Abstract
Lipid-based micelles provide an attractive option for therapeutic and diagnostic applications due to their small size (< 20 nm) and ability to self-assemble and improve the solubility of both hydrophobic drugs and dyes. Their use, however, has been challenged by the fact that these particles are inherently unstable in serum due to interactions with protein components, which favor the monomeric state. We have engineered serum stabilized micelles using short quadruplex forming oligonucleotide extensions as the lipid headgroup. Quadruplex formation on the surface of the particles, confirmed by 1H NMR, results in slight distortion of the otherwise spherical micelles and renders them resistant to disassembly by serum proteins for > 24 h. Using antisense oligonucleotides we demonstrated that disruption of the quadruplex leads to micelle destabilization and cargo release. The ability to use oligonucleotide interactions to control lipid particle stability represents a new approach in the design of programmed nanoscale devices.
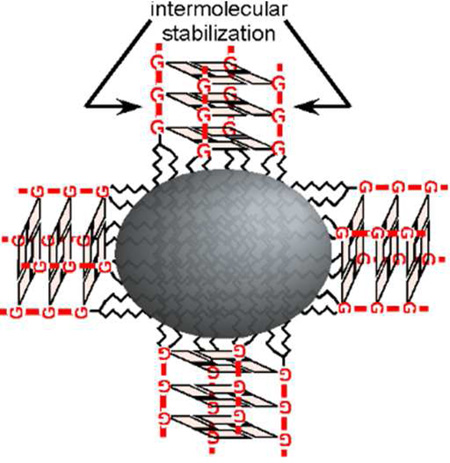
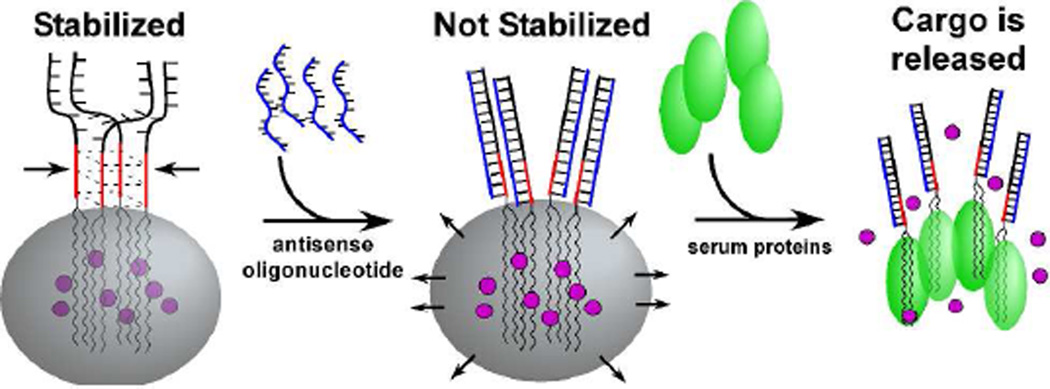
Graphical abstract
The structural characteristics of nucleic acids, specifically Watson-Crick base-pairing along with non-traditional hydrogen bonds between bases, make nucleic acids ideal building blocks for the design of complex nanostructures and nanodevices.1 Using oligonucleotide polymers and strand displacement by complementary hybridization, it is possible to create intricate circuits, logic-gated robots, and molecular automata for cellular recognition.2–4 Here, we have used similar strand displacement techniques to program the destabilization of nucleic acid modified micelles.
Lipid micelles generated by the self-assembly of monomers represent an important class of nanoparticle with the potential to enhance the solubility and delivery of hydrophobic drugs.5,6 Their small size (< 20 nm) makes them ideal candidates for both passive tumor uptake and deep tissue penetration.7 However, formation is a dynamic process in which lipid monomers exist in equilibrium with micelles. Destabilization of micelles in vivo occurs due to significant dilution upon systemic injection and is further affected by the presence of serum proteins such as BSA, which can interact with hydrophobic lipid tails.8,9 Thus, premature release of encapsulated components may occur before these particles reach their target (Figure 1A). The ability to stabilize lipid micelles but also allow for triggered cargo release remains a challenge.
Figure 1.
