Abstract
Objectives
Reliable monitoring of influenza seasons and pandemic outbreaks is essential for response planning, but compilations of reports on detection and prediction algorithm performance in influenza control practice are largely missing. The aim of this study is to perform a metanarrative review of prospective evaluations of influenza outbreak detection and prediction algorithms restricted settings where authentic surveillance data have been used.
Design
The study was performed as a metanarrative review. An electronic literature search was performed, papers selected and qualitative and semiquantitative content analyses were conducted. For data extraction and interpretations, researcher triangulation was used for quality assurance.
Results
Eight prospective evaluations were found that used authentic surveillance data: three studies evaluating detection and five studies evaluating prediction. The methodological perspectives and experiences from the evaluations were found to have been reported in narrative formats representing biodefence informatics and health policy research, respectively. The biodefence informatics narrative having an emphasis on verification of technically and mathematically sound algorithms constituted a large part of the reporting. Four evaluations were reported as health policy research narratives, thus formulated in a manner that allows the results to qualify as policy evidence.
Conclusions
Awareness of the narrative format in which results are reported is essential when interpreting algorithm evaluations from an infectious disease control practice perspective.
Keywords: influenza, detection algorithms, prediction algorithms, evaluation, meta-narrative review
Strengths and limitations of this study.
A metanarrative review of influenza detection and prediction algorithm evaluations was restricted to settings where authentic prospective data were used.
Application of a semiqualitative review method allowed attention to be paid to critical dissimilarities between narratives, for example, the learning period dilemma caused by the statistical models used in algorithms to detect or predict an influenza-related event must be determined in a preceding time interval.
Application of the review inclusion criteria resulted in the exclusion of a large number of papers. These papers may have contained additional narratives, but not on the appropriate topic.
Introduction
Experiences from winter influenza seasons1 and the pandemic pH1N1 outbreak in 20092 suggest that existing information systems used for detecting and predicting outbreaks and informing situational awareness show deficiencies when under heavy demand. Public health specialists seek more effective and equitable response systems, but methodological problems frequently limit the usefulness of novel approaches.3 In these biosurveillance systems, algorithms for outbreak detection and prediction are essential components.4 5 Regarding outbreak detection, characteristics influential for successful performance include representativeness of data and the type and specificity of the outbreak detection algorithm, while influential outbreak characteristics comprise the magnitude and shape of the signal and the timing of the outbreak.6 After detection, mathematical models can be used to predict the progress of an outbreak and lead to the identification of thresholds that determine whether an outbreak will dissipate or develop into an epidemic. However, it has been pointed out that present prediction models have often been designed for particular situations using the data that are available and making assumptions where data are lacking.7 8 In consequence, also biosurveillance models that have been subject to evaluation seldom produce output that fulfils standard criteria for operational readiness.9 For instance, a recent scoping review of influenza forecasting methods assessed studies that validated models against independent data.10 Use of independent data is vital for predictive model validation, because using the same data for model fitting and testing inflates estimates of predictive performance.11 The review concluded that the outcomes predicted and metrics used in validations varied considerably, which limited the possibility to formulate recommendations. Building on these experiences, we set out to perform a metanarrative review of evaluations of influenza outbreak detection and prediction algorithms. To ensure that the review results can be used to inform operational readiness, we restricted the scope to settings where authentic prospective surveillance data had been used for the evaluation.
