Highlight
Manipulation of phosphoenolpyruvate carboxykinase activity in rice leaves suggests that metabolism and transport participate in recycling of xylem amino acids and amides and that excess N modulates xylem hydraulics.
Key words: Amides, amino acids, asparagine, hydathode, phosphoenolpyruvate carboxykinase, rice (Oryza sativa), xylem.
Abstract
Measurements of amino acids in the guttation fluid and in the xylem exudates of cut leaves from intact plants provide evidence of the remarkable efficiency with which these nitrogenous compounds are reabsorbed from the xylem sap. This could be achieved by mechanisms involving intercellular transport and/or metabolism. Developmental changes in transcripts and protein showed that transcripts for phosphoenolpyruvate carboxykinase (PEPCK) increased from the base to the leaf tip, and were markedly increased by supplying asparagine. Supplying amino acids also increased the amounts of protein of PEPCK and, to a lesser extent, of pyruvate, Pi dikinase. PEPCK is present in the hydathodes, stomata and vascular parenchyma of rice leaves. Evidence for the role of PEPCK was obtained by using 3-mercaptopicolinic acid (MPA), a specific inhibitor of PEPCK, and by using an activation-tagged rice line that had an increase in PEPCK activity, to show that activation of PEPCK resulted in a decrease in N in the guttation fluid and that treatment by MPA resulted in an increase in amino acids in the guttation fluid and xylem sap towards the leaf tip. Furthermore, increasing PEPCK activity decreased the amount of guttation fluid, whereas decreasing PEPCK activity increased the amount of xylem sap or guttation fluid towards the leaf tip. The findings suggest the following hypotheses: (i) both metabolism and transport are involved in xylem recycling and (ii) excess N is the signal involved in modulating xylem hydraulics, perhaps via nutrient regulation of water-transporting aquaporins. Water relations and vascular metabolism and transport are thus intimately linked.
Introduction
The xylem not only supplies the aerial parts of the plant with water, but also transports nutrients from the root to the shoot. These include inorganic and organic forms of nitrogen, amino acids and amides. Therefore, there is a major flux of these solutes from the roots to the leaves via the transpiration stream. Since the xylem supplies both sources and sinks, it is unlikely that it always supplies the nitrogenous compounds that are required. For example, in the developing leaves of grasses, the developing sink tissues are at the base of the leaf and mature source tissues at the leaf tip. In the sink tissues of the leaf, it is probable that the majority of imported nitrogen is used to make proteins for photosynthesis and other processes, or for storage, whereas in the developed source tissues the potential input of nitrogen and other solutes via the transpiration stream is probably in excess of what is needed for biosynthesis. In these source tissues amino acids are recycled from the xylem into the leaf phloem for redistribution to developing sinks, such as flowers, fruits and seed (Pate and Gunning, 1972; Lalonde et al., 2003; Tegeder and Rentsch, 2010; Tegeder, 2012). In rice, even a young leaf plays a role as a supplier of remobilized nitrogen (Mae and Ohira, 1981). Therefore, there is a need for recycling of nitrogen and other solutes between the xylem and phloem that involves amino acid uptake from the xylem into the xylem parenchyma and transfer into the phloem (Tegeder and Rentsch, 2010; Okumoto and Pilot, 2011; Nagai et al., 2013). In addition, within leaves there is another pathway for the retrieval of solutes from the xylem sap via the hydathodes. The hydathodes allow the excretion of xylem sap (guttation) from the leaf margins, where they are positioned close to the ends of the conducting vessels. Guttation is driven by root pressure (Dieffenbach et al., 1980; Fisher et al., 1997). Possible functions of guttation might be the refilling of the xylem vessels after embolism (Ewers et al., 1997) and/or the exudation of toxic ions that are prevented from entering the mesophyll by, for example, the bundle sheath (Shatil-Cohen and Moshelion, 2012). In hydathodes, wall ingrowths typical of transfer cells are well developed in cells adjacent to the xylem (Maeda and Maedia, 1988), and may be regarded as a form of xylem parenchyma (Pate and Gunning, 1972). The hydathode functions as a selective reabsorption system with guttation fluid from the hydathode containing less K+ and NO3 −, and no Pi, compared to xylem exudates in barley (Nagai et al., 2013). Genes associated with the transport of a range of substances, including nitrate (Nazoa et al., 2003), sucrose (Schulze et al., 2000) and hexose (Schofield et al., 2009), as well as a glutamine dumper protein (Pilot et al., 2004; Pratelli et al., 2010), are expressed in hydathodes and the composition of guttation fluid is complex (Singh and Singh, 2013).
The question then arises as to how much of the excess solutes that arrive in the xylem are simply transported to the phloem via the intervening cells (xylem and phloem parenchyma) and how much they are metabolized (including by the hydathodes). Tegeder and Rentsch (2010) have provided an overview of transport in xylem–phloem transfer, with Hsu and Tsay (2013) studying evidence for the roles of the amino acid transporters AtAAP2 and AtAAP6, and nitrate transporters. Enzymes of nitrogen and carbon metabolism are located in the vasculature and in the hydathodes. Glutamine synthetase (GS1) protein has been detected in companion cells and vascular parenchyma cells of senescing leaf blades of rice, where it may generate Gln for transport of nitrogen to sink tissues (Kamachi et al., 1992; Sakurai et al., 1996; Yamaya and Kusano, 2014). In barley leaves, GS has similar locations, being prominent in the bundle sheath, mestome sheath and xylem parenchyma (Leegood, 2008). NADH-glutamate synthase (NADH-GOGAT) protein was found to accumulate in vascular parenchyma cells and the mestome sheath cells of developing young rice leaves (Hayakawa et al., 1994; Yamaya and Kusano, 2014). In the phloem parenchyma, an enzyme of carbohydrate metabolism, sucrose synthase, is also involved in the pathway of solutes to the phloem (Nolte and Koch, 1993). Cells surrounding the vascular system are enriched in enzymes that are not only key to photosynthesis in C4 plants, but also function in organic and amino acid metabolism in C3 plants (Hibberd and Quick 2002; Brown et al., 2010). In Arabidopsis all four NADP-malic enzyme (NADP-ME) genes are expressed in or around the leaf vasculature and in the hydathodes (Gerrard Wheeler et al., 2005). In barley leaves, NADP-ME protein is predominantly located within the xylem parenchyma (Leegood 2008). Both cytosolic and chloroplastic forms of pyruvate, orthophosphate dikinase (PPDK) accumulated preferentially in veins (Taylor et al., 2010). Phosphoenolpyruvate carboxykinase (PEPCK) protein is located within the the vascular tissues in a range of plants, such as grape, maize roots and leaves (Walker et al., 1999), in cucumber phloem companion cells and vascular parenchyma (Chen et al., 2004), in the vasculature of Arabidopsis (Chen et al., 2004; Malone et al., 2007; Brown et al., 2010; Penfield et al., 2012) and in Arabidopsis hydathodes (Penfield et al., 2012). In addition, there is evidence that PEPCK participates in the metabolism of Asn in tissues involved in transport in pea, presumably by metabolizing the oxaloacetate that is generated from Asn via aspartate (Delgado-Alvarado et al., 2007) and that cytosolic PPDK functions in nitrogen remobilization in senescing leaves (Taylor et al., 2010) and in gluconeogenesis during seedling establishment (Eastmond et al., 2015).
The aim of this study was to determine the role of metabolism, specifically of the ‘C4’ enzymes in rice leaves, in determining the amount and composition of guttation fluid and xylem sap in rice. This was done by determining how the amounts of transcripts and protein for these enzymes as well as the amount and composition of guttation fluid and xylem sap were influenced by N and by leaf development, and specifically what contribution is played by PEPCK to N recycling from the xylem in rice leaves.
