Abstract
Inhibitors of Apoptosis (IAP) family of genes encode BIR domain containing proteins with anti-apoptotic function. These proteins also contain RING or UBC domains and act by binding to major pro-apoptotic factors and ubiquitylating them. High levels of IAPs inhibit caspase-mediated apoptosis. For these cells to undergo apoptosis, IAP function must be neutralized by IAP-antagonists. Mammalian IAP knockouts do not exhibit obvious developmental phenotypes, but the cells are more sensitized to apoptosis in response to injury. Loss of the mammalian IAP-antagonist ARTS results in deficient stem cell apoptosis. In addition to the anti-apoptotic properties, IAPs regulate the innate immune response, and the loss of IAP function in humans is associated with immunodeficiency. The roles of IAPs in Drosophila apoptosis regulation is more apparent, where the loss of IAP1, or the expression of IAP antagonists in Drosophila cells, is sufficient to trigger apoptosis. In this organism, apoptosis as a fate is conferred by the transcriptional induction of the IAP antagonists. Many signaling pathways often converge on shared enhancer regions of IAP-antagonists. Cell death sensitivity is further regulated by post-transcriptional mechanisms, including those regulated by kinases, miRNAs and ubiquitin ligases. These mechanisms are employed to eliminate damaged or virus-infected cells, limit neuroblast (neural stem cell) numbers, generate neuronal diversity and sculpt tissue morphogenesis.
Keywords: Apoptosis, IAP-antagonist, REAPER, HID, ARTS, SMAC, DIAP1, XIAP, c-IAP1, BRUCE
Introduction
Apoptosis is one of the best-understood forms of cell death that is regulated through a combination of positive and negative factors. Among the negative regulators are anti-apoptotic proteins that share the Baculovirus IAP-Repeat (BIR) domains, which are now widely referred to as Inhibitors of Apoptosis (IAP) family of proteins.
The name of the BIR domain originates from the shared sequence of viral inhibitors of apoptosis found in the functional homologs of baculovirus p35, Cp-IAP and Op-IAP (Birnbaum, 1994; Clem, 1991, 1994; Crook, 1993). The family now includes X-linked IAP (XIAP), c-IAP1 and 2, Drosophila IAP1 and 2 (DIAP1 and 2), and BRUCE (BIR domain containing Ubiquitin Conjugating Enzyme) (Figure 1). Not all BIR domain-containing proteins regulate cell death, and certain BIR domain proteins are dedicated to the regulation of mitosis (Silke, 2001). The anti-apoptotic BIR domain proteins found in Drosophila and vertebrates mostly have C-terminal RING domains that have ubiquitin ligase activities (Yang, 2000). One exception to this is BRUCE, a potent anti-apoptotic protein that contains an Ubiquitin Conjugating Enzyme (UBC) motif instead of RING. These IAPs bind and ubiquitylate major pro-apoptotic proteins to exert their anti-apoptotic function. In addition, they are actively regulated in cells by their inhibitory molecules, referred to as IAP-antagonists. In this review, we will discuss the latest advances in the field, focusing on the roles of IAPs and their antagonists during animal development.
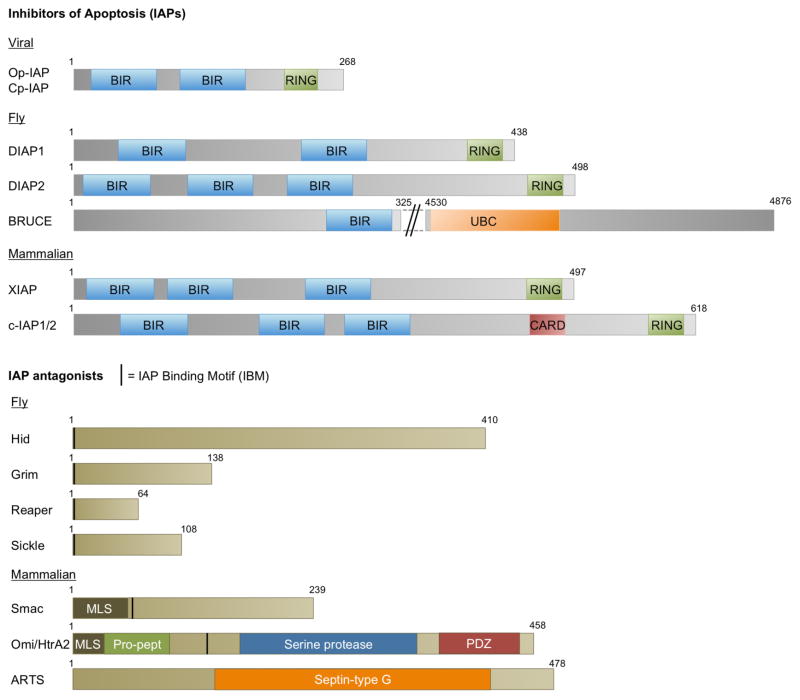
Figure 1. Domain maps of IAPs and their antagonists from various model systems.
All known IAPs contain at least one Baculovirus IAP-Repeat (BIR) domain. In addition, most have RING domains. BRUCE is the largest IAP (as indicated by the break in sequence in figure) and an exception in that it does not contain a RING domain but instead has an Ubiqutin Conjugation domain (UBC). Most IAP-antagonists contain a short 5–10 amino acid IAP-Binding Motif (IBM) at their N-terminii, usually immediately after the Methionine, which is cleaved to expose the IBM. The mammalian IAP antagonists, Smac, ARTS and Omi/HtrA2, localize to the mitochondria for their function and hence contain a Mitochondria Localization Sequence (MLS) amongst other domains. ARTS belongs is a non-canonical IAP-antagonist that does not have an N-terminal IBM, and instead uses the C-terminal sequences to bind IAPs. Domain maps to scale, source: www.uniprot.org.
IAP/antagonist interaction
In many cells, IAPs bind and inhibit active caspases to exert their anti-apoptotic function (Devereaux, 1997; Wang 1999; Goyal 2000). Caspases gain full catalytic activity after being proteolytically cleaved, so that the resulting small and large subunits of caspases can assemble to form active catalytic sites. IAPs can inhibit such proteolytically activated caspases (Srinivasula, 2001; Muro, 2002; Shapiro, 2008), and therefore, high levels of IAPs can block apoptosis at the last stage. However, cells with high levels of IAPs can undergo caspase-mediated apoptosis, if IAP antagonizing molecules are around to neutralize IAP function. The so-called IAP-antagonists were first discovered in Drosophila, and remains best characterized in this organism. Reflecting their important role in cell killing, these IAP-antagonists were named grim, reaper, hid and sickle (Chen, 1996; Christich, 2002; Grether, 1995; Srinivasula, 2002; White, 1994; Wing, 2002).
