disrupted by PTP1 and SnRK1.1 under submergence. The TXY (X-D/E/P) motif is a dual phosphorylation site and phosphorylation of both residues is essential for the activation of the MPKs (Taj et al., 2010). PTP1 is a negative regulator dephosphorylating the tyrosine phosphorylation of MPK6 (Gupta and Luan, 2003), suggesting that PTP1 regulated MPK6Y223 phosphorylation under submergence. To address how SnRK1.1 disrupted MPK6 signalling by phosphorylating PTP1, we demonstrated that MPK6 could interact with PTP1, but not with PTP1S7DS8D, which mimics the phosphorylated PTP1 (Fig. 6B). Although we could not directly examine the disruption of the interaction of PTP1 with MPK6 by the phosphorylation of PTP1 under submergence, the reduction of MPK6 phosphorylation in two PTP1 S7AS8A lines (Fig. 6C, D) and the maintenance of AMP in mpk6 and two PTP1 S7AS8A lines (Fig. 6E) supported that PTP1 phosphorylation by SnRK1.1 was required for MPK6 signalling in energy conservation under submergence. In addition, energy conservation (Fig. 2E), MPK6 phosphorylation, and signalling were also interrupted in the SnRK1.1 mutants (Fig. 8). Accordingly, we developed a possible signalling cascade that SnRK1 enhances the MPK6 signal through disrupting the interaction of PTP1 and MPK6 under submergence (Fig. 9).
Fig. 9.
Summary of SnRK1.1-mediated pathways under submergence. Solid lines indicate validated events and bold lines represent the new signalling pathways mediated by SnRK1 under submergence. Dotted lines indicate the predicted events.
SnRK1 regulates protein synthesis under submergence
Protein synthesis is an extremely energy-consuming process. In Arabidopsis cells under oxygen deprivation, in order to conserve energy, specific mRNAs are selected for translation into proteins (Branco-Price et al., 2008) or for storage in stress granules (Sorenson and Bailey-Serres, 2014). However, little is known about how the mRNAs are selected for translation in Arabidopsis. In animals, protein synthesis is regulated by the phosphorylation of specific translation initiation factors and ribosomal subunits. AMPK, the orthologue of SnRK1 in animals could phosphorylate mTOR which, in turn, phosphorylated the eIF4 complex to repress translation under hypoxia (Shimobayashi and Hall, 2014). In Arabidopsis, the TOR complex and S6K1 promote translation initiation (Moreau et al., 2010; Schepetilnikov et al., 2013). Notably, through identification of the targets of SnRK1.1, our data also revealed that SnRK1 modulates the phosphorylation of a specific eIF4 subunit under submergence (Fig. 4D; Supplementary Tables S1, S5). It is known that eIF4A is phosphorylated within 20min of anoxia in maize and that eIF4A is an ATP dependent RNA helicase and regulates the eIF4G–eIF3 complex turnover in yeast (Webster et al., 1991; Munoz and Castellano, 2012). In Arabidopsis under submergence, the cellular ATP content decreased dramatically (Fig. 2E) suggesting that the eIF4Fcomplex is involved in translation regulation under submergence. However, plants have two translation initiation complexes, eIF4F and eIFiso4F, to modulate differential translations of plant mRNAs (Mayberry et al., 2009; Patrick and Browning, 2012). It will be interesting to characterize the roles of these two eIF4F complexes and whether SnRK1.1 is involved in regulating translation initiation through phosphorylating these translation initiation factors under submergence.
On the other hand, two known RNA binding proteins, PENTA and GRP8, were found to be regulated by SnRK1. PENTA is known to bind to the 5′ UTR region of protochlorophyllide reductase (PORA) mRNA to regulate mRNA stability (Paik et al., 2012), and GRP8 is involved in the RNA spliceosome to alter the RNA splicing (Schoning et al., 2008). Among these candidates, eIFiso4G1 and PENTA were identified as the direct targets of SnRK1.1 (Fig. 4B, D). These findings suggest that SnRK1 may be involved in protein synthesis regulation under submergence.
SnRK1 synergistically regulates ABA signalling under submergence
SnRK1.1 is known to be a central regulator of the ABA and sugar signalling pathways. ABA is involved in systemic responses under flooding (Hsu et al., 2011), but is degraded under submergence in Arabidopsis (Benschop et al., 2007; Hsu et al., 2013). To examine whether SnRK1.1 is involved in ABA signalling under submergence, we compared the phosphoproteomic profiles of submergence with another phosphoproteomics analysis of ABA treatment (Fig. 3D; Kline et al., 2010). The phosphorylation of NHX1 and two members of the plasma membrane intrinsic protein (PIP) family of aquaporins were increased in Col-0, but not in SnRK1.1 K48M under submergence (Fig. 3D; Supplementary Table S5). PIP2 has been proposed to be an osmosensor (Maurel et al., 2008) and a cytosolic pH sensor in the roots (Tournaire-Roux et al., 2003). The phosphorylated PIP2S283 is transported to intracellular spherical bodies (Li et al., 2011) and controlled by ABA and salt stress (Kline et al., 2010). Here, our data also suggested that SnRK1.1 might trigger the phosphorylation of PIP2S283 to re-localize PIP2 in the intracellular spherical bodies in order to reduce water flow in cells under submergence.
