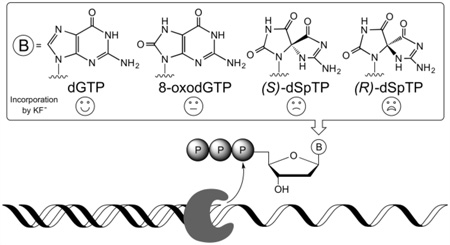
Abstract
Defining the biological consequences of oxidative DNA damage remains an important and ongoing area of investigation. At the foundation of understanding the repercussions of such damage is a molecular-level description of the action of DNA-processing enzymes, such as polymerases. In this work we focus on a secondary, or hyperoxidized, oxidative lesion of dG which is formed by oxidation of the primary oxidative lesion 2′-deoxy-8-oxo-7,8-dihydroguanosine (8-oxodG). In particular, we examine incorporation into DNA of the diastereomers of the hyperoxidized guanosine triphosphate lesion spiroiminodihydantoin-2′-deoxynucleoside-5′-triphosphate (dSpTP). Using kinetic parameters we describe the ability of the Klenow fragment of E. coli DNA polymerase I lacking 3'→5' exonuclease activity (KF−) to utilize (S)-dSpTP and (R)-dSpTP as building blocks during replication. We find that both diastereomers act as covert lesions, similar to a Trojan horse: KF− incorporates the lesion dNTP opposite dC, which is a non-mutagenic event, however, during the subsequent replication it is known that dSp is nearly 100% mutagenic. Nevertheless, using kpol/Kd to define the nucleotide incorporation specificity we find that the extent of oxidation of the dGTP-derived lesion correlates with its ability to be incorporated into DNA. KF− has the highest specificity for incorporation of dGTP opposite dC. The selection factors for incorporating 8-oxodGTP, (S)-dSpTP, and (R)-dSpTP are 1,700-, 64,000- and 850,000-fold lower respectively. Thus, KF− is rigorous in its discrimination against incorporation of the hyperoxidized lesion and these results suggest that the specificity of cellular polymerases provides an effective mechanism to avoid incorporating dSpTP lesions into DNA from the nucleotide pool.
Graphical Abstract
INTRODUCTION
Genomic DNA is exposed to a variety of endogenous and exogenous reactive oxygen species (ROS).1, 2 Due to its low redox potential,3 dG is the major target for ROS and a prototypic and well-studied oxidation product of dG is 2′-deoxy-8-oxo-7,8-dihydroguanosine (8-oxodG) (Figure 1).4–6 Indeed, in vivo 8-oxodG is found at steady-state levels of 0.3–4.2 per 106 dG.7 Studies performed in vitro have shown that 8-oxodG has a high miscoding frequency because it can base pair with both dC and dA, with the latter leading to G→T transversion mutations.8, 9 Despite its high miscoding frequency in vitro, 8-oxodG has a low mutation frequency in vivo yielding less than 10% G→T transversions.9–11 This low mutagenicity in vivo derives from the presence of an extensive repair system that counters the genetic effects of 8-oxodG.12
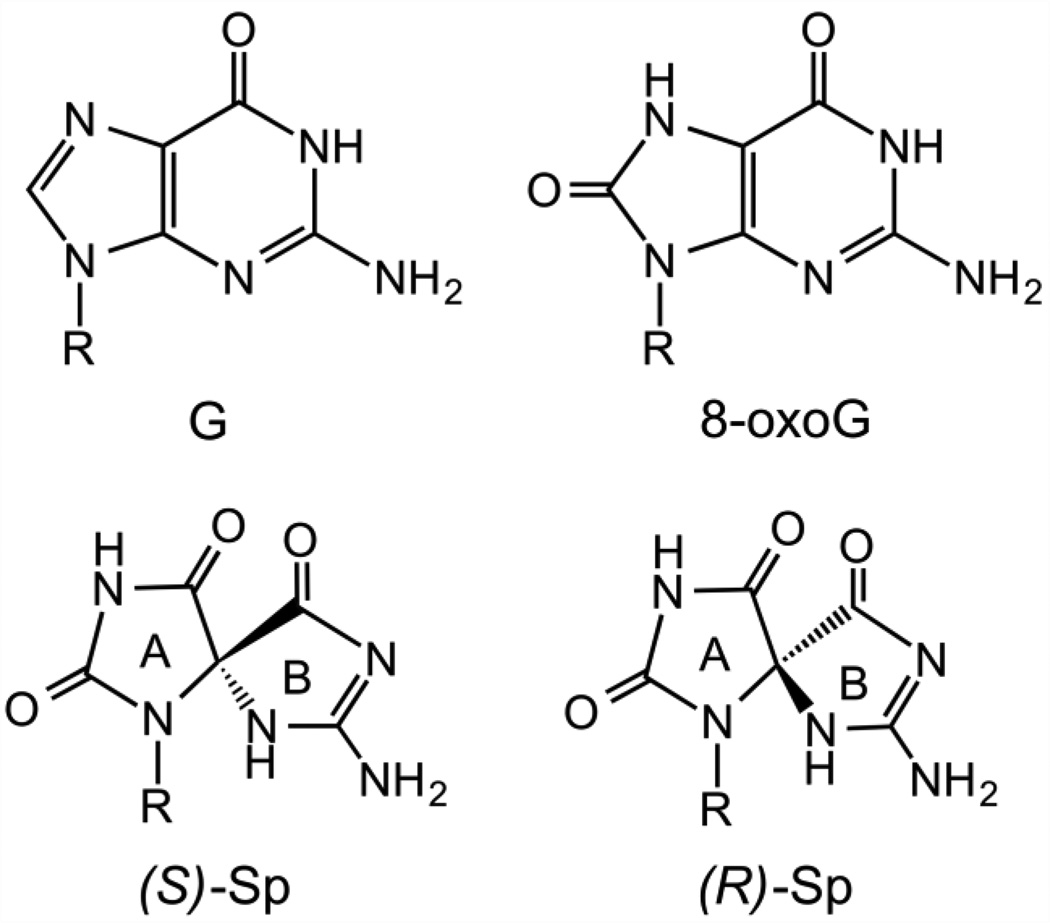
Figure 1.
Structures of dG, 8-oxodG, (S)-dSp, and (R)-dSp. The A and B rings of dSp are indicated.
In addition to the mutagenic potential of 8-oxodG, the lesion itself is chemically labile towards further oxidation.13 Several hyperoxidized lesions have been identified, including 2′-deoxy-spiroiminodihydantoin (dSp), which exists as a pair of diastereomers (Figure 1).14–20 Furthermore, the dSp lesion has been detected in genomic DNA from bacteria and mammalian cells. Hailer and co-workers detected the dSp lesion in E. coli treated with chromate,21 and more recently, Mangerich and co-workers identified dSp in Helicobacter hepaticus-infected mice, which develop inflammation-mediated carcinogenesis at levels of 1–5 per 108 nucleotides in genomic DNA.22 Several DNA glycosylases have been found to excise the dSp lesion from DNA in vitro and initiate the base excision repair (BER) process. The bacterial glycosylases FPG, Nei, and Nth remove dSp with differing preferences for the opposing base pair partner.23–25 The dSp lesion is not a substrate for the human enzyme OGG1 which is responsible for removing 8-oxodG, but the lesion is a substrate for yeast OGG1 and OGG2.26 Finally, eukaryotic27–30 and viral31 NEIL1 glycosylases can excise dSp from double-stranded DNA with some preference for the opposing base pair partner, and can also remove dSp from single-stranded DNA, bubble, and bulge structures. More recent results suggest that dSp can also be removed from DNA by nucleotide excision repair.32 If dSp is not removed prior to replication, in contrast to the mildly mutagenic 8-oxodG, the hyperoxidized dSp lesion is nearly 100% mutagenic in vitro33, 34 and in vivo,35–37 causing both G→T and G→C transversions.