Materials and methods
Plant material
Oryza sativa L. Indica cultivar IR72 seeds were sown on moist filter paper and incubated in sterile petri-dishes at 29 °C for 3 d then transferred to a growth chamber at a PPFD of 450 µmol m−2 s−1 with a 12h photoperiod (27 °C day, 24 oC night, 60% humidity). For the first amino acid measurements of xylem sap and guttation fluid, seedlings were sown in trays containing Levington M3 compost (Scotts, Ipswich, UK) for a further 4 d. On day 7, guttation fluid and xylem sap was collected from the cut base of the primary leaf using a clean, sterile tip attached to a pipette, flash frozen in liquid nitrogen and stored at −80 °C. The entire experiment was repeated three times with new seedlings. All subsequent guttation fluid and xylem sap samples were collected by this method. Guttation fluid at the tip was collected immediately after the 12h dark period and for all subsequent experiments. The first xylem sap sample was collected 1h into the light period for all experiments. All samples subsequently detailed were subject to flash freezing in liquid nitrogen and subsequent storage at −80 °C. For all the subsequent feeding experiments, seedlings were sown on moist filter paper and incubated in sterile petri-dishes at 29 °C for 3 d. Each seedling was transferred to an individual compartment of a black, plastic seed tray filled with washed perlite and placed in a propagator tray partially filled with deionized water so that the roots of the seedlings were kept moist for a further 4 d. For subsequent feeding experiments water was removed from the trays and immediately replaced with the appropriate metabolite solution. There were no other nutrients provided for the seedlings other than those detailed in any of the subsequent feeding experiments.
For the quantitative RT-PCR experiment on day 6, one tray of 24 seedlings was incubated with either water, 10mM L-Asn, or 10mM L-Gln for 24h with propagator lids in place. All solutions in this and subsequent experiments were adjusted to pH 6.0–7.0. On day 7, primary leaves were harvested and aligned with their bases on a pre-cooled glass plate. A sterile surgical blade was used to cut leaves into successive 0.5cm sections which were pooled into sterile RNase-free Eppendorf tubes and frozen in liquid nitrogen. Leaf section samples were subsequently analysed by quantitative RT-PCR. The entire experiment was repeated three times with new seedlings. All leaf samples for RT-PCR and subsequent experiments were taken 2h into the light period.
For the Western immunoblotting experiment on day 6, one tray of 24 seedlings was each incubated with water, 10mM Asn, 10mM Gln, 10mM L-Ala and 10mM L-malic acid for 24h with propagator lids in place. On day 7, primary leaves were harvested and aligned with their bases on a pre-cooled glass plate and harvested as detailed for the quantitative RT-PCR experiment. In addition, the FW of each sample was recorded. Leaf section samples were subsequently analysed by Western-immunoblotting.
For amino acid and xylem exudate volume determinations plants were fed as detailed above for Western-immunoblotting. For the 3-mercaptopicolinic acid (MPA) feeding experiments seedlings were grown as described for the control (water) and 10mM Asn feeding. Trays of 24 seedlings were fed for 24h with either water (control), or water with 350 µM MPA, 10mM Asn, or 10mM Asn with 350 µM MPA. On day 7 guttation fluid was collected from each tray of 24 seedlings, pooled and flash frozen in liquid nitrogen. A clean razor blade and ruler was used to remove 1.5cm from the tip of each in situ seedling and the propagator lid replaced for 1h to prevent drying out. After 1h exuded xylem sap was collected and pooled for each treatment tray and frozen. A further 1cm was cut cleanly from each primary leaf and exuded xylem sap collected again after 1h. Finally, the remaining leaf was cut off at the base for all seedlings and xylem sap collected after 1h as previously described. The entire experiment was repeated three times with new seedlings.
Protein concentrations in xylem exudate were determined by the Bradford method (Bio-Rad, Hemel Hempstead, UK) using samples from control (water fed) plants as described above for the control/MPA feeding experiments. For the investigation into the effect of MPA feeding on amount of PEPCK protein, intact leaves were taken from control (water-fed) and MPA-fed plants as described above and subject to SDS-PAGE and immunoblotting as detailed below.
For the immunohistochemistry investigations seedlings were grown as previously described for compost. At 28 d, 2mm slices of the first 4mm of young leaf tips were harvested into fixative using at least ten leaves.
Amino acid analysis
Amino acid analysis was performed using high performance liquid chromatography (HPLC). Guttation fluid samples were filtered directly through 0.20 µm Millipore filters whilst xylem sap samples were diluted with HPLC grade water prior to filtration. All samples (20 µl) were mixed with 200 µl of borate buffer (pH 10) and 30 µl of ortho-phthalaldehyde (OPA) for exactly 45s. A sample of 25 µl was then loaded after a further 60s onto a Luna C8 (250×4.6mm) column (Phenomenex, Macclesfield, UK) equilibrated with 25% (v/v) methanol, 75% (v/v) buffer [200mM Na acetate (pH 5.9), 1.5% (v/v) tetrahydrofuran]. Amino acids were eluted with a gradient of 25% (v/v) methanol, 75% (v/v) acetate buffer, to 90% (v/v) methanol, 10% (v/v) acetate buffer over 60min at a flow rate of 1.4ml min−1. The separated amino acids were detected by fluorescence using an excitation wavelength of 340nm and an emission wavelength of 455nm. Standard solutions of the amino acids were run under the same conditions to quantify each amino acid.
RTq-PCR
Total RNA from pooled leaf sections was purified from 7-d-old O. sativa using an RNeasy Plant Mini Kit (Qiagen, Crawley, UK). RNA was treated with DNase (Sigma-Aldrich, Poole, UK), cleaned with 1:1 (v/v) phenol:chloroform, ethanol precipitated and air-dried. RNA quantity was determined spectrophotometrically and the quality of RNA was validated by agarose gel electrophoresis [1.5% (w/v) agarose]. A 2 µg aliquot of each total RNA sample was used in reverse transcription reactions to make cDNA strands using random hexamers as primer and a Bioscript RTq-PCR kit (Bioline, London, UK). cDNA was diluted 10-fold with sterile water.
Primers were designed by QuantPrime programme (http://www.quantprime.de/) (Arvidsson et al., 2008) using an O. sativa Japonica database. Expression of target genes was normalized to U3, the small nucleolar RNA associated with protein 11 (SnU3-RNA). Target genes were PEPCK3, 10, NADP-ME and also genes for the Glutamine Dumper1 (GDU1), Glutamine synthetase (GS) and Asparaginase. The sequences of primers used to detect these are listed in Supplementary Table S1 at JXB online. Primers to all genes except GDU1 spanned an exon-exon junction further decreasing the possibility of amplification of genomic DNA. SnU3-RNA primers were selected from a range of reference gene primers after testing with RNA from all leaf sections. SnU3-RNA had a coefficient of variation of 28% under all conditions. All primer pairs for both reference and target genes were initially evaluated by checking PCR amplifications for every pair using at least five replicates. PCR amplifications from primer pairs that did not generate a single product and melt curves with a single uniform peak were rejected and new primers were selected. During analysis any amplification failing to meet this criterion was not included. A no-template control was analysed with all sample amplifications.
RTq-PCR was done using 1 µl of cDNA sample, 1 µl of primer mix (0.5 µM of each), 3 µl of water and 5 µl of 2× Sensimix (Quantace Sensimix NoRef Kit, Bioline, London, UK) following the manufacturer’s instructions using the RG6 000 (Corbett Research, UK). Parameters used were: 10min activation; followed by denaturation for 10s at 95 °C, annealing for 15s at 61 °C and extension for 25s at 72 °C for 45 cycles. The fluorescence intensity of SYBR green I was read and acquired at 72 °C after completion of the extension step of each cycle. Quantitation of individual transcripts was performed using the ‘Comparative Quantitation’ software supplied by Corbett Research for the Rotorgene. The mean efficiency of a group of cycling curves was calculated at the point that the cycling curves take off and was used to calculate a fold change according to the formula: fold change=efficiencyCt1−Ct2 where Ct1 and Ct2 were the take-off values of the cycling curves being compared. These fold-change values were expressed as the expression level relative to SnU3-RNA set to a value of 1 for each leaf section. SE values were calculated from the mean of two (technical) replicates for three independent experiments (biological replicates).
SDS-PAGE and western immunoblotting
Pooled leaf sections (10–20mg) were homogenized in a mortar containing 15 volumes of ice-cold 200mM Bicine-KOH (pH 9.8), 50mM dithiothreitol (DTT), then clarified by centrifugation at 14 000 ×g for 5min. Supernatants were added to an equal volume of SDS/PAGE solubilization buffer [62.5mM Tris/HCl (pH 6.8), 10% (v/v) glycerol, 5% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, 0.002% (w/v) bromophenol blue], placed at 100 °C for 3min, centrifuged at 14 000 ×g for 3min and supernatants analysed by SDS/PAGE.
