Abstract
Deficiency of carbamoyl phosphate synthetase I (CPSI) results in hyperammonemia ranging from neonatally lethal to environmentally induced adult-onset disease. Over 24 years, analysis of tissue and DNA samples from 205 unrelated individuals diagnosed with CPSI deficiency (CPSID) detected 192 unique CPS1 gene changes, of which 130 are reported here for the first time. Pooled with the already reported mutations, they constitute a total of 222 changes, including 136 missense, 15 nonsense, 50 changes of other types resulting in enzyme truncation, and 21 other changes causing in-frame alterations. Only ~10% of the mutations recur in unrelated families, predominantly affecting CpG dinucleotides, further complicating the diagnosis because of the “private” nature of such mutations. Missense changes are unevenly distributed along the gene, highlighting the existence of CPSI regions having greater functional importance than other regions. We exploit the crystal structure of the CPSI allosteric domain to rationalize the effects of mutations affecting it. Comparative modeling is used to create a structural model for the remainder of the enzyme. Missense changes are found to directly correlate, respectively, with the one-residue evolutionary importance and inversely correlate with solvent accessibility of the mutated residue. This is the first large-scale report of CPS1 mutations spanning a wide variety of molecular defects highlighting important regions in this protein.
Keywords: urea cycle, CPS1, hyperammonemia, structure, evolutionary
Introduction
Carbamoyl phosphate synthetase I (CPSI; HUGO-approved gene symbol, CPS1; MIM# 608307; E.C. 6.3.4.16) catalyzes the entry of ammonia into the urea cycle in the first and limiting step of this cycle [Krebs and Henseleit, 1932; Shambaugh, 1977] and its deficiency causes clinical hyperammonemia. High level of CPSI expression is limited to the liver [Neill et al., 2009], where the enzyme plays its essential role as a catalyst of the urea cycle, as well as the intestine, where the intramitochondrial part of the urea cycle is operative, generating citrulline for anabolic arginine synthesis [Windmueller and Spaeth, 1981]. Smaller but significant CPSI expression is also observed in the pancreas [Neill et al., 2009].
CPSI is a complex enzyme that makes carbamoyl phosphate (CP) from ammonia, bicarbonate, and two molecules of ATP in a three-step reaction (Fig. 1, bottom) in which bicarbonate is first phosphorylated by ATP, then the resulting carboxyphosphate reacts with ammonia to give carbamate and finally carbamate is phosphorylated by another ATP molecule yielding CP [Nyunoya et al., 1985; Oyanagi et al., 1980; Rubio, 1993; Rubio and Cervera, 1995; Rubio and Grisolia, 1981; Rubio et al., 1981; Ryall et al., 1985; Schofield, 1993; Summar et al., 2003]. Bicarbonate and carbamate are analogous compounds and in agreement with the chemical similarity of the two phosphorylation steps (Fig. 1, bottom) the enzyme, a polypeptide of 160 kDa, which is composed of N-terminal and C-terminal moieties of 40 and 120 kDa (Fig. 1, bottom), exhibits internal homology between the N- and C-terminal halves of its 120 kDa moiety [Nyunoya et al., 1985; Powers-Lee and Corina, 1986; Rubio, 1993; Schofield, 1993]. This moiety is homologous to the large subunit of Escherichia coli CPS, a subunit that catalyzes the whole reaction of CP synthesis from ammonia [Meister, 1989]. Each homologous half of the 120-kDa moiety is composed of N- and C-terminal domains of ~40 and ~20kDa [Alonso et al., 1992; Alonso and Rubio, 1995; Powers-Lee and Corina, 1986; Rodriguez-Aparicio et al., 1989]. The 40-kDa domains of the N- and of the C-half (labeled BPSD and CPSD in Fig. 1, bottom) catalyze, respectively, the phosphorylation of bicarbonate and of carbamate, whereas the C-terminal domain of 20 kDa (Fig. 1, bottom, ASD) hosts the site for the unique activator N-acetyl-L-glutamate, an essential allosteric effector of CPSI without which the enzyme is rendered inactive [Fresquet et al., 2000; Hart and Powers-Lee, 2009; Javid-Majd et al., 1996; Nyunoya et al., 1985; Oyanagi et al., 1980; Pierrat and Raushel, 2002; Rubio, 1993; Rubio and Cervera, 1995; Rubio and Grisolia, 1981; Rubio et al., 1981; Ryall et al., 1985; Schofield, 1993; Stapleton et al., 1996; Summar et al., 2003]. No function has been assigned thus far to the corresponding 20-kDa C-terminal domain of the first homologous half of the enzyme (labeled UFSD in Fig. 1, bottom). The CPSI polypeptide also has a 40-kDa N-terminal domain, which is homologous to the small subunit of bacterial CPS [Nyunoya et al., 1985], a subunit composed of two domains (labeled by homology in CPSI as ISD and GSD, Fig. 1 bottom) [Thoden et al., 1997]. In the bacterial enzyme the GSD domain binds and hydrolyzes the glutamine used as the source of ammonia [Meister, 1989]. However, human CPSI cannot use glutamine [Rubio et al., 1981]. Nevertheless, evidence obtained with the frog enzyme [Saeed-Kothe and Powers-Lee, 2003] suggests that the GSD domain of CPSI remains the entry site for the ammonia substrate and that this domain may contribute to the lower Km for ammonia seen in CPSI relative to other CPSs. CPSI is synthesized as a longer precursor having a 38-residue N-terminal mitochondrial targeting leader peptide (LP in Fig. 1) which is cleaved off upon mitochondrial entry [Ryall et al., 1985; Schofield, 1993].
Figure 1.
Distribution of mutations among the 38 CPS1 exons and correspondence with the domains of the protein. The CPSI coding sequence is schematized as a horizontal bar at the middle, with segments therein corresponding to individual exons (numbered; odd exons are shadowed gray). A parallel bar of the same length at the bottom schematizes the enzyme polypeptide, giving in different color the various enzyme domains, which are shown in exact correspondence with the exons encoding them: LP, mitochondrial targeting peptide (not present in mature CPSI). ISD and GSD, the two subdomains of the 40-kDa CPSI region corresponding to the small subunit of E. coli CPS. In the latter enzyme the ISD and the GSD are, respectively, the intersubunit interaction domain and a glutaminase domain. The glutaminase activity is lost in human CPSI. BPSD, UFSD, CPSD, and ASD, the four domains of the 120-kDa region of CPSI corresponding to the E. coli CPS large subunit. BPSD and CPSD, respective bicarbonate and carbamate phosphorylation domains. ASD, allosteric domain (the domain that binds NAG). UFSD, unknown function domain. The steps of the CPSI reaction are schematized below the protein, placing the two phosphorylation domains below the residues catalyzing them. Planted on the exons of the coding sequence, the banners in blue localize the missense mutations summarized in Supp. Table S1. Inverted banners below the coding sequence give the other types of mutations, in the color code as indicated at the figure foot. Novel mutations are boldface and italicized. Mutations with known neonatal-onset CPSID are shown in red font.
CPSI deficiency (CPSID; MIM# 237300), an autosomal recessive inborn error, can present as a devastating metabolic disease dominated by severe hyperammonemia in neonates or as a more insidious late-onset condition, generally manifesting as life-threatening hyperammonemic crises under catabolic situations. The frequency of CPSID is estimated to be 1:50,000–1:100,000 based on the observations in the Japanese and U.S. cohorts [Summar et al., 2008; Tuchman et al., 2008; Uchino et al., 1998], although the true incidence is still difficult to assess, because likely many patients die unreported, undiagnosed or both, with detection varying with the degree of awareness of the clinician and the access to testing, diagnostic, and life-supporting facilities. Genetic diagnosis is a key element in the diagnosis of CPSID and in the procurement of counseling, prenatal diagnosis, and eventually, of future procedures for disease-free embryo selection. Despite the fact that the human cDNA sequence has been known for nearly 20 years [Haraguchi et al., 1991] the path to genetic diagnosis has been slow. The large size of the CPS1 gene (4500-nucleotide coding sequence distributed among 38 exons separated by 37 intervening introns) [Häberle et al., 2003; Summar et al., 2003] and of the CPSI protein (1500 residues before cleavage of the 38-residue N-terminal signal peptide) and the complex multidomain/multiactive center/activator-dependent nature of the enzyme predict the existence of a large number of mutations associated with CPSID. However, <100 mutations have been reported thus far. Most of these reported mutations are small nucleotide changes, whereas large genomic rearrangements encompassing CPS1 have been reported infrequently [Aoshima et al., 2001a; Loscalzo et al., 2004].
We report here a wide repertoire of mutations, detected in tissues and DNA samples collected from 205 CPSID individuals over a period of 24 years. The samples were analyzed using a variety of diagnostic techniques revealing 192 unique changes of which the vast majority is reported here for the first time. These mutational data are merged with all previously reported mutations to provide a mutational database of sufficient size to allow assessing the frequency of different types of mutations in CPSID, the preference or lack of preference of missense changes for given amino acids or regions of the protein or of the gene and when clinical data permit for trying to connect the mutation to the disease presentation.