As the primary and secondary oxidation products of dG, the mutagenicity of both 8-oxodG and dSp have been well-studied when the lesion is present in DNA. However, in order to understand the full mutagenic potential of a lesion we must also understand the extent to which polymerases utilize the oxidized lesions when they are part of the nucleotide pool. Notably, it has been shown that a nucleobase is more susceptible to oxidation when it is part of a free nucleotide than when incorporated in duplex DNA. For instance, dATP in the nucleotide pool is 67-fold more easily oxidized than dA in duplex DNA.38 Although the preference is less pronounced for dGTP, it is still 9-fold easier to oxidize in the nucleotide pool relative to dG in duplex DNA.38 Therefore, it is important to consider the nucleotide pool as a source of oxidative damage. Indeed, it is known that 8-oxodGTP can be incorporated into DNA by several bacterial and mammalian polymerases.39–43 However, it remains unknown if polymerases can utilize dSpTP as a building block during DNA replication.
Here we determine the extent to which each diastereomer of dSpTP is a substrate for the Klenow Fragment of E. coli DNA polymerase I lacking 3'→5' exonuclease activity (KF−). We establish that KF− can incorporate each lesion into DNA, with a preference for (S)-dSpTP over (R)-dSpTP. We also define the kinetic parameters for incorporation of each lesion and discuss the incorporation efficiency relative to canonical dGTP and the primary oxidation product 8-oxodGTP.
EXPERIMENTAL PROCEDURE
Materials
Klenow Fragment 3'→5' exo− (KF−), calf intestinal alkaline phosphatase, and T4 polynucleotide kinase were purchased from New England Biolabs (Ipswich, MA). The canonical dNTPs were from Thermo Scientific (Waltham, MA) and 8-oxodGTP was from TriLink BioTechnologies (San Diego, CA).
Oligonucleotide Synthesis and Purification
Oligonucleotides were synthesized and purified according to methods published previously by our laboratory.44 Oligonucleotide concentrations were determined at 90 °C using molar extinction coefficients as determined by nearest-neighbor theory for single-stranded DNA45, 46 on a Beckman Coulter DU800 UV-visible spectrophotometer equipped with a peltier thermoelectric device. The identity of the oligonucleotides was confirmed by ESI-MS.
Synthesis, Purification, and Absolute Configuration of Spiroiminodihydantoin-2′-deoxynucleoside-5′-triphosphate
The hyperoxidized nucleotide triphosphate dSpTP was synthesized according to literature procedures from dGTP.47 The resulting solution was dried in vacuo, resuspended in a small amount of deionized water, and purified by ion-pairing HPLC on a Varian Microsorb C18 column (250 × 4.6 mm) (Agilent Technologies, Inc., Santa Clara, CA) using acetonitrile (mobile phase A) and 10 mM tetrabutylammonium hydroxide, 10 mM monopotassium phosphate, 0.25% methanol, pH = 7.0 (mobile phase B) as the mobile phases. As the gradient mobile phase A increased from 5 to 65% over 40 min at a flow rate of 1 mL/min. The dSpTP diastereomers were collected as a mixture and dried in vacuo; Figure S1A demonstrates that the mixture of dSpTP diastereomers was successfully separated from the dGTP starting material. The sample was resuspended in deionized water and the diastereomers were separated on the same HPLC column as above using mobile phase B (same as above) and mobile phase C (2.8 mM tetrabutylammonium hydroxide, 100 mM monopotassium phosphate, 30% methanol, pH = 5.5); as the gradient mobile phase C was increased from 50 to 70% over 40 min at flow rate 0.9 mL/min (Figure S1B). Removal of the ion-pairing reagent from the separated diastereomers was achieved by multiple rounds of purification on a Dionex DNAPac PA-100 anion exchange HPLC column (250 × 4.6 mm) (Thermo Scientific, Waltham, MA) using mobile phase D (25 mM ammonium bicarbonate, pH=8.5) and mobile phase E (800 mM ammonium bicarbonate, pH=8.5); as the gradient mobile phase E was increased from 0 to 50% over 15 min at a flow rate of 1 mL/min. Multiple rounds of anion exchange HPLC were performed to the point that the experimentally determined rate of nucleotide incorporation did not change. Notably, the diastereomers of dSpTP elute in the same order from the C18 and anion exchange HPLC columns. The volatile ammonium bicarbonate was removed by repetitive resuspension in deionized water and drying in vacuo. For all the HPLC analyses described above, absorbance was monitored at both 225 nm and 260 nm. The purified dSpTP diastereomers were characterized along with dGTP by anion exchange HPLC using mobile phase D (25 mM ammonium bicarbonate, pH=8.5) and mobile phase E (800 mM ammonium bicarbonate, pH=8.5); as the gradient mobile phase E was increased from 0 to 50% over 60 min at a flow rate of 1 mL/min (Figure S1C). The concentration of each diastereomer of dSpTP was determined by UV absorbance based on the extinction coefficient ε230nm = 4,900 M−1 cm−1.14 The molecular weights were verified by ESI-MS (Figure S2A, B).
The absolute configuration of the diastereomers of dSpTP was then assigned based on literature reports.48, 49 Each diastereomer of dSpTP (0.2 µmol) was resuspended in 1× Cutsmart buffer (20 mM Tris acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 0.1 mg/mL BSA, pH 7.9) (New England Biolabs, Ipswich, MA) and treated with 20 U calf intestinal alkaline phosphatase at 37 °C for 40 h. The phosphatase was removed by passing the solution through a 3,000 MWCO centrifugal filter (Sartorius Corp., Bohemia, NY). The resulting dSp nucleosides were then analyzed on a Thermo Scientific HyperCarb column (150 × 4.6 mm) (Thermo Scientific, Waltham, MA) with an isocratic flow of deionized water containing 0.1 % acetic acid at 1 mL/min; the absorbance was monitored at 240 nm (Figure S1D). The dSp nucleosides were then analyzed by ESI-MS (Figure S2C, D). Consistent with literature reports,49 removal of the triphosphate group switches the order of elution on the HyperCarb column relative to the C18 and anion exchange columns. Based on literature precedent,49 we assigned the absolute configurations of (S)-dSpTP (triphosphate elutes first from C18 and anion exchange columns, nucleoside elutes second from HyperCarb column) and (R)-dSpTP (triphosphate elutes second from C18 and anion exchange columns, nucleoside elutes first from HyperCarb column).
Preparation of DNA Primer/Template Assemblies
The primer strand was radiolabeled at the 5′-end with 32P using T4 polynucleotide kinase and annealed to the template in a 1:1.5 ratio of primer:template in a buffer containing 10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT, pH 7.9 by heating the solution at 90 °C for 5 min and allowing it to slowly return to room temperature over approximately 2.5 h.
Qualitative Single Nucleotide Triphosphate Incorporation Reactions
Primer/template DNA was incubated with KF− for 5 min on ice where the final sample contained 5 nM primer/template DNA, 25 nM KF−, 10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT and 0.04 mg/mL BSA, pH 7.9. The incorporation reaction was initiated by addition of nucleotide triphosphate to yield a final concentration of 20 nM dGTP, 5 µM 8-oxodGTP, 100 µM (S)-dSpTP, or 100 µM (R)-dSpTP and the samples were incubated for 15 min at room temperature. The reactions were quenched by addition of gel loading buffer (80% formamide, 1 mg/mL xylene cyanol, 1 mg/ml bromophenol blue, and 10 mM EDTA), electrophoresed through a 16% denaturing PAGE gel, and imaged by phosphorimagery.
Primer Extension after the Incorporation of Sp Nucleotide Triphosphate
To examine whether primer extension will continue after the incorporation of the dSpTP nucleotide triphosphates, DNA with template X = C was used. In addition to 100 µM of dSpTP, 20 nM of both dATP and dCTP were added to the reaction while the rest of conditions are the same as described above for the qualitative single nucleotide incorporation experiment.