Schematic of micelle stabilization. (A) Lipid monomers (1) form micelles above a critical micelle concentration (2), but this particle formation is disrupted by interactions with serum proteins (3) and dilution in vivo. (B) Intermolecular interactions between diC18-UGGGU headgroups result in G-quadruplex formation.
The use of oligonucleotides and even single nucleotides as lipid headgroups for the formation of micelles has previously been reported.10 However, while an oligonucleotide alone is unlikely to stabilize a micelle, we reasoned that we could engineer interactions between headgroups composed of short oligonucleotides which might enhance stability. Indeed, Edwardson et al. recently demonstrated that a self-assembled DNA nanocage could be used to precisely arrange C12 alkylchains to assemble a hydrophobic core within a tetrahedral structure.11
A variety of other approaches have previously been employed to stabilize micelles ranging from chemically crosslinking their hydrophobic tails12 to linking the hydrophilic head groups through covalent13,14 and reversible or reducible linkages. PEGylated peptides forming 3-helix bundles through intermolecular interactions at lipid headgroups have also been used to stabilize micelles.15,16 Drawing from this, we engineered lipids bearing short, highly stable 2’OMe RNA sequences capable of forming a parallel G-quadruplex which could bring four lipid monomers together (Figure 1B). In this way, micelles can be stabilized by intermolecular interactions that can be purposefully disrupted using antisense oligonucleotides designed to infiltrate the G-quadruplex.
Oligonucleotide-lipid conjugates were generated using standard solid phase DNA/RNA synthesis and a 1,2-dioctadecyl-sn-glycerol (diC18) phosphoramidite.17 We synthesized two short oligonucleotide-lipid conjugates composed of highly stable 2’OMe RNA bearing a 5’ lipid: (1) diC18-UUUUU, which is predicted to not possess any intermolecular interactions and (2) diC18-UGGGU, which is predicted to form a parallel G-quadruplex (Figure 1B).18,19 Following synthesis, deprotection, and purification, the conjugate identities were verified by LC/MS and used to generate micelles by thin-film hydration (see Supplemental Methods).
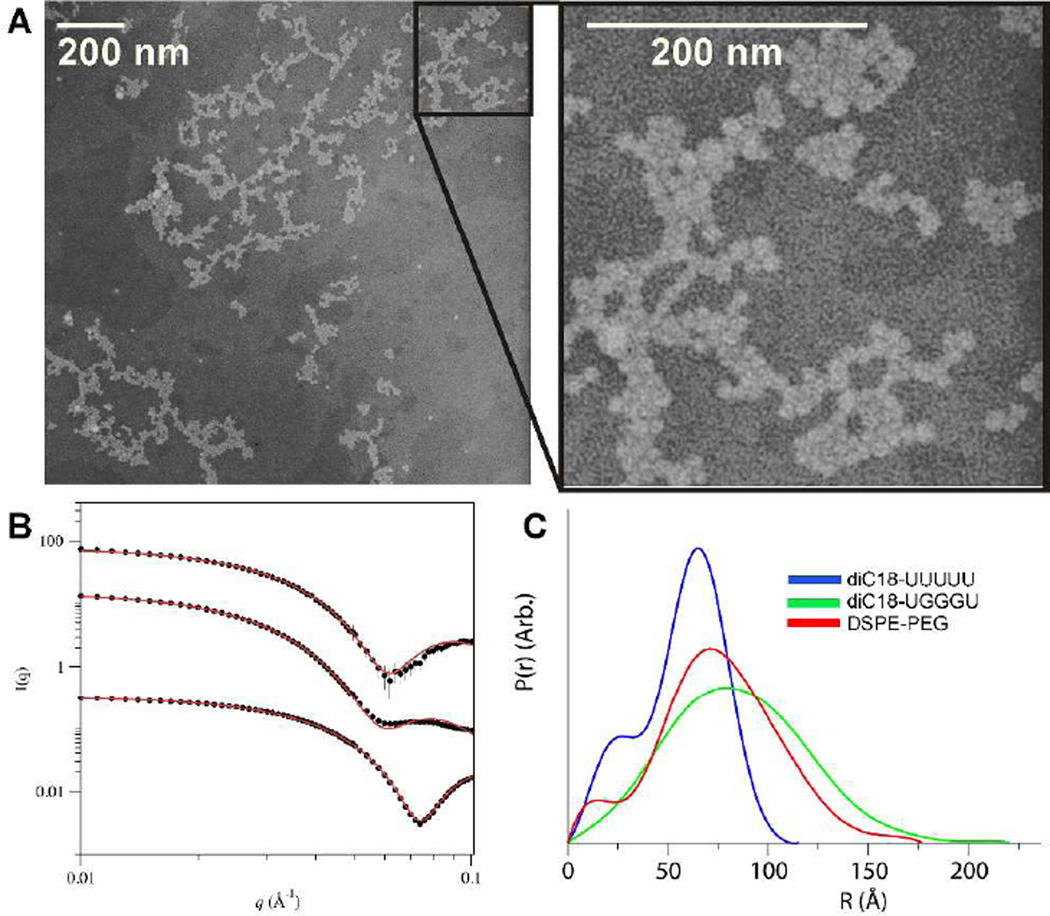
