Abstract
The long-term evolutionary interaction between the host immune system and symbiotic bacteria determines their cooperative rather than antagonistic relationship. It is known that commensal bacteria have evolved a number of mechanisms to manipulate the mammalian host immune system and maintain homeostasis. However, the strategies employed by the microbiome to overcome host immune responses in invertebrates still remain to be understood. Here, we report that the gut microbiome in mosquitoes utilizes C-type lectins (mosGCTLs) to evade the bactericidal capacity of antimicrobial peptides (AMPs). Aedes aegypti mosGCTLs facilitate colonization by multiple bacterial strains. Furthermore, maintenance of the gut microbial flora relies on the expression of mosGCTLs in A. aegypti. Silencing the orthologues of mosGCTL in another major mosquito vector (Culex pipiens pallens) also impairs the survival of gut commensal bacteria. The gut microbiome stimulates the expression of mosGCTLs, which coat the bacterial surface and counteract AMP activity. Our study describes a mechanism by which the insect symbiotic microbiome offsets gut immunity to achieve homeostasis.
Metazoans form symbioses with microorganisms to influence multiple aspects of host biology, such as nutrition, reproduction, metabolism and immunity. The insect commensal microbiome, which primarily resides in the gut, constitutes an intricate ecological environment1. Multiple immune systems have evolved in order to control unexpected microbial overgrowth or opportunistic infection in the insect gut2–4. A wide spectrum of antimicrobial peptides (AMPs) are constitutively expressed in mosquito gut epithelial cells and released into the lumen upon stimulation by the gut microbiome5,6. In contrast, the commensal gut microbiome might employ host proteins in order to conquer local immune responses, thereby enabling the gut microbiome to maintain homeostasis. Such intricate insect host–microbiome relationships in disease vector mosquitoes have lately received great attention due to their potential exploitation for disease transmission control; however, the survival strategy exploited by gut symbiotic microorganisms in mosquito gut ecosystems remains largely unknown to date.
C-type lectins are a large group of proteins in metazoans originally named for their ability to bind carbohydrate in a Ca2+ (C-type)-dependent manner7. C-type lectins bind microbial carbohydrate components8, thereby acting as key factors in both microbial pathogenesis and immune responses based on direct microbial binding. In mammals, a soluble mannose-binding lectin (MBL) acts as a pattern-recognition receptor that activates the complement cascade for the opsonization and clearance of microbial agents9. In contrast, multiple transmembrane C-type lectins, such as the mannose receptor (CD206), DC-SIGN (CD209) and its homolog L-SIGN (CD209L), have been identified as attachment factors for the cellular entry of Mycobacterium tuberculosis10 and many viruses11–14. Whole-genome analysis has revealed that a large number of C-type lectins are also present in invertebrates8,15. It has been shown that multiple C-type lectins act as susceptibility factors to facilitate infections of Plasmodium parasites16, the entomopathogenic fungi Beauveria bassiana17 and the West Nile18 and dengue19 viruses in mosquitoes, while a C-type lectin facilitates the entry of white spot syndrome virus in Marsupenaeus japonicus shrimp20. In contrast, other C-type lectins of mosquitoes, Drosophila and shrimps function as immune factors for the control of bacterial infections. Silencing these C-type lectins results in robust bacterial growth or shortening of the arthropod lifespan after infection21–23. Together, these findings highlight the pleiotropic effects of C-type lectins in invertebrate–microbial interactions.
In this study, we reveal that blockade of C-type lectins (mosGCTLs) suppresses gut bacterial survival and colonization in the mosquito Aedes aegypti and Culex pipiens pallens. We demonstrate that the expression of both mosGCTLs and AMPs is regulated by the gut microbiome via the immune deficiency (Imd) pathway, and that mosGCTLs directly coat the bacterial cells, neutralizing AMP-mediated elimination. We propose that the release of mosGCTLs is an immunoregulatory strategy to protect commensal bacteria from constitutive AMP-mediated elimination, thereby enabling eco-adaption of the gut microbiome in mosquitoes.
Results
A role for mosGCTLs in bacterial colonization of A. aegypti
Taking into account the pleiotropic roles of C-type lectins in host–microbe interactions, we examined the roles of mosquito C-type lectins (mosGCTLs) in Escherichia coli (ST515 strain) infection via dsRNA-mediated silencing in A. aegypti. The silencing efficiency of the mosGCTL dsRNAs was validated in our previous study19. Compared with the GFP dsRNA-treated control, knockdown of seven mosGCTL genes significantly reduced the number of E. coli cells by more than twofold in the colony-forming unit (c.f.u.) assay (Fig. 1a). No spectinomycin-resistant microbes were detected in the non-E. coli-inoculated mosquitoes (Supplementary Fig. 1). The most prominent opposite phenotype was exhibited by mosGCTL-24 (AAEL011607/AAEL014382) that shares 33.9 and 36.2% identity with CTL4 (AGAP005335) and CTLMA2 (AGAP005334) of the malaria vector mosquito Anopheles gambiae, respectively, which have been identified as anti-bacterial immune factors21. These data indicate that C-type lectins may play diverse roles in the bacteria–mosquito interaction.
Figure 1. The role of mosGCTLs in systemic bacterial inoculation in A. aegypti.
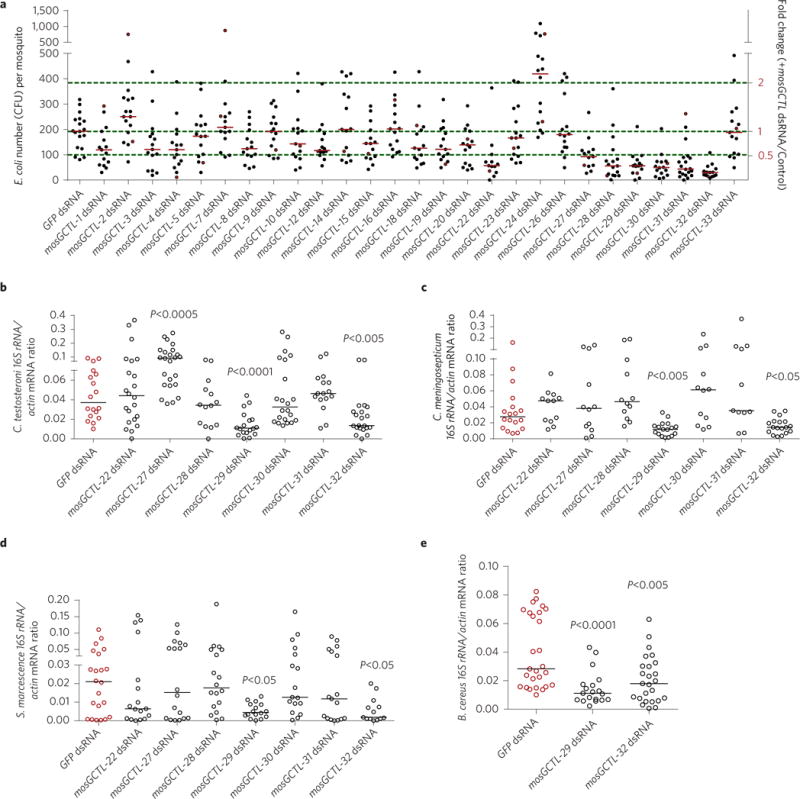
a, The role of mosGCTLs in systemic E. coli inoculation. Each mosGCTL gene was silenced through thoracic microinjection of dsRNA in A. aegypti. The number of E. coli was determined by a c.f.u. assay. Each dot represents one mosquito (n ≥ 12 in each group). b–d, The role of mosGCTLs in systemic inoculation of Gram-negative gut bacteria. Seven mosGCTLs, which act as susceptibility factors in E. coli infection (a), were silenced to investigate their roles in C. testosteroni (b), C. meningosepticum (c) and S. marcescens (d) infections in antibiotic-treated mosquitoes. The burden of these gut bacteria was determined by qPCR. e, The role of mosGCTL-29 and mosGCTL-32 in systemic infection with a Gram-positive gut bacterium B. cereus. Both mosGCTL-29 and mosGCTL-32 were knocked down by dsRNA inoculation in antibiotic-treated mosquitoes. The burden of the gut bacteria was determined by qPCR. b–e, The qPCR primers for gut bacteria 16S rDNA are described in Supplementary Table 5. One dot represents one mosquito gut. The horizontal lines represent the median values of the results. Data were analysed using the non-parametric Mann–Whitney test. a–e, The green fluorescent protein (GFP) dsRNA-treated mosquitoes served as mock controls. The results were combined from at least two biologically independent experiments.