SDS/PAGE was carried out using a 4.7% T/2.7% C stacking gel and a 7.5% T/2.7% C resolving gel. After electrophoresis, polypeptides were fixed in gels by immersion in 50% (v/v) methanol and 12% (v/v) acetic acid. Polypeptides were visualized by colloidal Coomassie Blue G-250 (Sigma-Aldrich, Poole, UK). For Western immunoblotting, transfer of polypeptides from an SDS/PAGE gel to Immunoblot PVDF membrane (Bio-Rad, Hemel Hempstead, UK) was done in a Bio-rad Tetra cell blotting apparatus. Immunoreactive polypeptides were visualized using a goat anti-rabbit IgG peroxidase-conjugated secondary antibody (Sigma-Aldrich Co. Ltd., Poole, UK) in conjunction with an enhanced chemiluminescence kit, and Hyperfilm ECL (GE Healthcare, Little Chalfont, UK) was used with an intensifying screen for up to 1min. For the various treatments on different Western immunoblots, adjustment to the same background brightness threshold was carried out using Photoshop.
Immunohistochemistry
Thin slices of leaf tissue (2mm) were cut and immediately immersed in fixative [7.5mg ml−1 sucrose, 30mg ml−1 formaldehyde in 100mM NaH2PO4 (pH 7.2)] and placed under vacuum (25 °C, five times for 30min each) and then left overnight at 4 °C. Samples were fixed, embedded and mounted as described by Walker et al. (1997) using the indicator substrate 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) tablets (Sigma-Aldrich, Poole, UK).
Antibodies
All antibodies were polyclonal and raised in rabbit. The antiserum specific for PEPC was raised against the enzyme purified from Amaranthus edulis L. (Dever et al., 1995). Antiserum specific for PEPCK raised against the enzyme purified from cucumber (Cucumis sativus L.) (Walker et al., 1995) was used in one of the immunolocalization studies. All other antibodies were affinity purified and derived from peptides designed to specific sequences in the corresponding enzymes in rice. See Supplementary Fig. S1 for details.
Measurement of PEPCK activity
Material (50mg) was homogenized in a mortar containing five volumes of ice-cold 200mM Bicine-KOH (pH 9.8), 50mM dithiothreitol (DTT), then clarified by centrifugation at 14 000 ×g for 5min. The carboxylase activity of PEPCK was measured in the supernatant (Walker et al., 2002). One unit of PEPCK activity produces 1 µmol product per min at 25 °C.
Results
Composition of xylem exudate and guttation fluid
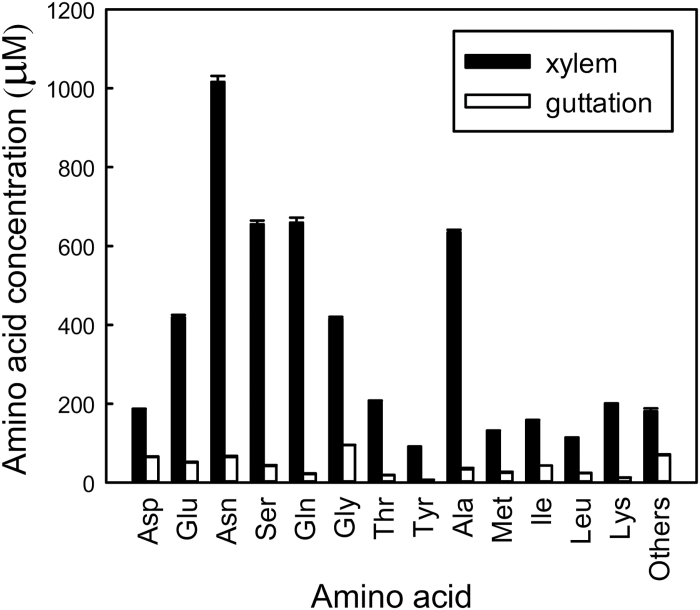
Figure 1 shows the concentrations of amino acid and amides (hereafter collectively referred to as amino acids) of xylem exudates, sampled from the leaf base and guttation fluid in young leaves of intact rice plants. It shows that the composition of the guttation fluid differs markedly from the xylem sap and that there is reabsorption of N across the whole range of amino acids, with particularly marked reductions in the concentrations of Asn, Gln, Ser and Ala.
Fig. 1.
Amino acid concentrations of xylem exudate and guttation sap in 7-d-old first leaves of O. sativa. The xylem exudate and guttation fluid measurements are from the combined volume of 24 plants. Values are the mean ±SE from three independent experiments. All measurements of amino acid contents in the xylem exudate are significantly different at the 5% level when compared to the respective guttation sap.
In addition to guttation fluid it was also possible to collect xylem exudate from cut leaves. There was no guttation or xylem sap exudation from detached leaves placed in water. Guttation and xylem sap exudation was collected from leaves still attached to the seedling.
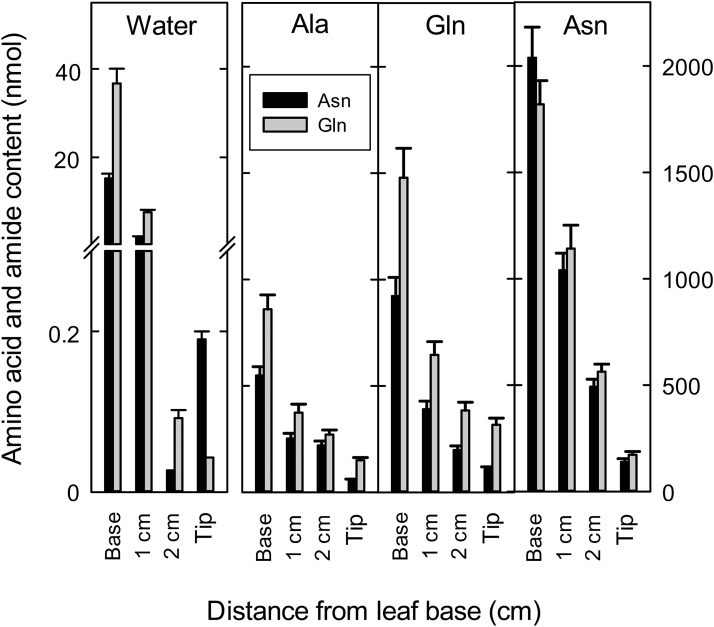
Figure 2 shows the contents of Asn and Gln in the xylem sap exuded from the leaf base, the cut leaf and in the guttation sap in young rice plants supplied with water, Asn, Gln and Ala. As in Fig. 1, there was a large decrease in Asn and Gln between the basal xylem exudate and the guttation fluid exuded from the tip of the leaf. Feeding with Asn induced the highest amounts of Asn and Gln in the xylem sap exuded from the leaf base. Supplying Gln and Ala increased the amounts of both Asn and Gln in the xylem exudate. Supply of Ala, Gln and Asn significantly increased the amounts of these amino acids in the guttation sap compared with supplying water (note the difference in Y-axis scales). However, for all treatments the amide content in the xylem was significantly lower than at the base of the leaf, and lower still in the guttation sap, indicating effective recycling of these substances even with the elevated xylem contents that resulted from supplying these substances to the root system. Note that although Fig. 1 shows concentrations, the remaining data are shown as amounts of amino acids because the volumes changed markedly. Ultimately it is the total amount of amino acids passing through the xylem to the leaf tip that relates to fluxes, whereas concentrations of amino acids in the xylem will tend to be regulated. Volume data are provided so that conversions can be made.
Fig. 2.
Amino acid and amide contents in guttation fluid at the tip (entire amount collected immediately after the 12h dark period) or xylem sap both at and various distances from the base (collected at 1h intervals in the light period) in 7-d-old primary leaves of O. sativa fed water (control), 10mM L-alanine, 10mM L-glutamine, or 10mM L-asparagine. The xylem exudate and guttation fluid measurements are from the combined volume of 24 plants. Values are the mean ±SE from three independent experiments. All xylem exudate and guttation sap amino acid contents are significantly different at the 5% level when compared to the control. Note break in Y-axis. Mean guttation and xylem exudate volumes (μl) were as follows: water, base to tip, 60, 11.5, 0.2, 41; alanine, 101, 58, 53, 135; glutamine, 112, 62, 45, 70; asparagine, 140, 111, 62, 103.