Micelles generated from diC18-UUUUU or diC18-UGGGU formed particles with ~10 nm diameters and low polydispersity (< 20%), similar in size to micelles generated using the lipid amphiphile DSPE-PEG(2000) as determined by dynamic light scattering (Supplemental Figure S1). Particle size was confirmed by negative-stain transmission electron microscopy (Figure 2A and Supplemental Figure S2). Based on the size similarity to DSPE-PEG, we estimate that the particles are composed of ~90 molecules each.20 Using small angle X-ray scattering (SAXS), we confirmed this estimation (Supplemental Table S1). SAXS data were fit to a scattering form factor for micelles with an ellipsoidal core with Gaussian chains on the surface (Figure 2B).21 The fit for the DSPE-PEG micelles using the model are in good agreement with previous SAXS characterization20 and display a slight ellipsoidal shape (ε = 1.16), with those formed from diC18-UUUUU proving more spherical (ε = 1.0) and those formed from diC18-UGGGU more oblate (ε = 1.95). For diC18-UGGGU, the fit suggests that the particles possess a large corona with a large radius of gyration (Rg) for the chains, consistent with the formation of a G-quadruplex.22 In contrast, the corona and Rg of the diC18-UUUUU micelles are smaller suggesting that headgroups are likely more diffuse and unstructured. Micelle shape was further investigated using GNOM22 to calculate the intraparticle distance distribution p(r) and the maximum dimension of the micelles, Dmax (Supplemental Table S2). A monodisperse spherical particle will display a bell-shaped p(r) curve whereas rod-like particles will display a bell-shape at small r, but would descend linearly at larger r values.23,24 The p(r) of each sample is closely bell-shaped with the DSPE-PEG and diC18-UGGGU micelles displaying more elongated distances, consistent with slight ellipsoidal character (Figure 2C). Furthermore, Dmax is in good agreement with micelle diameters determined by DLS.
Figure 2.
Micelle characterization. (A) TEM image of diC18-UGGGU micelles. (B) Experimental SAXS profiles of DSPE-PEG, diC18-UGGGU, and diC18-UUUUU micelles each at 2 mg/mL, from top to bottom, and fitted profiles from ellipsoid model fits (red lines). diC18-UGGGU and diC18-UUUUU data sets were vertically offset by scaling intensity values of 10−2 and 10−3, respectively. (C) Intraparticle distance distribution p(r) for diC18-UUUUU micelles (blue), diC18-UGGGU micelles (green), and DSPE-PEG micelles (red).