Methods
A metanarrative review12 was conducted to assess publications that prospectively evaluated algorithms for the detection or short-term prediction of influenza outbreaks based on routinely collected data. A metanarrative review was conducted because it is suitable for addressing the question ‘what works?’, and also to elucidate a complex topic, highlighting the strengths and limitations of different research approaches to that topic.13 Metanarrative reviews look at how particular research traditions have unfolded over time and shaped the kind of questions being asked and the methods used to answer them. They inspect the range of approaches to studying an issue, interpret and produce an account of the development of these separate ‘metanarratives’ and then form an overarching metanarrative summary. The principles of pragmatism (inclusion criteria are guided by what is considered to be useful to the audience), pluralism (the topic is illuminated from multiple perspectives; only research that lacks rigour is rejected), historicity (research traditions are described as they unfold over time), contestation (conflicting data are examined to generate higher order insights), reflexivity (reviewers continually reflect on the emerging findings) and peer review were applied in the analysis.12 Four steps were taken: an electronic literature search was carried out, papers were selected, data from these papers were extracted and qualitative and semiquantitative content analyses were conducted. For data extraction and analyses, researcher triangulation (involving several researchers with different backgrounds) was used as a strategy for quality assurance. All steps were documented and managed electronically using a database.
To be included in the review, an evaluation study had to apply an outbreak detection or prediction algorithm to authentic data prospectively collected to detect or predict naturally occurring influenza outbreaks among humans. Following the inclusive approach of the metanarrative review methodology, studies using clinical and laboratory diagnosis of influenza for case verification were included.14 For the evaluations of the prediction algorithms, correlation analyses were also accepted, because interventions could have been implemented during the evaluation period. In addition, studies were required to compare syndromic data with some gold standard data from known outbreaks. All studies published from 1 January 1998 to 31 January 2016 were considered.
PubMed was searched using the following search term combinations: ‘influenza AND ((syndromic surveillance) OR (outbreak detection OR outbreak prediction OR real-time prediction OR real-time estimation OR real-time estimation of R))’. The database searches were conducted in February 2016. Only articles and book chapters available in the English language were selected for further analysis. To describe the characteristics of the selected papers, information was documented regarding the main objective, the publication type, whether syndromic data were used, country, algorithm applied and context of application.
Information about the papers was analysed semiquantitatively by grouping papers with equal or similar characteristics and by counting the number of papers per group. In the next step, text passages, that is, sentences or paragraphs containing key terms (study aims, algorithm description and application context) were extracted and entered into the database. If necessary, sentences before and after a statement containing the key terms were added to ensure that the meaning and context were not lost. The documentation of data about the papers and the extraction of text were conducted by one reviewer and critically rechecked by a second reviewer. Next, content analysis of the extracted text was performed. The meaning of the original text was condensed. The condensed statements contained as much information as necessary to adequately represent the meaning of the text in relation to the research aim, but were as short and simple as possible to enable straightforward processing. If the original text contained several pieces of information, then a separate condensed statement was created for each piece of information. To analyse the information contained in the papers, a coding scheme was developed inductively. Also, a semantical system was developed to facilitate interpretation of algorithm performance. Values for the area under the curve (AUC) exceeding 0.90, 0.80 and 0.70, respectively, were chosen to denote very strong (outstanding), strong (excellent) and acceptable performance.15 The same limits are used to interpret the area under the weighted receiver operating characteristic curve (AUWROC) and volume under the time-ROC surface (VUTROC) metrics. Sensitivity, specificity and positive predictive value (PPV) limits were set at 0.95, 0.90 and 0.85, respectively, when weekly data were analysed, and 0.90, 0.85 and 0.80 when daily data were analysed, denoting very strong (outstanding), strong (excellent) and acceptable discriminatory performance. To interpret the strength of correlations, limit values were modified from the Cohen scale.16 This scale defines small, medium and large effect sizes as 0.10, 0.30 and 0.50, respectively. The limits for the present study were set at 0.90, 0.80 and 0.70 for analyses of weekly data, and 0.85, 0.75 and 0.65 for daily data, denoting very strong (outstanding), strong (excellent) and acceptable predictive performance. A summary of the sematic system is provided in table 1.
Table 1.