One very important drawback for rationalizing the effects of CPS1 mutations and for predicting the disease-causing potential of given mutations is the lack of direct structural CPSI data, because most structural information was derived from sequence analysis, limited proteolysis results, microcalorimetric data on protein unfolding, and protection by ligands and substrate-and cofactor-binding studies of the rat enzyme [Alonso et al., 1992; Alonso and Rubio, 1995; Cervera et al., 1993; Nyunoya et al., 1985; Powers-Lee and Corina, 1986; Rodriguez-Aparicio et al., 1989; Rubio, 1993; Rubio and Cervera, 1995; Schofield, 1993]. Rat CPSI has been used as a model for human CPSI [Pekkala et al., 2010] because both enzymes have identical length and exhibit 95% amino acid sequence identity but unhappily the structure of the rat enzyme or of any other CPSI has not been determined. Site-directed mutagenesis of CPSI could not be used to monitor the impact of mutations on enzyme stability and functionality until very recently, when insect cell and yeast cell systems have allowed the recombinant production of CPSI [Ahuja and Powers-Lee, 2008; Pekkala et al., 2009]. Although the crystal structure of Escherichia coli CPS has been known for some time already [Thoden et al., 1997], there was some uncertainty concerning its extrapolation to CPSI because of the differences between the two enzymes (in the case of the bacterial enzyme, presence of two subunits, use of glutamine as its substrate and activity in the absence of effectors) [Meister, 1989]. Nevertheless, the E. coli CPS structure has proved useful in the analysis of the impact of mutations affecting the catalytic domains, because these domains are shared by both enzymes [Yefimenko et al., 2005]. Only recently, the crystal structure of the C-terminal domain of human CPSI produced in a cell-free expression system was deposited in the Protein Databank (file 2yvq). This structure closely resembles that of the corresponding C-terminal domain of E. coli CPS [Pekkala et al., 2009]. Therefore, despite the absence of the remainder of the enzyme, the isolated C-terminal domain appears to be well folded. This structure was used to provide clues on the mechanism of NAG binding and of disease causation by mutations affecting that domain [Pekkala et al., 2009, 2010] but, in agreement with the limited size of this domain (~1/8 of the entire CPSI chain) the number of mutations falling on it is not large (see below). Nevertheless, the high similarity of the structures of this domain for CPSI and for E. coli CPS has placed on firm ground the view that the E. coli CPS structure can be used to model human CPSI, particularly because the sequence identity between the C-terminal domains of both enzymes is much lower than for the other domains. We have carried out this modeling, visualizing the mutations presented here on this structural context.
Methods
Sample Accrual
From 1985 to 2009, blood samples, DNA, or skin biopsies from 294 patients from North America, South America, Central America, Europe, the Middle East, Taiwan, Australia, and South Africa suspected of being affected by CPSID were investigated at either Vanderbilt University, at Münster or Zurich University or at Baylor College of Medicine. Patients were identified as those likely to have CPSID based on analysis of biochemical markers, liver enzyme testing, and/or a negative genetic study for defects of the N-acetylglutamate synthase (NAGS) gene or family history. Members of the same family with the same mutations are reported only once in this study to avoid overestimation of mutation rates and frequencies. Whenever available, parental DNA was investigated for confirmation of found mutations. Appropriate consent was obtained from all patients or their parents at the time at which each sample was collected in accordance with regulations at that time.
Mutation Detection and Characterization in CPSID Patients
We determined molecular changes in 205 unrelated CPSID patients both with neonatal and late-onset disease. While conducting this study, we utilized a wide range of mutation detection techniques. Our first series of patients were studied by reverse transcription and screening of cDNA derived from fibroblast or lymphoblastoid cell lines. Since that time we have developed the exon-flanking primers allowing direct analysis of the genomic DNA [Summar et al., 2003]. Whenever RNA was available, mutations affecting mRNA processing were assessed by screening the CPS1 message. Mutation detection methods included dideoxy-fingerprinting, restriction-endonuclease fingerprinting, single-strand conformation polymorphism analysis, fluorescent-detected melting, and direct-sequence analysis. Genomically screened sequences extended 500 bases into the sequence 5′ to the transcript start site and 50 bases past the 3′ end of the transcript in genomic DNA.
Mutation analyses at Münster and at Zurich University were performed using cultured skin fibroblasts as the source of RNA and DNA. Reverse transcriptase (RT)-PCR reactions using 1 µg of total RNA or direct sequencing of cDNA, followed by confirmation of identified mutations using genomic DNA were performed [Häberle et al., 2003; Rapp et al., 2001].
All changes have been confirmed by sequence analysis on fresh PCR products and were only classified as mutations if they were not identified in a group of 200 unrelated DNA samples of mixed ethnic background. Furthermore, a disease-causing role was assumed if the mutation was found on DNA and on mRNA, cosegregating with the disease or at least if the nature of the mutation suggested severity, that is, if a severe impact on the CPSI protein is expected. Polymorphic findings in the CPS1 gene have been reported elsewhere [Summar et al., 2004].
Nucleotide numbers correspond to GenBank entries NM_001875.2 for the cDNA and NC_000002.10 for DNA. Nucleotide numbering reflects cDNA numbering with 11 corresponding to A of the translation initiation codon in the reference sequence (according to journal guidelines under www.hgvs.org/mutnomen).
Molecular Modeling of Human CPSI Protein
A 3D structure of the human CPSI protein was obtained using comparative modeling of the E. coli CPS homologue bound to the inert ATP analogue AMPPNP (Protein Data Bank file 1bxr) [Thoden et al., 1999]. The 1bxr structure is the best available structural template found using PDBsum search [Laskowski et al., 1997]. It consists of two subunits that are fused in the human sequence. Chain A is the larger subunit homologous to the C-terminal moiety of CPSI and roughly maps 1100 amino acids. Chain B is the smaller subunit homologous to the N-terminus of CPSI and roughly maps 400 amino acids. The residue identity between the E. coli and human CPSI sequences is 41% for each chain, suggesting that the root mean square deviation between the human and E. coli CPSI structures should be within 2A [Chothia and Lesk, 1986]. Alignment was performed using MUSCLE according to a procedure described elsewhere [Edgar, 2004]. A protein structure homology-modeling server, SWISS-MODEL was used for the human CPSI modeling [Arnold et al., 2006] in the alignment mode for each domain/subunit. The derived models were combined into one structure with small ligands added in the original coordinates. Insertion of amino acids and sequence joining because of deletions was done using local geometric characteristics per SWISS-MODEL routine. These modifications could not be verified crystallographically.
The relative evolutionary importance (EI) of CPSI residues was estimated with the evolutionary trace (ET) method [Lichtarge et al., 1996; Mihalek et al., 2004, 2006; Morgan et al., 2006] using an alignment consisting of 182 eukaryotic sequences and the modeled structure. Using ET we computed importance rankings by systematically correlating residues variations with evolutionary divergences, in effect linking perturbations in sequence with perturbations in function as judged by phylogenetic distances. These rankings have been previously validated in large-scale retrospective analyses [Mihalek et al., 2003; Morgan et al., 2006] and from direct mutational studies that separated, rewired, or mimicked functional sites [Baameur et al., 2010; Lichtarge et al., 1997; Ribes-Zamora et al., 2007; Yao et al., 2003]. Here, the residues were ranked and then simply divided according to their percentile ranks in those of high, medium, and low EI as the top 30%, top 30–60%, and 60–100%, respectively. The ligand-binding sites and the interface were defined as the amino acids that are closer than 5Å to the ligand or domain in the atom–atom distances. The solvent accessibility (SA) was calculated as an absolute area using the DSSP algorithm [Kabsch and Sander, 1983]. Then the residues were categorized as high, medium, and low SA, if these values were less than 10, 10–50, and more than 50, respectively.
Results
Mutations Found in Our Cohort of 294 Patients of CPSID
Within the cohort of 294 patients investigated from 1985–2009, 205 patients were confirmed to have CPSID (102 patients at Vanderbilt University, 81 patients at Münster or Zurich University, and 22 patients at Baylor College of Medicine). During the same period, the diagnosis of CPSID could not be confirmed in 5 patients at Vanderbilt, in 30 patients at Münster or Zurich, and in 54 patients at Baylor College of Medicine.