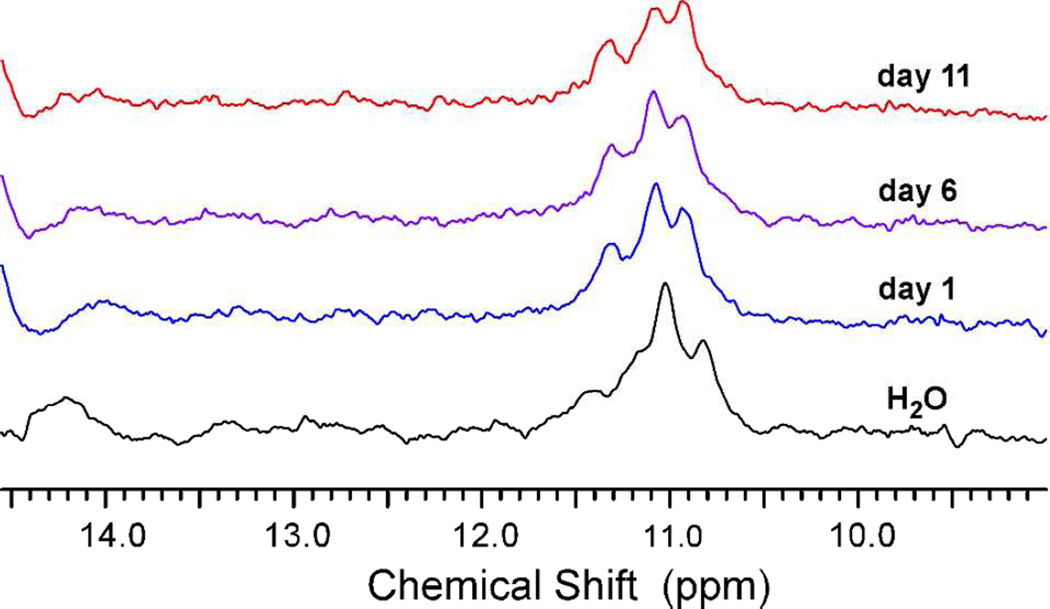
The alteration in the shape of micelles generated with diC18-UUUUU vs. diC18-UGGGU suggested that the engineered headgroup interactions within the diC18-UGGGU micelles lead to a change in the overall micelle structure. To confirm quadruplex formation we preformed 1H NMR on these micelles. The imino protons in a G-quadruplex show diagnostic chemical shifts of 10–12 ppm vs. 13–14 ppm in a standard Watson-Crick duplex and are much more resistant to deuterium exchange in D2O, with half-times of days to weeks.18,25 Consistent with this, the 1H NMR spectra for the diC18-UGGGU micelles displayed three imino protons between 10.8 and 11.3 ppm, all of which remained resistant to deuterium exchange for >> 24 h (Figure 3). A similar NMR analysis of diC18-UUUUU micelles does not show these characteristic imino proton shifts (data not shown) and that of diC18-UGUUU micelles, which shows a predicted 10.8 ppm imino proton shift characteristic of G-G interactions, does not show resistance to deuterium exchange (Supplemental Figure S3).26
Figure 3.
Overlay of NMR spectra for diC18-UGGGU micelles after D2O exchange. diC18-UGGGU micelles were incubated in D2O for 24 h (blue), 6 days (purple), and 11 days including one 24 h incubation at 37 °C (red). Similar to the H2O spectra (black), the 1H NMR spectra for diC18-UGGGU micelles display characteristic imino proton shifts for the three guanine bases which remain resistant to deuterium exchange > 24 h.