E. coli is not part of the commensal microbiome of A. aegytpi. To characterize the role of mosGCTLs in the survival of commensal bacterial in mosquitoes, we isolated cultivable bacteria from the female A. aegypti midgut and categorized them as members of 14 species based on 16S rDNA sequencing, Comamonas testosteroni, Chryseobacterium meningosepticum, Serratia marcescens and Bacillus cereus were found to be the most abundant species (Supplementary Table 1). Many reports have described the pathogenic nature of S. marcescens in mosquitoes and Drosophila24, however, other studies have identified S. marcescens as an indigenous gut bacterium in mosquitoes25,26. Additionally, the other three bacteria have been well established as gut commensal microbes in mosquitoes26–28. Specific 16S rDNA primers were designed and validated for qPCR detection of each of these four gut microbes (Supplementary Fig. 2). We first assessed the role of the seven mosGCTL genes, found to facilitate systemic E. coli infections, in colonization of three Gram-negative gut bacteria. Compared with the GFP mock group, knockdown of mosGCTL-29 and mosGCTL-32, also designated as CTLGA8 and CTL15, respectively15, significantly reduced the burdens of thoracically inoculated C. testosteroni (Fig. 1b), C. meningosepticum (Fig. 1c) and S. marcescens (Fig. 1d) in antibiotic-treated mosquitoes. Intriguingly, mosGCTL-27 exhibited a role in resistance to infection by C. testosteroni (Fig. 1b), which is consistent with the role of mosGCTL-24 in E. coli infection. We next determined the role of mosGCTL-29 and mosGCTL-32 in the infection of a Gram-positive bacterium, B. cereus. Like the Gram-negative bacteria, the members of the Bacillus genus have peptidoglycans that activate the Imd pathway in Drosophila29,30. Silencing of mosGCTL-29 and mosGCTL-32 also significantly reduced the load of B. cereus (Fig. 1e). Taken together, our results indicate that these two mosGCTLs are critical host factors for bacterial colonization of A. aegypti.
mosGCTLs facilitate the colonization of commensal bacteria in the gut of mosquitoes
Commensal bacteria have evolved a number of mechanisms to manipulate the mammalian host immune system and maintain homeostasis, such as recruitment of regulatory T cells31,32. In invertebrates, which lack such complex immune regulatory machineries, the strategies employed by the microbiome to overcome host immune responses are unknown. We therefore investigated whether commensal bacteria might exploit mosGCTLs to achieve homeostasis. Both mosGCTL-29 and mosGCTL-32 are highly expressed in the haemolymph but are also found in the midgut of A. aegypti at both the mRNA and protein levels (Fig. 2a,b). Although both mosGCTLs are predicted to be soluble proteins, immunofluorescence staining indicated that they are also localized to the lumenal surface of the gut epithelial layer (Supplementary Fig. 3). Therefore, we determined the role of these mosGCTLs in the gut colonization of commensal bacteria. In antibiotic-treated mosquitoes, both knockdown of mosGCTL-29 and mosGCTL-32 (Fig. 2c) and immuno-blockade by feeding mosGCTL antisera (Fig. 2d) significantly suppressed colonization of C. testosteroni, C. meningosepticum, S. marcescens and B. cereus in the mosquito midguts.
Figure 2. mosGCTLs facilitate the colonization of the A. aegypti midgut by gut bacteria.
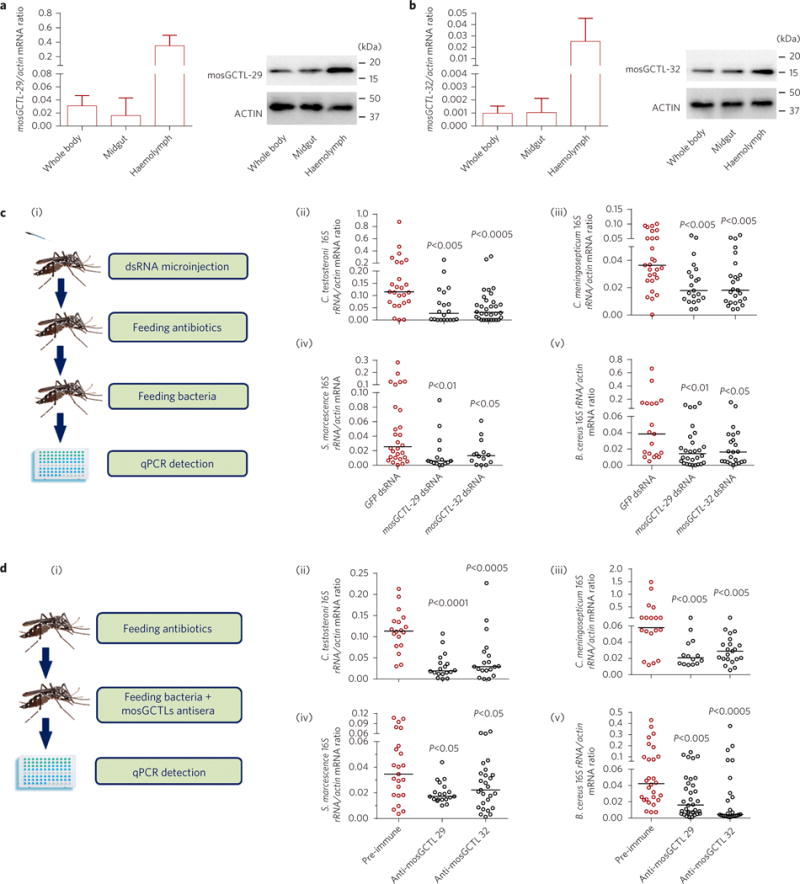
a,b, Distribution of mosGCTL-29 (a) and mosGCTL-32 (b) in A. aegypti. The abundance of mosGCTLs was detected in the whole bodies, midguts and haemolymph via qPCR (left panels) and western blotting (right panels). For the western blotting experiment, the mosquito whole bodies, midguts and haemolymph were dissected and lysed for detection. The polyclonal antibodies against mosGCTL-29 and mosGCTL-32 were generated in mice. The detection of actin served as an internal control. A total of 100 μg of protein lysate was loaded in each lane. Data in a,b are represented as mean±s.d. in each group. c, Knockdown of mosGCTLs impaired colonization of gut bacteria in the A. aegypti midgut. (i) Schematic representation of the study design. mosGCTL-29 and mosGCTL-32 were silenced in antibiotic-treated mosquitoes. The GFP dsRNA-treated mosquitoes served as mock controls. Subsequently, 1 optical density (OD) of C. testosteroni (ii), C. meningosepticum (iii), S. marcescens (iv) and B. cereus (v) was fed to the mosquitoes 3 days after dsRNA inoculation, respectively. d, Immuno-blockade of mosGCTLs reduced colonization of gut bacteria in the A. aegypti midgut. (i) Schematic representation of the study design. The antibodies against mosGCTL-29 or mosGCTL-32 were fed together with 1 OD of C. testosteroni (ii), C. meningosepticum (iii), S. marcescens (iv) and B. cereus (v) to the antibiotic-treated mosquitoes. Mosquitoes fed a pre-immune antibody served as the mock control. c,d, The bacterial burden was determined by SYBR Green qPCR. The qPCR primers for bacterial 16S rDNA are described in Supplementary Table 5. One dot represents one mosquito gut. The horizontal line represents the median value of the results. Data were analysed using the non-parametric Mann–Whitney test. The results were combined from at least two biologically independent experiments.