IAP-antagonists play particularly visible roles in Drosophila apoptosis regulation: Virtually all apoptosis is abolished in the absence of these genes, whereas their overexpression is sufficient to kill cells (White, 1994; Chen, 1996; Grether, 1995; White, 1996). Genetic interaction screens have identified DIAP1, DIAP2 and BRUCE as downstream targets (Hay, 1995; Wang, 1999; Goyal, 2000; Lisi, 2000; Vernooy, 2002; Arama, 2003). In living cells of Drosophila, DIAP1 normally inhibits both, initiator and effector caspases (Hawkins, 1999; Meier, 2000; Yan, 2004; Tenev, 2005). DIAP1 uses its ubiquitin ligase activity to directly ubiquitylate the initiator caspase DRONC (Lee, 2011; Ryoo, 2004; Wilson, 2002) and also helps to destabilize the upstream adaptor, the apoptosome holoenzyme, which is a protein complex that serves to activate DRONC (Akdemir, 2006; Shapiro, 2008). DIAP1 is a key player as demonstrated by the observation that virtually all somatic cells undergo apoptosis in diap1 mutant embryos (Goyal, 2000; Lisi, 2000; Wang, 1999). DIAP2 has a more confined role in inhibiting a specific effector caspase (Ribeiro, 2007), and while overexpression of DIAP2 can inhibit IAP-antagonist-induced apoptosis (Hay, 1995), the loss of this gene does not show the dramatic apoptosis phenotype as seen in diap1 mutants (Huh, 2007; Ribeiro, 2007). BRUCE is also a potent anti-apoptotic gene, and this protein exerts its effect by using its UBC domain to ubiquitylate IAP-antagonists for proteasomal degradation (Arama, 2003; Bartke, 2004; Domingues, 2012; Hao, 2004; Vernooy, 2002). Mammalian IAP antagonists, Smac and Omi/HtrA2, were also identified based on its ability to physically bind to XIAP (Du, 2000; Verhagen, 2000). However, mouse genetics studies indicate that IAP antagonists primarily target c-IAP1 in vivo (Vince, 2007).
IAP-antagonists share a conserved N-terminal 4 – 8 residues that directly bind to a groove within the IAP BIR domain, allowing caspases to be liberated from IAPs (Wu, 2000; Wu, 2001). Furthermore, they promote the auto-ubiquitination and degradation of IAPs (Li, 2011; Ryoo, 2002; Yoo, 2002). Notable in this interaction is the fact that the first methionine of the N-terminal IAP binding motif must be lost, and the new N-terminus must start with an alanine residue, in order to fit into an IAP BIR groove (Wu, 2000).
Mitochondrial Association of IAP Antagonists
How can cells make peptides that do not start with an N-terminal methionine residue? In mammals, the IAP antagonist Smac encodes an N-terminal mitochondrial localization motif followed by an IAP-binding motif that is similar to the N-terminal residues of GRIM, REAPER, HID and SICKLE (Du, 2000; Verhagen, 2000) (Figure 1). Omi/HtrA2 is a mitochondrial protease that has a similar IAP-binding motif (Hegde, 2002; Martins, 2002; van Loo, 2002) – although in Drosophila, Omi/HtrA2 does not appear to regulate IAPs and downstream caspases (Yacobi-Sharon, 2013). The N-terminal mitochondrial localization sequences of Smac and Omi/HtrA2 are cleaved off while being trafficked into the mitochondrial intermembrane space, generating new N-terminal motifs that start with alanine residues and with the ability to bind to BIR domain grooves. Because mammalian IAP-antagonists are sequestered into the mitochondrial intermembrane space, they do not inhibit IAPs that reside in the cytoplasm during non-apoptotic conditions (Du, 2000; Verhagen, 2000; Hegde, 2002; Martins, 2002; van Loo, 2002). Only when Bcl-2 family proteins help release the mitochondrial proteins into the cytoplasm during apoptosis do these proteins get into contact with IAPs in the cytoplasm, neutralizing their target IAPs.
In case of the Drosophila IAP-antagonists, there are no N-terminal mitochondrial localization sequences, and it remains unclear how these Drosophila proteins lose their N-terminal methionine residues. Drosophila IAP-antagonists do not enter the mitochondrial intermembrane space, but localize to the mitochondrial outer membrane. Hid contains a C-terminal tail anchor sequence that inserts into the mitochondrial outer membrane, with the IAP-binding motif facing the cytoplasm (Abdelwahid, 2007; Haining, 1999). GRIM and REAPER each contain an amphipathic helix that are required for their mitochondrial outer membrane localization (Claveria, 2002; Olson, 2003a; Sandu, 2010). These proteins form multimers with each other, and such association is important for their mitochondrial localization (Sandu, 2010). Mutating their mitochondrial localization sequences disrupt their pro-apoptotic function (Abdelwahid, 2007; Claveria, 2002; Olson, 2003a). More recently, it was found that cdk7 mutants block the mitochondrial localization of Hid, and such conditions abolished Hid’s cell killing activity (Morishita, 2013). Why should these proteins localize to the mitochondrial outer membrane to trigger apoptosis? Certain studies have implicated pro-apoptotic roles of IAP-antagonists that are independent of DIAP1 (Abdelwahid, 2007; Thomenius, 2011; Thress, 1999). On the other hand, DIAP1 overexpression almost completely blocks IAP-antagonist-induced apoptosis in vivo (Hay, 1995), suggesting that DIAP1 independent effects of IAP-antagonists are likely to be subtle, at best. A different explanation was proposed recently, suggesting that the IAP-binding activity of the antagonists is linked to the mitochondrial localization. Specifically, subcellular fractionation studies indicate that only the mitochondria-associated pool of Hid, but not the cytoplasmic pool, has the ability to bind to recombinant DIAP1 proteins (Morishita, 2013).
The role of mammalian IAP-antagonists
Genetic analysis of mammalian IAP-antagonists have shown varying outcomes. Perhaps Smac has drawn the most attention, but a knockout study brings into question its biological significance: The Smac-deficient mice grow normally, and the knockout cell lines respond normally to the apoptotic stimuli that were tested (Okada, 2002). Omi/HtrA2 mutations are associated with Parkinson’s disease (Strauss, 2005), which is more consistent with its role in mitochondrial homeostasis, but not with IAP-antagonist function. However, there is another mammalian IAP-antagonist, ARTS. This is a splice isoform of Septin4, originally identified in a retroviral insertion screen (Larisch, 2000). Subsequent studies have revealed that this protein binds to XIAP, but not through an N-terminal sequence as found in other IAP-antagonists. Instead, the nine C-terminal end residues of ARTS serve as the XIAP1 binding motif (Reingewertz, 2011). Upon binding, ARTS promotes the ubiquitin-mediated degradation of XIAP, similar to the Drosophila IAP antagonists (Gottfried, 2004). ARTS knockout mice are have elevated levels of XIAP, resulting in enhanced cell death resistance. Perhaps as a result, these animals have increased numbers of hematopoietic stem cells and hair follicle stem cells. On the negative side, these mice are more prone to develop tumors (Garcia-Fernandez, 2010). On the positive side, they display marked improvement in wound healing and regeneration (Fuchs, 2013).
Transcriptional regulation of Drosophila IAP-antagonists
Unlike mammalian IAP-antagonists, which are initially segregated into the mitochondrial intermembrane space, transcriptional induction of IAP-antagonists in Drosophila is sufficient to trigger apoptosis. In fact, the transcription of grim, reaper and sickle foreshadows apoptosis induction in this organism (White, 2004; Chen, 2006). As a result, there is much interest in understanding the transcriptional regulation mechanisms of Drosophila IAP-antagonists.