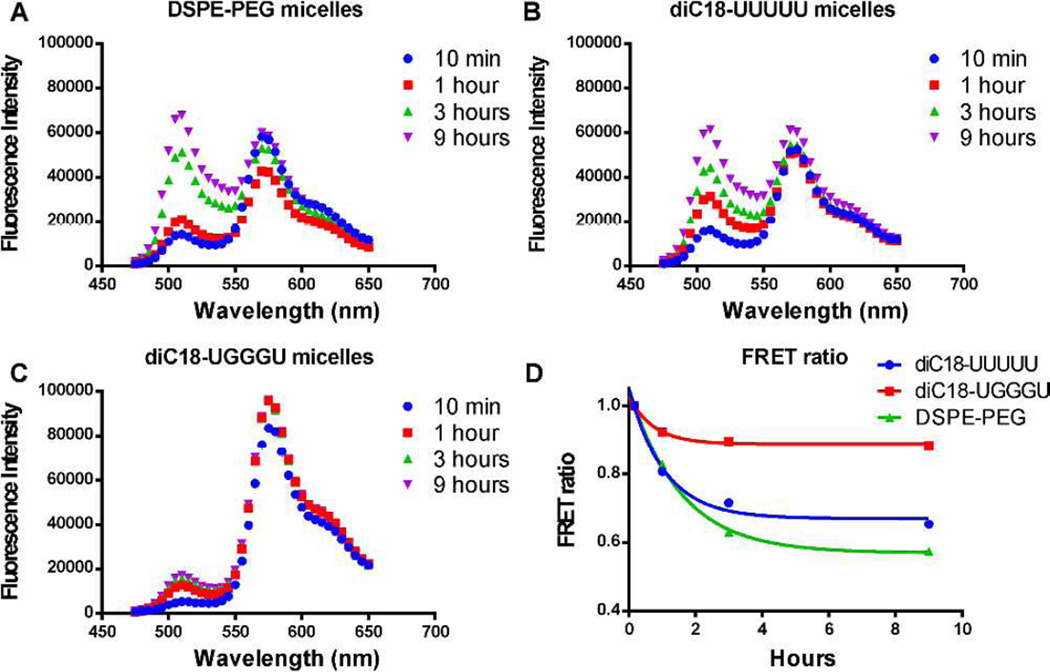
In order to assess the effects of quadruplex stabilization on micelle stability, we encapsulated a lipophilic FRET pair (DiO/DiI) within our micelles and monitored the release of the dyes over time.5,6,16 When co-encapsulated within an intact micelle, excitation at 450 nm results in FRET, and fluorescence emission is observed at 565 nm. Dye release results in separation of the FRET pair and an increase in fluorescence intensity at 505 nm. When both DSPE-PEG micelles and diC18-UUUUU micelles are diluted in 90% fetal bovine serum (FBS), there is an increase in fluorescence intensity at 505 nm as these lipids exchange with serum proteins (Figure 4A and 4B; t1/2 = 1.06 ± 0.06 h and 0.74 ± 0.17 h, respectively). This destabilization is also marked by a decrease in the FRET ratio (I565/[I565+I505]), which approaches an experimentally determined value of ~0.5 in 9 h indicating almost complete release of encapsulated cargo (Figure 4D and Supplemental Figure S4). Destabilization is FBS dependent (Supplemental Figure S5) predominantly due to interactions with α/β globulins (Supplemental Figure S6), which have previously been shown to contribute to micelle destabilization in vivo.5
Figure 4.
FRET assay to measure micelle stability. (A) FRET spectra of DSPE-PEG micelles and (B) diC18-UUUUU micelles show an increase in fluorescence intensity at 505 nm indicating micelle destabilization. (C) FRET spectra of diC18-UGGGU micelles. (D) Graph of normalized FRET ratio (I565/[I565+I505]) over 9 h.
In contrast, diC18-UGGGU micelles, remained stable for > 24 h (Figures 4C and 4D). Micelle stability did not rely on the presence of potassium (Supplemental Figures S6 and S7) suggesting that like other parallel quadruplexes, Na+ alone was sufficient for stabilization.27
Micelle stability was also assessed by measuring the kinetics of lipid exchange using fluorescently labeled lipids.16 Fluorescein labeled diC18-UUUUU and diC18-UGGGU lipids assembled into micelles with diameters similar to those of their unlabeled counterparts (~10 nm; data not shown). Due to the close proximity of the dyes in the head-group, fluorescence is quenched. However, to ensure that fluorescence quenching was not due solely to the formation of quadruplexes, micelles were generated using a 5:1 ratio of unlabeled to labeled lipid. These FITC-labeled micelles were diluted with a 600-fold excess of unlabeled micelles, and the rate of fluorescence recovery was measured (Supplemental Figure S8). For diC18-UGGGU micelles, there is no increase in fluorescence, indicating that particles remain quenched overtime. On the other hand, consistent with the data from our dye release assays, diC18-UUUUU micelles dequench over the course of 3 h (t1/2 = 0.25 ± 0.01 h), suggesting exchange of fluorescently labeled lipids with unlabeled lipids. Similar results are observed when the FITC-labeled micelles were diluted in FBS (Supplemental Figure S9).