Table 1 shows the volumes of guttation fluid (at the tip) or of xylem sap exuded from cut leaves at varying distances from the base collected over a period of 1h. Of note is that guttation fluid was exuded more slowly than xylem exudate and the volume shown was the entire amount of guttation fluid at the beginning of the photoperiod from intact plants. It should be emphasized that the exudate and guttation fluid measurements are not directly comparable as exudation is an experimentally controlled process occurring in the light whereas guttation relies upon leaf processes occurring in the dark that cannot be experimentally manipulated. We checked that there was no contamination of xylem exudates with phloem sap by assessing protein concentration in the exudates. The mean protein concentration at the base and tip (guttation fluid) was 9.9 and 5.62ng μl−1 respectively, comparable to that in the xylem sap, but 10 000 times lower than in the phloem (Richardson et al., 1982; Atkins et al., 2011). In the control, amounts of xylem exudate decreased from the leaf base to the tip. In order to modify the composition of the xylem sap we supplied metabolites to the roots of intact plants. Supplying Ala, Gln and Asn greatly increased the volumes of xylem exudate and guttation fluid, from a doubling at the base to as much as a several hundred-fold increase at 2cm. By contrast, feeding malate (which was also taken up and led to a 4.5-fold increase in malate in the basal xylem exudate and a 14-fold increase in the guttation fluid) (Supplementary Table S2) had no significant effect on the volume except for a 3-fold increase in xylem exudate at 1cm (Table 1; P=0.038).
Table 1.
Volumes of guttation fluid at the tip (entire amount collected) or xylem exudate both at and various distances from the base (collected after 1h) in 7-d-old first leaves of rice plants fed various compounds at a concentration of 10mM, or 350 µM MPA
Values are the mean ±SE of three independent experiments of 24 plants.
| Distance from base (cm) | Mean volume of xylem exudate/guttation fluid per compound ±SE (µl) | ||||||
|---|---|---|---|---|---|---|---|
| Control | Malate | Ala | Gln | Asn | Control+MPA | Asn+MPA | |
| Base | 60±4.04 | 71±8.90 | 101±8.69 | 112±11.00 | 140±14.29 | 61±4.06 | 130±12.90 |
| 1 | 11.5±0.76 | 33±4.41 | 58±5.29 | 62±6.11 | 111±11.26 | 20±2.52 | 92±8.51 |
| 2 | 0.2±0.06 | 0.4±0.12 | 53±4.58 | 45±5.13 | 62±7.06 | 21±2.60 | 95±10.02 |
| Tip | 41±2.08 | 62±6.69 | 135±11.37 | 70±8.02 | 103±10.49 | 67±4.36 | 96±8.54 |
Changes in gene expression after feeding Asn and Gln
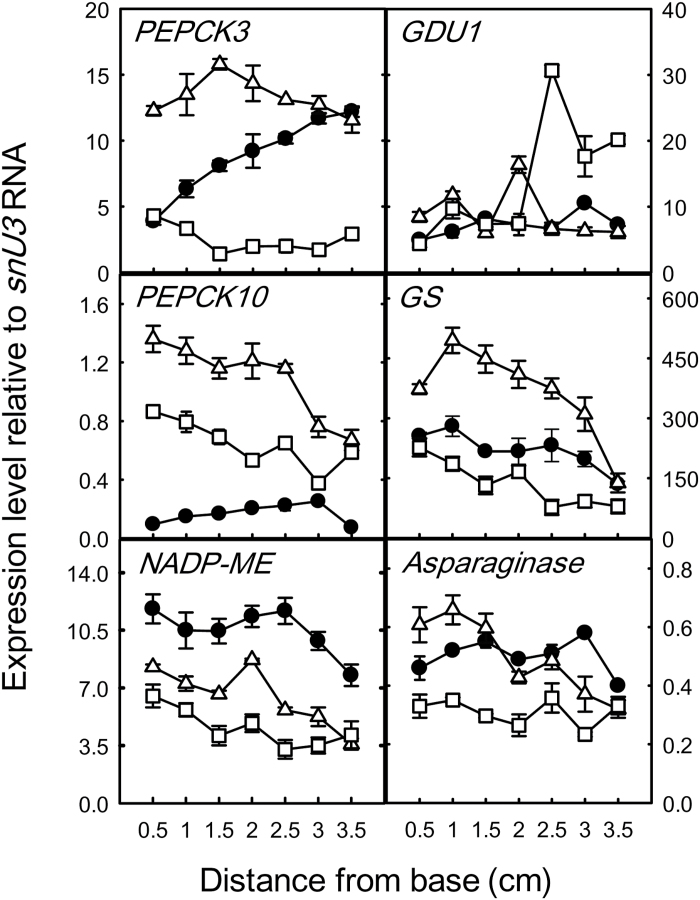
Figure 3 shows the relative expression of selected genes compared to the control, small nuclear RNA U3 (Os01g59500) (snU3; Brown et al., 2003), in leaves of young rice plants supplied with water, Asn or Gln. In leaves of plants supplied with water, expression of PEPCK3 (Os03g15050) increased from base to tip while expression of PEPCK10 (Os10g13700) increased only slightly from the base until just below the tip (the numbers refer to the two PEPCK genes in rice that are found within chromosomes 3 and 10, respectively; Kawahara et al., 2013). Expression of NADP-ME (Os01g52500) and GS (chloroplastic GS2; Os04g56400) tended to decrease from base to tip, while expression of ASPARAGINASE (Os04g46370) (Grant and Bevan, 1994) and glutamine dumper 1 (Os08g34700) (GDU1, known to be expressed in the vascular tissues and in hydathodes of Arabidopsis; Pilot et al., 2004) remained relatively constant. Supplying Asn generally increased expression of PEPCK3, PEPCK10 and GS, although with overall declines of PEPCK10 and GS along the leaf, it induced a decline in ASPARAGINASE along the leaf but had less effect on GDU1, and it decreased the expression of NADP-ME along the leaf. Supplying Gln decreased expression of PEPCK3 (except at the base), GS, NADP-ME and ASPARAGINASE, but overall increased expression of PEPCK10 (although decreasing along the leaf) and increased expression of GDU1 towards the leaf tip.
Fig. 3.
Relative expression of various genes compared to snU3 RNA in 7-d-old control (●), asparagine-fed (∆) and glutamine-fed (□) first leaves of O. sativa. Results are expressed as means ±SE of three biological replicates.
Immunoblots for key enzymes
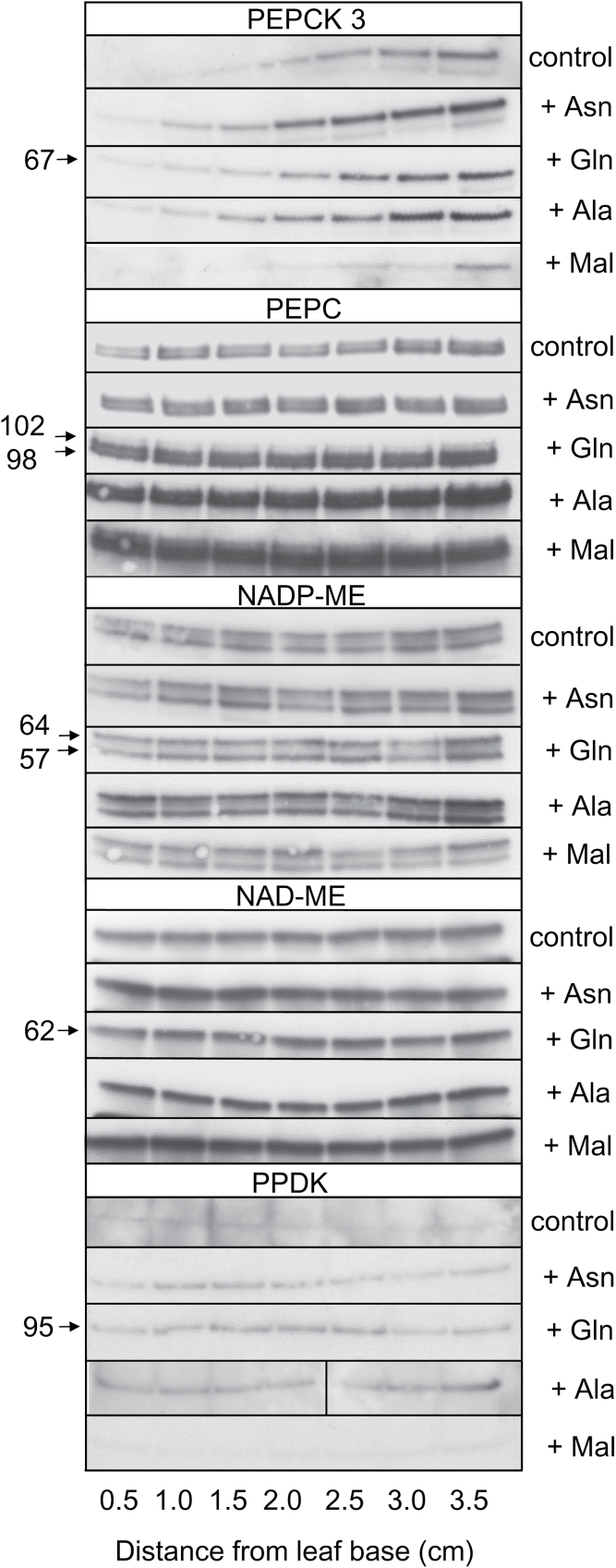
Figure 4 shows that PEPCK3 protein increased from the base to the tip of the leaf. Feeding with Asn, Gln and Ala increased the amount of PEPCK3 protein, particularly in the middle region of the leaf (1.5–2.5cm from the base), while supplying malate decreased PEPCK3 in the middle region of the leaf. The amount of PEPC increased marginally from base to tip, and was increased along the entire length of the leaf by Gln, Ala and malate. The amounts of NAD-malic enzyme (NAD-ME) and two isoforms of NADP-ME were constant from the base to the tip of the leaf, with the amount of NADP-ME being slightly increased by the supply of Ala. The amount of PPDK was constant from the base to tip of the leaf but was slightly increased by supplying Asn and Gln, and notably by Ala at the leaf tip. Note that there was a good correspondence between transcripts (Fig. 3) and protein for control and Asn-fed PEPCK3, but not for Gln-fed PEPCK3 or for NADP-ME.