Investigations of the 205 patients revealed a total of 192 different sequence alterations of the CPS1 gene, which are either certain or likely to be pathogenic. All possible types of mutations were detected. Of the 192 mutations identified, the majority (n = 115) were missense changes (Fig. 1 and Supp. Table S1), 82 of which are described here for the first time (in bold type in Supp. Table S1), and 33 had been reported already, 25 of them in patients of this cohort [Eeds et al., 2006; Häberle et al., 2003; Rapp et al., 2001; Summar, 1998] and another 8 in other patient cohorts [Finckh et al., 1998; Ihara et al., 1999; Khayat, 2009; Kurokawa et al., 2007; Ono et al., 2009; Pekkala et al., 2010; Wakutani et al., 2001]. Double-mutated alleles carrying each of them two missense mutations (p.Ile986Thr together with p.Ala304Val as well as p.Lys875Glu together with p.Lys843Ser) in addition to a third missense mutation on the second allele were identified in two of our patients and were confirmed by parental testing, raising the question regarding which of the two mutations in the same allele is disease-causing. One novel missense mutation (mutation 94; Supp. Table S1) changes the G at the last base of exon 24 by a T, and thus it may also decrease splicing efficiency.
Protein truncation mutations accounted for 58 of the 192 mutations (Supp. Tables S2 and S3), with 37 truncation mutations described here for the first time and 21 reported before in patients of this cohort [Eeds et al., 2006; Häberle et al., 2003; Rapp et al., 2001; Summar, 1998]. Only 13 of the mutations causing protein truncation were true nonsense changes (Supp. Table S2), with the remainder being due to frameshift of different origins (Supp. Table S3), including small deletions or insertions (22 deletions, 11 insertions, 2 deletions+insertions, and 1 nucleotide duplication), one large genomic deletion (2,559 nucleotides) of exon 24 resulting in frameshift, and 7 splice site changes causing frameshift (3 causing exon skipping, and 4 resulting in smaller deletions or insertions).
A third type of mutations was represented by 19 in-frame insertions or deletions (Supp. Table S4), of which 11 are unreported and 7 reported already in patients of this cohort [Eeds et al., 2006; Häberle et al., 2003; Summar, 1998], mostly due to splice aberrations (n = 12, of which 9 were proven to induce exon-skipping), but also including four small deletions, 1 deletion of intermediate size (90 nucleotides), and 1 duplication.
Overall, 148 mutations were single-nucleotide substitutions, with the remainder 42 being either insertions or deletions, of which only 1 was a relatively large genomic deletion. Of the 129 single-nucleotide substitutions falling in the coding sequence, 32 fall on CpG dinucleotides, yielding a rate of mutation in these dinucleotides ~13-fold higher than in the remainder of the coding sequence. The vast majority of mutations were private, which was in agreement with previous reports [Aoshima et al., 2001a,b; Eeds et al., 2006; Häberle et al., 2003; Hoshide et al., 1993]. Of the 192 mutations found here, 2 recurred in two patients (1 affecting a CpG dinucleotide) and another 1 (c.2148T>A) in four patients of this cohort. This low frequency of recurrence (< 2%) contrasts drastically with the ~20% recurrence found among the 27 mutations reported in the largest CPSID cohort studied thus far [Kurokawa et al., 2007], suggesting that the largely homogeneous genetic pool of Japanese patients was the reason for the high recurrence in the study. Further evidence for low recurrence of the mutations in CPSID is provided by the observation that only 12 (8.1%) of the 148 single nucleotide substitutions in our cohort have been reported in patients from other cohorts (Supp. Tables S1–S4), and that these affect mostly CpG dinucleotides (underlined): c.130C>T (p.Q44X), c.1631C>T (p.T544M), c.1760G>A (p.R587H9, c.2339G>A (p.R780H), c.2359C>T (p. p.R787X), 2407C>G (p.R803G), c.2429A>G (p.Q810R), c.2549G>A (p.R850H), c.2945G>A (p.G982D), c.3265C>T (p.R1089C), c.3784C>T (p.R1262X), and c.4357C>T (p.R1453W). Alternative changes affecting the same base were observed also for nine mutations within our cohort: c.1592T>A/G (p.V531E/G); c.1760G>A/T (p.R587H/L), c.2407C>A/G/T (p.803S/G/C), c.2732G>A/T (p.6911E/V), c.2945G>A/T (p.G982D/V), c.3464C>A/T (pA1155E/V), c.3785G>A/C (p.R1262Q/P), c.3969_3970insC/insCC (Supp. Tables S1 and S3).
The Mutation Database for CPSID
The new mutations reported presently, together with those that had been reported already, yield a total of 222 mutations (Fig. 1 and Supp. Tables S1–S4), comprising 136 missense changes, 15 nonsense mutations, 50 insertions, deletions, duplications, indels and splice aberrations that introduce stop codons and thus truncate the enzyme and 21 changes of these last types that do not affect the reading frame and therefore do not cause enzyme truncation. Thus, this expanded mutational database confirms and provides broader support to the main inferences made in our cohort, including the conclusions that the majority of the mutations are single-nucleotide substitutions (77%) with predominance of missense changes (61%) mostly of “private” nature, with increased tendency of mutations affecting CpG pairs (~22%) making up a large fraction (~65%) of the few mutations that recur (< 10%) in different families. Next in the frequency to the nucleotide substitutions are small deletions, which represent ~13% of the mutational database, followed by small insertions or duplications (~7%), indels (~2%), and large deletions (only 2, ~1%).
The nonsense changes (n = 15) are necessarily disease-causing. They represent a larger fraction (~3%) of all the possible nonsense single-nucleotide substitutions (n = 516) for the CPS1 gene than the fraction of all the possible missense single nucleotide changes in this gene (n = 8,974) represented in the present database (n = 136, corresponding to a fraction of ~1.5%), possibly reflecting the fact that only a fraction of the missense changes is expected to cause CPSID, highlighting the importance of providing experimental data or sound in silico proof of the pathogenic potential of given missense changes.
Uneven Distribution of the Missense Mutations along CPSI
The distribution of missense mutations among different exons (normalized per 100 exon nucleotides) (Fig. 2A and B) is highly uneven, with many more mutations clustering in some exons than in other exons, even after removing from the analysis the mutations falling on CpG dinucleotides (Fig. 2B, thick line and triangles). The peak of highest density of missense mutations is observed at exon 24 and the surrounding exons (Fig. 2B), at the boundary between both homologous halves of the region encoding the 120-kDa catalytic moiety of CPSI. In both homologous halves, peaks of high mutation density are observed in exons encoding portions of the corresponding phosphorylation domain (peaks at exons 13 and 17 for the bicarbonate phosphorylation domain and at exons 30 and 33 for the carbamate phosphorylation domain). In line with the functional importance of CPSI activation by NAG [Pekkala et al., 2010], there are also missense mutation peaks at exons 35 and 37, corresponding to the regulatory C-terminal domain where NAG binds. The occurrence of mutations in exons encoding parts of the small subunit-like region supports the functional importance of this region [Ahuja and Powers-Lee, 2008].
Figure 2.
Density of missense mutations per CPS1 exon or CPSI domain and correlation with the domain structure of the enzyme. A: Bar representation of the CPS1 coding sequence/CPSI protein to show the exon and domain composition and the different sizes of the various exons and domains, superimposing on them vertical lines corresponding to the exact locations of the different missense changes summarized in Table 1. The bar has the same length as the 4500-base coding sequence or 1500-residue amino acid sequence at the X-axes of the plots below. It is divided into the various exons, which are represented to scale and numbered. The different domains along the CPSI protein are superimposed on the exon bar, again in exact representation of sizes and boundaries (given as the number of the amino acid residue in the protein sequence), coloring each domain as in the bar at the bottom of Figure 1. The full domain name is shown above the bar in its appropriate color. Question marks indicate unknown function. The span in the polypeptide of the 40-kDa and 120-kDa moieties of the protein and of the two homologous repeats that make up the 120-kDa moiety are shown above the bar. The setting is such that there is exact physical correspondence between the exons and the domains in the bar and the two plots below it. B: The number of missense mutations (from Table 1) per exon, normalized per 100 nucleotides, is plotted along the coding sequence, with the symbols placed at the middle point of each exon (some representative exon numbers are given). Thin line, all missense mutations. Thick line and large filled triangles, missense mutations that do not fall on CpG dinucleotides. C: Bar plot of the number of missense mutations per domain or subdomain, normalized per 100 amino acids, along the amino acid sequence of CPSI. The width of each bar corresponds to the number of residues in the domain. The colors and names of the bars correspond to the naming and coloring of the domains that are given at the bottom of Figure 1. The shadowed bars that are superimposed on both phosphorylation domains, BPSD and CPSD, correspond to the frequency of mutations in the three subdomains A, B, and C, that compose each of these phosphorylation domains. Horizontal broken lines, number of missense mutations that do not fall on CpG dinucleotides.