Next, we investigated whether the bacteria-protecting property of mosGCTLs could be generalized to the whole microbial flora. Knockdown of mosGCTL-29 and mosGCTL-32 significantly reduced the total bacterial burden in A. aegypti midguts 3 and 6 days post-dsRNA inoculation (Fig. 3a), while an immuno-blockade accomplished by feeding mosquitoes mosGCTL antisera repressed the growth of gut microorganisms at 24 h and 48 h post-blood feeding (Fig. 3b). We next wished to examine if mosGCTL-29 and mosGCTL-32 have conserved functions in Culex pipiens pallens, another disease-vector mosquito that is phylogenetically closely related to A. aegypti. Since there is no clear orthology between mosGCTL-29 and mosGCTL-32 and Culex C-type lectins, we selected the top five hits in the genome of Culex quinquefasciatus (Supplementary Table 2), a mosquito species that is very closely related to C. pipiens pallens33, for functional characterization. Each of the five genes was knocked down via thoracic microinjection of dsRNA into C. pipiens pallens. Then, E. coli (ST515 strain) was thoracically inoculated into the mosquitoes, and the resultant bacterial loads were quantified by the c.f.u. assay 4 h post-infection. Silencing CPIJ007869, the Culex homologue with the highest identity to mosGCTL-32, significantly reduced bacterial viability after systemic infection (Fig. 3c). We then investigated the role of CPIJ007869 in maintenance of the Culex gut microbiota ecosystem. Knockdown of CPIJ007869 significantly impaired the microbial flora burden in the C. pipiens pallens midgut 3 and 6 days after gene silencing (Fig. 3d), thereby confirming that certain C-type lectins are key factors maintaining mosquito gut microbiome homeostasis.
Figure 3. mosGCTLs contribute to the maintenance of gut microbiota in A. aegypti and C. pipiens pallens.
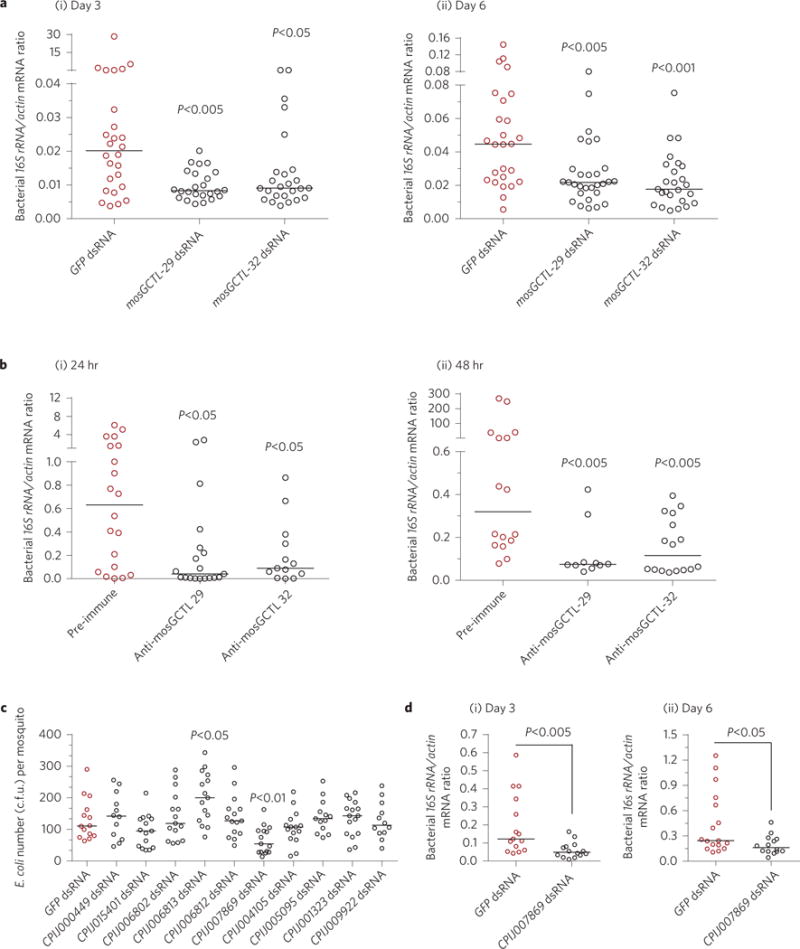
a, Knockdown of mosGCTLs reduced the abundance of gut microbiota in A. aegypti. The microbial load in the midgut was determined 3 days (i) and 6 days (ii) after gene silencing by qPCR using universal bacterial primers. b, Immuno-blockade of mosGCTLs repressed the enormous growth of the gut microbiota after blood feeding. Antisera against mosGCTL-29 and mosGCTL-32 were fed together with fresh human blood. Mosquitoes fed a pre-immune antibody served as the mock controls. The microbial load in the midgut was determined at 24 h (i) and 48 h (ii) by qPCR. c, The role of mosGCTL orthologues in systemic E. coli inoculation of C. pipiens pallens. Ten mosGCTL orthologues to mosGCTL-29/mosGCTL-32 were individually silenced through thoracic microinjection of dsRNA in C. pipiens pallens. At least 12 mosquitoes were included in each group. d, Knockdown of a mosGCTL orthologue (CPIJ007869) reduced the burden of the gut microbiota in C. pipiens pallens. The microbial load in the midgut was determined 3 days (i) and 6 days (ii) after dsRNA inoculation by qPCR. One dot represents one mosquito gut. The horizontal line represents the median of the results. The results were combined from at least two biologically independent experiments. Data were analysed using the non-parametric Mann–Whitney test.
Gut microbiome induces expression of mosGCTLs and AMPs via the Imd pathway
We assessed the expression of mosGCTLs in adult A. aegypti mosquitoes following systemic infection with E. coli (ST515 strain) and these four gut bacteria. Expressions of 8 mosGCTLs in E. coli-, 7 mosGCTLs in C. testosteroni-, 19 mosGCTLs in C. meningosepticum-, 12 mosGCTLs in S. marcescens-, and 9 mosGCTLs in B. cereus-inoculated mosquitoes were increased by more than twofold (Supplementary Table 3). Both mosGCTL-29 and mosGCTL-32 were significantly induced by these bacterial infections (Fig. 4a). Insects are equipped with multiple immune signaling pathways that respond to microbial invasion, including the immune deficiency (Imd), Toll and JAK–STAT pathways that are best known to respond to microbial invasion with production of AMPs2. Knockdown of Imd pathway components (Imd and Rel2), but not factors of any of the other pathways (Supplementary Fig. 4a,b), significantly impaired the induction of mosGCTL-29 and mosGCTL-32 at 4 h after E. coli (Fig. 4b) and gut bacterial (Supplementary Fig. 5a,c,e,g) infections. At the same time, the expression of AMP genes is also highly induced (Fig. 4c), but this induction is dramatically impaired by silencing modulators of the Imd pathway (Fig. 4d and Supplementary Fig. 5b,d,f,h). Both mosGCTLs and AMPs are constitutively expressed in the mosquito midguts, and their expression is suppressed following feeding with antibiotics, indicating that it is triggered by commensal bacteria residing in the gut lumen (Fig. 4e). Indeed, silencing the Imd and Rel2 genes significantly reduced the constitutive expression of mosGCTLs and AMPs in the midguts (Fig. 4f). Taken together, our data indicate that constitutive expression of mosGCTLs and AMPs is simultaneously triggered by the gut microbiome via the Imd pathway.