In addition, AKS1 and AKS2, which are involved in ABA-dependent stomata closure (Takahashi et al., 2013), were dephosphorylated in Col-0, but remained phosphorylated in SnRK1 K48M under submergence (Supplementary Table S5), indicating that SnRK1 might be involved in regulating stomatal closure under submergence. Stomatal closure has been proposed to be a part of the hypoxia-triggered immunity response (Hsu et al., 2013) and to be involved in O2 influx into the shoots under submergence. Under submergence, there is a balance between O2 influx and consumption in the shoots. Plants can maintain the O2 level in the shoots by taking up O2 from stomata during dark periods or photosynthesis; it is then diffused to the roots through gas space diffusion (Lee et al., 2011). These findings, together with our results, raise the possibility that SnRK1 and ABA synergistically regulate stomatal opening to diffuse O2 into the shoots in the early stage of submergence.
In summary, through our phosphoproteomic analysis, we discovered many SnRK1.1 downstream targets that participate in various biological processes (Fig. 9). Among these, proteins involved in the hypoxic response and the sugar metabolism enzymes, TPS and F2KP, have been well characterized in the SnRK1-mediated energy starvation response. The majority of these proteins still need to be examined further to dissect the significance of their phosphorylation under submergence. We started to investigate the potential significance of phosphorylation of the ROS signal regulator, PTP1, which is a negative regulator of MPK6. Examination of downstream targets of PTP1 indicated that SnRK1 might be involved in PTP1-MPK6 signalling and the downstream transcriptional regulation. Considering that the energy starvation response and ROS are critical signals of oxygen deprivation, our results show that SnRK1-mediated phosphorylation is important in the early stage of submergence.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The heterocomplex profile of SnRK1 under submergence.
Figure S2. The profile of SnRK1 transgene lines.
Figure S3. Flowchart of iTRAQ sample preparation and prediction of SnRK1.1 targets.
Figure S4. The recombinant proteins of SnRK1.1 substrates and the phosphorylation pattern of IP-SnRK1.1 under submergence.
Figure S5. Immunoprecipitation of SnRK1 in Col-0 and inactive SnRK1K48M in cMYC-SnRK1.1 K48M.
Figure S6. The profile of two PTP1 transgene lines.
Figure S7. Examination of the interactions between PTP1S7AS8A and PTP1S7DS8D with negative controls.
Table S1. Quantification of phosphoproteins in Col-0 and SnRK1.1 K48M under submergence.
Table S2. Quantification of proteins in Col-0 and SnRK1.1 K48M under submergence.
Table S3. Phosphorylation peptides up-regulated in Col-0 under submergence.
Table S4. Phosphorylation peptides down-regulated in Col-0 under submergence.
Table S5. Differential phosphorylation of selected submergence- responsive genes in SnRK1.1 K48M mutants.
Table S6. Protein abundance of differential phosphorylation responsive genes in SnRK1.1 K48M mutants.
Table S7. Phosphorylation motifs identified in whole seedings of Arabidopsis under submergence.
Table S8. Primer list.
Acknowledgements
We thank Dr Chih-Yu Lin and Ms Chen-Yun Ting for UPLC-MS/MS parameter optimization and the Metabolomics Core Facility for technical support. We also thank Dr Choun-Sea Lin, Ms Fu-Hui Wu, and Ms Shu-Chen Shen for assistance with the protoplast BiFC assays and confocal microscopy. This work was supported by the Ministry of Science and Technology (grant numbers NSC 101-2321-B-001-036 and MOST 104-2321-B-001-053) and Academia Sinica, Taiwan.