The uneven distribution of the mutations is also patent when the localization of these in the different enzyme domains was examined (Fig. 2C): missense mutations were found more frequently in domains composing the large subunit-like region, which performs substrate binding, catalysis, and NAG regulation, than in the domains composing the small subunit-like portion of CPSI (domains ISD and GSD; Fig. 2C). Irrespective of whether mutations falling on CpG dinucleotides are considered (Fig. 2C, bars) or not (Fig. 2C, dashed horizontal lines), the highest occurrence of missense mutations (normalized per 100 amino acids) is found in the bicarbonate phosphorylation domain (Fig. 2C, bar labeled BPSD), which is a key catalytic and substrate binding domain. In the structure of E. coli CPS [Thoden et al., 1997] this domain occupies a central position, interacting with all other enzyme domains. Interestingly, the second most frequent localization of clinical missense mutations is (Fig. 2C) in the 20-kDa C-terminal domain of the first homologous half of the enzyme (the domain labeled UFSD in Fig. 1, bottom), a domain of unascertained function. In E. coli CPS this domain interacts nearly exclusively (it also makes limited contacts with the small subunit) with the C and A subdomains of, respectively, the bicarbonate and the carbamate phosphorylation domains (both phosphorylation domains are composed of three well-defined consecutive structural domains called A, B, and C) [Thoden et al., 1997]. These subdomains are easily identified in the sequence of human CPSI, and they are found here to exhibit the highest frequency of missense mutations for the entire protein (Fig. 2B; the subdomain bars are shaded within the BPSD and CPSD bars), suggesting that these two subdomains and the intervening UFSD constitute a continuous highly eloquent region for missense changes, at the center of the large subunit-like moiety.
Structure-Based Rationalization of the Effects of the Missense Mutations Affecting the C-Terminal Domain of CPSI
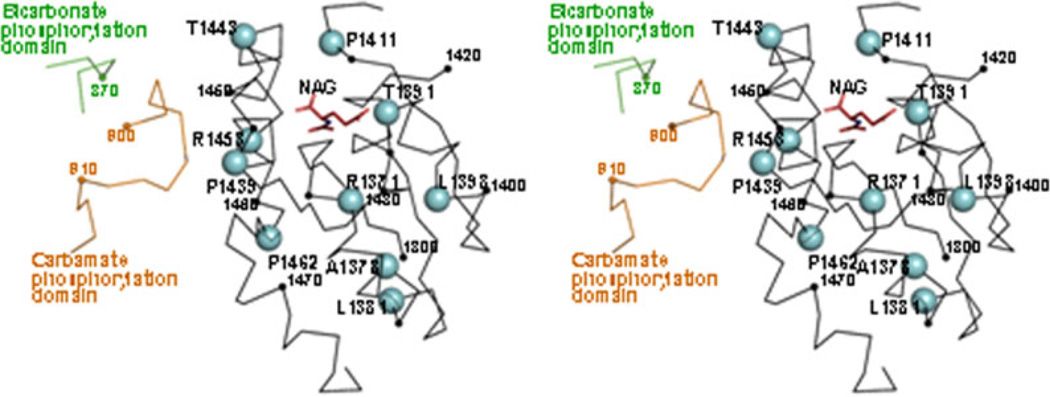
The only CPSI domain for which the crystal structure was determined is the C-terminal or allosteric domain (PDB file 2yvq), where the site of NAG binding was identified [Pekkala et al., 2009] and where three missense changes (one of them a polymorphism, p.G1376S) have been localized previously [Pekkala et al., 2010]. We now localize in this structure (Fig. 3) eight additional mutations from the present database. Of these, T1391 directly interacts with NAG, and its substitution by methionine in the p.T1391M mutation is highly likely to hamper NAG binding. The mild effect of the p.P1411L mutation was attributed [Pekkala et al., 2010] to the nature of the amino acid change in an apparently quite important position close to the NAG site [Pekkala et al., 2010]. Residues P1439, T1443, and R1453 directly shape the NAG binding site, suggesting that structural changes on this site may be responsible for the effects of the p.P1439L, p.T1443A, p.R1453W, and p.R1453Q mutations (proven experimentally for the p.R1453W and p.R1453Q mutations) [Pekkala et al., 2010]. In addition, P1439 and R1453 are also in a potential surface of transmission to the remainder of the enzyme of the activating signal of NAG. Finally, p.P1462R, p.R1371L, p.A1378T, and p.L1381S most likely have an important structural role, disturbing the domain fold, given the nature and location of the substitutions in important structural elements of the domain. The clustering of p.R1371L, p.A1378T, and p.L1381S in a single helix (α1) of the five belonging to this α3β5α2 sandwich-folded domain possibly reflects the important structural role of this helix, which occupies the central position among the three helices of the a3 layer of the sandwich (Fig. 3). Indeed, the potential effect of the p.L1398V mutation (if any, given the mildness of the L>V substitution) should stem from the disturbance of interactions within the α3 layer with this central organizing helix.
Figure 3.
Structure of the C-terminal domain of CPSI, with indication of the missense mutations found within this domain. Stereo view of the crystal structure of this domain (Cα trace in black) produced in a cell-free system (Protein DataBank file 2yvq), with fitting in the structure of the bound NAG molecule (in sticks and colored) [Pekkala et al., 2009]. The green and brown loops belong, respectively, to the bicarbonate and carbamate phosphorylation domains of the superimposed structure of E. coli CPS. Small spheres mark every tenth residue (numbering some of them, in small type). Large cyan spheres correspond to the indicated allosteric domain residues, which are those at which missense mutations have been found (Supp. Table S1). With the purpose of comparison, the projection and mode of representation are as similar as possible to those in Figure 1D of [Pekkala et al., 2010].
Locating Mutations in a Model of the Human CPSI Structure
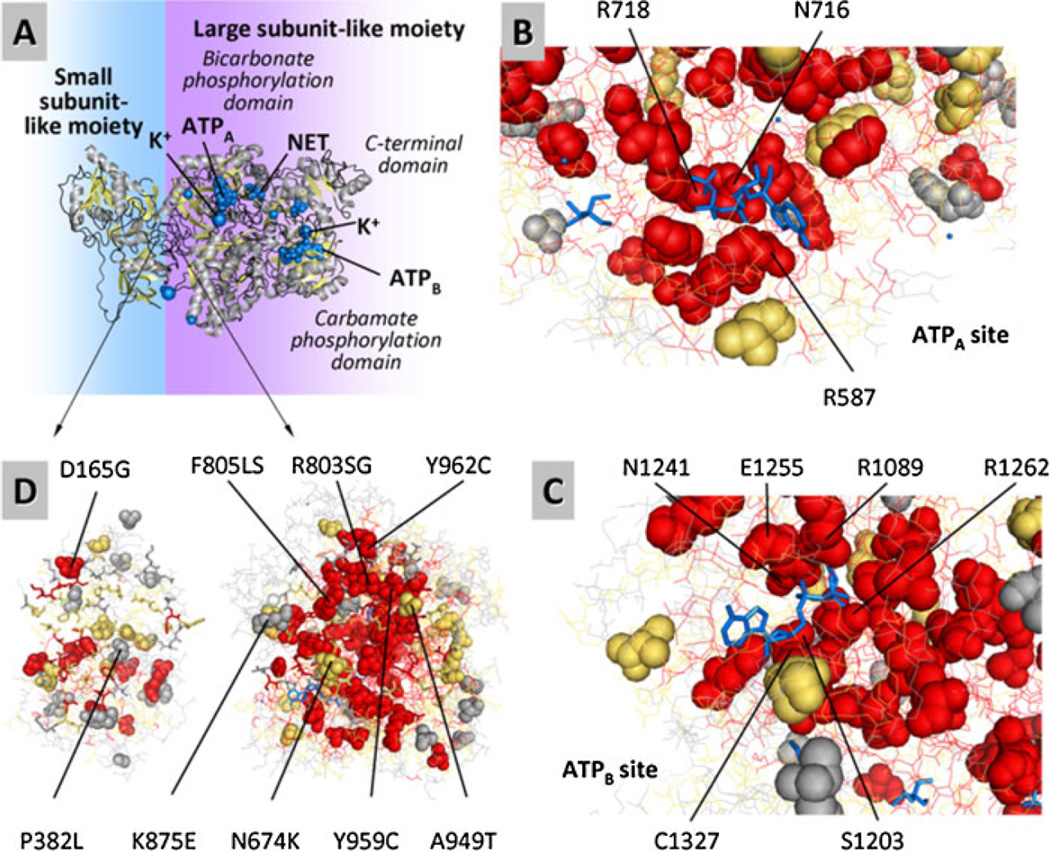
The lack of direct structural information for CPSI except for the C-terminal domain led us to model the structure on the basis of that reported for E. coli CPS (Fig. 4A – D). This modeled structure, enhanced with evolutionary importance (EI) scores for each residue, revealed the functional sites and helped to visualize the possible impact of each missense mutation (Fig. 4B–D). Because all reported mutations come from CPSID patients, it is expected that they will affect either a functional site or the folding stability of CPSI. Indeed, these mutations highly correlated with the EI of the mutated residues with a Fisher’s exact test probability of p = 5 · 10−14. Notably, putative ligand-binding sites (for both ATPs, both Mn ions, both K+ ions, the ornithine activator of E. coli CPS and one Cl− ion) harbored 21 mutations on 16 residues, the majority of which have high EI (13 high, 2 medium, 1 low) suggesting that these mutations either affect the ability of CPSI for binding similar ligands (both nucleotide molecules and the metal atoms) or alter sites that are important functionally in as yet unidentified ways (the cases for the regions corresponding to the ornithine and Cl−-binding sites of E. coli CPS) (Fig. 4B and C). Moreover, the interface of small and large CPSI moieties harbored 12 mutations on 9 residues, most of which have high-medium EI (4 high, 4 medium, 1 low; Figs. 4D) suggesting that the interactions between both moieties could be very important and negatively affected in these patients, in agreement with the mutation density plot (Fig. 2), which also led to the same conclusion, given the peak of mutations in exon 24, at the boundary between both moieties. Not surprisingly, the majority of the missense mutations (94) correspond in the model to internal residues, suggesting that they affect the folding of the protein, which agrees with studies on other proteins [Wang and Moult, 2001]. This is supported by the significant correlation of the CPSI mutations with solvent inaccessibility that results in a Fisher’s exact test probability of P = 9 · 10−9 . The distribution of the CPSI mutations in functional sites and among levels of EI and SA of the residues are summarized in Table 1.