Figure 4. mosGCTLs and AMPs are simultaneously regulated by the Imd pathway.
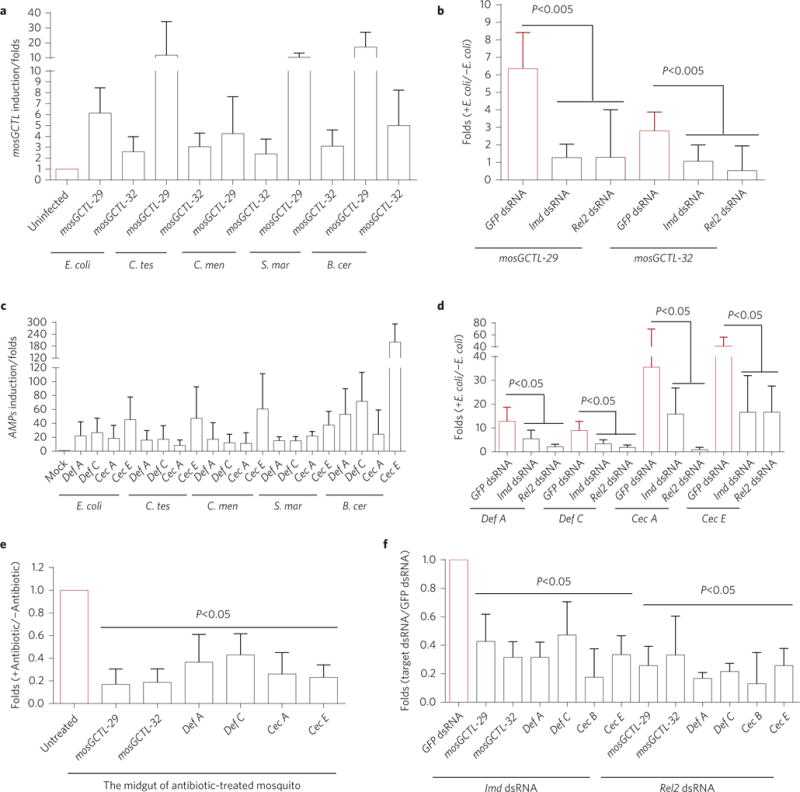
a,c, The expression of mosGCTLs and AMPs were simultaneously induced by systemic inoculation with E. coli, C. testosteroni (C. tes), C. meningosepticum (C. men), S. marcescens (S. mar) and B. cereus (B. cer). The mock-treated mosquitoes received inoculations of PBS buffer. The mRNA abundance of mosGCTLs (a) and AMPs (c) was assessed 4 h after bacterial inoculation. The induction is presented as the fold-change relative to that in the mock mosquitoes without bacteria treatment. b,d, The role of the Imd signaling pathway in mosGCTL and AMP induction. The key factors in the Imd pathway (Imd and Rel2) were silenced in A. aegypti. A 0.005 OD aliquot of E. coli in PBS was sequentially inoculated 3 days later, and the expression of inducible mosGCTLs (b) and AMPs (d) was assessed 4 h after bacterial inoculation. Mosquitoes inoculated with GFP dsRNA and subsequently infected with E. coli were included as controls. The induction is presented as the fold-change relative to that in the uninfected mosquitoes. e, Removal of gut commensal bacteria suppressed the expression of mosGCTLs and AMPs in the midguts. The expression of mosGCTLs and AMPs was determined in the midguts of antibiotic-treated mosquitoes. Mosquitoes without antibiotic treatment served as mock controls. f, Knockdown of the Imd and Rel2 genes reduced the expression of mosGCTLs and AMPs in the midguts. The Imd and Rel2 genes were silenced by dsRNA-mediated thoracic microinjection. Mosquitoes inoculated with GFP dsRNA were included as controls. After 3 days, the midguts of the treated mosquitoes were isolated to determine the mRNA abundance of mosGCTLs and AMPs. The result was read through qPCR and normalized to A. aegypti actin (AAEL011197). The qPCR primers are described in Supplementary Table 5. Data are represented as mean±s.d. in each group and analysed using the nonparametric Mann–Whitney test. The experiment was biologically repeated three times with similar results.
mosGCTLs function as antagonists of AMP-mediated bacterial elimination
Insects employ multiple and diverse immune reactions to control bacterial infections, including production of AMPs2, thioester (TE)-containing proteins (TEPs)34 and reactive oxygen species (ROS)3 or prophenoloxidase (Pro-PO)-mediated melanization35. Therefore, we evaluated whether these mosGCTLs facilitated bacterial survival by regulating the established immune machineries. Knockdown of mosGCTL-29 and mosGCTL-32 failed to regulate these immune reactions and effectors (Supplementary Fig. 6 and Supplementary Table 4), indicating that mosGCTLs facilitate bacterial colonization via mechanisms not involving regulation of the above-mentioned immune responses in A aegypti.
AMPs are immune effectors that electrostatically or hydrophobically interact with bacterial surfaces orchestrating their elimination via different mechanisms including lysis, disruption of proton gradient or other membrane perturbations. The 17 AMPs discovered to date in A. aegypti are categorized into several independent groups, including Defensin (Def), Cecropin (Cec), Diptericin (Dpt), Attacin (Att) and Gambicin (Gam)36. We expressed each of these AMPs in Drosophila S2 cells (Supplementary Fig. 7a). Incubation of E. coli cells with supernatants of these AMP-expressing S2 cells revealed that four Defs, seven Cecs, Att and Dpt significantly suppress the bacterial growth (Supplementary Fig. 7b). We have shown that the mosGCTLs are simultaneously regulated with the AMPs via the Imd pathway (Fig. 4 and Supplementary Fig. 5). Intriguingly, incubation of mosGCTL-29 or mosGCTL-32 in the S2 supernatant enabled the neutralization of AMP-mediated E. coli elimination (Fig. 5a).
Figure 5. mosGCTLs are antagonists for AMP-mediated bacterial elimination.
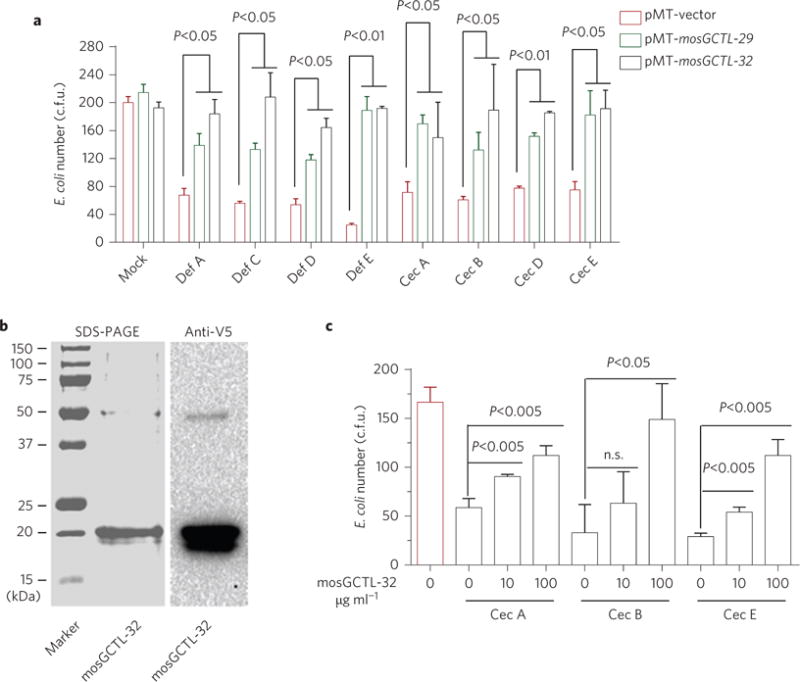
a, mosGCTL-29 and mosGCTL-32 expressed in S2 supernatant interrupted AMP-mediated bacterial elimination. The empty-vector-transfected S2 supernatant with AMPs served as the mock control. The S2 supernatant with mosGCTLs and/or AMPs was individually mixed with the E. coli cells. b, Expression and purification of mosGCTL-32 using a Drosophila S2 cell expression system. c, The purified mosGCTL-32 protein antagonized Cec-mediated bacterial killing. The purified recombinant mosGCTL-32 protein was pre-incubated with the E. coli cells, and subsequently the synthesized Cec peptides were added to the cells. The same concentration of BSA was used as the mock control. a,c, After 2 h of incubation, E. coli viability was assessed by the c.f.u. assay. Data are represented as mean±s.d. in each group and analysed using the non-parametric Mann–Whitney test. All experiments were biologically repeated three times with similar results.