Single-Turnover Kinetics Reactions
Reactions were performed at 25 °C using a Rapid Quench Flow apparatus (RQF-3) from KinTek Corporation (Austin, TX). A solution consisting of 10 nM primer/template DNA, 50 nM KF−, 10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.08 mg/mL BSA, pH 7.9 was incubated for 5 min on ice. The incorporation reaction was initiated by adding various concentrations of each nucleotide triphosphate (listed in each figure legend) in buffer containing 10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT, pH 7.9. For each reaction in the RQF, 15 µL DNA·KF− and 15 µL dNTP were loaded into the reaction loops, rapidly mixed and incubated for the designated time before being quenched by 85 µL of 100 mM EDTA. The final reaction concentrations were 5 nM primer/template DNA, 25 nM KF−, dNTP (various concentrations), 10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.04 mg/mL BSA, pH 7.9. Gel loading buffer was added and the samples were analyzed by 16% denaturing PAGE gel and imaged by phosphorimagery.
Data Analysis
KinTek Explorer software (KinTek Corporation, Austin, TX) was used to globally fit the kinetic data.50, 51 The data were iteratively fit until a best fit was obtained according to a specific enzyme model. Because binding of a canonical nucleotide triphosphate to a polymerase is diffusion limited, k1 was fixed at 150 µM−1 s−1 for dGTP.52 However, for the oxidized and hyperoxidized nucleotide triphosphates 8-oxodGTP and dSpTP, k1 was fixed at 10 µM−1 s−1 due to their reduced binding.51 The lower and upper limits associated with each kinetic parameter were defined using the FitSpace function of KinTek Explorer. The best fit curves and experimental data were exported from KinTek Explorer and plotted in KaleidaGraph (Synergy software, Reading, PA).
RESULTS AND DISCUSSION
Qualitative Analysis of Incorporation of Spiroiminodihydantoin Nucleotide Triphosphate by Klenow Fragment
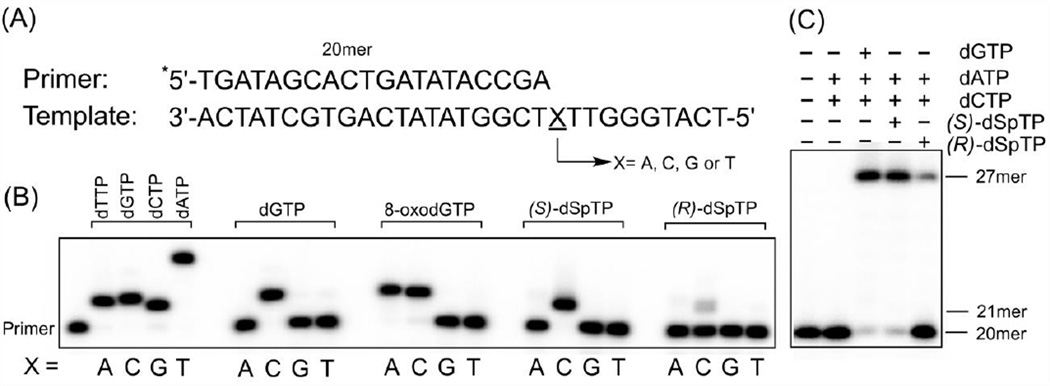
In order to study incorporation of the hyperoxidized nucleotide triphosphate dSpTP into DNA, templates were designed with either X = dA, dC, dG, or dT at the site of incorporation (Figure 2A). The diastereomers of dSpTP were resolved and examined separately. Primer extension reactions were catalyzed by KF− and were initiated by the addition of either dGTP, 8-oxodGTP, (S)-dSpTP, or (R)-dSpTP. The results are shown in Figure 2B. As expected, dGTP is incorporated exclusively opposite dC, and 8-oxodGTP is incorporated opposite both dC and dA.8 Both diastereomers of dSpTP are incorporated opposite dC, with more incorporation observed for (S)-dSpTP relative to (R)-dSpTP. Notably, in order to observe sufficient amounts of the extended primer product, the concentration of 8-oxodGTP was 250-fold greater than dGTP, and the concentration of the dSpTP diastereomers was 5,000-fold greater than dGTP. The experiments were also performed with dSpTP that was prepared using a different synthetic strategy (using 8-oxodGTP as the starting material47 rather than dGTP), and the same results were obtained (data not shown). Obtaining the same results with these different preparations of dSpTP indicates that these polymerase incorporation results are not due to trace contaminants of the dGTP starting material that may remain after synthesis and purification, but rather to dSpTP itself. That the product observed in Figure 2A corresponds to incorporation of (S)- and (R)-dSpTP rather than potential contaminating dGTP starting material is further supported by the difference in the migration between the single nucleotide incorporation products when dGTP or our preparation of dSpTP are added to the primer (Figure S3).
Figure 2.
(A) Sequence of primer and template strands. Primer was 32P-radiolabeled at 5'-end as indicated by the asterisk. (B) Primer extension on different template DNA (X = A, C, G or T) with either dGTP, 8-oxodGTP, (S)-dSpTP, or (R)-dSpTP. The five lanes on the left in panel B are the primer and the positive controls with correct nucleotide for each template DNA. (C) Primer extension after the incorporation of dSpTP. See Experimental Procedures for reaction conditions.
Having observed that both (S)-dSpTP and (R)-dSpTP can be incorporated into DNA from the nucleotide pool, we next examined whether the polymerase can extend beyond dSpTP or if incorporation of the hyperoxidized nucleotide would terminate replication. To avoid competition between dSpTP and dGTP, the dGTP was excluded from the reactions. Both dATP and dCTP were included and under these experimental conditions the full extension product would be a 27-mer. As shown in Figure 2C, KF− can extend past both diastereomers of dSpTP to yield the 27-mer product, suggesting that if incorporated from the nucleotide pool the lesions are not absolute blocks to replication. It is noteworthy that these qualitative experiments were performed using an excess of polymerase, and that under conditions limiting in polymerase stalling at the lesion site could occur.
In previous work, it was shown that when dSp is located in the template strand, KF− incorporates dATP and dGTP opposite the lesion both in vitro and in vivo.33–37 In fact, the correct base dCTP is almost never incorporated opposite dSp, meaning that the lesion is nearly 100% mutagenic if formed in genomic DNA. Based on these prior observations, we expected the dSpTP diastereomers to be incorporated into templates that contain X = dA and dG. Consistent with this logic, 8-oxodG is paired with dA and dC regardless of whether the lesion is in the template or is incorporated from the nucleotide pool.8, 33, 40, 42 Our observation that KF− only incorporates the dSpTP diastereomers opposite of dC in the template provides an intriguing example where a polymerase exhibits a different preference based on whether the lesion is in the template or in the nucleotide pool.
It is of note that because the hyperoxidized lesion is a dG-derived lesion, the incorporation of dSpTP opposite dC does not immediately represent a mutagenic event. However, in a subsequent round of replication the hyperoxidized lesion will serve as the template and in this context is known to be nearly 100% mutagenic. Thus, one could liken the dNTP form of the hyperoxidized lesion to a Trojan horse; the dNTP is incorporated into DNA by polymerase but will not exhibit mutagenicity until a G → C or G → T mutation occurs in the next round of replication.