Figure 4.
Structural model of human CPSI and its binding sites, predicting the localization of the mutated residues and providing an indication of the evolutionary importance (EI) of each residue. A: A ribbon representation of the enzyme, with ligands represented by blue spheres. The model was built from the structure of E. coli CPS using SWISS-MODEL (see Methods and Supporting Information). Similarly to the E. coli template structure, the BPSD and CPSD domains are shown binding AMPPNP, an inert ATP analogue labeled as ATPA and ATPB in the BPSD and CPSD subdomains, respectively. The NH3 analogue, a tetraethylammonium ion, is labeled as NET and is located in the BPSD subdomain. In the human CPSI ornithine is not considered to be an effector, but it is an activator of E. coli CPS. It has been placed here at the equivalent location in the model, between the CPSD and ASD subdomains. B–D: Detailed view of the CPSD and BPSD subdomains. Residues with high, medium, and low evolutionary importance are colored as red, orange, and gray, respectively. Ligands are shown blue, in sticks representation. The majority of the missense mutations affect evolutionary important residues and occur internally (low solvent accessibility). Sticks and lines represent nonmutated residues at and away from the binding site of interest, respectively. B: Bicarbonate phosphorylation subdomain binding ATPA. C: Carbamate phosphorylation subdomain binding ATPB. D: Missense mutation affecting amino acid residues predicted to play role in the interaction between the small subunit-like and large subunit-like moieties of CPSI.
Table 1.
The Relationship of CPSI Mutations, Functional Sites, Evolutionary Importance, and Solvent Accessibility
| Evolutionary importance of a residue |
|||||
|---|---|---|---|---|---|
| High | Medium | Low | |||
| Binding amino acid residues | Residues forming small-ligand binding sites | ANP | 14 (11)† | – | – |
| ORN | 1 (1) | – | 1 (1) | ||
| K+ | 6 (5) | 1 (1) | – | ||
| Cl | 1(1) | – | – | ||
| All small ligands | 21 (18) | 1 (1) | 1 (1) | ||
| Domain interface residues | N-terminus interface | 1 (1) | 1 (1) | 1 (1) | |
| C-terminus interface | 5 (3) | 6 (4) | 1 (1) | ||
| All interfacing residues | 6 (4) | 7 (5) | 2 (2) | ||
| Nonbinding amino acid residues | Solvent accessibility of residues | Low accessibility (0–10) | 37 (32) | 12 (11) | 10 (10) |
| Medium accessibility (11–50) | 15 (12) | 8 (7) | 9 (9) | ||
| High accessibility (>50) | 1 (1) | 1 (1) | 8 (8) | ||
This table summarizes the distribution of mutations in relation to the CPSI functional sites, evolutionary importance, and solvent accessibility. The number outside of parenthesis refers to the number of mutations. The number in parenthesis refers to the number of mutated residues. For example, 14 (11) in the upper left column (denoted by †) would refer to 14 mutations affecting 11 different amino acid residues, which form the ANP-binding site of CPSI. The table illustrates (highlighted in bold) that mutations primarily affect (a) evolutionary important residues clustering around small-ligand binding sites and (b) residues with low solvent accessibility.
Discussion
The current report summarizes our experience over a period of 24 years. Molecular analysis of 205 CPSID patients enrolled in this study revealed 192 sequence alterations of the CPS1 gene considered causative of CPSID. This report adds up to a total of 222 mutations in the CPS1 gene. This number encompasses isolated reports and small groups of CPSID patients [Aoshima et al., 2001a,b; Finckh et al., 1998; Funghini et al., 2003; Hoshide et al., 1993; Ihara et al., 1999; Khayat, 2009; Kurokawa et al., 2007; Ono et al., 2009; Wakutani et al., 2001] as well as previously described mutations within this cohort [Eeds et al., 2006, 2007; Falik-Zaccai et al., 2008; Häberle et al., 2003; Klaus et al., 2009; Mitchell et al., 2009; Rapp et al., 2001; Summar, 1998; Wakutani et al., 2001; Wong et al., 1994]. Our study illustrates the broad genetic heterogeneity at the CPS1 locus with private mutations being the rule and recurrence of mutations only found as an exception, occurring >10-fold more frequently on CpG dinucleotides than on other nucleotides. Indeed, of the whole database of 222 mutations only 16 mutations appear to have been found in more than one family (see Supp. Tables S1–S4). Nevertheless, if the mutations at the same nucleotide that cause different amino acid changes are included also, then the number of families with mutational recurrence is increased to 23, approximately 10% of all the mutations pooled in the present study. Double mutated alleles were found in two patients but we do not know whether all of the involved nucleotide changes would cause disease if present on one allele as the only sequence alteration.
Mutations in the CPS1 gene are distributed along the entire coding region, but missense changes, which represent the vast majority of the mutations (Fig. 5), are distributed unevenly across all exons, with exons 1, 4, 6, 27, 29, and 34 not hosting any of these mutations. This is not merely a stochastic effect resulting from the differences in size of the various exons and different distribution of the hypermutable CpG dinucleotides among the exons. When both these effects are eliminated, the distribution of the mutations remains highly uneven (Fig. 2). Most likely, the heterogeneous distribution reflects the fact that some enzyme regions are more important than others for enzyme folding and functionality. Therefore, mutations in these regions would be expected to have a higher functional repercussion, thus being more represented in cases of CPSID. This is supported by localizing the mutations in the protein, because this localization analysis reveals that the region at and around the boundary between the two repeats that make up the large subunit-like moiety has strong mutational representation (Fig. 2B and C), most likely reflecting the need for extremely well-orchestrated interaction and architecture of the two repeats to fulfill their role in catalyzing cooperatively the multistep CPS reaction [Yefimenko et al., 2005]. In addition to this boundary region, the regions involved in both phosphorylation steps are shown by the structural modeling (Fig. 4B and C) and mutation density studies (Fig. 2) to be well represented in the mutational database collected, as expected, given their high functional importance in the catalysis of the reaction. The present structural modeling studies strongly suggest (Fig. 4D) that missense mutations cluster also in the putative regions of interaction between the large subunit-like and the small subunit-like moieties of the enzyme (Fig. 2), pointing to the importance of the interactions between both enzyme moieties. Such importance explains the conservation through evolution of the small subunit-like region of CPSI despite the fact that this part of the enzyme has lost its role as a catalyst, because CPSI cannot use glutamine as a substrate. In E. coli CPS, addition of the small subunit to the active isolated large subunit (with ammonia as substrate, used with low affinity) strongly increases the thermal stability of both subunits [Cervera et al., 1993], suggesting that the corresponding region of CPSI could play a similar stabilizing role. Furthermore, removal of the small subunit-like region of CPSI decreased the Vmax of the enzyme and increased the Km for ATP ~200-fold and ~12-fold, respectively [Ahuja and Powers-Lee, 2008]. The present data on CPS1 mutations further support the role of the small subunit-like domain by demonstrating that a number of clinically ascertained missense mutations fall within this domain, in fact occurring with high density in exon 2 at the N-terminal part of this moiety. Therefore, the present mutational spectrum provide evidence that mutations in both sequence-predicted domains (ISD and GSD, Fig. 1 bottom) of the small subunit-like moiety are important, in as yet uncharacterized ways, for CPSI functionality. A negative control confirming that the differences in mutation density among various exons are significant is the observation of no missense mutations in exon 1, an exon that encodes the mitochondrial targeting peptide, which is absent from the mature enzyme and thus is unimportant for activity. Previous studies with other proteins such as ornithine transcarbamylase have proven that the mitochondrial targeting function of N-terminal signaling sequences might be only little affected by missense mutations [Horwich et al., 1986], as reflected also in the lack of mutations in this region causing CPSID.
Figure 5.
The summary of all CPS1 mutations reported to date.