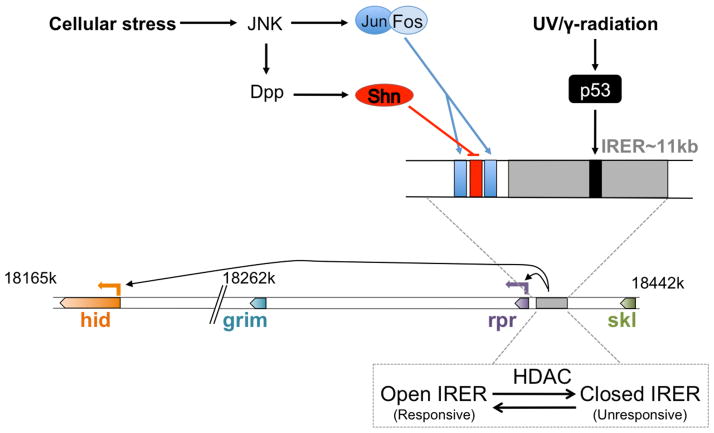
Stress-activated pathways mediated by the tumor suppressor gene p53 respond to a number of distinct stress conditions, including DNA damage (Brodsky, 2000; Ollmann, 2000) and viral infection (Liu, 2011, 2013). p53 directly binds to a regulatory sequence that lies between reaper and sickle (Brodsky, 2000) (Figure 2). This site is part of a broader irradiation-responsive element that controls the induction of multiple IAP-antagonists, including hid that lies more than 250kbp away (Zhang, 2008). Interestingly, this locus is active only during early embryogenesis, and subsequently becomes silenced through epigenetic regulation, thereby making cells insensitive to irradiation-induced apoptosis (Zhang, 2008).
Figure 2. A schematic showing the Drosophila H99 locus and its regulation.
The 3L chromosome arm contains all four of the IAP-antagonists, hid, grim, reaper (rpr) and sickle (skl), clustered in the H99 locus. A ~12kbp region upstream of rpr is the hub of most transcriptional regulation and controls transcription of rpr and hid, although the hid locus is more than 250kbp away. It contains a ~11kbp Irradiation-Responsive Enhancer Region (IRER) containing a 20bp p53 binding site critical for radiation induced apoptosis. The IRER region is heavily epigenetically regulated during development by histone deacetylases (HDAC) and methylases. The rpr upstream promoter region contains sites for binding several transcription factors, notably Jun/Fos and Schnurri (Shn), which mediate context dependent JNK-driven apoptosis.
Jun N-terminal Kinase (JNK) signaling is also tightly associated with IAP-antagonist induction in Drosophila (Figure 2). The heterodimeric transcription factors Jun and Fos form the AP-1 complex to mediate the transcriptional response to this pathway. In fact, IAP-antagonist expression triggers a transcriptional feed back loop that induces other IAP-antagonists to augment their pro-apoptotic effects, and this is mediated by JNK signaling and p53 (Kuranaga, 2002; Shlevkov, 2012). Analysis of the reaper upstream sequence in fact shows functionally significant binding sites. However, JNK is also involved in cell migration and other physiological events without the induction of apoptosis. Why certain cells evade JNK-induced apoptosis has been a mystery up until recently. It has now been found that JNK-mediated apoptosis signal is integrated at the reaper regulatory sequences, together with other anti-apoptotic signals. DPP signaling, mediated by the transcription factor Schnurri, represses JNK-mediated reaper expression through a binding site located in between those of AP-1 (Beira, 2014) (Figure 2). During embryonic development, dorsal closure is regulated by JNK signaling, but these cells do not undergo apoptosis, due to Dpp-Schnurri mediated repression of IAP-antagonist gene expression.
The steroid hormone ecdysone, which triggers the onset of metamorphosis, is also well known to induce massive cell death during metamorphosis. The effect is, in part, due to the transcriptional induction of grim, reaper and hid by the Ecdysone Receptor (EcR), which binds to a reaper-upstream enhancer sequence (Jiang, 2000). In addition to the control of IAP-antagonist, a recent study shows that Ecdysone signaling is required for the proper induction of downstream caspases. That study noted that the resulting increase in caspase expression renders cells sensitive to reaper and hid-induced apoptosis during the late 3rd instar stage of Drosophila development, as ecdysone signaling becomes active at this stage (Kang, 2014). In addition, loss of EGF Receptor/MAP Kinase survival signaling pathway induces hid transcription (Kurada, 1998).
Post-transcriptional regulation of IAP-antagonists
When IAP-antagonists were first discovered, it had been noted that the transcripts of hid are distributed more broadly than the actual pattern of apoptosis, suggestive of post-transcriptional regulatory mechanisms (Grether, 1995). Ever since, distinct types of regulatory mechanisms have been elucidated.
As in many other cellular mechanisms, kinases play roles in IAP-antagonist regulation. A well-characterized example is MAP Kinase, which phosphorylates Hid to inhibit its cell killing activity (Bergmann, 1998; Bergmann A, 2002). This EGF Receptor/MAP Kinase survival signaling pathway also regulates hid transcription (Kurada, 1998). More recently, CDK7 was identified as a gene required for IAP-antagonist-induced cell death. However, it remains unclear whether CDK7 directly phosphorylates IAP-antagonists (Morishita, 2013).
Many IAPs and their antagonists are regulated through ubiquitylation. As introduced earlier, both mammalian and insect IAPs undergo auto-ubiquitylation when bound by IAP-antagonists (Yang, 2000; Yoo, 2002; Ryoo, 2002). IAP-antagonists are ubiquitylatied by IAPs (Olson, 2003b). Lysine-deficient REAPER is more stable, but it was recently demonstrated that BRUCE can add ubiquitins on REAPER even at non-lysine residues and target it for degradation (Domingues, 2012). In the developing eye discs of Drosophila, sensitivity to apoptosis changes as cells transition from an unspecified state to differentiated photoreceptors. Unspecified cells are more vulnerable to apoptosis, as DIAP1 is kept low through ubiquitylation by the Cullin-3 complex in these cells. Differentiated photoreceptors accumulate DIAP1 to gain apoptotic resistance (Fan, 2014).
The role of micro RNAs (miRs) in various cellular processes has been intensely studied in the past decade. Not surprisingly, IAP-antagonists are also targets of miRs in Drosophila. One of the first to be discovered was the miR, bantam, which targets hid for translational suppression (Brennecke, 2003). bantam mutants are homozygous viable but are smaller in size due to reduced tissue growth. Consistently, overexpression leads to overgrowth of tissue due to an increase in cell numbers. GFP-reporter studies show that bantam binds to at least five sites in the hid 3′UTR region and target it for degradation by RNAi. Similar reporter studies show miR-2 to target sites in reaper, grim and sickle transcripts (Stark, 2003) and miR-14 to target reaper (Xu, 2003). More recent studies have found miR-6 and 11 as additional regulators of all four IAP-antagonists (Ge, 2012; Truscott, 2011). Though single mutants are viable, miR-6/miR-11 double mutants are embryonic lethal and show defects in the CNS, suggesting that they may have overlapping roles. Broadly, spatial expression patterns of miRNAs and their targets contribute to regulation of cell death, but an additional layer of complexity is added by competition amongst miRNA for the same targets.