References
- Avila J, Gregory OG, Su D, Deeter TA, Chen S, Silva-Sanchez C, Xu S, Martin GB, Devarenne TP. 2012. The beta-subunit of the SnRK1 complex is phosphorylated by the plant cell death suppressor Adi3. Plant Physiology 159, 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, Gonzalez Besteiro MA, Carreri A, Hirt H, Buchala A, Metraux JP, Peck SC, Ulm R. 2009. MAP kinase phosphatase 1 and protein tyrosine phosphatase 1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis . The Plant Cell 21, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Millenaar FF, Smeets ME, van Z, anten M, Voesenek LA, Peeters AJ. 2007. Abscisic acid antagonizes ethylene-induced hyponastic growth in Arabidopsis. Plant Physiology 143, 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. 2008. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana . The Plant Journal 56, 743–755. [DOI] [PubMed] [Google Scholar]
- Chang R, Jang CJ, Branco-Price C, Nghiem P, Bailey-Serres J. 2012. Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis . Plant Molecular Biology 78, 109–122. [DOI] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD. 2012. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiology 158, 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleries R, Galvez J, Espino M, Ribes J, Nunes V, de Heredia ML. 2012. BootstRatio: a web-based statistical analysis of fold-change in qPCR and RT-qPCR data using resampling methods. Computers in Biology and Medicine 42, 438–445. [DOI] [PubMed] [Google Scholar]
- Confraria A, Martinho C, Elias A, Rubio SI, Baena-Gonzalez E. 2013. miRNAs mediate SnRK1-dependent energy signalling in Arabidopsis . Frontiers in Plant Science 4, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, Elias CA, Baena-Gonzales E. 2014. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Frontiers in Plant Science 5, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Mentzen WI, de la Fuente A, Hirt H. 2008. Towards functional phosphoproteomics by mapping differential phosphorylation events in signalling networks. Proteomics 8, 4453–4465. [DOI] [PubMed] [Google Scholar]
- Delatte TL, Sedijani P, Kondou Y, Matsui M, de Jong GJ, Somsen GW, Wiese-Klinkenberg A, Primavesi LF, Paul MJ, Schluepmann H. 2011. Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signalling pathway. Plant Physiology 157, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Luan S. 2003. Redox Control of Protein Tyrosine Phosphatases and Mitogen-Activated Protein Kinases in Plants. Plant Physiology 132, 1149–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hardie DG. 1998. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Molecular Biology 37, 765–748. [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey SJ. 2009. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochemical Journal 419, 247–259. [DOI] [PubMed] [Google Scholar]
- Hardie DG. 2011. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes & Development 25, 1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harthill JE, Meek SEM, Morrice N, Peggie MW, Borch J, Wong BHC, MacKintosh C. 2006. Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. The Plant Journal 47, 211–223. [DOI] [PubMed] [Google Scholar]
- Hsu FC, Chou MY, Chou SJ, Li YR, Peng HP, Shih MC. 2013. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis . The Plant Cell 25, 2699–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FC, Chou MY, Peng HP, Chou SJ, Shih MC. 2011. Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis . PLoS One 6, e28888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame H, ardie D, Thomas M. 2009. SnRK1 (snf1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana . The Plant Journal 59, 316–328. [DOI] [PubMed] [Google Scholar]
- Kline KG, Barrett-Wilt GA, Sussman MR. 2010. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proceedings of the National Academy of Sciences, USA 107, 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Schmidt W. 2012. Quantitative phosphoproteome profiling of iron-deficient Arabidopsis roots. Plant Physiology 159, 403–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LA, Bailey-Serres J. 2011. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytologist 190, 457–471. [DOI] [PubMed] [Google Scholar]
- Li X, Wang X, Yang Y, Li R, He Q, Fang X, Luu DT, Maurel C, Lin J. 2011. Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. The Plant Cell 23, 3780–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM. 2007. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. The Plant Cell 19, 2484–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Verdoucq L, Luu DT, Santoni V. 2008. Plant aquaporins: membrane channels with multiple integrated functions. Annual Review of Plant Biology 59, 595–624. [DOI] [PubMed] [Google Scholar]
- Mayberry LK, Allen ML, Dennis MD, Browning KS. 2009. Evidence for variation in the optimal translation initiation complex: plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs. Plant Physiology 150, 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens E, Larondelle Y, Hers HG. 1990. Induction of pyrophosphate: fructose 6-phosphate 1-phosphotransferase by anoxia in rice seedlings. Plant Physiology 93, 584–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y, Dolferus R, Ismond KP, Good AG. 2007. Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana . The Plant Journal 49, 1108–1121 [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Good AG. 2008. Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana . Plant Cell Physiology 49, 92–102. [DOI] [PubMed] [Google Scholar]
- Moreau M, Sormani R, Menand B, Veit B, Robaglia C, Meyer C. 2010. The TOR complex and signaling pathway in plants. In: Hall M, Tamanoi F, eds. The enzymes , Vol. 27 Academic Press/Elsevier: Oxford, UK, 285–302. [Google Scholar]