Finally, some mutations map in the allosteric domain, where mutations are expected to impact enzyme activity because of the essentiality of NAG activation for CPSI activity. The crystal structure of this domain produced in a cell-free system appears to represent that of the well-folded domain despite the absence of the remainder of the enzyme. Thus, this structure has been highly instrumental in allowing to rationalize the effects of the mutations affecting this domain, of which a number of them appear to affect overall folding of the domain, others to influence NAG binding and a third group having impact in the transmission of the allosteric signal to nearby domains.
We also demonstrate here a strong statistical evidence of significant dependence of CPS1 mutations on the EI (P = 5 · 10−14 ) of the amino acid residue in the sequence of CPS and on the SA (P = 9 · 10−9 ) of the corresponding residues in the 3D structure of E. coli CPS1bxr. As already indicated, missense mutations fall on the small-ligand binding sites of this enzyme (both ATP sites, Mn, K, and curiously in sites corresponding to the ornithine and putative Cl− sites of the E. coli CPS structure), the interface of the small and large domains and internally at significantly higher frequencies than expected by chance, suggesting that they impact the binding, the intramolecular interaction and the folding of the enzyme, all important traits of the CPSI function. These observations are important for two reasons. First, even in the absence of a structural model of human CPSI, the E. coli structure can be used as a model, as previously shown experimentally [Yefimenko et al., 2005], to suggest putative pathogenicity of novel CPS1 mutations exploiting EI and SA analysis. Second, the high dependence of the CPSID missense mutations on the EI and SA scores in the E. coli CPS structural model suggests that a plausible 3D structure of the human CPSI protein can be created using comparative homology modeling, as shown in Figure 4 and in the Supporting Information. [Supp. Figs. S1 and S2 store the CPSI model in standard pdb format as well as in pse format, respectively; Supp. Fig. S1 and S2 show the CPSI model with all information presented in this manuscript, namely, missense mutations, EI, binding sites, and subdomains. The file CPS1_model.pdb opens with any protein structure software and gives the atomic coordinates for the CPS1 model in Protein Databank format (Supp. Fig. S1). The file CPS1_model.pse should be opened with the program Pymol and presents the same model highlighting the residues hosting the missense mutations included in the present manuscript as well as evolutionary importance, binding sites, and subdomains (Supp. Fig. S2).)
The CPS1 locus is also known to be affected by a high proportion of mutations causing RNA instability [Eeds et al., 2006]. A total of 65 protein truncation mutations are now known with 37 novel mutations reported here. As with the missense mutations, protein truncation mutations can be found in almost all exons (hitherto not yet in exons 6, 10, 13, 21, 28, 30, 36). The high proportion of protein truncation mutations can cause difficulties for the RNA-based genetic diagnostic methods, because many of the affected alleles become subject to nonsense-mediated decay and might thus escape their detection [Eeds et al., 2006]. Taken together, the group of missense and protein truncation mutations makes up for 201 of 222 total changes in the CPS1 gene.
In comparison to the number of missense and of protein truncation mutations, only a small fraction of mutant alleles was found to be affected by splice site alterations, small deletions, insertions, deletion insertions, or duplications. Except for one mutation (c.4102-239A>G), we did not find single nucleotide changes deep within intronic regions affecting the CPS1 transcript. This surprising low incidence of sequence alterations affecting intronic parts of the CPS1 gene is also substantiated with respect to small deletions and can be regarded as a genuine representation of the true occurrence of these mutations because 147 of 205 patients were investigated primarily at the mRNA level.
The effect of splice site mutations on the CPS1 transcript has been proven in most cases by demonstrating the loss of respective exons in mRNA from lymphoblasts or fibroblasts. In addition, use of an alternative intronic splice site resulting in an insertion has been found in patients affected by a splice site mutation of exon 34 (c.4101+2T>C) [Klaus et al., 2009]. There are further mutations affecting splice sites and hereby leading to frameshift because of an exon loss (c.1981+2T>G and c.4404 + 1G>A) or leading to use of cryptic exonic (c.1549 + 1G>T) or intronic (c.1837−8A>G) splice sites. Because these latter mutations result in premature termination of translation, we classified them as protein truncation mutations. As with small deletions, only few patients are affected by small insertions or duplications. Most of these changes as well as the combined occurrence of deletion and insertion were found to result in a frameshift with premature stop of translation and are therefore also classified as protein truncation mutations.
Most of the deletions found are small and result in the loss of single or few amino acids. Only three deletions, namely, c.622_711del90, c.711+686_1164 + 136del4260, and c.2895 + 429_c.2960−281del2559, lead to loss of single exons and could be amenable to detection using MLPA or oligo-based array CGH. In turn, c.711+686_1164 + 136del4260 and c.2895 + 429_c.2960−281del2559 are the only large intragenic deletions in the CPS1 gene described until now. The last of the two was classified here as a protein truncation mutation since it results in a frameshift. In addition to these, there are 24 deletions of single or few nucleotides resulting in frameshift and early stop of translation.
Certain limitations apply to this study. We did not aim for a comprehensive study of genotypes of patients suffering from CPSID. Many patients are affected by homozygous missense mutations, possibly due to consanguinity. For this group of patients, the time of onset of CPSID is provided whenever known (Supp. Table S1). Most of the patients affected by a homozygous mutation presented at neonatal age and carried a high risk of mortality. It is known that in at least half of the patients CPSID first manifests itself outside the neonatal period. Therefore, it would be interesting to examine whether patients that are compound heterozygous are more likely to exhibit late presentations. More comprehensive phenotypic profiling on well-studied genotypes of CPSID patients will be necessary to get insight on this question and also to further characterize putative genotype-phenotype correlations.
Supplementary Material
Acknowledgments
Contract grant sponsor: The National Institutes of Health; Contract grant numbers: GM066099; GM079656 (to O.L.); Contract grant sponsor: Prometeo of the Spanish and Valencian Governments; Contract grant number: BFU2008-05021 (to V.R.); Contract grant sponsor: NIH; Contract grant numbers: R01CA92313 (to M.S., L.H., M.Y., S.R.); ES09915 (to M.S., L.H., M.Y., S.R.); U54RR019453 (to M.S., S.R.); Contract grant sponsor: O’Malley Foundation (to M.S.).
The authors thank clinical colleagues from many countries for sending patient’s samples. Also, the enthusiasm of our lab technicians performing investigations over the many years of this study is acknowledged. ET servers can be found at: http://mammoth.bcm.tmc.edu/. J.H. acknowledges Orphan Europe for defraying part of the cost of mutation determination in Münster and Zurich.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Ahuja V, Powers-Lee SG. Human carbamoyl-phosphate synthetase: insight into N-acetylglutamate interaction and the functional effects of a common single nucleotide polymorphism. J Inherit Metab Dis. 2008;31:481–491. doi: 10.1007/s10545-008-0913-y. [DOI] [PubMed] [Google Scholar]
- Alonso E, Cervera J, Garcia-Espana A, Bendala E, Rubio V. Oxidative inactivation of carbamoyl phosphate synthetase (ammonia). Mechanism and sites of oxidation, degradation of the oxidized enzyme, and inactivation by glycerol, EDTA, and thiol protecting agents. J Biol Chem. 1992;267:4524–4532. [PubMed] [Google Scholar]