The role of IAP-antagonists in nervous system development
Arguably, a majority of apoptosis observed during development occurs in the nervous system. Reinforcing this view, recent studies on the Drosophila mutants for grim, reaper and hid, revealed their intricate roles in neuronal development. In the ventral nerve cord of the central nervous system, most neuroblasts (neural stem cells) in the abdominal segments undergo apoptosis during late embryogenesis, and a few additional dying cells are observed in the late third instar larval stage. grim and reaper double mutants show a dramatic impairment of neuroblast cell death in this tissue, leading to a significantly enlarged ventral nerve cord (Peterson, 2002; Tan, 2011). This indicates that, in wild type animals, developmental cues confer death as a fate to the abdominal segment neuroblasts. Apoptosis is induced specifically in the abdominal segment neuroblast due to the regulation of grim, reaper and hid by the Hox gene expressed in that region, AbdA (Bello, 2003). A temporal series of transcription factors that are expressed in these cells make up a combinatorial code to determine the timing of apoptosis (Maurange, 2008). In addition, there appears to be a cell death signal originating from the progeny: Those progeny express the Notch ligand, delta, and the resulting Notch signaling contributes to AbdA induction to express IAP-antagonists (Arya, 2015).
Analogous to the example of cell death induction in the ventral nerve cord, a recent study has found that a combinatorial transcription code induces grim and reaper to confer death as a fate to certain differentiating cells. In the region of the optic lobe Outer Proliferation Center (tOPC), neuroblasts temporally express a series of transcription factors to confer different fates to the differentiating neurons. In one of the early lineages, a specific transcription factor helps the cells interpret Notch signal as an apoptotic signal. In a later lineage, a different transcription factor helps the cells to perceive Notch signaling in an opposite way – as a survival signal (Bertet, 2014). Such a strategy allows a diverse array of neuronal subtypes to be established in this tissue.
IAPs and their antagonists in sculpting morphogenesis
In Drosophila, mutant alleles of DIAP1 are referred to as thread, as one of the hypomorphic alleles, thread1 causes the fly antenna tip to appear as thin as a thread. Normally, that region of the antenna, which is called the arista, has many branches, which disappear in thread1 mutants due to excessive death of cells in the larval antennal discs. Conversely, loss of the IAP-antagonist hid, causes excessive branches to appear in the arista (Cullen, 2004). These observations indicate that apoptosis is regulated in antennal discs to regulate the morphogenesis of the arista during development.
One of the important functions of apoptosis is to sculpt body structures. An example is the role of the Drosophila Hox gene Deformed (Dfd) in inducing reaper to kill cells along the segment boundary (Lohmann, 2002). A more recent study has found that Drosophila joint boundaries also undergo apoptosis. How do the tissue know where to make the joint boundary? The data seem to indicate that the Dpp morphogenic gradient is a key determinant. Dpp is secreted from the organizers to form a gradient, but when there is a sharp discontinuity in the gradient, it is known to activate JNK signaling and the induction of IAP-antagonists. This is what happens at the joints, leading to the death of those boundary cells (Manjon, 2007). Notch pathway also contributes to tarsal joint development. This pathway induces the transcription factor Dys, which in turn induce reaper and hid to help sculpt joints (Cordoba, 2014).
Recent work on Drosophila IAP-antagonists has revealed a surprising role of apoptosis in unexpected morphogenic processes, such as tissue rotation. It had been noted that hid mutants can survive to adulthood, and the surviving males have their genitalia rotated in abnormal angles. A live imaging study of the developing male genitalia in the pupal stage helped elucidate this rotation process in detail. Two distinct domains, each rotating 180 °C, have the incremental effect of rotating 360 °C. These two domains are initially part of the same epithelial layer, and the investigators found that hid-induced cell death allows the two domains to separate and rotate away from each other. A failure to separate the two domains leads to a rotational defect (Suzanne, 2010).
Non-apoptotic roles of IAPs in morphogenesis, cell migration and proliferation
Caspases were primarily studied as proteins dedicated to apoptosis induction, but now, there are increasing numbers of studies implicating caspases in diverse non-apoptotic roles. Since caspases in Drosophila are tightly regulated by DIAP1, IAP-antagonists and DIAP1 are also involved in those non-apoptotic processes. The examples include their roles in regulating non-autonomous cell proliferation, cell migration and dendritic morphology regulation.
The loss of DIAP1, or activation of IAP-antagonists, not only induces caspase activation, but also activates a pathway that triggers mitogen expression and the proliferation of neighboring cells. This phenotype is dramatically augmented to cause tissue overgrowth if effector caspases are blocked with p35. It turns out p35 does not inhibit the initiator caspase DRONC, and latter has an apoptosis-independent function in activating the JNK pathway to promote the expression of mitogenic genes such as wingless and dpp (Ryoo, 2004; Huh, 2004; Perez-Garijo, 2004. Kondo, 2006; Fan 2008).
DIAP1’s target DRONC is involved in a number of other non-apoptotic roles. One of the non-apoptotic cellular processes that resemble apoptosis is neuronal dendrite pruning. In order to make, or break, proper synaptic connections, certain dendrites have to undergo dramatic morphological changes. Certain neurons have adopted the caspases regulatory network to eliminate, not the entire cell, but specific dendrites. For example, the Drosophila initiator caspase DRONC can promote dendrite pruning during development, and this process is inhibited by DIAP1 (Kuo, 2006). Other regulators of dendrite pruning have been discovered, and in one example, it was discovered that the AAA ATPase, VCP, regulates dendrite pruning by binding to DIAP1 and facilitating its degradation (Rumpf, 2011).
Sensory organ development of Drosophila also involves caspases in a non-apoptotic mechanism. During the formation of the precursor cells, caspases cleave Shaggy, a negative regulator of Wnt signaling. The cleavage by caspase converts the substrate to an active kinase, promoting the formation of sensory organ precursors (Kanuka, 2005). Not surprisingly, such caspase activity is under the control of DIAP1. DIAP1 is regulated in these cells, not by IAP-antagonists, but through phosphorylation by by IKK epsilon. Loss of this kinase results in the stabilization of DIAP1, which in turn, blocks caspase-mediated shaggy cleavage and activation (Kuranaga, 2006). The IKK epsilon/DIAP1/caspase cascade is also involved in F-actin turnover at the cellular margin and contributes to the morphogenic changes of cultured cells (Oshima, 2006).
Such effects of DIAP1 on the cytoskeleton are not limited to morphological changes, but can also affect cell migration. In the Drosophila ovary, border cells migrate during ovary development to a specific position, and this process requires the GTPase Rac that helps to rearrange the cytoskeleton during this process. Evidence indicates that DIAP1 and DRONC regulate Rac, and without DIAP1, border cells fail to migrate properly (Geisbrecht, 2004). In an analogous mechanism, XIAP1 and c-IAP-1/2 regulate mammalian cell motility by ubiquitylating C-RAF (Dogan, 2008). The non-apoptotic roles of DIAP1 bring up an important question. How can some cells regulate DIAP1 without triggering apoptosis? Live imaging of DIAP1 in the sensory organ precursor cells of Drosophila indicates that the turnover rate of DIAP1 varies between cell types, and such temporal regulation of DIAP1 may determine whether the downstream caspases are utilized for apoptotic or non-apoptotic roles (Koto, 2009).