To validate the antagonistic role of mosGCTLs, we expressed and purified a recombinant mosGCTL-32 protein in the Drosophila S2 expression system (Fig. 5b). Expression of mosGCTL-29 in S2 cells was too low for successful purification (data not shown). The expression of mosGCTL-32 was probed by immuno-staining with anti-V5 targeting a tag on the recombinant protein (Fig. 5b). We also commercially synthesized the AMPs Cec-A, Cec-B and Cec-E, which are inducible by the gut commensal bacteria and antagonized by mosGCTLs. In vitro synthesis of Def peptides was unsuccessful due to their mispaired disulfide bond. The bacterial cells were dramatically killed by incubation with the synthesized Cec peptides. However, incubation of purified mosGCTL-32 recombinant protein prevented the Cec-mediated elimination (Fig. 5c), indicating that mosGCTLs act as immune antagonists to protect microbial flora in mosquitoes.
mosGCTLs interrupt the deposition of antimicrobial peptides onto bacterial cells
The carbohydrate recognition domain of C-type lectins is capable of binding polysaccharides on the surface of microorganisms37. Next, we assessed whether the identified mosGCTL directly interfaced with bacterial cells, thereby protecting them from mosquito immune responses. E. coli cells were incubated with purified mosGCTL-32 protein; the coating of mosGCTL-32 on the bacterial surface was clearly detected using immunofluorescence staining (Fig. 6a) and flow cytometry (Fig. 6b). In a sugar competition assay, mosGCTL-32 bound on the bacterial surface was specifically eluted using a buffer containing galactose (Fig. 6c,i), while incubation of mosGCTL-32 with galactose impaired mosGCTL-32 binding to E. coli cells (Fig. 6c,ii). The coating of the bacterial surface with mosGCTLs was determined by western blotting with an anti-V5 antibody (Fig. 6c). These data indicate that surface polysaccharides, presumably involving galactose, mediate the interaction between mosGCTL-32 and microorganisms. Using immunofluorescence microscopy (Fig. 6d) and flow cytometry (Fig. 6e), we showed that mosquito Cecropins, a well-characterized group of AMPs, directly deposit on the bacterial cells. However, preincubation of bacterial cells with mosGCTL-32 significantly reduced the amount of Cec-associated E. coli cells detected by both immunofluorescence and flow cytometry, indicating that mosGCTLs protect bacteria cells by directly impairing the deposition of AMPs onto bacterial cells. We next assessed the role of mosGCTLs in maintenance of commensal gut microorganisms in A. aegypti. Compared with the GFP dsRNA-treated controls, knockdown of mosGCTL-29 and mosGCTL-32 enhanced the number of gut microorganisms coated by Cec peptides that were orally introduced by sugar feeding (Fig. 6f,g). Altogether, mosGCTLs maintain survival of gut microbiome by directly impairing the deposition of AMPs onto the bacterial surface.
Figure 6. mosGCTLs interrupt the deposition of AMPs onto bacteria cells.
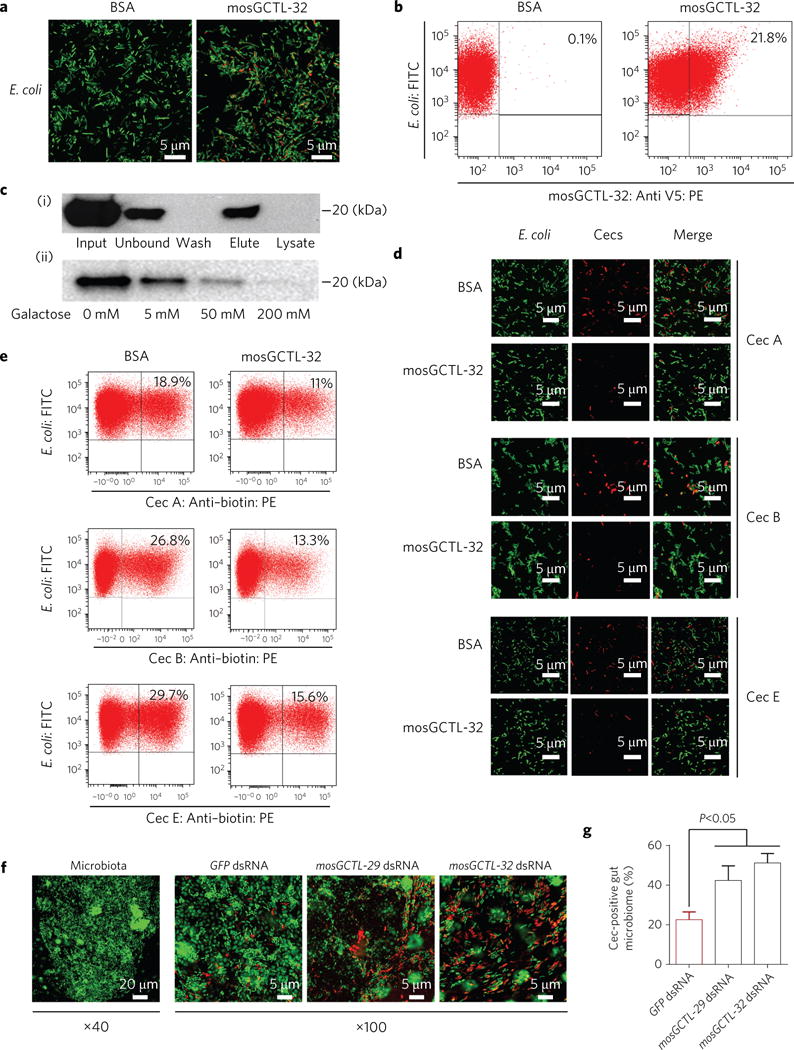
a,b, Determination of the interaction between mosGCTL-32 and E. coli cells by confocal microscopy (a) and flow cytometry (b). c, The interaction between mosGCTL-32 and bacterial cells was mediated by surface polysaccharides. (i) Elution assay. (ii) Competitive assay. All samples with mosGCTLs was determined by western blotting with an anti-V5 antibody. d,e, mosGCTLs interrupt the deposition of AMPs onto bacterial cells. The cells with AMPs were assessed using an immunofluorescence assay (d) or numbered using a flow cytometry assay (e). The E. coli cells (ST515 strain) were equipped with a GFP reporter with constitutive expression. f,g, The role of mosGCTLs in maintenance of commensal bacteria in the mosquito midguts. The mosGCTLs-silencing mosquitoes were fed with the Alexa 546-labelled Cec mixture (with an equal amount of Cec A, B and E). Mosquitoes inoculated with GFP dsRNA were used as negative controls. The midguts of fed mosquitoes were dissected at 4 h post oral feeding. (f) SYTO 16 green fluorescent nucleic acid dye was used to stain gut microbiome DNA. (g) The percentage (%) of Cec-coated cells was calculated as red-stained cells/total green-stained cells. The result was independently measured in the six different midguts. Data are represented as mean±s.d. in each group and analysed using the non-parametric Mann–Whitney test. a,d,f, The bacteria were imaged with the multiple track mode of a Zeiss LSM780 meta confocal microscopy. The scale bars represent 5 μm in a, b, and the right panel of f (×100), and represent 20 μm in the left panel of f (×40). a–g, The experiments were biologically repeated three times with similar results.