It is possible to rationalize how a polymerase could use different base pairing for a lesion, depending on whether the lesion serves as the template or is incorporated from the nucleotide pool. Prior to making a covalent linkage between the primer and incoming nucleotide, a polymerase probes the template nucleobase for favorable electrostatic interactions. It has been proposed that for KF− the electrostatic profile is a significant determinant in nucleotide selectivity.53 Interestingly, the B-ring of the lowest energy tautomer of the diastereomers of dSpTP49 can form two of the same hydrogen bonds as the Watson-Crick face of dGTP (using a lone pair on the carbonyl and a hydrogen of the exocyclic amine) suggesting a means by which the hyperoxidized lesion pairs with dC. Other tautomers of dSpTP (i.e., the imino tautomer or the amino unconjugated tautomer49), although energetically less favorable, can form three hydrogen bonds with dC. Furthermore, a nucleotide triphosphate in solution has more conformational freedom than a nucleobase in a template strand because it is not confined by base stacking, base pairing, and covalent attachment to the sugar-phosphate backbone. This conformational freedom could allow incoming dSpTP to adopt the anti conformation and present the B-ring to base pair with dC in the template. However, characterizations of dSp have shown that the A- and B-rings are perpendicular to one another.54 Due to this structural constraint, when the lesion is located in the template, the A-ring of dSp could be presented for base pairing with an incoming nucleotide, which would explain the lack of pairing with dCTP when the lesion is in the template. Indeed, a recent crystal structure of (S)-dSp as the templating nucleobase in the active site of DNA polymerase β (pol β) reveals that the lesion is in the syn conformation and uses the A-ring as the base pairing face.54
While the information is not yet available for Sp, crystal structures of primer/template-polymerase-dNTP ternary complexes where dSp is either the templating base or dSpTP is the incoming nucleotide would greatly inform our results. Previous ternary structures of pol β bound to DNA containing 8-oxodG as the templating base or as the incoming 8-oxodGTP revealed the molecular basis for different base pairing. When 8-oxodG served as the templating base, the lesion was observed while base pairing with an incoming dCTP. Both 8-oxodG and dCTP were in the anti conformation and Watson-Crick base pairing was observed.55 In the ternary structure where 8-oxodGTP was the incoming nucleotide the templating base was dA.56 While dA remained anti the 8-oxodGTP was in the syn conformation. The syn conformation was stabilized by Hoogsteen hydrogen-bonding with the templating dA. In other work with human DNA polymerase η, which replicates a templating 8-oxodG in a nearly error-free manner, ternary complexes revealed that an Arg residue from a finger domain prevents formation of the 8-oxodG:A mispair.57 Similar structural studies would reveal the molecular basis for why a templating dSp base pairs with dATP and dGTP, whereas the nucleotide version dSpTP base pairs with a templating dC.
Kinetic Analyses of Nucleotide Triphosphate Incorporation
To evaluate the biological relevance of incorporation of dSpTP into DNA from the nucleotide pool, we performed experiments to compare the kinetic parameters kpol and Kd of the hyperoxidized nucleotide to dGTP and 8-oxodGTP. To avoid the complications that arise from multiple turnovers of a processive polymerase, i.e., the rate being dominated by dissociation of the DNA·KF− complex, the incorporation assays were performed under single-turnover conditions in which [KF−]>>[DNA]. Because single-turnover experiments using DNA polymerases occur rapidly and are too fast to be mixed and quenched manually, we used a rapid quench flow (RQF) apparatus for these experiments. Furthermore, because dGTP, 8-oxodGTP, (S)-dSpTP, and (R)-dSpTP share the ability to be incorporated opposite C, the kinetic analyses were performed using the template where X = dC.
Single nucleotide incorporation assays were performed using varying concentrations of dNTP in order to examine the concentration dependence of the rate of incorporation and amplitude of product formation. Under these conditions, the concentration dependence of the rate of incorporation reports kpol and Kd. kpol is the rate of the slowest step up to and including the phosphoryl transfer step in which the nucleotide is covalently attached to the primer, and Kd is the ground state nucleotide dissociation constant Kd. The DNA and KF− were pre-incubated prior to initiating the reaction by addition of dNTP. In this manner, the measured Kd is for nucleotide binding to the KF−·DNA complex rather than binding of KF− to the DNA. In this report, we use kpol/Kd to define the nucleotide incorporation specificity.58
Single-Turnover Kinetic Analysis of dGTP and 8-oxodGTP Incorporation
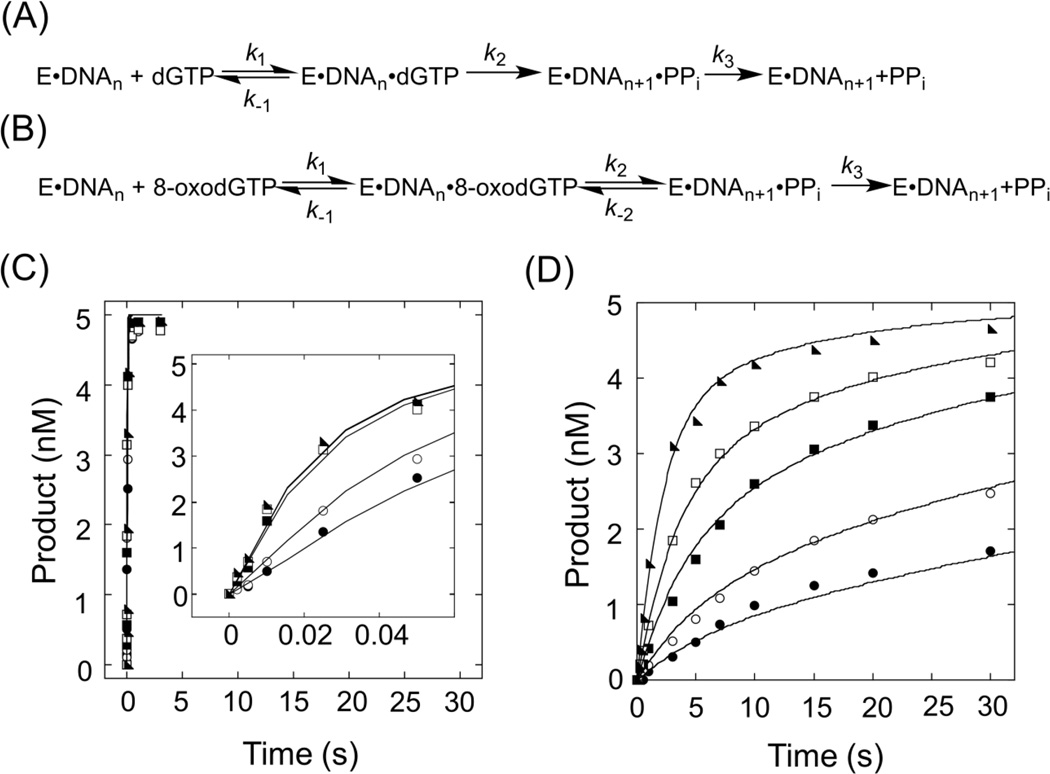
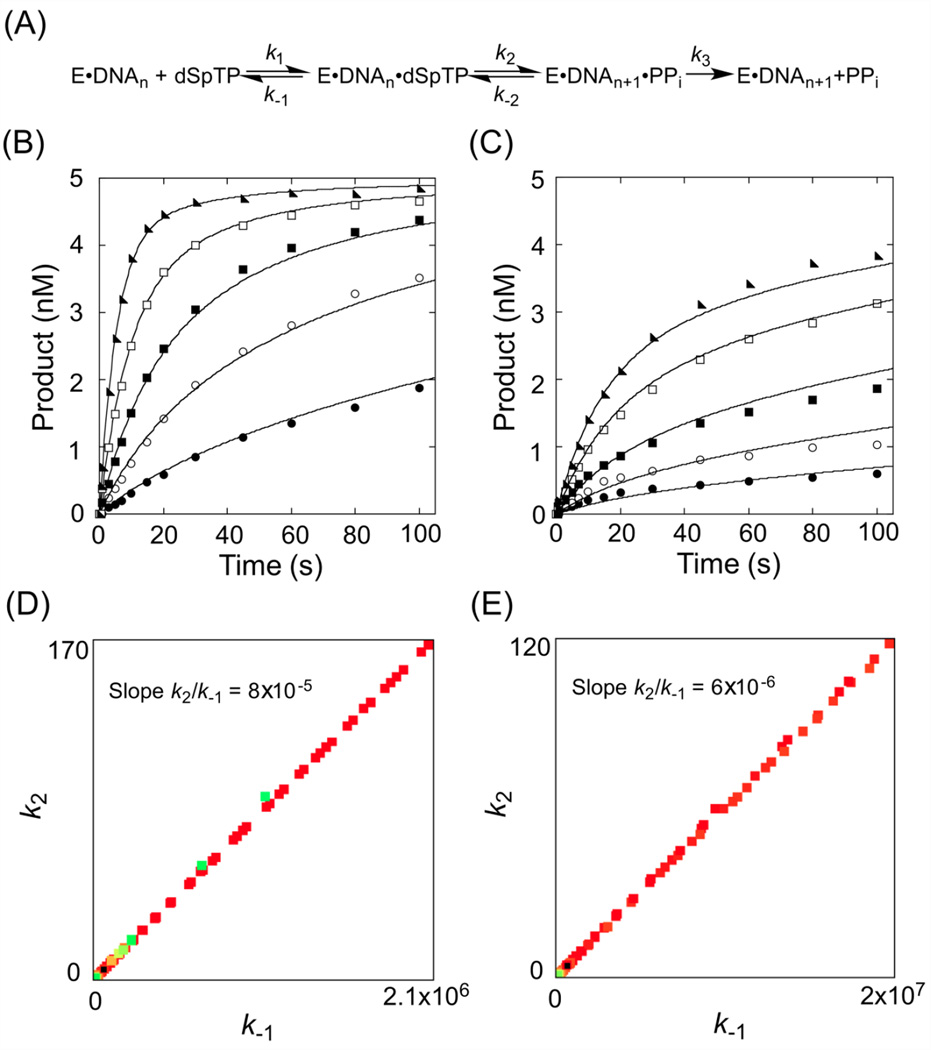
As the canonical nucleotide to incorporate opposite dC, dGTP was employed as a control for the incorporation experiments. A time course was performed at several dGTP concentrations and the data were fit globally to the model shown in Figure 3A. In this model Kd is defined by k−1/k1, k2 is kpol, and k3 is release of pyrophosphate. For KF− incorporating a canonical dNTP, it is generally accepted that kpol includes a rate limiting conformational change59 that controls nucleotide specificity,60 followed by phosphoryl transfer; however, other results suggest that the phosphoryl transfer step might limit the rate.61 It is also known that for incorporation of canonical nucleotides k3 is faster than k2.62 In our analysis, a Kd of 0.8 µM and kpol of 41 s−1 define a specificity constant kpol/Kd (or k2/Kd) of 51 µM−1 s−1 for the incorporation of dGTP opposite dC (Table 1). It is notable that the final amplitude of 21-mer product is the same at each concentration of dGTP and the slope of the rise converge with increasing concentration (Figure 3C). These observations show that kpol is fast and there are no slower steps after incorporation that would cause incorporation to be reversible. Both of these conditions are accounted for in the model in Figure 3A.