Fig. 4.
Immunoblots showing the effect of nitrogenous compounds or malate on the abundance of the PEPCK 3, PEPC, NADP-ME, NAD-ME and PPDK proteins in 0.5cm leaf sections taken from base to tip of the first leaf of 7-d-old leaves of O. sativa. Loadings on gels contained the soluble protein content of 0.33mg of FW of tissue. Numbers on the left hand Y-axis refer to molecular masses (kDa). Note that the image is constructed from different immunoblots.
Localization of PEPCK
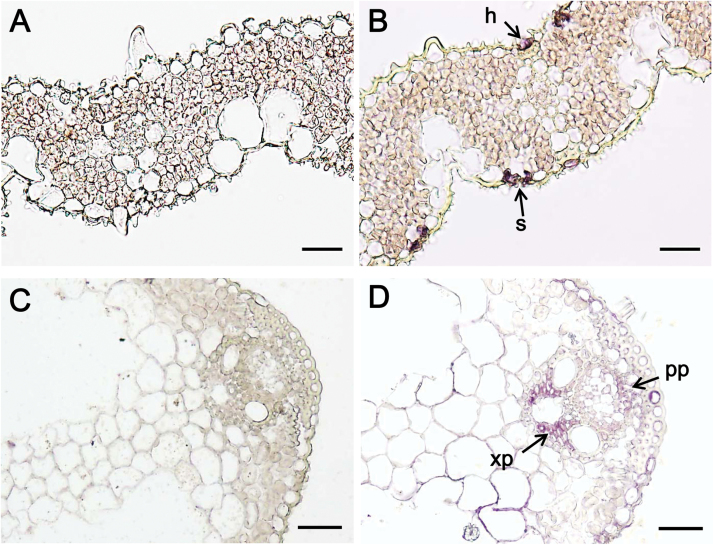
PEPCK was shown by immunolocalization to be present in the stomata, hydathodes and parenchyma cells close to the xylem and phloem (Fig. 5). Pre-immune controls showed no comparable staining in these cell types. A peptide antibody designed specifically to the chromosome 3 isoform of PEPCK in rice was used (Fig. 5B). This clearly showed staining in the hydathode and stomata, however staining in the vascular tissue was not as clear. This was due to the relatively weak expression of PEPCK in the vasculature and the high background coloration in the surrounding tissue. The peptide antibody was very specific because of affinity purification but as a consequence had to be used at a high concentration, leading to a high background coloration. We therefore used a different PEPCK antibody (Fig. 5D). This antibody was made using purified cucumber PEPCK as the antigen. Since it was a protein antibody (not affinity purified) it worked at a much more dilute concentration and unspecific background staining was largely eliminated. Staining of the vasculature was clearly visible, however this antibody was less reactive to PEPCK located in the stomata and hydathode of rice.
Fig. 5.
PEPCK is present in the stomata, s, hydathode, h, and vascular tissue (xylem parenchyma, xp; phloem parenchyma, pp). Transverse sections of rice after immunodiazostaining with pre-immune (A and C) and PEPCK serum (B and D). Serum used in panel B was a peptide antibody to the chromosome 3 isoform in rice. Serum used in panel D was a protein antibody to PEPCK in cucumber that was more sensitive to PEPCK located in the vascular tissue but less sensitive to PEPCK located in the stomata and hydathode of rice. Bars: 20 μm for A, B; 30 μm for C, D.
Influence of decreased PEPCK activity on sap amount and composition
In order to test if PEPCK and its associated metabolism influenced the utilization and recycling of amino acids delivered in the xylem, we carried out two experiments to alter the activity of PEPCK. We supplied the intact plants with a specific inhibitor of PEPCK, 3-mercaptopicolinic acid (Leegood and ap Rees, 1978). In order to supply different compounds and MPA to seedlings it was also necessary to grow them in Perlite, rather than compost. One major effect of this change was that Asn was much less prominent as a solute transported in the xylem. Since PEPCK is implicated in Asn metabolism (Delgado et al., 2007), we therefore did an additional treatment in which MPA was supplied to Asn-fed, as well as control plants. We checked, by immunoblotting, that supplying MPA for 24h did not affect the amount of PEPCK protein in rice leaves (Supplementary Fig. S2). Supplying MPA decreased xylem recycling in the leaf as there was an increase in the amount of amino acids, 2cm from the base and at the tip, either with or without an additional Asn supply. However, MPA made no difference to the total amino acids in the xylem exudate from the base of the leaf, with or without Asn, showing that there was no overall effect of MPA on the supply of amino acids from the roots (Table 2). Feeding MPA increased the volume of the xylem exudate and guttation fluid in the control leaves, except at the base, but had no additional effect when it was supplied with Asn (Table 1). Feeding MPA clearly inhibited Asn metabolism and resulted in an increase in amino acids and amides in the xylem. For full sets of data see Supplementary Tables S3, S4.
Table 2.
Total amounts of amino acids in guttation fluid at the tip (entire amount collected) or xylem sap both at and various distances from the base (collected after 1h) in 7-d-old primary leaves of rice plants fed water (control) and water with 350 μM MPA or 10mM Asn (control) and Asn with 350 μM MPA
Measurements are the mean ±SE of the combined volume from three independent measurements each of 24 plants. Mean guttation fluid and xylem exudate volumes (μl) required for conversion to μM are as follows: −MPA, base to tip, 60, 11.5, 0.2, 41; +MPA, 61, 20, 21, 67; –MPA+Asn, 140, 111, 62, 103; +MPA+Asn, 130, 92, 95, 96.
|
Distance from base
(cm) |
Total amount of amino acids (nmol) | |||
|---|---|---|---|---|
| -MPA | +MPA | -MPA+Asn | +MPA+Asn | |
| Base | 225±18 | 222±19 | 4900±340 | 5343±492 |
| 1 | 20.54±1.48 | 43.58±4.76 | 2813±241 | 2963±250 |
| 2 | 0.207±0.02 | 14.91±1.34 | 1280±86 | 2085±200 |
| Tip | 2.77±0.2 | 10.60±1.14 | 404±38 | 704±41 |
We did not find any activation-tagged lines in which PEPCK activity was reduced. However, we used an activation-tagged line of rice (in a different variety from the rest of the experiments) that had an elevated expression and amount of PEPCK (Hsing et al., 2007). Table 3 shows that, compared with the wild type, leaves of the heterozygous and homozygous mutants had an increase in both PEPCK transcripts and up to a doubling of PEPCK activity. Increases in PEPCK activity in these plants led to a decrease in the total amount of amino acids and amides in the guttation fluid and a decrease in the guttation fluid volume.