Spermatid differentiation is a process that involves dramatic morphological changes, including the removal of the bulk of cytoplasm along the elongating spermatids through a process termed “spermatid individualization”. Caspases have been adopted in these cells to mediate the massive cytoplasmic removal in this differentiation process (Arama, 2003). In Drosophila, caspases form a gradient in their activity to regulate spermatid differentiation, so that the regions of the spermatids that are the last to individualize have the lowest caspase activity. This gradient is formed by a counter gradient of the IAP protein, BRUCE (Kaplan, 2011). Consistently, Drosophila bruce mutants show male sterility (Arama, 2003). An analogous regulation of caspases occurs during mammalian spermatid differentiation through XIAP1 and its antagonist, ARTS. In fact, mice lacking the ARTS/Septin 4 locus show defects in sperm cell maturation (Kissel, 2005).
The roles of IAPs in the innate immune response
Early studies of mammalian IAPs, XIAP and c-IAP-1/2 focused on their ability to regulate effector caspases and apoptosis, but in vivo studies of these IAPs increasingly point to their important roles in TNFα signaling and the innate immune response. Upon infection by virus or pathogenic bacteria, various cells in our body produce TNFα to initiate immune response signaling. There are at least three distinct pathways that can be activated downstream of TNFα receptors: The extrinsic cell death pathway mediated by caspase-8 and -3; NF-κB signaling that leads to cytokine production; and RIP (receptor interacting protein)-1 and -3/MLKL mediated “ necroptotic cell death (Silke, 2011). As introduced earlier, c-IAP1 and 2 are mammalian IAP proteins initially identified based on their physical association with TNFα receptor 2 (Rothe, 1995; Uren, 1996). c-IAP1/2 specifically bind to RIP1 while in a complex with TNFα receptor. Upon TNFα stimulation, c-IAP1 and 2 ubiquitylate RIP1 to activate NK-κB and MAP kinase signaling (Bertrand, 2008). Loss of c-IAP1/2, or deubiquitination of RIP1 triggers the formation of a different TNF receptor signaling complex that, instead of promoting NF-κB signaling, activates the caspase-8 mediated apoptosis or RIP3/MLKL-dependent necroptosis (Tenev., 2011; Vince, 2007). c-IAP1 and 2 also ubiquitylate a different, yet related protein, RIP2, and such ubiquitination promotes NF-κB signaling and cytokine production (Bertrand, 2009). In Drosophila, DIAP1 and 2 are the closest homologs of c-IAP1/2, and while DIAP1 primarily regulates apoptosis, DIAP2 promotes the activation of the Drosophila NF-κB homolog, Relish, as part of an innate immune response to gram positive bacteria infection (Huh, 2007).
XIAP was originally characterized as an anti-apoptotic protein that primarily inhibits at least two effector caspases, caspase-3 and -7 (Deveraux, 1997). There was a slight disappointment to the field when it was first reported that XIAP1−/− mice do not show obvious developmental abnormalities (Olayioye, 2005). Subsequent studies revealed subtle cell death phenotypes: XIAP1 deficient sympathetic neurons are more vulnerable to apoptosis after cytochrome c injection (Potts, 2003), and the mutant fibroblasts are sensitized to TNFα-induced apoptosis (Schile, 2008).
Interestingly, more recent studies also implicate XIAP in TNFα signaling and immune response. Mutations in human XIAP (also referred to as BIRC4) have been found to underlie immunodeficiency with aberrant activation of macrophages and dendritic cells, and the accumulation of activated T lymphocytes after viral infection (Rigaud, 2006; Damgaard, 2013; Marsh, 2010; Pachlopnik Schmid, 2011). Similarly, XIAP knockout mice show reduced ability to clear infectious pathogens (Bauler, 2008; Prakash, 2010). These immunodeficiency phenotypes are difficult to explain through XIAP’s ability to inhibit effector caspases. Recent studies have found that XIAP1 has an inhibitory effect on TNFα signaling, a process that is also regulated by c-IAP1/2. However, XIAP1 and c-IAP1/2 have different mechanisms of action: Whereas c-IAP1/2 ubiquitylate RIP1 while bound with TNF receptor to activate NF-κB signaling, XIAP1 appears to ubiquitylate RIP1 at a later stage of signaling, within a distinct complex. The loss of such XIAP activity results in abnormally high inflammasome activity, caspase-1 activation, and IL-1β secretion from dendritic cells (Yabal, 2014). Human patients with XIAP mutation suffer from hyperinflammation, and XIAP’s effect on inflammasome provides a molecular explanation.
Invertebrate IAPs and their antagonists also respond to viral infection as part of an innate immune response. Mosquitos and Drosophila induce IAP-antagonists when infected with DNA or RNA viruses, which helps to kill cells infected with virus and block their propagation (Liu, 2013). Certain virus have evolved to inhibit such innate immune response by evolving IAPs in their genome (Clem, 2005), and the best characterized examples of viral IAPs include Orgyia pseudotsugata Op-IAP, which primarily inhibit initiator caspase activity (Birnbaum, 1994; LaCount, 2000). It appears that these viral IAPs have a more stable anti-apoptotic activity than their cellular homologs: Whereas XIAP, c-IAP1/2 and DIAP1 have short half-lives and undergo auto-ubiquitination and degradation upon binding to IAP-antagonists (Yang, 2000; Ryoo, 2002), or after cleavage by caspases near the N-terminus (Yokokura, 2004; Ditzel, 2003), Op-IAPs lack the N-terminal degrons found in cellular IAPs and exhibit more stability (Cerio, 2010; Vandergaast, 2015). Restoring apoptosis by introducing Drosphila reaper into Sindbis virus impaired their ability to infect mosquitoes, and a gradual negative selection against reaper expression in the recovered virus (O’Neill, 2015). Together, these observations indicate that IAPs and IAP-antagonists regulate the degree of viral propagation in insect hosts.
Concluding Remarks
It has been more than two decades since IAPs and their antagonists were first discovered, but dramatic new discoveries continue to be made in this field. A number of them are particularly notable: For example, although it had been thought that IAP antagonists play no obvious roles in mammalian development, knockout of ARTS revealed defects in stem cell death. Exciting biological roles of IAPs and IAP-antagonists in innate immune response, in contributing to neuronal diversity and numbers, and playing unexpected roles in morphogenesis, have been discovered only recently. There are still many unanswered questions. We still do not fully understand why Drosophila IAP-antagonists must localize to the mitochondrial outer membrane, and how it is trafficked to that site. The intricate regulatory mechanisms that converge on transcription, translation, and post-translational levels, are only beginning to be understood. We hope to see major advances in these areas in coming years.
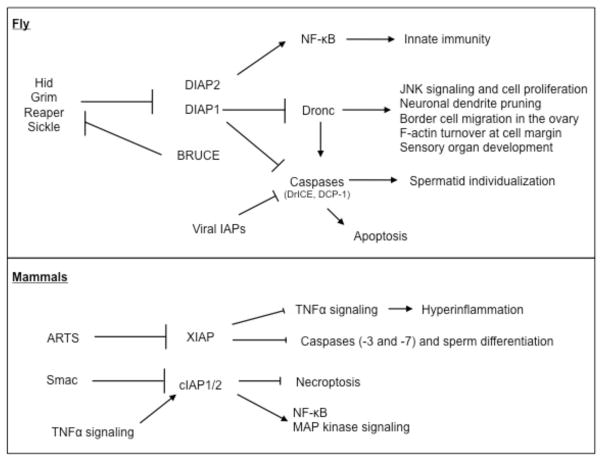
Figure 3. A schematic diagram of IAP-antagonists and their targets.