Finally, since the CMC is an important factor in defining micelle stability, we performed experiments to determine the CMC value for both diC18-UUUUU and diC18-UGGGU micelles. Using multiple approaches the CMC values proved too low to detect (< 10 pM; Supplemental Figure S10). It is interesting to note that although diC18-UUUUU micelles display a very low CMC, this value is not sufficient to maintain stability in the presence of serum proteins. Thus, a low CMC is not the only prerequisite for stability in biological fluids.
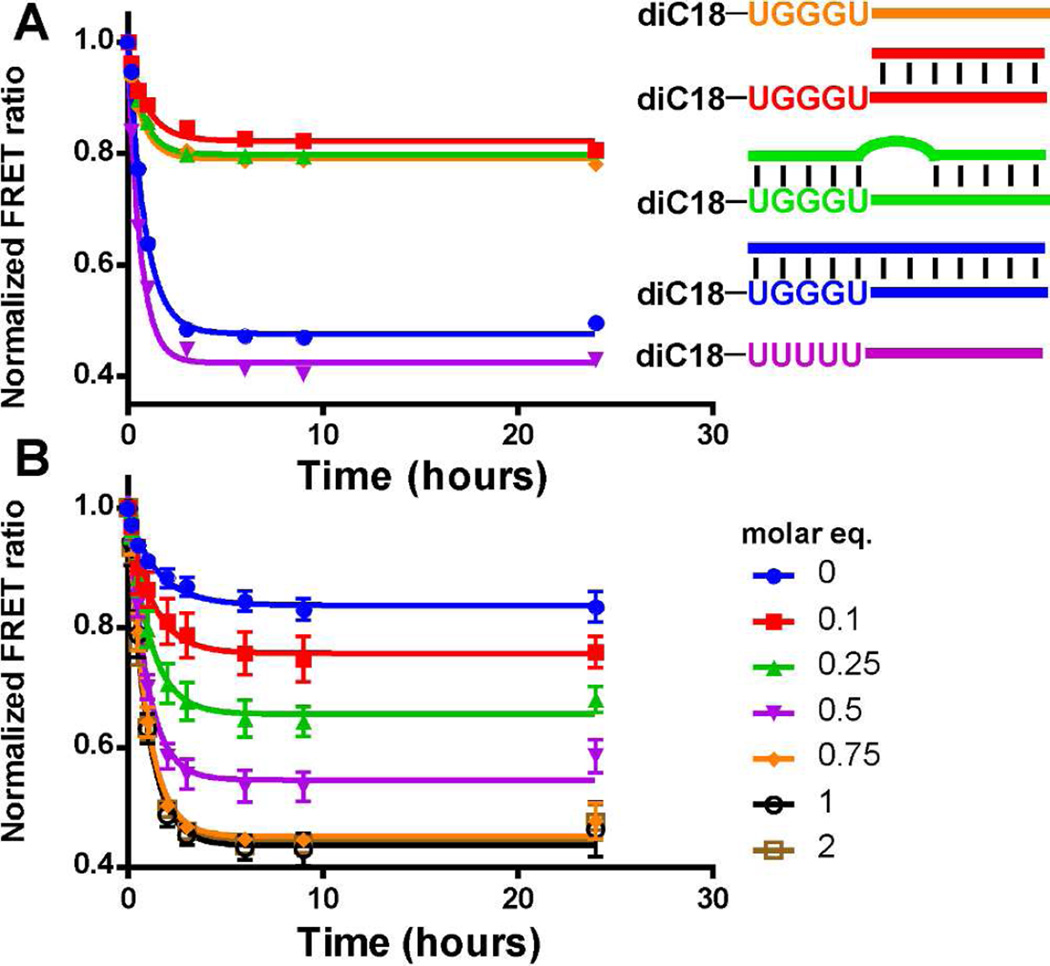
The marked difference in stability between the diC18-UUUUU micelles and the diC18-UGGGU micelles suggested that modifications at the lipid headgroup might be used to alter micelle stability and subsequently induce cargo release. We synthesized diC18-UGGGU micelles bearing a 3’ oligonucleotide extension (diC18-UGGGU-ext) such that the addition of an antisense oligonucleotide would disrupt the stabilizing G-quadruplex (Figure 5). Micelles formed from diC18-UGGGU-ext were ~10 nm in diameter (data not shown) and were stable in serum for > 24 h in the presence of FBS according to the FRET stability assay (Figure 6A; orange). However, upon addition of equimolar amounts of a perfectly complementary oligonucleotide, the stability is lost (Figure 6A; blue, t1/2 = 0.60 ± 0.03 h), and the FRET ratio decreased at a rate similar to that of diC18-UUUUU-ext micelles (Figure 6A; purple, t1/2 = 0.44 ± 0.03 h). The onset of triggered release is rapid and can be induced at a desired time point following pre-incubation (Supplemental Figure S11). On the other hand, micelles remain intact when an antisense oligonucleotide is introduced that does not hybridize with the G-quadruplex forming sequence, UGGGU (Figure 6A, red) or contains a 3-base mismatch (Figure 6A, green). Hybridization of antisense oligonucleotides with micelles was confirmed by gel electrophoresis (Supplement Figure S12).