Table 3.
Characteristics of Oryza sativa L. Japonica cultivar Taichung wild-type and PEPCK transgenic lines (TRIM line M0035095) (Hsing et al., 2007)
Relative expression of PEPCK compared to glutaredoxin and PEPCK enzyme activity were measured in leaves. Copy number was determined to be one for the inserted gene. Guttation fluid was pooled from leaves of wild-type, heterozygous and homozygous plants, the volume measured and amino acid contents determined for each genotype. Values are the mean of three measurements from six plants ±SE. See Supplementary Tables S5–S7.
| Plant |
Gene expression
mean ±SE (% wild type) |
Enzyme activity mean ±SE
(µmol min −1 g −1 FWt) (% wild type) |
Total amino
acid content (nmol) |
Guttation fluid volume
(µl) |
|---|---|---|---|---|
| Wild type | 1.98±0.05 (100) | 0.062±0.005 (100) | 9.70±0.85 | 33±3 |
| Heterozygote | 3.01±0.22 (152) | 0.091±0.006 (147) | 4.92±0.46 | 27±3 |
| Homozygote | 5.71±0.42 (288) | 0.135±0.019 (218) | 2.51±0.13 | 13±1 |
Discussion
The measurements of amino acids and amides in the guttation fluid and in the xylem exudates of cut leaves, when compared with the xylem exudate from the base of the shoot, provide striking evidence of the efficiency with which these nitrogenous compounds are reabsorbed from the xylem sap during its passage through the leaf. Feeding with Gln increased the contents of both Asn and Gln in the xylem exudate and also significantly increased the contents of these amino acids in the guttation sap perhaps indicating that Gln is not as effectively recycled as Asn.
Of course, amino acid recycling is essential to maintaining the nitrogen economy of the plant, but it then raises the question of whether this is achieved wholly by mechanisms involving intercellular transport or whether metabolism might also play a role. The fact that hydathodes are enriched in many enzymes of primary metabolism, as outlined in the introduction, led us to investigate whether metabolism might also be involved in N recycling. PEPCK is located in secretory tissues such as Arabidopsis hydathodes (Penfield et al., 2012), and Arabidopsis nectaries and the stigma (Malone et al., 2007), as well as in the xylem and phloem parenchyma cells associated with the vasculature, as outlined in the introduction and shown in Fig. 5. A survey of developmental changes in transcripts and protein for some of these key enzymes revealed that only transcripts for PEPCK3 increased from the base to the leaf tip and that they were markedly increased by supplying Asn, while transcripts for GLN DUMPER1 increased at the leaf tip when Gln was supplied. Supplying N also increased the amounts of protein of PEPCK and, to a much lesser extent, of PPDK. The pattern of PEPCK3 transcript and PEPCK protein abundance in rice is similar to that observed for transcript abundance in the developing maize leaf (Li et al., 2010). The observation that PEPCK protein is induced by a range of nitrogenous compounds (Chen et al., 2004), and particularly by Asn in developing seeds (Walker et al., 1999; Delgado-Alvarado et al., 2007), suggests that it is involved in the metabolism of oxaloacetate deriving from Asn (via aspartate) in tissues that import or recycle nitrogen arriving as Asn (Delgado-Alvarado et al., 2007). Thus in the hydathodes and vascular cells PEPCK may play a role in retrieving Asn from the xylem (Fig. 6).
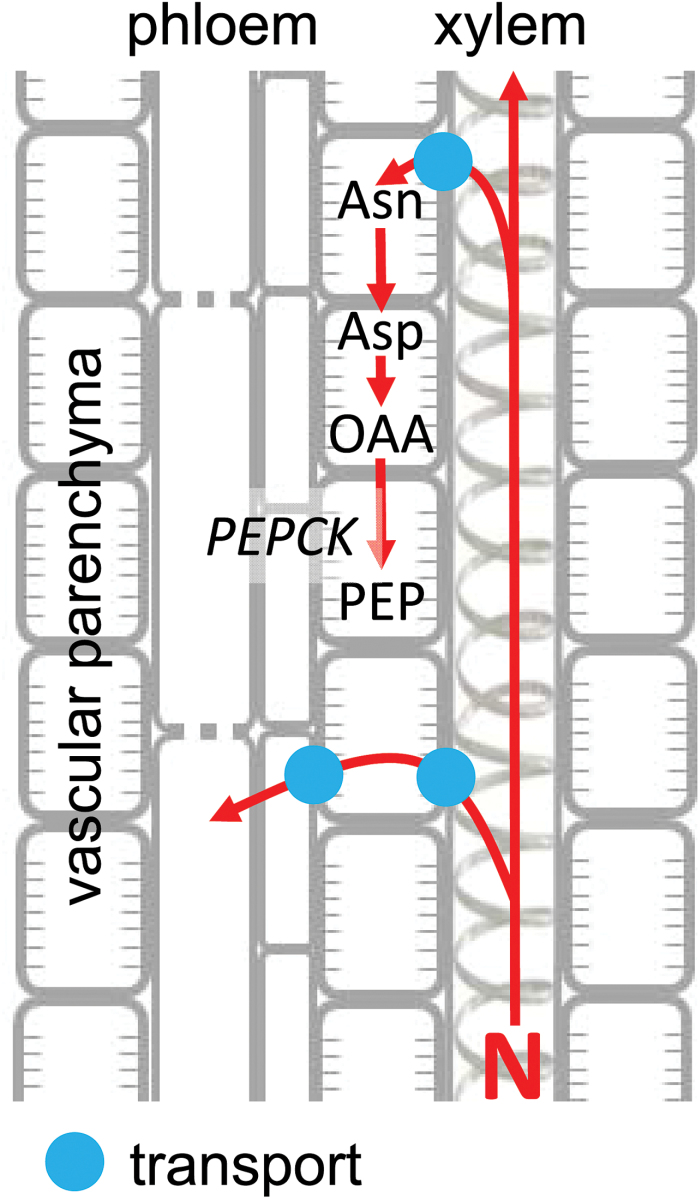
Fig. 6.
Schematic presentation of the roles of transport and the possible metabolic role of PEPCK in nitrogen recycling from the xylem. Transport involves unloading of amino acids and amides (N) from the xylem, passage through the vascular parenchyma between the xylem and phloem, and loading into the phloem. Metabolism involves similar transport processes in conjunction with metabolism in the vascular parenchyma. Specifically, the role of PEPCK would involve the metabolism of OAA (oxaloacetate) deriving from Asp and, ultimately, Asn. N released by Asn and Asp metabolism could then be also transported into the phloem.
Evidence for a role for PEPCK in xylem recycling in rice leaves was obtained by using MPA, a specific inhibitor of PEPCK in animals and plants (DiTullio et al., 1974; Jomain-Baum et al., 1976; Leegood and ap Rees, 1978; Wingler et al., 1999) to show that inhibition of PEPCK resulted in an increase in amino acids in the guttation fluid as well as in the xylem exudate, except at the base. This finding was corroborated by using an activation-tagged rice line that had an increase in PEPCK activity showing that activation of PEPCK resulted in a decrease in amino acids in the guttation fluid. Taken together these results support the hypothesis that PEPCK is involved in N recycling. Furthermore, decreasing PEPCK increased the amount of xylem leaf exudate and guttation fluid whereas increasing PEPCK decreased the amount of guttation fluid. The simplest interpretation is that altering the efficiency of this recycling feeds back on xylem hydraulic conductivity.
There is previous evidence that transfer of N from the xylem to phloem involves metabolism as well as direct transfer of amino acids. Sharkey and Pate (1975) fed 14C-labelled amino compounds singly to fruiting shoots of white lupin through the transpiration stream and measured the distribution of 14C in solutes of the phloem sap. The carbon of Asn, the major nitrogenous solute of phloem, was derived from five amino compounds in the xylem. Although the most important of these transfers was the direct throughput of Asn from xylem to phloem, significant amounts of carbon in phloem Asn were also exchanged from aspartate, glutamate, Gln and γ-aminobutyrate supplied to the xylem. On the other hand, carbon in aspartate and glutamate in the phloem was derived from a wide number of xylem amino acids, with little direct transfer of either amino acid from the xylem to the phloem (Sharkey and Pate, 1975). Differences between the xylem and phloem sap composition of maize leaves might also have a similar explanation (Martin et al., 2006).