Upper panel shows the relationships between Drosophila genes, whereas the lower panel shows mammalian IAPs and their antagonists.
Acknowledgments
This work was supported by the NIH grant R01 EY020866 to H.D.R.
References
- Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved’ova L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006;113:1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Arya R, Sarkissian T, Tan Y, White K. Neural stem cell progeny regulate stem cell death in a Notch and Hox dependent manner. Cell Death Differ. 2015 doi: 10.1038/cdd.2014.235. in press. [DOI] [PMC free article] [PubMed]
- Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14:801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Bauler LD, Duckett CS, O’Riordan MX. XIAP regulates cytosol-specific innate immunity to Listeria infection. PLoS Pathog. 2008;4:e1000142. doi: 10.1371/journal.ppat.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beira JV, Spinghorn A, Gunther S, Hufnagel L, Pyrowolakis G, Vincent JP. The Dpp/TGFβ-dependent corepressor Schnurri protects epithelial cells from JNK-induced apoptosis in Drosophila embryos. Dev Cell. 2014;31:240–247. doi: 10.1016/j.devcel.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Bergmann AAJ, McCall K, Steller H. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev Cell. 2002;2:159–170. doi: 10.1016/s1534-5807(02)00116-8. [DOI] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell. 2014;158:1173–1186. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801. doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gilard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Birnbaum MJ, Clem RJ, Miller LK. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. doi: 10.1128/jvi.68.4.2521-2528.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Cerio RJ, Vandergaast R, Friesen PD. Host insect inhibitor-of-apoptosis SfIAP functionally replaces baculovirus IAP but is differentially regulated by its N-terminal leader. J Virol. 2010;84:11448–11460. doi: 10.1128/JVI.01311-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Christich A, Kauppila S, Chen P, Sogame N, Ho SI, Abrams JM. The damage-responsive Drosophila gene sickle encodes a novel IAP binding protein similar to but distinct from reaper. Curr Biol. 2002;12:137–140. doi: 10.1016/s0960-9822(01)00658-3. [DOI] [PubMed] [Google Scholar]
- Claveria C, Caminero E, Martinez AC, Campuzano S, Torres M. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J. 2002;21:3327–3336. doi: 10.1093/emboj/cdf354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RJ. The role of apoptosis in defense against baculovirus infection in insects. Curr Top Microbiol Immunol. 2005;289:113–129. doi: 10.1007/3-540-27320-4_5. [DOI] [PubMed] [Google Scholar]
- Clem RJ, Fechneimer M, Miller LK. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- Clem RJ, Miller LK. Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba S, Estella C. The bHLH-PAS transcription factor dysfusion regulates tarsal joint formation in response to Notch activity during drosophila leg development. PLoS Genet. 2014;10:e1004621. doi: 10.1371/journal.pgen.1004621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen K, McCall K. Role of programmed cell death in patterning the Drosophila antennal arista. Dev Biol. 2004;275:82–92. doi: 10.1016/j.ydbio.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Damgaard RB, Fiil BK, Speckmann C, Yabal M, zur Stadt U, Bekker-Jensen S, Jost PJ, Ehl S, Mailand N, Gyrd-Hansen M. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signaling. EMBO Mol Med. 2013;5:1278–1295. doi: 10.1002/emmm.201303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, Meier P. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- Dogan T, Harms GS, Hekman M, Karreman C, Oberoi TK, Alnemri ES, Rapp UR, Rajalingam K. X-linked and cellular IAPs modulate the stability of C-RAF kinase and cell motility. Nat Cell Biol. 2008;10:1447–1455. doi: 10.1038/ncb1804. [DOI] [PubMed] [Google Scholar]
- Domingues C, Ryoo HD. Drosophila BRUCE inhibits apoptosis through non-lysine ubiquitination of the IAP-antagonist REAPER. Cell Death Differ. 2012;19:470–477. doi: 10.1038/cdd.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a Mitochondrial Protein that Promotes Cytochrome c-Dependent Caspase Activation by Eliminating IAP Proteins. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev Cell. 2008;14:399–410. doi: 10.1016/j.devcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Multiple mechanisms modulate distinct cellular susceptibilities toward apoptosis in the developing Drosophila. Dev Cell. 2014;30:48–60. doi: 10.1016/j.devcel.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Brown S, Gorenc T, Rodriguez J, Fuchs E, Steller H. Sept4/ARTS regulates stem cell apoptosis and skin regeneration. Science. 2013;341:286–289. doi: 10.1126/science.1233029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez M, Kissel H, Brown S, Gorenc T, Schile AJ, Rafii S, Larisch S, Steller H. Sept4/ARTS is required for stem cell apoptosis and tumor suppression. Genes Dev. 2010;24:2282–2293. doi: 10.1101/gad.1970110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Chen YW, Weng R, Lim SF, Buescher M, Zhang R, Cohen SM. Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell Death Differ. 2012;19:839–846. doi: 10.1038/cdd.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht ER, Montell DJ. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell. 2004;118:111–125. doi: 10.1016/j.cell.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Gottfried Y, Rotem A, Lotan R, Steller H, Larisch S. The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. EMBO J. 2004;23:1627–1635. doi: 10.1038/sj.emboj.7600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Haining WN, Carboy-Newcomb C, Wei CL, Steller H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:4936–4941. doi: 10.1073/pnas.96.9.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, Katayama R, Hashimoto C, Zhang X, Noda T, Tsuruo T, Naito M. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6:849–860. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- Hawkins CJ, Wang SL, Hay BA. A cloning method to identify caspases and their regulators in yeast: identification of Drosophila IAP1 as an inhibitor of the Drosophila caspase DCP-1. Proc Natl Acad Sci USA. 1999;96:2885–2890. doi: 10.1073/pnas.96.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukattash R, Cilenti L, DuBois G, Lazebnik Y, Zervos AS, Fernandes-Alnemri T, Alnemri ES. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts IAP-caspase interaction. J Biol Chem. 2002;277:432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Huh JR, Foe I, Muro I, Chen CH, Seol JH, Yoo SJ, Guo M, Park JM, Hay BA. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- Jiang C, Lamblin AF, Steller H, Thummel CS. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol Cell. 2000;5:445–455. doi: 10.1016/s1097-2765(00)80439-6. [DOI] [PubMed] [Google Scholar]