- Alonso E, Rubio V. Affinity cleavage of carbamoyl-phosphate synthetase I localizes regions of the enzyme interacting with the molecule of ATP that phosphorylates carbamate. Eur J Biochem. 1995;229:377–384. doi: 10.1111/j.1432-1033.1995.tb20478.x. [DOI] [PubMed] [Google Scholar]
- Aoshima T, Kajita M, Sekido Y, Kikuchi S, Yasuda I, Saheki T, Watanabe K, Shimokata K, Niwa T. Novel mutations (H337R and 238-362del) in the CPS1 gene cause carbamoyl phosphate synthetase I deficiency. Hum Hered. 2001a;52:99–101. doi: 10.1159/000053360. [DOI] [PubMed] [Google Scholar]
- Aoshima T, Kajita M, Sekido Y, Mimura S, Itakura A, Yasuda I, Saheki T, Watanabe K, Shimokata K, Niwa T. Carbamoyl phosphate synthetase I deficiency: molecular genetic findings and prenatal diagnosis. Prenat Diagn. 2001b;21:634–637. doi: 10.1002/pd.123. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Baameur F, Morgan DH, Yao H, Tran TM, Hammitt RA, Sabui S, McMurray JS, Lichtarge O, Clark RB. Role for the regulator of G-protein signaling homology domain of G protein-coupled receptor kinases 5 and 6 in beta 2-adrenergic receptor and rhodopsin phosphorylation. Mol Pharmacol. 2010;77:405–415. doi: 10.1124/mol.109.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera J, Conejero-Lara F, Ruiz-Sanz J, Galisteo ML, Mateo PL, Lusty CJ, Rubio V. The influence of effectors and subunit interactions on Escherichia coli carbamoyl-phosphate synthetase studied by differential scanning calorimetry. J Biol Chem. 1993;268:12504–12511. [PubMed] [Google Scholar]
- Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeds AM, Hall LD, Yadav M, Willis A, Summar S, Putnam A, Barr F, Summar ML. The frequent observation of evidence for nonsense-mediated decay in RNA from patients with carbamyl phosphate synthetase I deficiency. Mol Genet Metab. 2006;89:80–86. doi: 10.1016/j.ymgme.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Eeds AM, Mortlock D, Wade-Martins R, Summar ML. Assessing the functional characteristics of synonymous and nonsynonymous mutation candidates by use of large DNA constructs. Am J Hum Genet. 2007;80:740–750. doi: 10.1086/513287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falik-Zaccai TC, Kfir N, Frenkel P, Cohen C, Tanus M, Mandel H, Shihab S, Morkos S, Aaref S, Summar ML, Khayat M. Population screening in a Druze community: the challenge and the reward. Genet Med. 2008;10:903–909. doi: 10.1097/GIM.0b013e31818d0e0f. [DOI] [PubMed] [Google Scholar]
- Finckh U, Kohlschütter A, Schafer H, Sperhake K, Colombo JP, Gal A. Prenatal diagnosis of carbamoyl phosphate synthetase I deficiency by identification of a missense mutation in CPS1 . Hum Mutat. 1998;12:206–211. doi: 10.1002/(SICI)1098-1004(1998)12:3<206::AID-HUMU8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Fresquet V, Mora P, Rochera L, Ramón-Maiques S, Rubio V, Cervera J. Site-directed mutagenesis of the regulatory domain of Escherichia coli carbamoyl phosphate synthetase identifies crucial residues for allosteric regulation and for transduction of the regulatory signals. J Mol Biol. 2000;299:979–991. doi: 10.1006/jmbi.2000.3794. [DOI] [PubMed] [Google Scholar]
- Funghini S, Donati MA, Pasquini E, Zammarchi E, Morrone A. Structural organization of the human carbamyl phosphate synthetase I gene (CPS1) and identification of two novel genetic lesions. Hum Mutat. 2003;22:340–341. doi: 10.1002/humu.9184. [DOI] [PubMed] [Google Scholar]
- Häberle J, Schmidt E, Pauli S, Rapp B, Christensen E, Wermuth B, Koch HG. Gene structure of human carbamylphosphate synthetase 1 and novel mutations in patients with neonatal onset. Hum Mutat. 2003;21:444. doi: 10.1002/humu.9118. [DOI] [PubMed] [Google Scholar]
- Haraguchi Y, Uchino T, Takiguchi M, Endo F, Mori M, Matsuda I. Cloning and sequence of a cDNA encoding human carbamyl phosphate synthetase I: molecular analysis of hyperammonemia. Gene. 1991;107:335–340. doi: 10.1016/0378-1119(91)90336-a. [DOI] [PubMed] [Google Scholar]
- Hart EJ, Powers-Lee SG. Role of Cys-1327 and Cys-1337 in redox sensitivity and allosteric monitoring in human carbamoyl phosphate synthetase. J Biol Chem. 2009;284:5977–5985. doi: 10.1074/jbc.M808702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Kalousek F, Fenton WA, Pollock RA, Rosenberg LE. Targeting of pre-ornithine transcarbamylase to mitochondria: definition of critical regions and residues in the leader peptide. Cell. 1986;44:451–459. doi: 10.1016/0092-8674(86)90466-6. [DOI] [PubMed] [Google Scholar]
- Hoshide R, Matsuura T, Haraguchi Y, Endo F, Yoshinaga M, Matsuda I. Carbamyl phosphate synthetase I deficiency. One base substitution in an exon of the CPS I gene causes a 9-basepair deletion due to aberrant splicing. J Clin Invest. 1993;91:1884–1887. doi: 10.1172/JCI116405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara K, Nakayama H, Hikino S, Hara T. Mutation in CPS1 . Hum Genet. 1999;105:375. [Google Scholar]
- Javid-Majd F, Stapleton MA, Harmon MF, Hanks BA, Mullins LS, Raushel FM. Comparison of the functional differences for the homologous residues within the carboxy phosphate and carbamate domains of carbamoyl phosphate synthetase. Biochemistry. 1996;35:14362–14369. doi: 10.1021/bi961184q. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Khayat M. Novel human pathological mutations. Gene symbol: CPS1. Disease: carbamoyl phosphate synthetase I deficiency. Hum Genet. 2009;125:336. [PubMed] [Google Scholar]
- Klaus V, Vermeulen T, Minassian B, Israelian N, Engel K, Lund AM, Roebrock K, Christensen E, Häberle J. Highly variable clinical phenotype of carbamylphosphate synthetase 1 deficiency in one family: an effect of allelic variation in gene expression? Clin Genet. 2009;76:263–269. doi: 10.1111/j.1399-0004.2009.01216.x. [DOI] [PubMed] [Google Scholar]
- Krebs H, Henseleit K. Untersuchungen über die Harnstoffbildung im Tierkörper. Hoppe-Seyler’s Zeitschrift für Physiologische Chemie. 1932;210:325–332. [Google Scholar]
- Kurokawa K, Yorifuji T, Kawai M, Momoi T, Nagasaka H, Takayanagi M, Kobayashi K, Yoshino M, Kosho T, Adachi M, Otsuka H, Yamamoto S, Murata T, Suenaga A, Ishii T, Terada K, Shimura N, Kiwaki K, Shintaku H, Yamakawa M, Nakabayashi H, Wakutani Y, Nakahata T. Molecular and clinical analyses of Japanese patients with carbamoylphosphate synthetase 1 (CPS1) deficiency. J Hum Genet. 2007;52:349–354. doi: 10.1007/s10038-007-0122-9. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Hutchinson EG, Michie AD, Wallace AC, Jones ML, Thornton JM. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- Lichtarge O, Yamamoto KR, Cohen FE. Identification of functional surfaces of the zinc binding domains of intracellular receptors. J Mol Biol. 1997;274:325–337. doi: 10.1006/jmbi.1997.1395. [DOI] [PubMed] [Google Scholar]
- Loscalzo ML, Galczynski RL, Hamosh A, Summar M, Chinsky JM, Thomas GH. Interstitial deletion of chromosome 2q32–34 associated with multiple congenital anomalies and a urea cycle defect (CPS I deficiency) Am J Med Genet A. 2004;128A:311–315. doi: 10.1002/ajmg.a.30105. [DOI] [PubMed] [Google Scholar]
- Meister A. Mechanism and regulation of the glutamine-dependent carbamyl phosphate synthetase of Escherichia coli . Adv Enzymol Relat Areas Mol Biol. 1989;62:315–374. doi: 10.1002/9780470123089.ch7. [DOI] [PubMed] [Google Scholar]
- Mihalek I, Res I, Lichtarge O. A family of evolution-entropy hybrid methods for ranking protein residues by importance. J Mol Biol. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- Mihalek I, Res I, Lichtarge O. Evolutionary trace report-maker: a new type of service for comparative analysis of proteins. Bioinformatics. 2006;22:1656–1657. doi: 10.1093/bioinformatics/btl157. [DOI] [PubMed] [Google Scholar]
- Mihalek I, Res I, Yao H, Lichtarge O. Combining inference from evolution and geometric probability in protein structure evaluation. J Mol Biol. 2003;331:263–279. doi: 10.1016/s0022-2836(03)00663-6. [DOI] [PubMed] [Google Scholar]
- Mitchell S, Ellingson C, Coyne T, Hall L, Neill M, Christian N, Higham C, Dobrowolski SF, Tuchman M, Summar M. Genetic variation in the urea cycle: a model resource for investigating key candidate genes for common diseases. Hum Mutat. 2009;30:56–60. doi: 10.1002/humu.20813. [DOI] [PubMed] [Google Scholar]
- Morgan DH, Kristensen DM, Mittelman D, Lichtarge O. ET viewer: an application for predicting and visualizing functional sites in protein structures. Bioinformatics. 2006;22:2049–2050. doi: 10.1093/bioinformatics/btl285. [DOI] [PubMed] [Google Scholar]