Figure 5.
Micelle destabilization using antisense oligonucleotides. diC18-UGGGU-ext micelles remain intact in the presence of serum proteins. Addition of antisense oligonucleotides that disrupt G-quadruplex formation results in micelle destabilization and cargo release.
Figure 6.
Programmable micelle stability. (A) Plot of FRET ratios upon addition of antisense oligonucleotides. The perfectly complementary oligonucleotide (blue) disrupts the stability of diC18-UGGGU-ext micelles (orange), which looks similar to the diC18-UUUUU-ext micelles (purple). Antisense strands that do not disrupt the G-quadruplex (red) or contain a 3-base mutation (green) do not affect micelle stability. (B) Change in FRET ratio upon titration of the perfectly complementary antisense oligonucleotide. “Molar eq.” indicates molar equivalent of antisense oligonucleotide as compared to the concentration of the diC18-UGGGU-ext micelles.
When titrating in molar equivalents of the perfectly complementary oligonucleotide to diC18-UGGGU-ext micelles, micelle stability is completely disrupted in the presence of 0.75:1 or greater (1:1 or 2:1) amounts of the antisense oligonucleotide (Figure 6B). The addition of lower levels (< 0.75:1) resulted in intermediate levels of dye release. Interestingly, all micelles decay with similar rates (t1/2 0.69 ± 0.09 h) suggesting that only a subset of micelles is loaded with enough antisense oligonucleotide to destabilize and release cargo. Thus, it appears that hybridization to the extension and infiltration of the quadruplex may proceed in a cooperative manner in which hybridization ‘primes’ a given particle for additional hybridization events. Cooperative transitions have been observed with other oligonucleotide functionalized nanoparticle systems.28 A more detailed mechanistic analysis to confirm this is currently underway.
In summary, we have designed a lipid-based micelle bearing a RNA modification at the lipid headgroup that forms a G-quadruplex which stabilizes the micelle in serum. By extending the RNA sequence at the 3’ end, micelle destabilization can be programmed by introduction of a complementary oligonucleotide which infiltrates the quadruplex leading to cargo release. Finally, while our work here has focused on the use of a simple G-quadruplex as a stabilizing element, it is easy to envision how this approach could be incorporated into much more complex systems and nanoscale devices.
Supplementary Material
Acknowledgments
This work was supported by the Training Program in Cellular and Molecular Biology and Genetics, T32 GM007491. Thanks to Dr. Sean Cahill and Dr. Richard Harris for assistance with NMR. Thanks to Leslie Cummins and Frank Macaluso at Einstein’s Analytical Imaging Facility for collecting TEM images. SAXS data at NSLS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental methods and additional experimental data. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Sources
No competing financial interests have been declared.