Figure 3.
Kinetic analysis of dGTP and 8-oxodGTP incorporation by KF−. Enzyme model used for (A) dGTP and (B) 8-oxodGTP. (C) Global fitting of incorporation of dGTP opposite of template dC. The concentrations of dGTP are 0.5 µM (closed circles), 1 µM (open circles), 10 µM (closed squares), 50 µM (open squares), and 100 µM (closed triangles). (D) Global fitting of incorporation of 8-oxodGTP opposite of template dC. The concentrations of 8-oxodGTP are 1 µM (closed circles), 2 µM (open circles), 5 µM (closed squares), 10 µM (open squares), and 50 µM (closed triangles). The curves superimposed with the experimental data were generated by KinTek Explorer fitting.
Table 1.
Kinetic Parameters for the Incorporation of dNTPs Opposite Template dC.
| Nucleotide | Kd (µM) | k2 (s−1) | k2/Kd (µM−1 s−1) | selection factora |
|---|---|---|---|---|
| dGTP | 0.8 | 41 | 51 | 1 |
| 8-oxodGTP | 15.2 | 0.45 | 0.030 | 1,700 |
| (S)-dSpTP | 1,700b | 1.4b | 8×10−4 | 64,000 |
| (R)-dSpTP | 7,000b | 0.45b | 6×10−5 | 850,000 |
Selection factor is the ratio of the specificity of incorporation (k2/Kd) of dGTP relative to the dNTP of interest.
Values are the lower bound provided by KinTek Explorer global fitting.
In the case of 8-oxodGTP incorporation opposite of dC, the single-turnover plots at each nucleotide concentration are noticeably biphasic (Figure 3D). Additionally, the amplitude is dependent on nucleotide concentration suggesting that after incorporation there is a slow step that allows phosphoryl transfer to be reversible. Indeed, similar results were obtained for the incorporation of 8-oxodGTP opposite dC by the human mitochondrial DNA polymerase.42 In accordance with Hanes and co-workers,42 we accounted for the differences observed for incorporation of 8-oxodGTP relative to dGTP by fitting the data to the model shown in Figure 3B, which allows for reversibility of the phosphoryl transfer. A Kd of 15.2 µM describes nucleotide binding while the incorporation step has a forward rate (k2) of 0.45 s−1 and a reverse rate (k−2) of 0.08 s−1. In contrast to canonical nucleotides, for modified nucleotides it is not known whether k2 is limited by the conformational change, but it has been shown that when KF incorporates a mismatched nucleotide, the rate of phosphoryl transfer is not limited by the preceding conformational change.63 Pyrophosphate release is indeed slower than incorporation and occurs at a rate of 0.09 s−1, which allows k2 to be reversible. Therefore, unlike for dGTP, kpol ≠ k2. When taking into account the slow pyrophosphate release, the kpol/Kd value (0.016 µM−1 s−1, where kpol = k2k3/(k−2+k3))42 is slightly smaller than the k2/Kd (0.03 µM−1 s−1) for 8-oxodGTP. Interestingly, it has been proposed that slow pyrophosphate release is an additional factor that limits incorporation of 8-oxodGTP by mitochondrial polymerase.42
Single-Turnover Kinetic Analysis of dSpTP Incorporation
Compared to dGTP and 8-oxodGTP, experiments using the diastereomers of dSpTP required significantly higher nucleotide triphosphate concentrations and extended incubation times to observe a sufficient amount of product for analysis (Figure 4B, 4C). We also observed differences in incorporation between the two dSpTP diastereomers. Specifically, the amount of product accumulated after a particular reaction time for (S)-dSpTP is significantly greater than for (R)-dSpTP at the same nucleotide concentration, consistent with the results of the qualitative incorporation presented in Figure 2B. Global fitting of the data to the models in Figure 3A and 3B reveal that the data are described better by a model (Figure 4A) that allows reversibility of the incorporation step (data not shown), similar to the results observed for 8-oxodGTP. However, unlike what we observed for incorporation of dGTP or 8-oxodGTP, for the diastereomers of dSpTP the parameters in the kinetic scheme were not well constrained (Figure 4D, E and Figure S4). As a consequence, we could not obtain individual quantitative values for Kd and kpol. Because kpol is limited by the amount of available DNA·KF−·dNTP complex, poor binding of the hyperoxidized nucleotide triphosphate to the DNA·KF− complex likely causes the lack of parameter constraint. Importantly, these data indicate that the Kd for both diastereomers of dSpTP is greater than the nucleotide concentrations used here and these values are at least on the millimolar scale. Even though k−1 and k2 are not constrained and can vary across a wide range for both diastereomers of dSpTP, analysis by KinTek Explorer FitSpace reveals there is a linear correlation between k2 and k−1 (Figure 4D, E). Since k1 was fixed at 10 µM−1 s−1 during the fitting (see Experimental Procedures), there is also a linear correlation between k2 and Kd (Kd = k−1/k1 = k−1/10). Therefore we use k2/Kd to compare the incorporation specificities of the dNTPs used in this study, rather than kpol/Kd. From the slope of FitSpace analysis k2/k−1 (or k2/(10×Kd)), we defined the incorporation specificity k2/Kd for (S)-dSpTP and (R)-dSpTP as 8×10−4 µM−1 s−1 and 6×10−5 µM−1 s−1, respectively.