Discussion
Commensal bacteria maintain homeostasis in the insect gut4. The release of antimicrobial peptides (AMPs), which is precisely regulated by NF-κB-like signals, balances the growth of gut commensal bacteria in mosquitoes5,6. Conversely, the symbiotic microbiome might exploit some host factors to offset the adverse effects of gut immunity activation by themselves, thereby allowing their adaptation to the harsh gut environment for long-term colonization. In this study, we found that multiple mosGCTLs and AMP peptides were simultaneously upregulated by the Imd pathway. These mosGCTLs acted as vital players in colonization of commensal bacteria in the mosquito midgut. They directly interface with the bacterial cells and interrupt AMP deposition, exhibiting a role for immune antagonists in protecting commensal bacteria from AMP-mediated elimination. The commensal microbial flora includes many species of microorganisms, most of which are Gram-negative bacteria6. We identified seven mosGCTLs that functioned as susceptibility factors in E. coli colonization of A. aegypti. However, two out of these seven mosGCTLs played a role in the colonization of multiple gut bacteria. Carbohydrate recognition is a predominant property of C-type lectins7,8. Different C-type lectins have variable specificities for different carbohydrate moieties. Considering the diversity of bacterial species in the mosquito gut and the variation of carbohydrate moieties on the bacterial surface, we speculate that different bacteria might exploit a different set of mosGCTLs to combat AMP-mediated elimination. Induction of a variable spectrum of mosGCTLs by different bacterial species might be a strategy to shape population diversity of the gut symbiotic microbes, thereby enforcing gut homeostasis. Future studies are needed to determine the mosGCTL binding affinity with other bacterial carbohydrate moieties, thus elucidating the association between mosGCTLs and the gut commensal bacteria. In addition, we should note that both AMPs and mosGCTLs are constitutively expressed at a basal level in microbiome–gut homeostasis. However, little is known about the actual physiological concentrations of the AMPs and mosGCTLs in the mosquito gut and other tissues. We recognize that knowledge of their physiological concentration may provide a basis for fully interpreting the results presented in this study.
The long-term evolutionary interaction of the host immune system with symbiotic bacteria determines their cooperative rather than antagonistic relationship. Indeed, many immune regulatory strategies that allow for the survival of symbiotic bacteria have been described for both mammals31,32 and insects. In the insect guts, a wide spectrum of AMPs are constitutively released into the lumen via gut-microbiome-mediated activation of Imd signaling5,38,39. However, the Imd pathway is largely repressed by multiple negative regulatory mechanisms, to avoid potential detrimental effects on resident symbiotic microbes39–42. The Drosophila amidase peptidoglycan recognition proteins (PGRPs), such as PGRP-LB and PGRP-SC, can scavenge immunostimulatory peptidoglycan and reduce its abundance, thereby enabling hosts to tolerate commensal bacterial colonization in the gut lumen40,42,43. PGRP-LC interacting inhibitor of Imd signaling (PIMS), which is inducible by the Imd pathway, modulates Imd signaling by re-localizing of PGRP-LC from the plasma membrane to an intracellular compartment39. In addition, there are several intracellular negative regulators that restrict the Imd signaling in the Drosophila gut41,44. The presence of multiple non-redundant immunoregulatory mechanisms in the gut helps hosts to tolerate residential microbes and thus achieve commensal bacteria–gut homeostasis.
In haematophagous insects, commensal bacteria largely proliferate during blood meal digestion. Therefore, maintenance of gut homeostasis is a challenge for the insect gut epithelia6. In many blood-feeding insects such as mosquitoes, the midgut can form a dityrosine peritrophic matrix (PM) in response to blood feeding5. In the gut of mosquitoes, rapid bacterial growth following a blood meal induces the expression of an immunomodulatory peroxidase, which associates with dual oxidase to mediate protein cross-linking through forming dityrosine bonds. Indeed, the formation of the PM reduces the rate of diffusion of immune elicitors and decreases microbial recognition by immune receptors on the gut epithelia, thereby playing an important role in prevention of immune overreaction against the symbiotic microbiome during blood feeding5. In this study, we found that several soluble C-type lectins coat bacterial surfaces via polysaccharide. It is plausible that the mosGCTL coat may not only prevent AMPs from obtaining access to bacteria, but it may also mask the bacterial ligands from being recognized by pattern recognition receptors (PRRs) of the gut epithelial cells, a similar scenario to the PM. However, genetic manipulation of mosGCTLs failed to alter the expression of AMPs and many other key immune genes, indicating that mosGCTLs do not interfere with PRR-mediated signaling.
C-type lectins demonstrate different functions after binding particular oligosaccharides on the microbial surface, even when they share similar sequences. Both MBLs and surfactant proteins (SPs) are members of the mammalian collectin family. However, MBLs but not SPs bind mannosylated components on microbial surfaces and subsequently associate with MBL-associated serine proteases (MASPs) to activate the complement cascades9,45. Results of a previous study indicated that some insect C-type lectins recognized bacterial cells to facilitate haemocyte-mediated phagocytosis22. In agreement with the previous observation, we identified a member of the C-type lectins (mosGCTL-24) that played a role in the resistance to E. coli infection in A. aegypti. Knockdown of mosGCTL-27 (AAEL011612) enhanced the burden of C. testosteroni in mosquitoes. However, our dsRNA-mediated screening of the mosGCTL family identified multiple C-type lectins in A. aegypti and C. pipiens pallens that conversely facilitated bacterial survival by interrupting AMP deposition on bacterial cells, further implicating the pleiotropic roles of C-type lectins in the interaction between invertebrates and microorganisms.
Mosquitoes transmit multiple human pathogens of medical importance throughout the world. The mosquito gut is a pivotal pathogen entry site that determines pathogen colonization and survival. The gut harbors a microbiome that interacts with midgut cells and is important for vector physiology46. Recently accumulated evidence indicated that the gut microbiome influenced vector competence6,26,47,48, implying that midgut microbial flora would be a promising intervention target for the control and prevention of mosquito-borne diseases. Our investigation elucidated an unappreciated role for C-type lectins in gut ecology, uncovered a mechanism by which the commensal microbiome conquered constitutive immune activation, and furthermore might offer intervention targets for the control of vector-borne diseases in nature.
Methods
Mosquitoes and bacteria
Aedes aegypti (the Rockefeller strain) and Culex pipiens pallens (the Beijing strain) were maintained in the laboratory in a low-temperature illuminated incubator (model 818, Thermo Electron Corporation) at 26 °C and 80% humidity according to standard rearing procedures18. The E. coli ST515 strain, which is equipped with a GFP reporter and spectinomycin resistance, was cultured on LB plates with 100 μg ml−1 spectinomycin at 37 °C. C. testosteroni, C. meningosepticum, S. marcescens and B. cereus were isolated from the A. aegypti midgut and cultured on LB plates without any antibiotics at 37 °C.
Antibody generation
The mosGCTL-29 and mosGCTL-32 genes were amplified from A. aegypti cDNA and cloned into a pET-28a (+) expression vector. The cloning primers are presented in Supplementary Table 5. The recombinant mosGCTL proteins were expressed in the E. coli BL21 DE3 strain, with the insoluble form in inclusion bodies. The proteins were resolved by 8 M urea and purified using a purification kit (Clontech, Cat. No 635515). Polyclonal antibodies were produced by three boosting immunizations in C57BL/6 mice. The antibody against Drosophila actin (CST, Cat. No 4967) was used for detection of A. aegypti actin. The antibodies for the tags were purchased from the Medical & Biological Lab (MBL).