- Kang Y, Bashirullah A. A steroid-controlled global switch in sensitivity to apoptosis during Drosophila development. Dev Biol. 2014;386:34–41. doi: 10.1016/j.ydbio.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanuka H, Kuranaga E, Takemoto K, Hiratou T, Okano H, Miura M. Drosophila caspase transduces Shaggy/GSK-3beta kinase activity in neural precursor development. EMBO J. 2005;24(21):3793–3806. doi: 10.1038/sj.emboj.7600822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel H, Georgescu M, Larisch S, Manova K, Hunnicutt GR, Steller H. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev Cell. 2005;8:353–364. doi: 10.1016/j.devcel.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol. 2006;26:7258–7268. doi: 10.1128/MCB.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koto A, Kuranaga E, Miura M. Temporal regulation of Drosophila IAP1 determines caspase functions in sensory organ development. J Cell Biol. 2009;182:219–231. doi: 10.1083/jcb.200905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Igaki T, Sawamoto K, Ichijo H, Okano H, Miura M. Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nat Cell Biol. 2002;4:705–710. doi: 10.1038/ncb842. [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, Kobayashi M, Hayashi S, Miura M. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell. 2006;126(3):583–596. doi: 10.1016/j.cell.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- LaCount DJ, Hanson SF, Schneider CL, Friesen PD. Caspase inhibitor p35 and inhibitor of apoptosis Op-IAP block in vivo proteolytic activation of an effector caspase at different steps. J Biol Chem. 2000;275:15657–15664. doi: 10.1074/jbc.M000791200. [DOI] [PubMed] [Google Scholar]
- Larisch S, Yi Y, Lotan R, Kerner H, Eimerl S, Tony Parks W, Gottfried Y, Birkey Reffey S, de Caestecker MP, Danielpour D, Book-Melamed N, Timberg R, Duckett CS, Lechleider RJ, Steller H, Orly J, Kim SJ, Roberts AB. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat Cell Biol. 2000;2:915–921. doi: 10.1038/35046566. [DOI] [PubMed] [Google Scholar]
- Lee TV, Fan Y, Wang S, Srivastava M, Broemer M, Meier P, Bergmann A. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet. 2011;7:e1002261. doi: 10.1371/journal.pgen.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang J, Shi Y. Structural mechanisms of DIAP1 auto-inhibition and DIAP1-mediated inhibition of drICE. Nat Commun. 2011;2:408. doi: 10.1038/ncomms1418. [DOI] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Becnel JJ, Zhang Y, Zhou L. Induction of reaper ortholog mx in mosquito midgut cells following baculovirus infection. Cell Death Differ. 2011;18:1337–1345. doi: 10.1038/cdd.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Behura SK, Clem RJ, Schneemann A, Becnel J, Severson DW, Zhou L. p53-mediated rapid induction of apoptosis conveys resistance to viral infection in Drosophila melanogaster. PLoS Pathog. 2013;9:e1003137. doi: 10.1371/journal.ppat.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann I, McGinnis N, Bodmer M, McGinnis W. The Drosophila Hox gene Deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell. 2002;110:457–466. doi: 10.1016/s0092-8674(02)00871-1. [DOI] [PubMed] [Google Scholar]
- Manjon C, Sanchez-Herrero E, Suzanne M. Sharp boundaries of Dpp signalling trigger local cell death required for Drosophila leg morphogenesis. Nat Cell Biol. 2007;9:57–63. doi: 10.1038/ncb1518. [DOI] [PubMed] [Google Scholar]
- Marsh RA, Madden L, Kitchen BJ, Mody R, McClimon B, Jordan MB, Bleesing JJ, Zhang K, Filipovich AH. XIAP1 deficiency: a unique primary immunodeficiency best classified as X-linked familial hemophagocytic lymphohistiocytosis and not as X-linked lymphoproliferative disease. Blood. 2010;116:1079–1082. doi: 10.1182/blood-2010-01-256099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins LM, Iaccarino I, Tenev T, Gschmeissner S, Totty NF, Lemoine NR, Sabopoulos J, Gray CW, Creasy CL, Dingwall C, Downward J. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a Reaper-like motif. J Biol Chem. 2002;277:439–444. doi: 10.1074/jbc.M109784200. [DOI] [PubMed] [Google Scholar]
- Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita J, Kang MJ, Fidelin K, Ryoo HD. CDK7 regulates the mitochondrial localization of a tail-anchored proapoptotic protein, Hid. Cell Rep. 2013;5:1481–1488. doi: 10.1016/j.celrep.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I, Hay BA, Clem RJ. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated processed form of the apical caspase DRONC. J Biol Chem. 2002;20:49644–49650. doi: 10.1074/jbc.M203464200. [DOI] [PubMed] [Google Scholar]
- Okada H, Suh WK, Jin J, Woo M, Du C, Elia A, Duncan GS, Wakeham A, Itie A, Lowe SW, Wang X, Mak TW. Generation and characterization of Smac/DIABLO-deficient mice. Mol Cell Biol. 2002;22:3509–3517. doi: 10.1128/MCB.22.10.3509-3517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye MA, Kaufmann H, Pakusch M, Vaux DL, Lindeman GJ, Visvader JE. XIAP-deficiency leads to delayed lobuloalveolar development in the mammary gland. Cell Death Differ. 2005;12:87–90. doi: 10.1038/sj.cdd.4401524. [DOI] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, Duyk G, Friedman L, Prives C, Kopczynski C. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Olson MR, Holley CL, Gan EC, Colon-Ramos DA, Kaplan B, Kornbluth S. A GH3-like domain in reaper is required for mitochondrial localiztion and induction of IAP degradation. J Biol Chem. 2003a;278:44758–44768. doi: 10.1074/jbc.M308055200. [DOI] [PubMed] [Google Scholar]
- Olson MR, Holley CL, Yoo SL, Huh JR, Hay BA, Kornbluth S. Reaper is regulated by IAP-mediated ubiquitination. J Biol Chem. 2003b;278:4028–4034. doi: 10.1074/jbc.M209734200. [DOI] [PubMed] [Google Scholar]
- O’Neill K, Olson BJ, Huang N, Unis D, Clem RJ. Rapid selection against arbovirus-induced apoptosis during infection of a mosquito vector. Proc Natl Acad Sci USA. 2015;112:E1152–1161. doi: 10.1073/pnas.1424469112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Takeda M, Kuranaga E, Ueda R, Aigaki T, Miura M, Hayashi S. IKK epsilon regulates F actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr Biol. 2006;16(15):1531–1537. doi: 10.1016/j.cub.2006.06.032. [DOI] [PubMed] [Google Scholar]
- Pachlopnik Schmid J, Canioni D, Moshous D, Touzot F, Mahlaoui N, Hauck F, Kanegane H, Lopez-Granados E, Mejstrikova E, Pelier I, Galicier L, Galambrun C, Barlogis V, Bordigoni P, Fourmaintraux A, Hamidou M, Dabadie A, Le Deist F, Haerynck F, Ouachee-Chardin M, Rohrlich P, Stephan JL, Lenoir C, Rigaud S, Lambert N, Milili M, Schiff C, Chapel H, Picard C, de Saint Basile G, Blanche S, Fischer A, Latour S. Clinical similarities and differences of patients with X-linked lymphoproliferative syndrome type 1 (XLP-1/SAP deficiency) versus type 2 (XLP-2/XIAP deficiency) Blood. 2011;117:1522–1529. doi: 10.1182/blood-2010-07-298372. [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Martin FA, Morata G. Caspase inhibition during apoptosis causes abnormal signaling and developmental aberrations in Drosophila. Development. 2004;131:5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- Peterson C, Carney GE, Taylor BJ, White K. reaper is required for neuroblast apoptosis during Drosophila development. Development. 2002;129:1467–1476. doi: 10.1242/dev.129.6.1467. [DOI] [PubMed] [Google Scholar]