Figure 4.
Kinetic analysis of (S)-dSpTP and (R)-dSpTP incorporation by KF−. (A) Enzyme model used for global fitting. (B) Global fitting of incorporation of (S)-dSpTP opposite of template dC. (S)-dSpTP concentrations are 10 µM (closed circles), 20 µM (open circles), 50 µM (closed squares), 100 µM (open squares), and 200 µM (closed triangles). (C) Global fitting of incorporation of (R)-dSpTP opposite of template dC. (R)-dSpTP concentrations are 50 µM (closed circles), 100 µM (open circles), 200 µM (closed squares), 400 µM (open squares), and 600 µM (closed triangles). The curves superimposed with the experimental data were generated by KinTek Explorer fitting. FitSpace analysis by KinTek Explorer showing the results of the initial excursions to map the boundaries of a good fit for (D) (S)-dSpTP and (E) (R)-dSpTP.
Our kinetic results demonstrate that KF− exhibits a stereochemical preference and more readily incorporates (S)-dSpTP over (R)-dSpTP. A general stereochemical preference for the (S) diastereomer has been reported previously for several other DNA-processing enzymes. The Burrows laboratory showed that KF− was more efficient at inserting dATP opposite dSp1 relative to dSp2, where the diastereomers were first described based on their order of elution from an anion exchange HPLC column.34 Subsequent studies revealed that dSp1 and dSp2 are (S)-dSp and (R)-dSp, respectively.48, 49 The Essigmann laboratory similarly showed that pol II and pol IV more efficiently bypass dSp1 in vivo64 and that the relative amount of G → C and G → T transversions can vary for dSp1 and dSp2 depending on the sequence context.35–37 Furthermore, nuclease P165 and the BER glycosylase human NEIL128, 30, 32 also have a preference for the dSp1 configuration. Molecular dynamics simulations showed that hNEIL1 makes better contacts with (S)-dSp in its binding pocket, justifying the stereochemical preference of the enzyme.66
Comparing the Kinetic Parameters for dGTP, 8-oxodGTP, (S)-dSpTP, and (R)-dSpTP
Our results show that the extent of oxidation of the dGTP-derived lesion is correlated with its ability to be incorporated into DNA. The specificity of incorporation is highest for dGTP opposite dC by KF−. Compared to dGTP, the selection factors for incorporating 8-oxodGTP, (S)-dSpTP, and (R)-dSpTP are 1,700-, 64,000- and 850,000-fold lower respectively (Table 1). Notably, since pyrophosphate release is slow for 8-oxodGTP, (S)-dSpTP, and (R)-dSpTP, kpol/Kd must be smaller than k2/Kd, so these selection factors are lower limits for specificity.
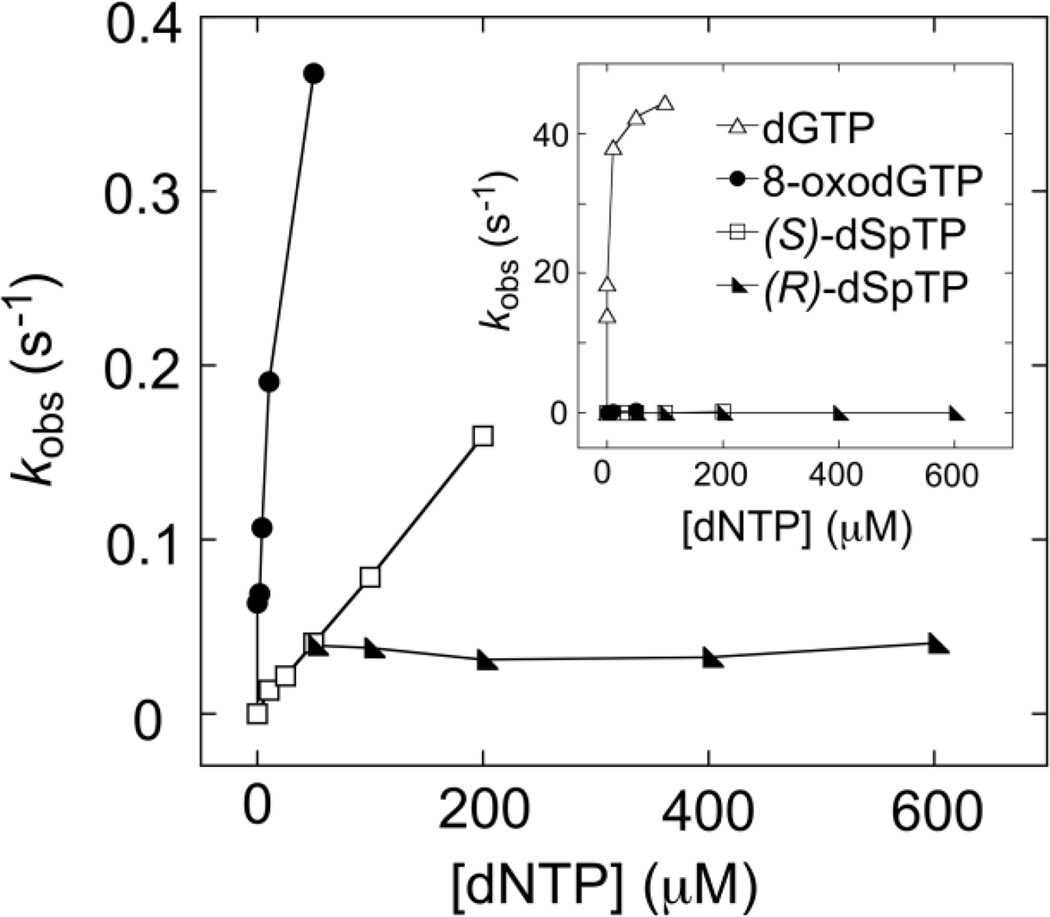
In order to provide a graphical representation of the difference in specificity for incorporation of the canonical, oxidized, and hyperoxidized guanine lesions we also analyzed our kinetic data using traditional Michaelis-Menten methods, in which the observed rate (kobs) is plotted against nucleotide triphosphate concentration (Figure 5). The slope of the rise can be used to define incorporation specificity kpol/Kd. In fact, the kpol/Kd from this Michaelis-Menten analysis (data not shown) is comparable to the k2/Kd obtained by global fitting. Clearly, the incorporation kinetics for dGTP are significantly different from those of oxidized nucleotides (Figure 5 inset, open triangles). For the lesion dNTPs, a rise in kobs as a function of dNTP concentrations is observed for 8-oxodGTP and (S)-dSpTP, with 8-oxodGTP more steep than (S)-dSpTP. However, for (R)-dSpTP, kobs was extremely slow across the entire concentration range, and there was no obvious dependence of the rate on nucleotide concentration. These results are entirely consistent with the quantitative parameters obtained by global fitting.
Figure 5.
Analysis of dGTP (open triangles), 8-oxodGTP (closed circles), (S)-dSpTP (open squares), and (R)-dSpTP (closed triangles) kinetic results using Michaelis-Menten techniques where the observed rates are plotted against nucleotide concentrations. The insert is a zoomed out view of the main panel and allows for comparison of dGTP to the three oxidized nucleotides.