Generation of antibiotic-treated mosquitoes and bacterial oral feeding
Mosquitoes were provided cotton balls moistened with a 10% sucrose solution including 20 units of penicillin and 20 μg of streptomycin ml−1 for 3 days26. Then, the mosquitoes were starved for 24 h to allow the antibiotics to be metabolized prior to bacterial challenge. The removal of the microbes was confirmed by qPCR using a universal bacteria primer5 after the mosquitoes were decontaminated in 70% ethanol, rinsed in sterile PBS, and the midguts dissected under aseptic conditions. After antibiotic treatment, the mosquitoes were fed a mixture containing C. testosterone, C. meningosepticum, S. marcescens and B. cereus (OD600 = 1) respectively, with fresh human blood (1/1 v/v) via the Hemotek feeding system (6W1). Subsequently, the mosquitoes were cold-anesthetized, and the fully fed mosquitoes were separated into new containers and maintained for further investigation.
Gene silencing and bacterial systemic inoculation in mosquitoes
We have described the detailed procedures used for gene silencing in mosquitoes elsewhere18,19. Briefly, the mosquitoes were cold-anesthetized on a cold tray (BioQuip), and subsequently 1 μg 300 nl−1 of dsRNA was microinjected into the mosquito thoraxes. The mosquitoes were allowed to recover for 3 days under standard rearing conditions. Then, the mosquitoes received a thoracic microinjection of with bacteria suspended in PBS (OD600 = 0.005 for the E. coli ST515 strain; OD600 = 0.05 for C. testosteroni; OD600 = 0.05 for C. meningosepticum; OD600 = 0.05 for S. marcescens; OD600 = 0.05 for B. cereus). After 4 h of rearing, the treated mosquitoes were killed and homogenized for the quantitative assays. For the E. coli-treated mosquitoes, the mosquito lysates in PBS buffer were plated onto agar plates containing 100 μg spectinomycin ml−1. The number of cells was counted after overnight incubation at 37 °C (the colony-forming unit assay). For the mosquitoes inoculated with C. testosterone, C. meningosepticum, S. marcescens and B. cereus, the whole bodies of the mosquitoes were homogenized in Buffer I of an RNeasy Mini Kit (Qiagen, Cat. No 74106) with a Pestle Grinder System (Fisher Scientific, Cat. No 03-392-106). The detailed procedure for total RNA isolation was described in the RNeasy Kit manual. Complementary DNA (cDNA) was randomly reverse-transcribed using an iScript cDNA Synthesis Kit (Bio-Rad, Cat. No 1708891). The burden of C. testosterone, C. meningosepticum, S. marcescens and B. cereus was measured with qPCR with the primers listed in Supplementary Table 5 and subsequently normalized to A. aegypti actin (AAEL011197).
Measurement of gut microbiota by 16S rDNA qPCR
The surface of a mosquito was sterilized with 70% ethanol and washed twice with sterile PBS. The midgut was carefully removed from the abdomen under aseptic conditions to reduce contamination from bacteria. Midgut total RNA was extracted from the sample with the RNeasy Mini Kit (Qiagen, Cat. No 74106). cDNA was randomly reverse-transcribed using an iScript cDNA Synthesis Kit (Bio-Rad, Cat. No 1708891). The 16S rDNA was amplified using a pair of universal primers5. The burden of gut microbiota was normalized to A. aegypti actin (AAEL011197).
Protein generation in a Drosophila expression system
The mosGCTL-32 gene was amplified and inserted into the pMT/BiP/V5-His A vector (Invitrogen, Cat. No V4130-20), and then the recombinant plasmids were transfected into Drosophila S2 cells in combination with a hygromycin selection vector (pCo-Hygro) for construction of stable cells. The primers for PCR and gene cloning are shown in Supplementary Table 5. The stable-cell screening and purification have been described in our previous study18,19. Briefly, the transfected cells were selected using 300 μg hygromycin-B ml−1 (Invitrogen, Cat. No 10687-010) for 4 weeks. The resistant cells were grown in spinner flasks, switched to Express Five serum-free medium (GIBCO, Invitrogen, Cat. No 10486025) for 3 days, and induced with copper sulfate at a final concentration of 500 μM for 4 days. The culture medium was collected for protein purification with a metal affinity resin (Clontech, Cat. No PT1320-1). The protein was eluted with 100 mM imidazole, extensively dialysed against PBS (pH 7.5), and concentrated via centrifugal filtration through a 10-kDa filter (Millipore, Cat. No pLCC07610). The protein concentration was measured using Protein Assay Dye (Bio-Rad, Cat. No 500-0006) and a Nanodrop 2000c spectrophotometer (Thermo Scientific). The protein purity was verified with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), and the specificity of purification was confirmed by immunoblotting.
Sugar competitive assay
We determined mosGCTL-32 binding to the polysaccharides of the bacterial surface using two separate approaches. (1) Elution assay. The E. coli cells were pelleted by centrifugation at 12,000 g for 1 min, washed three times and resuspended with PBS. The purified mosGCTL-32 protein was incubated with the cells in PBS for 1 h at 4 °C. The microorganisms were washed four times with PBS. Then, the binding proteins were released with 50 μl elution buffer containing 50 mM galactose. Samples of the unbound proteins, the wash buffer, the elution buffer with galactose and the bacterial lysates were subjected to detection by immunoblotting. (2) Competitive assay. The purified mosGCTL-32 protein was incubated with different concentrations of galactose in PBS (0, 5, 50, and 200 mM) for 30 min at 4 °C. Then, the E. coli cells were added to the mixture above and incubated for 1 h at 4 °C. The microorganisms were washed four times with PBS. The bacterial lysates were detected by immunoblotting.
Detection of AMP-mediated anti-bacterial activity
The A. aegypti AMP genes without signal sequences were cloned into the pMT/BiP/V5-His vector (Invitrogen, Cat. No V4130-20), and subsequently the recombinant plasmids were transfected into Drosophila S2 cells cultured in medium without antibiotics. The supernatants were collected 48 h after transfection. The expression of these AMP peptides was determined by western blotting with an anti-V5 antibody. For the anti-bacterial assay, the E. coli cells were washed three times with PBS and diluted to 2.5 × 102 cells per 100 μl. Subsequently, 100 μl of the supernatants with AMPs were added to 100 μl of bacteria suspensions for a 1 h incubation at room temperature. The mixture was plated onto LB plates with spectinomycin, and the E. coli colonies were counted for the c.f.u. assay.
Antagonistic role of mosGCTL in Cec-mediated bacterial elimination
The E. coli cells were washed three times with PBS and diluted to 2.5 × 102 cells per 100 μl. 10 μg ml−1 or 100 μg ml−1 of purified mosGCTL-32 protein (final concentration) was added to the bacteria suspension and incubated for 30 min at 4 °C. Then, the synthesized Cec peptides (Lifetein), including 2 μg Cec A, 0.2 μg Cec B and 0.2 μg Cec E per 100 μl, were separately added and incubated with the bacteria for 1 h at room temperature. The viability of the bacterial cells was determined by the c.f.u. assay.