- Potts PR, Singh S, Knezek M, Thompson CB, Deshmukh M. Critical function of endogenous XIAP in regulating caspase activation during sympathetic neuronal apoptosis. J Cell Biol. 2003;163:789–799. doi: 10.1083/jcb.200307130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash H, Albrecht M, Becker D, Kuhlmann T, Rudel T. Deficiency of XIAP leads to sensitization for Chlamydophila pneumoniae pulmonary infection and dysregulation of innate immune response in mice. J Biol Chem. 2010;285:20291–20302. doi: 10.1074/jbc.M109.096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reingewertz TH, Shalev DE, Sukenik S, Blatt O, Rotem-Bamberger S, Lebendiker M, Larisch S, Friedler A. Mechanism of the interaction between the intrinsically disordered C-terminus of the pro-apoptotic ARTS protein and the Bir3 domain of XIAP. PLoS One. 2011;6:e24655. doi: 10.1371/journal.pone.0024655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P. Diap2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol. 2007;31:7. doi: 10.1083/jcb.200706027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, Fischer A, de Saint Basile G, Latour S. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNF-R2-TRAF signaling complex contains two novel proteins related to baculoviral-inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- Rumpf S, Lee SB, Jan LY, Jan YN. Neuronal remodeling and apoptosis require VCP-dependent degradation of the apoptosis inhibitor DIAP1. Development. 2011;138:1153–1160. doi: 10.1242/dev.062703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Sandu C, Ryoo HD, Steller H. Drosophila IAP antagonists form multimeric complexes to promote cell death. J Cell Biol. 2010;190:1039–1052. doi: 10.1083/jcb.201004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro PJ, Hsu HH, Jung H, Robbins ES, Ryoo HD. Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol. 2008;10:1440–1446. doi: 10.1038/ncb1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov E, Morata G. A dp53/JNK-dependant feedback amplification loop is essential for the apoptotic response to stress in Drosophila. Cell Death Differ. 2012;19:451–460. doi: 10.1038/cdd.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J. The regulation of TNF signalling: what a tangled web we weave. Curr Opin Immunol. 2011;23:620–626. doi: 10.1016/j.coi.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Silke J, Vaux DL. Two kinds of BIR-containing protein - inhibitors of apoptosis, or required for mitosis. J Cell Sci. 2001;114:1821–1827. doi: 10.1242/jcs.114.10.1821. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandez-Alnemri T, Shi Y, Alnemri ES. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- Srinivasula S, Datta P, Kobayashi M, Wu J-W, Fujioka M, hedge R, Zhang Z, Mukattash R, Fernandes-Alnemri T, Shi Y, Jaynes JB, Alnemri ES. sickle, a novel Drosophila death gene in the reaper/hid/grim region, encodes an IAP-inhibitory protein. Curr Biol. 2002;12:125–130. doi: 10.1016/s0960-9822(01)00657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:e60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, Gasser T, Wszolek Z, Muller T, Bornemann A, Wolburg H, Downward J, Riess O, Schulz JB, Kruger R. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum Mol Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- Suzanne M, Petzoldt AG, Speder P, Coutelis JB, Steller H, Noselli S. Coupling of apoptosis and L/R patterning controls stepwise organ looping. Curr Biol. 2010;20:1773–1779. doi: 10.1016/j.cub.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Yamada-Mabuchi M, Arya R, St Pierre S, Tang W, Tosa M, Brchmann C, White K. Coordinated expression of cell death genes regulates neuroblast apoptosis. Development. 2011;138:2197–2206. doi: 10.1242/dev.058826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, Meier P. The ripoptosome, a signaling platform that assembles in response to genotyoxic stress and loss of IAPs. Mol Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Thomenius M, Freel CD, Horn S, Krieser R, Abdelwahid E, Cannon R, Balasundaram S, White K, Kornbluth S. Mitochondrial fusion is regulated by Reaper to modulate Drosophila programmed cell death. Cell Death Differ. 2011;18:1640–1650. doi: 10.1038/cdd.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress K, Evans EK, Kornbluth S. Reaper-induced dissociation of a Scythe-sequestered cytochrome c-releasing activity. EMBO J. 1999;18:5486–5493. doi: 10.1093/emboj/18.20.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott M, Islam AB, Lopez-Bigas N, Frolov MV. mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes Dev. 2011;25:1820–1834. doi: 10.1101/gad.16947411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Pakusch M, Hawkins CJ, Puls KL, Vaux DL. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc Natl Acad Sci USA. 1996;1996:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loo G, van Gurp M, Depuydt B, Srinivasula SM, Rodriguez I, Alnemri ES, Gevaert K, Vandekerckhove J, Declercq W, Vandenabeele P. The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity. Cell Death Differ. 2002;9:20–26. doi: 10.1038/sj.cdd.4400970. [DOI] [PubMed] [Google Scholar]
- Vandergaast R, Mitchell JK, Byers NM, Friesen PD. Insect inhibitor-of-Apoptosis (IAP) proteins are negatively regulated by signal-induced N-terminal degrons absent within viral IAP proteins. J Virol. 2015;89:4481–4493. doi: 10.1128/JVI.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen A, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a Mammalian Protein that Promotes Apoptosis by Binding to and Antagonizing IAP Proteins. Cell. 2000;102:43–54. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- Vernooy SY, Chow V, Su J, Verbrugghe K, Yang J, Cole S, Olson MR, Hay BA. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr Biol. 2002;12:1164–1168. doi: 10.1016/s0960-9822(02)00935-1. [DOI] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, Brink R, Leverkus M, Tergaonkar V, Schneider P, Callus BA, Koentgen F, Vaux DL, Silke J. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Wing JP, Karres JS, Ogdahl JL, Zhou L, Schwartz LM, Nambu JR. Drosophila sickle is a novel grim-reaper cell death activator. Curr Biol. 2002;12:131–135. doi: 10.1016/s0960-9822(01)00664-9. [DOI] [PubMed] [Google Scholar]
- Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Wu JW, Cocina AE, Chai J, Hay BA, Shi Y. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Yabal M, Muller N, Adler H, Knies N, Bross CJ, Damgaard RB, Kanegane H, Ringelhan M, Kaufmann T, Heikenwalder M, Strasser A, Gross O, Ruland J, Peschel C, Gyrd-Hansen M, Jost PJ. XIAP restricts TNF- and RIP3-dependent cell death and inflammasome activation. Cell Rep. 2014;7:1796–1808. doi: 10.1016/j.celrep.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Yacobi-Sharon K, Namdar Y, Arama E. Alternative germ cell death pathway in Drosophila involves HtrA2/Omi, lysosomes, and a caspase-9 counterpart. Dev Cell. 2013;25:29–42. doi: 10.1016/j.devcel.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Yan N, Wu JW, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol. 2004;11:420–428. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P, White K. Dissection of DIAP1 functional domains via a mutant replacement strategy. J Biol Chem. 2004;279:52603–52612. doi: 10.1074/jbc.M409691200. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang S, Feldman RMR, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lin N, Carroll PM, Chan G, Guan B, Xiao H, Yao B, Wu SS, Zhou L. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression. Dev Cell. 2008;14:481–493. doi: 10.1016/j.devcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]