Biological Considerations
In addition to the kinetic parameters describing incorporation of lesion dNTPs, one must also consider the action of enzymes responsible for cleansing the nucleotide pool. The importance of removing 8-oxodGTP from the nucleotide pool is underscored by the fact that E. coli lacking MutT, a phosphatase that converts 8-oxodGTP to 8-oxodGMP,39 have a 100- to 10,000-fold higher mutation rate compared to wild type E. coli.67 This dramatic increase in mutation rate in the absence of MutT indicates that the nucleotide pool represents a biologically significant source of 8-oxodGTP.68, 69 However, it has been demonstrated that the diastereomers of dSpTP are not good substrates for MutT or other known MutT-type nucleotide pool sanitization enzymes in E. coli.70 Therefore, preventing the incorporation of dSpTP into DNA may depend solely on the ability of a polymerase to discriminate the hyperoxidized nucleotide triphosphate from dGTP. Indeed, consistent with our kinetic results, in vivo studies demonstrate that introduction of dSpTP1 or dSpTP2 into E. coli cells does not significantly increase the mutation frequency.70 This result is in contrast to the high mutagenicity observed in E. coli when the Sp lesions are in the DNA template.35–37, 64 Our results suggest that the lack of mutagenicity of dSpTP is because the E. coli cellular polymerases effectively discriminate the hyperoxidized nucleotides and do not use them as building blocks during replication. It is notable, however, that although it is difficult to force dSpTP into DNA from the nucleotide pool, if incorporation occurs, the lesion is a powerful source of mutations. Indeed, the “Trojan horse” quality of dSpTP makes it an intriguing player in the field of lethal mutagenesis71, 72 where a miscoding nucleotide is used as an antiviral agent to accelerate viral mutation rates and drive a viral population to extinction. In order to be effective in this manner, however, the incorporation efficiency of dSpTP would likely need to be higher than that observed for KF−.
In this work, we used KF− as a model polymerase. Similar incorporation efficiencies have been observed for KF− and some mammalian replicative polymerases incorporating 8-oxodGTP into DNA,73 although mammalian polymerases have a higher preference to incorporate 8-oxodGTP opposite the correct dC instead of the mismatched dA template.74 Therefore, we expect the selection factor for mammalian polymerases incorporating the hyperoxidized dNTP to be even more rigorous than KF− and to prevent incorporation of dSpTP from the nucleotide pool.
As a final consideration the in vivo concentrations of (S)-dSpTP and (R)-dSpTP will also influence the extent to which the hyperoxidized lesion can be incorporated into DNA. These values remain to be determined but are most certainly well below the millimolar Kd values suggested by our experiments. Thus, we conclude that while dSpTP can be described as a Trojan horse, the specificity of cellular polymerase provides an effective defense against using the hyperoxidized nucleotide as a building block during DNA replication, and that the dSp lesion is a much more potent mutagen when it is formed in genomic DNA.
Supplementary Material
Acknowledgments
FUNDING SOURCES
This work was supported by an Institutional Development Award (IDeA) from the National Institutes of Health National Center for Research Resources (NIH/NCRR) (P20RR016457).
We thank Prof. Kenneth Johnson (Univ. of Texas, Austin) for helpful discussions including data analysis using KinTek Explorer software. We also thank Dr. Eric Olmon, Ms. Katharina Bilotti, and Ms. Erin Kennedy for helpful discussion.
ABBREVIATIONS
- 8-oxodGTP
2'-deoxy-8-oxo-7,8-dihydroguanosine-5'-triphosphate
- BER
base excision repair
- dGTP
2'-deoxyguanosine-5'-triphosphate
- dSpTP
spiroiminodihydantoin-2'-deoxynucleoside-5'-triphosphate
- ESI-MS
electrospray ionization mass spectrometry
- KF−
Klenow Fragment 3'→5' exo−
- RQF
rapid quench flow
- ROS
reactive oxygen species
Footnotes
SUPPORTING INFORMATION AVAILABLE
HPLC and ESI-MS characterization of the Sp nucleosides and nucleotide triphosphates, denaturing PAGE characterization of different migration for single nucleotide incorporation product of either dGTP or (S)-dSpTP, 3-D confidence contours from KinTek Explorer analysis for dGTP and 8-oxodGTP. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Gates KS. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadet J, Douki T, Ravanat JL. Oxidatively generated base damage to cellular DNA. Free Radical Biol. Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 3.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 4.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 5.Cadet J. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Delaney S, Jarem DA, Volle CB, Yennie CJ. Chemical and biological consequences of oxidatively damaged guanine in DNA. Free Radical Res. 2012;46:420–441. doi: 10.3109/10715762.2011.653968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moller P, Cooke MS, Collins A, Olinski R, Rozalski R, Loft S. Harmonising measurements of 8-oxo-7,8-dihydro-2'-deoxyguanosine in cellular DNA and urine. Free Radical Res. 2012;46:541–553. doi: 10.3109/10715762.2011.644241. [DOI] [PubMed] [Google Scholar]
- 8.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 9.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J. Biol. Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 10.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 11.Klein JC, Bleeker MJ, Saris CP, Roelen HC, Brugghe HF, van den Elst H, van der Marel GA, van Boom JH, Westra JG, Kriek E, et al. Repair and replication of plasmids with site-specific 8-oxodG and 8-AAFdG residues in normal and repair-deficient human cells. Nucleic Acids Res. 1992;20:4437–4443. doi: 10.1093/nar/20.17.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steenken S, Jovanovic SV, Bietti M, Bernhard K. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2'deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J. Am. Chem. Soc. 2000;122:2373–2374. [Google Scholar]
- 14.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org. Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 15.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem. Res. Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 16.Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin and guanidinohydantoin are the dominant products of 8-oxoguanosine oxidation at low fluxes of peroxynitrite: mechanistic studies with 18O. Chem. Res. Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- 17.Munk BH, Burrows CJ, Schlegel HB. An exploration of mechanisms for the transformation of 8-oxoguanine to guanidinohydantoin and spiroiminodihydantoin by density functional theory. J. Am. Chem. Soc. 2008;130:5245–5256. doi: 10.1021/ja7104448. [DOI] [PubMed] [Google Scholar]
- 18.Fleming AM, Muller JG, Dlouhy AC, Burrows CJ. Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2'-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing. J. Am. Chem. Soc. 2012;134:15091–15102. doi: 10.1021/ja306077b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rokhlenko Y, Geacintov NE, Shafirovich V. Lifetimes and reaction pathways of guanine radical cations and neutral guanine radicals in an oligonucleotide in aqueous solutions. J. Am. Chem. Soc. 2012;134:4955–4962. doi: 10.1021/ja212186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui L, Ye W, Prestwich EG, Wishnok JS, Taghizadeh K, Dedon PC, Tannenbaum SR. Comparative analysis of four oxidized guanine lesions from reactions of DNA with peroxynitrite, singlet oxygen, and γ-radiation. Chem. Res. Toxicol. 2013;26:195–202. doi: 10.1021/tx300294d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem. Res. Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 24.Hazra TK, Muller JG, Manuel RC, Burrows CJ, Lloyd RS, Mitra S. Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 2001;29:1967–1974. doi: 10.1093/nar/29.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnamurthy N, Muller JG, Burrows CJ, David SS. Unusual structural features of hydantoin lesions translate into efficient recognition by Escherichia coli Fpg. Biochemistry. 2007;46:9355–9365. doi: 10.1021/bi602459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Recognition and removal of oxidized guanines in duplex DNA by the base excision repair enzymes hOGG1, yOGG1, and yOGG2. Biochemistry. 2003;42:11373–11381. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- 27.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair. 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20715–20719. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair. 2007;6:1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, David SS. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J. Am. Chem. Soc. 2013;135:13851–13861. doi: 10.1021/ja4059469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow Fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 35.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 36.Delaney S, Delaney JC, Essigmann JM. Chemical-biological fingerprinting: probing the properties of DNA lesions formed by peroxynitrite. Chem. Res. Toxicol. 2007;20:1718–1729. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 38.Kamiya H, Kasai H. Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steady-state kinetics of the incorporation. J. Biol. Chem. 1995;270:19446–19450. doi: 10.1074/jbc.270.33.19446. [DOI] [PubMed] [Google Scholar]