Interruption of the deposition of Cec peptides onto bacteria cells by mosGCTL
The cells of the E. coli ST515 strain (with the GFP reporter) were incubated with mosGCTL-32 protein (50 μg ml−1 final concentration) for 1 h at 4 °C. The same amount of BSA was added to the bacterial cells as a negative control. After five washes, 2 μg Cec A, 0.2 μg Cec B and 0.2 μg Cec E peptides labeled with biotin were incubated with the bacterial cells for 1 h at room temperature, respectively. Subsequently, the cells were fixed with 4% paraformaldehyde (USB) after five washes. For the flow cytometry assay, the cells were stained with an anti-biotin rabbit antibody (CST, Cat. No 5597) and an Alexa 546-conjugated anti-rabbit IgG antibody (Invitrogen, Cat. No A-11010). Then, the treated cells were examined using a FACS Calibur flow cytometer (BD Biosciences). Dead cells were excluded on the basis of forward and side light scatter. The data were analysed using the FlowJo software. For the confocal imaging, the bacterial cells were stained by an Alexa 546-conjugated anti-rabbit IgG and placed on sialylated slides, and subsequently imaged with the multi-track mode of a Zeiss LSM 780 meta confocal microscope.
Supplementary Material
Acknowledgments
This work was funded by grants from the National Natural Science Foundation of China (81301412, 81422028, 81571975 and 61472205), the National Key Basic Research Program of the Chinese Ministry of Science and Technology (MOST) (2013CB911500), the Excellent Young Scientist Foundation of Beijing (2013D009004000002), Grand Challenges Explorations of the Bill & Melinda Gates Foundation (OPP1021992), and the National Institute of Health of the United States (AI103807 and AI099625). We thank the Professor George K. Christophides from Imperial College London, who provided critical suggestions for the manuscript. G.C. is a Newton Advanced Fellow awarded by the Academy of Medical Sciences and the Newton Fund, and a Janssen Investigator of Tsinghua University. We thank the technical supports from the Core Facility of Center for Life Sciences and Center of Biomedical Analysis (Tsinghua University).
Footnotes
Accession codes. The nucleotide sequences of the mosquitoes Aedes aegypti and Culex quinquefasciatus used in this study were deposited in the gene sets AaegL3.3 and CpipJ2.2 of Vectorbase database (https://www.vectorbase.org).
Author contributions
G.C. designed the experiments and wrote the manuscript; X.P. performed the majority of the experiments and analysed data; X.X., R.Z., Y. L. and J.L. helped with the RNA isolation and qPCR detection; Q.L. provided Culex pipiens pallens and contributed to the discussion. P.W. contributed experimental suggestions and strengthened the writing of the manuscript. All authors reviewed, critiqued and provided comments to the text.
Additional information
Supplementary information is available online.
Competing interests
The authors declare no competing financial interests.
References
- 1.Valiente Moro C, Tran FH, Raharimalala FN, Ravelonandro P, Mavingui P. Diversity of culturable bacteria including Pantoea in wild mosquito Aedes albopictus. BMC Microbiol. 2013;13:70–80. doi: 10.1186/1471-2180-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 3.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 4.Buchon N, Broderick NA, Lemaitre B. Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nature Rev Microbiol. 2013;11:615–626. doi: 10.1038/nrmicro3074. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science. 2010;327:1644–1648. doi: 10.1126/science.1184008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drickamer K. Two distinct classes of carbohydrate-recognition domains in animal lectins. J Biol Chem. 1988;263:9557–9560. [PubMed] [Google Scholar]
- 8.Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 9.Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 10.Tailleux L, et al. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. 2003;197:121–127. doi: 10.1084/jem.20021468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halary F, et al. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity. 2002;17:653–664. doi: 10.1016/s1074-7613(02)00447-8. [DOI] [PubMed] [Google Scholar]
- 12.Klimstra WB, Nangle EM, Smith MS, Yurochko AD, Ryman KD. DC-SIGN, and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J Virol. 2003;77:12022–12032. doi: 10.1128/JVI.77.22.12022-12032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tassaneetrithep B, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JL, et al. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterhouse RM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 17.Wang YH, et al. A critical role for CLSP2 in the modulation of antifungal immune response in mosquitoes. PLoS Pathog. 2015;11:e1004931. doi: 10.1371/journal.ppat.1004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng G, et al. A C-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell. 2010;142:714–725. doi: 10.1016/j.cell.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, et al. Transmission-blocking antibodies against mosquito C-type lectins for dengue prevention. PLoS Pathog. 2014;10:e1003931. doi: 10.1371/journal.ppat.1003931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang XW, Xu YH, Xu JD, Zhao XF, Wang JX. Collaboration between a soluble C-type lectin and calreticulin facilitates white spot syndrome virus infection in shrimp. J Immunol. 2014;193:2106–20117. doi: 10.4049/jimmunol.1400552. [DOI] [PubMed] [Google Scholar]
- 21.Schnitger AK, Yassine H, Kafatos FC, Osta MA. Two C-type lectins cooperate to defend Anopheles gambiae against Gram-negative bacteria. J Biol Chem. 2009;284:17616–17624. doi: 10.1074/jbc.M808298200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanji T, Ohashi-Kobayashi A, Natori S. Participation of a galactose-specific C-type lectin in Drosophila immunity. Biochem J. 2006;396:127–138. doi: 10.1042/BJ20051921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XW, Xu JD, Zhao XF, Vasta GR, Wang JX. A shrimp C-type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. J Biol Chem. 2014;289:11779–11790. doi: 10.1074/jbc.M114.552307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suryawanshi RK, et al. Mosquito larvicidal and pupaecidal potential of prodigiosin from Serratia marcescens and understanding its mechanism of action. Pestic Biochem Physiol. 2015;123:49–55. doi: 10.1016/j.pestbp.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Bahia AC, et al. Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ Microbiol. 2014;16:2980–2994. doi: 10.1111/1462-2920.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez JL, et al. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl Trop Dis. 2012;6:e1561. doi: 10.1371/journal.pntd.0001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dada N, et al. Comparative assessment of the bacterial communities associated with Aedes aegypti larvae and water from domestic water storage containers. Parasit Vectors. 2014;7:391. doi: 10.1186/1756-3305-7-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaio AO, et al. Use of the checkerboard DNA–DNA hybridization technique for bacteria detection in Aedes aegypti (Diptera: Culicidae) (L.) Parasit Vectors. 2011;4:237. doi: 10.1186/1756-3305-4-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leulier F, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nature Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 30.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 32.Palm NW, de Zoete MR, Flavell RA. Immune–microbiota interactions in health and disease. Clin Immunol. 2015;159:122–127. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou D, et al. Cloning and characterization of prophenoloxidase A3 (proPOA3) from Culex pipiens pallens. Comp Biochem Physiol B Biochem Mol Biol. 2012;162:57–65. doi: 10.1016/j.cbpb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moita LF, Wang-Sattler R, Michel K, Zimmermann T, Blandin S. In vivo identification of novel regulators and conserved pathways of phagocytosis in Anopheles gambiae. Immunity. 2005;23:65–73. doi: 10.1016/j.immuni.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Yassine H, Kamareddine L, Osta MA. The mosquito melanization response is implicated in defense against the entomopathogenic fungus Beauveria bassiana. PLoS Pathog. 2012;8:e1003029. doi: 10.1371/journal.ppat.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao XP, et al. Complement-related proteins control Flavivirus infection of Aedes aegypti by inducing antimicrobial peptides. PLoS Pathog. 2014;10:e1004027. doi: 10.1371/journal.ppat.1004027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, et al. The roles of direct recognition by animal lectins in antiviral immunity and viral pathogenesis. Molecules. 2015;20:2272–2295. doi: 10.3390/molecules20022272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lhocine N, et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity. 2011;35:770–779. doi: 10.1016/j.immuni.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Ryu JH, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 42.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaidman-Rémy A, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 44.Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc Natl Acad Sci USA. 2006;103:16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartshorn KL, et al. Mechanism of binding of surfactant protein D to influenza A viruses: importance of binding to haemagglutinin to antiviral activity. Biochem J. 2000;351:449–458. [PMC free article] [PubMed] [Google Scholar]
- 46.Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 47.Meister S, et al. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5:e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cirimotich CM, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