Why metabolism is involved in xylem recycling remains an open question. One possibility is that metabolism is acting as a sink to promote N transfer from the xylem, another is that metabolism reconfigures the N profile to suit the immediate needs of the phloem, for example, to supply seeds (Sharkey and Pate, 1975) or developing tissues. If PEPCK were involved, then the pathway is likely to involve the release of ammonia from Asn, which could then be reassimilated via GS and GOGAT, enzymes which are present in the vasculature (Kamachi et al., 1992; Hayakawa et al., 1994; Sakurai et al., 1996).
The other important finding from these studies was that the various experimental treatments modulated the amount of guttation and xylem exudate, suggesting that the amount of amino acids and amides in the xylem exudate modulated the hydraulic conductivity of the xylem and that this might change along the length of the leaf. It should be noted that malate had a much smaller effect, suggesting that N could be a key factor. Water relations and vascular metabolism and transport are thus intimately linked. An excess of amino acids in the xylem may be the signal involved in modulating xylem hydraulic conductivity. There is already evidence that nutritional information can be translated into a hydraulic response, whereby a plant’s transpiration, stomatal conductance and root hydraulic conductivity can be influenced by the supply of certain mineral nutrients (Clarkson et al., 2000). The xylem is a finely regulated water transport system (Nardini et al., 2011). Vascular bundle-sheath cells have been hypothesized to control the solute and hydraulic conductivity of the leaf vascular system (Fricke 2002), with the latter altered via the specific activity of bundle sheath cell aquaporins (Shatil-Cohen et al., 2011). It is also established that water-transporting aquaporins are regulated in response to N (Gaspar et al., 2003; Loqué et al., 2005, Hacke et al., 2010), probably by phosphorylation that is mediated both by N (Engelsberger and Schulze, 2012) and, specifically in the leaf veins, by light (Prado et al., 2013).
The above findings allow us to propose the following hypotheses: (i) both metabolism and transport are involved in xylem recycling and (ii) excess N is a signal involved in modulating xylem hydraulic conductivity, whether in the roots or shoots. Thus feeding N increases hydraulic conductivity, and feeding MPA inhibits Asn metabolism and results in an increase in amino acids and amides in the xylem. This also appears to increase conductivity. These observations are not restricted to rice and we have made similar findings in both barley and maize (in which PEPCK also functions in the bundle sheath in C4 photosynthesis).
Supplementary data
Supplementary data can be found at JXB online.
Fig. S1. Immunoblot showing the difference in specificity between antibodies specific to the chromosome 3 and the chromosome 10 isoform of PEPCK.
Fig. S2. Western immunoblot of PEPCK3 in MPA or water fed leaves.
Table S1. Sequences of primers used for gene expression.
Table S2. Concentration of malate in guttation fluid and xylem exudate in water- or malate-fed plants.
Table S3. Amounts of amino acids in guttation fluid and xylem exudate in plants fed water or water+MPA.
Table S4. Amounts of amino acids in guttation fluid and xylem exudate in plants fed Asn or Asn+MPA.
Table S5. Sequences of primers used for genotyping.
Table S6. Sequences of primers used for copy number determination.
Table S7. Sequences of primers used for PEPCK3 gene expression.
Acknowledgements
This work was supported by the Bill and Melinda Gates Foundation as part of the C4-Rice programme administered by the International Rice Research Institute, and also by the EU Framework 7 Programme, 3to4.
Glossary
Abbreviations:
- GOGAT
glutamate synthase
- GS
glutamine synthetase
- MPA
3-mercaptopicolinic acid
- NADP-ME
NADP-malic enzyme
- PEPCK
phosphoenolpyruvate carboxykinase
- PPDK
pyruvate, orthophosphate dikinase.
References
- Arvidsson S, Kwasniewski M, Riano-Pachou DM, Mueller-Roeber B. 2008. Quantprime- a flexible tool for reliable high throughput primer design for quantitative PCR. BioMedCentral Bioinformatics doi: 10.1186/1471-2105-9-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CA., Smith PMC, Rodriguez-Medina C. 2011. Macromolecules in phloem exudates-a review. Protoplasma 248, 165–172. [DOI] [PubMed] [Google Scholar]
- Bartlett JG, Alves SC, Smedley M, Snape JW, Harwood WA. 2008. High-throughput Agrobacterium-mediated barley transformation. Plant Methods doi: 10.1186/1746-4811-4-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JWS, Echeverria M, Qu L-H. 2003. Plant snoRNAs: functional evolution and new modes of gene expression. Trends in Plant Science 8, 42–49. [DOI] [PubMed] [Google Scholar]
- Brown NJ, Palmer BG, Stanley S, et al. 2010. C4 acid decarboxylases required for C4 photosynthesis are active in the mid-vein of the C3 species Arabidopsis thaliana, and are important in sugar and amino acid metabolism. Plant Journal 61, 122–133. [DOI] [PubMed] [Google Scholar]
- Chen Z-H, Walker RP, Técsi LI, Lea PJ, Leegood RC. 2004. Phosphoenolpyruvate carboxykinase in cucumber plants is increased both by ammonium and by acidification and is present in the phloem. Planta 219, 48–58. [DOI] [PubMed] [Google Scholar]
- Clarkson DT, Carvajal M, Henzler T, Waterhouse RN, Smyth AJ, Cooke DT, Steudle E. 2000. Root hydraulic conductance: diurnal aquaporin expression and the effects of nutrient stress. Journal of Experimental Botany 51, 61–70. [PubMed] [Google Scholar]
- Delgado-Alvarado A, Walker RP, Leegood RC. 2007. Phosphoenolpyruvate carboxykinase in developing pea seeds is associated with tissues involved in solute transport and is nitrogen responsive. Plant, Cell & Environment 30, 225–235. [DOI] [PubMed] [Google Scholar]
- Dever LV, Blackwell RD, Fullwood NJ, Lacuesta M, Leegood RC, Onek LA, Pearson M, Lea PJ. 1995. The isolation and characterisation of mutants of the C4 photosynthetic pathway. Journal of Experimental Botany 46, 1363–1376. [Google Scholar]
- Dieffenbach H, Kramer D, Lüttge U. 1980. Release of guttation fluid from passive hydathodes of intact barley plants. I. Structural and cytological aspects. Annals of Botany 45, 397–401. [Google Scholar]
- DiTullio NW, Berkoff CE, Blank B, Kostos V, Stack EJ, Saunders HL. 1974. 3-mercaptopicolinic acid, an inhibitor of gluconeogenesis. Biochemical Journal 138, 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Astley HM, Parsley K, Aubry S, Williams BP, Menard GN, Craddock CP, Nunes-Nesi A, Fernie AR, Hibberd JM. 2015. Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nature Communications 6, 6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX. 2012. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen starved Arabidopsis seedlings. Plant Journal 6, 978–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers FW, Cochard H, Tyree MT. 1997. A survey of root pressures in vines of a tropical lowland forest. Oecologia 11, 191–196. [DOI] [PubMed] [Google Scholar]
- Fisher B, Guillermo Angeles I, Ewers FW, Lopez-Portillo J. 1997. Survey of root pressure in tropical vines and woody species. International Journal of Plant Science 158, 44–50. [Google Scholar]
- Fricke W. 2002. Biophysical limitation of cell elongation in cereal leaves. Annals of Botany 90, 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar M, Bousser A, Sissoeff I, Roche O, Hoarau J, Mahe A. 2003. Cloning and characterization of ZmPIP1-5b, an aquaporin transporting water and urea. Plant Science 16, 21–31. [Google Scholar]
- Gerrard Wheeler MC, Tronconi MA, Drincovich MF, Andreo CS, Flügge U-I, Maurino VG. 2005. A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiology 13, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Bevan MW. 1994. Asparaginase gene expression is regulated in a complex spatial and temporal pattern in nitrogen-sink tissues. Plant Journal 5, 695–704. [Google Scholar]