- 39.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 40.Pavlov YI, Minnick DT, Izuta S, Kunkel TA. DNA replication fidelity with 8-oxodeoxyguanosine triphosphate. Biochemistry. 1994;33:4695–4701. doi: 10.1021/bi00181a029. [DOI] [PubMed] [Google Scholar]
- 41.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase δ. Steady-state and pre-steady-state kinetic analysis. J. Biol. Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 42.Hanes JW, Thal DM, Johnson KA. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J. Biol. Chem. 2006;281:36241–36248. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 43.Brown JA, Duym WW, Fowler JD, Suo Z. Single-turnover kinetic analysis of the mutagenic potential of 8-oxo-7,8-dihydro-2'-deoxyguanosine during gap-filling synthesis catalyzed by human DNA polymerases λ and β. J. Mol. Biol. 2007;367:1258–1269. doi: 10.1016/j.jmb.2007.01.069. [DOI] [PubMed] [Google Scholar]
- 44.Jarem DA, Wilson NR, Delaney S. Structure-dependent DNA damage and repair in a trinucleotide repeat sequence. Biochemistry. 2009;48:6655–6663. doi: 10.1021/bi9007403. [DOI] [PubMed] [Google Scholar]
- 45.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 46.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye Y, Muller JG, Burrows CJ. Synthesis and characterization of the oxidized dGTP lesions spiroiminodihydantoin-2'-deoxynucleoside-5'-triphosphate and guanidinohydantoin-2'-deoxynucleoside-5'- triphosphate. J. Org. Chem. 2006;71:2181–2184. doi: 10.1021/jo052484t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karwowski B, Dupeyrat F, Bardet M, Ravanat JL, Krajewski P, Cadet J. Nuclear magnetic resonance studies of the 4R and 4S diastereomers of spiroiminodihydantoin 2'-deoxyribonucleosides: absolute configuration and conformational features. Chem. Res. Toxicol. 2006;19:1357–1365. doi: 10.1021/tx060088f. [DOI] [PubMed] [Google Scholar]
- 49.Fleming AM, Orendt AM, He Y, Zhu J, Dukor RK, Burrows CJ. Reconciliation of chemical, enzymatic, spectroscopic and computational data to assign the absolute configuration of the DNA base lesion spiroiminodihydantoin. J. Am. Chem. Soc. 2013;135:18191–18204. doi: 10.1021/ja409254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson KA, Simpson ZB, Blom T. FitSpace explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal. Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 51.Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 52.Johnson KA. Chapter 23 Fitting enzyme kinetic data with KinTek global kinetic explorer. Methods Enzymol. 2009;467:601–626. doi: 10.1016/S0076-6879(09)67023-3. [DOI] [PubMed] [Google Scholar]
- 53.Paul N, Nashine VC, Hoops G, Zhang P, Zhou J, Bergstrom DE, Davisson VJ. DNA polymerase template interactions probed by degenerate isosteric nucleobase analogs. Chem. Biol. 2003;10:815–825. doi: 10.1016/j.chembiol.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Eckenroth BE, Fleming AM, Sweasy JB, Burrows CJ, Doublie S. Crystal structure of DNA polymerase beta with DNA containing the base lesion spiroiminodihydantoin in a templating position. Biochemistry. 2014;53:2075–2077. doi: 10.1021/bi500270e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batra VK, Shock DD, Beard WA, McKenna CE, Wilson SH. Binary complex crystal structure of DNA polymerase β reveals multiple conformations of the templating 8-oxoguanine lesion. Proc. Natl. Acad. Sci. U. S. A. 2012;109:113–118. doi: 10.1073/pnas.1112235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batra VK, Beard WA, Hou EW, Pedersen LC, Prasad R, Wilson SH. Mutagenic conformation of 8-oxo-7,8-dihydro-2'-dGTP in the confines of a DNA polymerase active site. Nat. Struct. Mol. Biol. 2010;17:889–890. doi: 10.1038/nsmb.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patra A, Nagy LD, Zhang Q, Su Y, Muller L, Guengerich FP, Egli M. Kinetics, structure, and mechanism of 8-oxo-7,8-dihydro-2'-deoxyguanosine bypass by human DNA polymerase η. J. Biol. Chem. 2014;289:16867–16882. doi: 10.1074/jbc.M114.551820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee HR, Helquist SA, Kool ET, Johnson KA. Importance of hydrogen bonding for efficiency and specificity of the human mitochondrial DNA polymerase. J. Biol. Chem. 2008;283:14402–14410. doi: 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 60.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bakhtina M, Roettger MP, Kumar S, Tsai MD. A unified kinetic mechanism applicable to multiple DNA polymerases. Biochemistry. 2007;46:5463–5472. doi: 10.1021/bi700084w. [DOI] [PubMed] [Google Scholar]
- 62.Hanes JW, Johnson KA. A novel mechanism of selectivity against AZT by the human mitochondrial DNA polymerase. Nucleic Acids Res. 2007;35:6973–6983. doi: 10.1093/nar/gkm695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eger BT, Benkovic SJ. The minimal kinetic mechanism for misincorporation by DNA polymerase I (Klenow fragment) Biochemistry. 1992;31:9227–9236. doi: 10.1021/bi00153a016. [DOI] [PubMed] [Google Scholar]
- 64.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 65.Chen X, Fleming AM, Muller JG, Burrows CJ. Endonuclease and exonuclease activities on oligodeoxynucleotides containing spiroiminodihydantoin depend on the sequence context and the lesion stereochemistry. New J. Chem. 2013;37:3440–3449. doi: 10.1039/C3NJ00418J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia L, Shafirovich V, Geacintov NE, Broyde S. Lesion specificity in the base excision repair enzyme hNeil1: modeling and dynamics studies. Biochemistry. 2007;46:5305–5314. doi: 10.1021/bi062269m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yanofsky C, Cox EC, Horn V. The unusual mutagenic specificity of an E. coli mutator gene. Proc. Natl. Acad. Sci. U. S. A. 1966;55:274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tajiri T, Maki H, Sekiguchi M. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res., DNA Repair. 1995;336:257–267. doi: 10.1016/0921-8777(94)00062-b. [DOI] [PubMed] [Google Scholar]
- 69.Kamiya H, Ishiguro C, Harashima H. Increased A:T→C:G mutations in the mutT strain upon 8-hydroxy-dGTP treatment: direct evidence for MutT involvement in the prevention of mutations by oxidized dGTP. J. Biochem. 2004;136:359–362. doi: 10.1093/jb/mvh132. [DOI] [PubMed] [Google Scholar]
- 70.Hori M, Suzuki T, Minakawa N, Matsuda A, Harashima H, Kamiya H. Mutagenicity of secondary oxidation products of 8-oxo-7,8-dihydro-2'-deoxyguanosine 5'-triphosphate (8-hydroxy-2'- deoxyguanosine 5'-triphosphate) Mutat. Res. 2011;714:11–16. doi: 10.1016/j.mrfmmm.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 71.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perales C, Martin V, Domingo E. Lethal mutagenesis of viruses. Curr. Opin. Virol. 2011;1:419–422. doi: 10.1016/j.coviro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 73.Macpherson P, Barone F, Maga G, Mazzei F, Karran P, Bignami M. 8-oxoguanine incorporation into DNA repeats in vitro and mismatch recognition by MutSα. Nucleic Acids Res. 2005;33:5094–5105. doi: 10.1093/nar/gki813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katafuchi A, Nohmi T. DNA polymerases involved in the incorporation of oxidized nucleotides into DNA: their efficiency and template base preference. Mutat. Res. 2010;703:24–31. doi: 10.1016/j.mrgentox.2010.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.