- Neill MA, Aschner J, Barr F, Summar ML. Quantitative RT-PCR comparison of the urea and nitric oxide cycle gene transcripts in adult human tissues. Mol Genet Metab. 2009;97:121–127. doi: 10.1016/j.ymgme.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyunoya H, Broglie KE, Widgren EE, Lusty CJ. Characterization and derivation of the gene coding for mitochondrial carbamyl phosphate synthetase I of rat. J Biol Chem. 1985;260:9346–9356. [PubMed] [Google Scholar]
- Ono H, Suto T, Kinoshita Y, Sakano T, Furue T, Ohta T. A case of carbamoyl phosphate synthetase 1 deficiency presenting symptoms at one month of age. Brain Dev. 2009;31:779–781. doi: 10.1016/j.braindev.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Oyanagi K, Nakamura K, Sogawa H, Tsukazaki H, Minami R, Nakao T. A study of urea-synthesizing enzymes in prenatal and postnatal human liver. Pediatr Res. 1980;14:236–241. doi: 10.1203/00006450-198003000-00012. [DOI] [PubMed] [Google Scholar]
- Pekkala S, Martinez AI, Barcelona B, Gallego J, Bendala E, Yefimenko I, Rubio V, Cervera J. Structural insight on the control of urea synthesis: identification of the binding site for N-acetyl-L-glutamate, the essential allosteric activator of mitochondrial carbamoyl phosphate synthetase. Biochem J. 2009;424:211–220. doi: 10.1042/BJ20090888. [DOI] [PubMed] [Google Scholar]
- Pekkala S, Martinez AI, Barcelona B, Yefimenko I, Finckh U, Rubio V, Cervera J. Understanding carbamoyl-phosphate synthetase I (CPS1) deficiency by using expression studies and structure-based analysis. Hum Mutat. 2010;31:801–808. doi: 10.1002/humu.21272. [DOI] [PubMed] [Google Scholar]
- Pierrat OA, Raushel FM. A functional analysis of the allosteric nucleotide monophosphate binding site of carbamoyl phosphate synthetase. Arch Biochem Biophys. 2002;400:34–42. doi: 10.1006/abbi.2002.2767. [DOI] [PubMed] [Google Scholar]
- Powers-Lee SG, Corina K. Domain structure of rat liver carbamoyl phosphate synthetase I. J Biol Chem. 1986;261:15349–15352. [PubMed] [Google Scholar]
- Rapp B, Häberle J, Linnebank M, Wermuth B, Marquardt T, Harms E, Koch HG. Genetic analysis of carbamoylphosphate synthetase I and ornithine transcarbamylase deficiency using fibroblasts. Eur J Pediatr. 2001;160:283–287. doi: 10.1007/s004310100725. [DOI] [PubMed] [Google Scholar]
- Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat Struct Mol Biol. 2007;14:301–307. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Aparicio LB, Guadalajara AM, Rubio V. Physical location of the site for N-acetyl-L-glutamate, the allosteric activator of carbamoyl phosphate synthetase, in the 20-kilodalton COOH-terminal domain. Biochemistry. 1989;28:3070–3074. doi: 10.1021/bi00433a050. [DOI] [PubMed] [Google Scholar]
- Rubio V. Structure-function studies in carbamoyl phosphate synthetases. Biochem Soc Trans. 1993;21:198–202. doi: 10.1042/bst0210198. [DOI] [PubMed] [Google Scholar]
- Rubio V, Cervera J. The carbamoyl-phosphate synthase family and carbamate kinase: structure-function studies. Biochem Soc Trans. 1995;23:879–883. doi: 10.1042/bst0230879. [DOI] [PubMed] [Google Scholar]
- Rubio V, Grisolia S. Human carbamoylphosphate synthetase I. Enzyme. 1981;26:233–239. doi: 10.1159/000459184. [DOI] [PubMed] [Google Scholar]
- Rubio V, Ramponi G, Grisolia S. Carbamoyl phosphate synthetase I of human liver. Purification, some properties and immunological cross-reactivity with the rat liver enzyme. Biochim Biophys Acta. 1981;659:150–160. doi: 10.1016/0005-2744(81)90279-5. [DOI] [PubMed] [Google Scholar]
- Ryall J, Nguyen M, Bendayan M, Shore GC. Expression of nuclear genes encoding the urea cycle enzymes, carbamoyl-phosphate synthetase I and ornithine carbamoyl transferase, in rat liver and intestinal mucosa. Eur J Biochem. 1985;152:287–292. doi: 10.1111/j.1432-1033.1985.tb09196.x. [DOI] [PubMed] [Google Scholar]
- Saeed-Kothe A, Powers-Lee SG. Gain of glutaminase function in mutants of the ammonia-specific frog carbamoyl phosphate synthetase. J Biol Chem. 2003;278:26722–26726. doi: 10.1074/jbc.M303774200. [DOI] [PubMed] [Google Scholar]
- Schofield JP. Molecular studies on an ancient gene encoding for carbamoyl-phosphate synthetase. Clin Sci (Lond) 1993;84:119–128. doi: 10.1042/cs0840119. [DOI] [PubMed] [Google Scholar]
- Shambaugh 3rd GE. Urea biosynthesis I. The urea cycle and relationships to the citric acid cycle. Am J Clin Nutr. 1977;30:2083–2087. doi: 10.1093/ajcn/30.12.2083. [DOI] [PubMed] [Google Scholar]
- Stapleton MA, Javid-Majd F, Harmon MF, Hanks BA, Grahmann JL, Mullins LS, Raushel FM. Role of conserved residues within the carboxy phosphate domain of carbamoyl phosphate synthetase. Biochemistry. 1996;35:14352–14361. doi: 10.1021/bi961183y. [DOI] [PubMed] [Google Scholar]
- Summar ML. Molecular genetic research into carbamoyl-phosphate synthase I: molecular defects and linkage markers. J Inherit Metab Dis. 1998;21(Suppl 1):30–39. doi: 10.1023/a:1005349306311. [DOI] [PubMed] [Google Scholar]
- Summar ML, Dobbelaere D, Brusilow S, Lee B. Diagnosis, symptoms, frequency and mortality of 260 patients with urea cycle disorders from a 21-year, multicentre study of acute hyperammonaemic episodes. Acta Paediatr. 2008;97:1420–1425. doi: 10.1111/j.1651-2227.2008.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summar ML, Hall L, Christman B, Barr F, Smith H, Kallianpur A, Brown N, Yadav M, Willis A, Eeds A, Cermak E, Summar S, Wilson A, Arvin M, Putnam A, Wills M, Cunningham G. Environmentally determined genetic expression: clinical correlates with molecular variants of carbamyl phosphate synthetase I. Mol Genet Metab. 2004;81(Suppl 1):S12–S19. doi: 10.1016/j.ymgme.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Summar ML, Hall LD, Eeds AM, Hutcheson HB, Kuo AN, Willis AS, Rubio V, Arvin MK, Schofield JP, Dawson EP. Characterization of genomic structure and polymorphisms in the human carbamyl phosphate synthetase I gene. Gene. 2003;311:51–57. doi: 10.1016/s0378-1119(03)00528-6. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Holden HM, Wesenberg G, Raushel FM, Rayment I. Structure of carbamoyl phosphate synthetase: a journey of 96 A from substrate to product. Biochemistry. 1997;36:6305–6316. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Wesenberg G, Raushel FM, Holden HM. Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding. Biochemistry. 1999;38:2347–2357. doi: 10.1021/bi982517h. [DOI] [PubMed] [Google Scholar]
- Tuchman M, Lee B, Lichter-Konecki U, Summar ML, Yudkoff M, Cederbaum SD, Kerr DS, Diaz GA, Seashore MR, Lee HS, McCarter RJ, Krischer JP, Batshaw ML Urea Cycle Disorders Consortium of the Rare Diseases Clinical Research Network. Cross-sectional multicenter study of patients with urea cycle disorders in the United States. Mol Genet Metab. 2008;94:397–402. doi: 10.1016/j.ymgme.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino T, Endo F, Matsuda I. Neurodevelopmental outcome of long-term therapy of urea cycle disorders in Japan. J Inherit Metab Dis. 1998;21(Suppl 1):151–159. doi: 10.1023/a:1005374027693. [DOI] [PubMed] [Google Scholar]
- Wakutani Y, Nakayasu H, Takeshima T, Mori N, Kobayashi K, Endo F, Nakashima K. A case of late-onset carbamoyl phosphate synthetase I deficiency, presenting periodic psychotic episodes coinciding with menstrual periods. Rinsho Shinkeigaku. 2001;41:780–785. [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Windmueller HG, Spaeth AE. Source and fate of circulating citrulline. Am J Physiol. 1981;241:E473–E480. doi: 10.1152/ajpendo.1981.241.6.E473. [DOI] [PubMed] [Google Scholar]
- Wong LJ, Craigen WJ, O’Brien WE. Postpartum coma and death due to carbamoyl-phosphate synthetase I deficiency. Ann Intern Med. 1994;120:216–217. doi: 10.7326/0003-4819-120-3-199402010-00007. [DOI] [PubMed] [Google Scholar]
- Yao H, Kristensen DM, Mihalek I, Sowa ME, Shaw C, Kimmel M, Kavraki L, Lichtarge O. An accurate, sensitive, and scalable method to identify functional sites in protein structures. J Mol Biol. 2003;326:255–261. doi: 10.1016/s0022-2836(02)01336-0. [DOI] [PubMed] [Google Scholar]
- Yefimenko I, Fresquet V, Marco-Marin C, Rubio V, Cervera J. Understanding carbamoyl phosphate synthetase deficiency: impact of clinical mutations on enzyme functionality. J Mol Biol. 2005;349:127–141. doi: 10.1016/j.jmb.2005.03.078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.