- Hacke UG, Plavcová L, Almeida-Rodriguez A, King-Jones S, Zhou W, Cooke JEK. 2010. Influence of nitrogen fertilization on xylem traits and aquaporin expression in stems of hybrid poplar. Tree Physiology 3, 1016–1025. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Nakamura T, Hattori F, Mae T, Ojima K, Yamaya T. 1994. Cellular localization of NADH-dependent glutamate-synthase protein in vascular bundles of unexpanded leaf blades and young grains of rice plants. Planta 193, 455–460. [Google Scholar]
- Hibberd JM, Quick WP. 2002. Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415, 451–454. [DOI] [PubMed] [Google Scholar]
- Hsing Y-I, Chern C-C, Fan M-J, et al. 2007. A rice gene activation/knock-out mutant resource for high throughput functional genomics. Plant Molecular Biology 63, 351–364. [DOI] [PubMed] [Google Scholar]
- Hsu P-K, Tsay Y-F. 2013. Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiology 163, 844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomain-Baum M, Schramm VL, Hanson RW. 1976. Mechanism of 3-mercaptopicolinic acid inhibition of hepatic phosphoenolpyruvate carboxykinase (GTP). Journal of Biological Chemistry 251, 37–44. [PubMed] [Google Scholar]
- Kamachi K, Yamaya T, Hayakawa T, Mae T, Ojima K. 1992. Vascular bundle-specific localization of cytosolic glutamine synthetase in rice leaves. Plant Physiology 99, 1481–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, De-La Bastide M, Hamilton JP, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Tegeder M, Throne-Holst M, Frommer WB, Patrick JW. 2003. Phloem loading and unloading of sugars and amino acids. Plant, Cell & Environment 26, 37–56. [Google Scholar]
- Leegood RC. 2008. Roles of the bundle sheath cells in leaves of C3 plants. Journal of Experimental Botany 59, 1663–1673. [DOI] [PubMed] [Google Scholar]
- Leegood RC, ap Rees T. 1978. Identification of regulatory steps in gluconeogenesis in cotyledons of Cucurbita pepo . Biochimica et Biophysica Acta 524, 207–218. [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Loqué D, Ludewig U, Yuan LX, von Wirén N. 2005. Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiology 137, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mae T, Ohira K. 1981. The remobilization of nitrogen related to leaf growth and senescence in rice plants (Oryza sativa L.). Plant and Cell Physiology 22, 1067–1074. [Google Scholar]
- Maeda E, Maeda K. 1988. Ultrastructural studies of leaf hydathodes. II. Rice (Oryza sativa) leaf tips. Japanese Journal of Crop Science 57, 733–742. [Google Scholar]
- Malone S, Bahrami AR, Walker RP, Gray JE, Leegood RC. 2007. Phosphoenolpyruvate carboxykinase in Arabidopsis thaliana: Changes in isoforms and location during vegetative and floral development. Plant and Cell Physiology 48, 441–450. [DOI] [PubMed] [Google Scholar]
- Martin A, Lee J, Kichey T, et al. 2006. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Ohnishi M, Uehara T, et al. 2013. Ion gradients in xylem exudate and guttation fluid related to tissue ion levels along primary leaves of barley. Plant, Cell & Environment 36, 1826–1837. [DOI] [PubMed] [Google Scholar]
- Nardini A, LoGullo MA, Salleo S. 2011. Refilling embolized xylem conduits: Is it a matter of phloem unloading? Plant Science 180, 604–611. [DOI] [PubMed] [Google Scholar]
- Nazoa P, Vidmar JJ, Tranbarger TJ, Mouline K, Damiani I, Tillard P, Zhuo D, Glass ADM, Touraine B. 2003. Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Molecular Biology 52, 689–703. [DOI] [PubMed] [Google Scholar]
- Nolte KD, Koch KE. 1993. Companion-cell specific localization of sucrose synthase in zones of phloem loading and unloading. Plant Physiology 101, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S, Pilot G. 2011. Amino acid export in plants: A missing link in nitrogen cycling. Molecular Plant 4, 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate JS, Gunning BES. 1972. Transfer cells. Annual Review of Plant Physiology 23, 173–196. [Google Scholar]
- Penfield S, Clements S, Bailey KJ, Gilday AD, Leegood RC, Gray JE, Graham IA. 2012. Expression and manipulation of PHOSPOENOLPYRUVATE CARBOXYKINASE 1 identifies a role for malate metabolism in stomatal closure. Plant Journal 69, 679–688. [DOI] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VPM, Frommer WB. 2004. Overexpression of GLUTAMINE DUMPER 1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16, 1827–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado K, Boursiac Y, Tournaire-Roux C, Monneuse J-M, Postaire O, Da Ines O, Schäffner AR, Hem S, Santoni V, Maurel C. 2013. Regulation of Arabidopsis leaf hydraulics involves light-dependent phosphorylation of aquaporins in veins. Plant Cell 25, 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Voll LM, Horst RJ, Frommer WB, Pilot G. 2010. Stimulation of nonselective amino acid export by glutamine dumper proteins. Plant Physiology 152, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson PT, Baker DA, Ho LC. 1982. The chemical composition of cucurbit vascular exudates. Journal of Experimental Botany 33, 1239–1247. [Google Scholar]
- Sakurai N, Hayakawa T, Nakamura T, Yamaya T. 1996. Changes in the cellular localization of cytosolic glutamine synthetase protein in vascular bundles of rice leaves at various stages of development. Planta 200, 306–311. [Google Scholar]
- Schofield RA, Bi Y-M, Kant S, Rothstein SJ. 2009. Over-expression of STP13, a hexose transporter, improves plant growth and nitrogen use in Arabidopsis thaliana seedlings. Plant, Cell & Environment 32, 271–285. [DOI] [PubMed] [Google Scholar]
- Schulze W, Weise A, Frommer WB, Ward JM. 2000. Function of the cytosolic N-terminus of sucrose transporter AtSUT2 in substrate affinity. Federation of European Biochemical Societies Letters 485, 189–194. [DOI] [PubMed] [Google Scholar]
- Sharkey PJ, Pate JS. 1975. Selectivity in xylem to phloem transfer of amino acids in fruiting shoots of white lupin (Lupinus albus L.). Planta 127, 251–62. [DOI] [PubMed] [Google Scholar]
- Shatil-Cohen A, Moshelion M. 2012. Smart Pipes. The bundle sheath role as xylem-mesophyll barrier. Plant Signaling Behavior 7, 1088–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatil-Cohen A, Attia Z, Moshelion M. 2011. Bundle-sheath cell regulation of xylem-mesophyll water transport via aquaporins under drought stress: a target of xylem-borne ABA? Plant Journal 67, 72–80. [DOI] [PubMed] [Google Scholar]
- Singh S, Singh TN. 2013. Guttation 1: chemistry, crop husbandry and molecular farming. Phytochemistry Reviews 12, 147–172. [Google Scholar]
- Taylor L, Nunes-Nesi A, Parsley K, Leiss A, Leach G, Coates S, Wingler A, Fernie AR, Hibberd JM. 2010. Cytosolic pyruvate, orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. Plant Journal 62, 641–652. [DOI] [PubMed] [Google Scholar]
- Tegeder M. 2012. Transporters for amino acids in plant cells: some functions and many unknowns. Current Opinions in Plant Biology 15, 315–321. [DOI] [PubMed] [Google Scholar]
- Tegeder M, Rentsch D. 2010. Uptake and partitioning of amino acids and peptides. Molecular Plant 3, 997–1011. [DOI] [PubMed] [Google Scholar]
- Walker RP, Acheson RM, Tesci LI, Leegood RC. 1997. Phosphoenolpyruvate carboxykinase in C4 plants: its role and regulation. Australian Journal of Plant Physiology 24, 459–468. [Google Scholar]
- Walker RP, Chen Z-H, Acheson RM, Leegood RC. 2002. Effects of phosphorylation on phosphoenolpyruvate carboxykinase from the C4 plant Guinea Grass. Plant Physiology 12, 165–172. [PMC free article] [PubMed] [Google Scholar]
- Walker RP, Chen Z-H, Técsi L, Famiani F, Lea PJ, Leegood RC. 1999. Phosphoenolpyruvate carboxykinase plays a role in interactions of carbon and nitrogen metabolism during grape seed development. Planta 210, 9–18. [DOI] [PubMed] [Google Scholar]
- Walker RP, Trevanion SJ, Leegood RC. 1995. Phosphoenolpyruvate carboxykinase from higher plants: purification from cucumber and evidence for rapid proteolytic cleavage in extracts from a range of plant tissues. Planta 196, 58–63. [Google Scholar]
- Wingler A, Walker RP, Chen Z-H, Leegood RC. 1999. Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle-sheath of maize. Plant Physiology 120, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaya T, Kusano M. 2014. Evidence supporting distinct functions of three cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. Journal of Experimental Botany 65, 5519–5525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.