- Munoz A, Castellano MM. 2012. Regulation of translation initiation under abiotic stress conditions in plants: Is it a conserved or not so conserved process among Eukaryotes? Comparative and Functional Genomics 2012, 406357–406365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. 2009. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis . Proceedings of the National Academy of Sciences, USA 106, 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TH, Rung JH, Villadsen D. 2004. Fructose-2,6-bisphosphate: a traffic signal in plant metabolism. Trends in Plant Science 9, 556–563. [DOI] [PubMed] [Google Scholar]
- Nunes C, Primavesi LF, Patel MK, Martinez-Barajas E, Powers SJ, Sagar R, Fevereiro PS, Davis BG, Paul MJ. 2013. Inhibition of SnRK1 by metabolites: tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiology and Biochemistry 63, 89–98. [DOI] [PubMed] [Google Scholar]
- Paik I, Yang S, Choi G. 2012. Phytochrome regulates translation of mRNA in the cytosol. Proceedings of the National Academy of Sciences, USA 109, 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick RM, Browning KS. 2012. The eIF4F and eIFiso4F Complexes of Plants: An Evolutionary Perspective. Comparative and Functional Genomics 2012, 287814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. 2008. Trehalose metabolism and signaling. Annual Review of Plant Biology 59, 417–441. [DOI] [PubMed] [Google Scholar]
- Polge C, Thomas M. 2007. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends in Plant Science 12, 20–28. [DOI] [PubMed] [Google Scholar]
- Pucciariello C, Banti V, Perata P. 2012. a. ROS signaling as common element in low oxygen and heat stresses. Plant Physiology and Biochemistry 59, 3–10. [DOI] [PubMed] [Google Scholar]
- Pucciariello C, Parlanti S, Banti V, Novi G, Perata P. 2012. b. Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiology 159, 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, De S, met I, Vandesteene L, Naudts M, Leyman B, Van D, ijck P, Rolland F, Beeckman T, Thevelein JM. 2009. Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana . Plant, Cell & Environment 32, 1015–1032. [DOI] [PubMed] [Google Scholar]
- Ramon M, Rolland F, Sheen J. 2008. Sugar sensing and signaling. In: The Arabidopsis book. The American Society of Plant Biologists, 6, e0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F. 2013. The hybrid four-CBS-domain KINbetagamma subunit functions as the canonical gamma subunit of the plant energy sensor SnRK1. The Plant Journal 75, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A, Adamo M, Crozet P, et al. 2013. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis . The Plant Cell 25, 3871–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG. 1999. Regulation of spinach snf1-related (SNRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. The Plant Journal 19, 433–439. [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martinez E, Geldreich A, Keller M, Ryabova LA. 2013. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. The EMBO Journal 32, 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoning JC, Streitner C, Meyer IM, Gao Y, Staiger D. 2008. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Research 36, 6977–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP. 2005. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nature Biotechnology 23, 1391–1398. [DOI] [PubMed] [Google Scholar]
- Shen W, Reyes MI, Hanley-Bowdoin L. 2009. Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation Loop. Plant Physiology 150, 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN. 2014. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nature Reviews Molecular Cell Biology 15, 155–162. [DOI] [PubMed] [Google Scholar]
- Sorenson R, Bailey-Serres J. 2014. Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis . Proceedings of the National Academy of Sciences, USA 111, 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taj G, Agarwal P, Grant M, Kumar A. 2010. MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signaling & Behavior 5, 1370–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Ebisu Y, Kinoshita T, Doi M, Okuma E, Murata Y, Shimazaki K. 2013. bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Science Signaling 6, ra48. [DOI] [PubMed] [Google Scholar]
- Tournaire-Roux C, Sutka M, Javot H, Gout E, Gerbeau P, Luu DT, Bligny R, Maurel C. 2003. Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425, 393–397. [DOI] [PubMed] [Google Scholar]
- Tsai AYL, Gazzarrini S. 2012. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. The Plant Journal 69, 809–821. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Deutsch EW, Wang R, et al. 2014. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature Biotechnology 32, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C, Gaut RL, Browningfl KS, RavellI JM, Roberts JKM. 1991. Hypoxia enhances phosphorylation of Eukaryotic Initiation Factor 4A in maize root tips. Journal of Biological Chemistry 266, 23341–23346. [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. 2009. Tape–Arabidopsis sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HY, Liu KH, Wang YC, Wu JF, Chiu WL, Chen CY, Wu SH, Sheen J, Lai EM. 2014. AGROBEST: an efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 10, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY. 2014. Hydrogen peroxide controls transcriptional responses of ERF 73 /HRE1 and ADH1 via modulation of ethylene signaling during hypoxic stress. Planta 239, 877–885. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. 2009. Inhibition of Snf1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiology 149, 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.