REFERENCES
- 1.Zhang F, Nangreave J, Liu Y, Yan H. J Am Chem Soc. 2014;136:11198. doi: 10.1021/ja505101a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amir Y, Ben-Ishay E, Levner D, Ittah S, Abu-Horowitz A, Bachelet I. Nat Nanotechnol. 2014;9:353. doi: 10.1038/nnano.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 4.Rudchenko M, Taylor S, Pallavi P, Dechkovskaia A, Khan S, Butler VP, Rudchenko S, Stojanovic MN. Nat Nanotechnol. 2013;8:580. doi: 10.1038/nnano.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Kim S, He W, Wang H, Low PS, Park K, Cheng JX. Langmuir. 2008;24:5213. doi: 10.1021/la703570m. [DOI] [PubMed] [Google Scholar]
- 6.Chen HT, Kim SW, Li L, Wang SY, Park K, Cheng JX. P Natl Acad Sci USA. 2008;105:6596. doi: 10.1073/pnas.0707046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torchilin VP. Journal of Controlled Release. 2001;73:137. doi: 10.1016/s0168-3659(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 8.Castelletto V, Krysmann M, Kelarakis A, Jauregi P. Biomacromolecules. 2007;8:2244. doi: 10.1021/bm070116o. [DOI] [PubMed] [Google Scholar]
- 9.Kastantin M, Missirlis D, Black M, Ananthanarayanan B, Peters D, Tirrell M. J Phys Chem B. 2010;114:12632. doi: 10.1021/jp1001786. [DOI] [PubMed] [Google Scholar]
- 10.Raouane M, Desmaele D, Urbinati G, Massaad-Massade L, Couvreur P. Bioconjug Chem. 2012;23:1091. doi: 10.1021/bc200422w. [DOI] [PubMed] [Google Scholar]
- 11.Edwardson TG, Carneiro KM, McLaughlin CK, Serpell CJ, Sleiman HF. Nat Chem. 2013;5:868. doi: 10.1038/nchem.1745. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Xiao K, Luo J, Xiao W, Lee JS, Gonik AM, Kato J, Dong TA, Lam KS. Biomaterials. 2011;32:6633. doi: 10.1016/j.biomaterials.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joralemon MJ, O'Reilly RK, Hawker CJ, Wooley KL. J Am Chem Soc. 2005;127:16892. doi: 10.1021/ja053919x. [DOI] [PubMed] [Google Scholar]
- 14.Sun X, Rossin R, Turner JL, Becker ML, Joralemon MJ, Welch MJ, Wooley KL. Biomacromolecules. 2005;6:2541. doi: 10.1021/bm050260e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong H, Dube N, Shu JY, Seo JW, Mahakian LM, Ferrara KW, Xu T. ACS Nano. 2012;6:5320. doi: 10.1021/nn301142r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong H, Shu JY, Dube N, Ma Y, Tirrell MV, Downing KH, Xu T. J Am Chem Soc. 2012;134:11807. doi: 10.1021/ja3048128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selden NS, Todhunter ME, Jee NY, Liu JS, Broaders KE, Gartner ZJ. J Am Chem Soc. 2012;134:765. doi: 10.1021/ja2080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheong C, Moore PB. Biochemistry. 1992;31:8406. doi: 10.1021/bi00151a003. [DOI] [PubMed] [Google Scholar]
- 19.Petraccone L, Erra E, Randazzo A, Giancola C. Biopolymers. 2006;83:584. doi: 10.1002/bip.20591. [DOI] [PubMed] [Google Scholar]
- 20.Arleth L, Ashok B, Onyuksel H, Thiyagarajan P, Jacob J, Hjelm RP. Langmuir. 2005;21:3279. doi: 10.1021/la047588y. [DOI] [PubMed] [Google Scholar]
- 21.Pedersen JS. J Appl Crystallogr. 2000;33:637. [Google Scholar]
- 22.Svergun DI. J Appl Crystallogr. 1992;25:495. [Google Scholar]
- 23.Small Angle X-ray Scattering. London: Academic Press; 1982. [Google Scholar]
- 24.Neutron, X-ray and light scattering: introduction to an investigative tool for colloidal and polymeric systems. North-Holland: 1991. [Google Scholar]
- 25.Adrian M, Heddi B, Phan AT. Methods. 2012;57:11. doi: 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharya PK, Cha J, Barton JK. Nucleic Acids Res. 2002;30:4740. doi: 10.1093/nar/gkf601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marathias VM, Bolton PH. Biochemistry. 1999;38:4355. doi: 10.1021/bi982604+. [DOI] [PubMed] [Google Scholar]
- 28.Jin R, Wu G, Li Z, Mirkin CA, Schatz GC. J Am Chem Soc. 2003;125:1643. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.