Summary of semantic system used to interpret algorithm performance
| Performance |
|||
|---|---|---|---|
| Measurement | Outstanding | Excellent | Acceptable |
| Outbreak detection and prediction | |||
| AUC, AUWROC, VUTROC | 0.90 | 0.80 | 0.70 |
| Sensitivity, specificity, PPV (weekly) | 0.95 | 0.90 | 0.85 |
| Sensitivity, specificity, PPV (daily) | 0.90 | 0.85 | 0.80 |
| Only outbreak prediction | |||
| Pearson's correlation (weekly) | 0.90 | 0.80 | 0.70 |
| Pearson's correlation (daily) | 0.85 | 0.75 | 0.65 |
AUC, area under the curve; AUWROC, area under the weighted receiver operating characteristic curve; PPV, positive predictive value; VUTROC, volume under the time-ROC surface.
Condensed statements could be labelled with more than one code. The creation of the condensed statements and their coding was carried out by one reviewer and rechecked by a second reviewer. Preliminary versions were compared and agreed upon, which resulted in final versions of the condensed statements and coding. The information about the detection and prediction algorithms was summarised qualitatively in tables and analysed semiquantitatively on the basis of the coding. Next analysis phase consisted of identifying the key dimensions of algorithm evaluations, providing a narrative account of the contribution of each dimension and explaining conflicting findings. The resulting two narratives (biodefence informatics and health policy research) are presented using descriptive statistics and narratively without quantitative pooling. In the last step, a wider research team and policy leaders (n=11) with backgrounds in public health, computer science, statistics, social sciences and cognitive science were engaged in a process of testing the findings against their expectations and experience, and their feedback was used to guide further reflection and analysis. The final report was compiled following this feedback.
Results
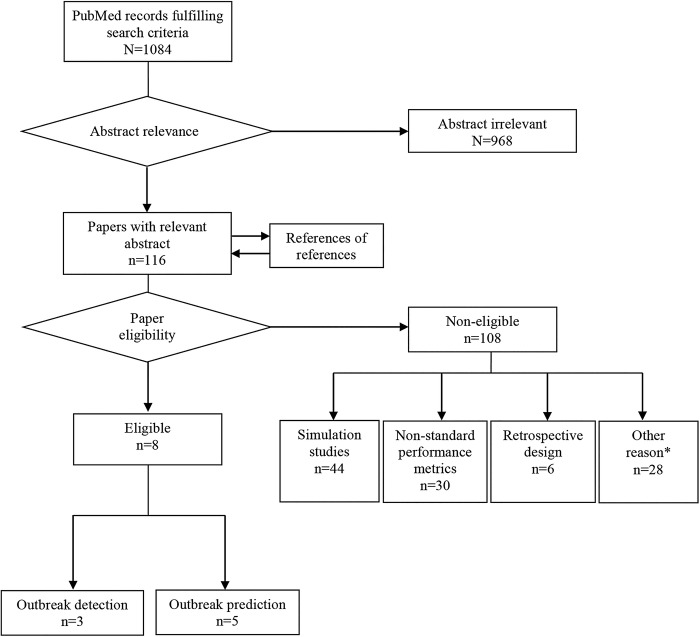
The search identified eight studies reporting prospective algorithm performance based on data from naturally occurring influenza outbreaks: three studies17–19 evaluated one or more outbreak detection algorithms and five20–24 evaluated prediction algorithms (figure 1).
Figure 1.
Flow chart of the paper selection process. Additional reasons for exclusion (*) included that the case definition did not comprise at least a clinical diagnosis of influenza or influenza-like illness.
Regarding outbreak detection, outstanding algorithm performance was reported from a Spanish study18 for two versions of algorithms based on hidden Markov models and Serfling regression (table 2). Simple regression was reported to show poor performance in this study. The same technique displayed excellent performance on US influenza data in a study comparing algorithm performances on data from two continents, as did time-series analysis and the statistical process control method based on cumulative sum (CUSUM).19 However, the performance of these three algorithms was found to be poor to acceptable when applied on Hong Kong data in the latter study.
Table 2.
Evaluation algorithms include in the metanarrative review and their absolute and relative performance
| Study | Algorithm | Modification | Temporal | Absolute performance | Relative performance |
|---|---|---|---|---|---|
| Outbreak detection | |||||
| Closas et al17 | Kolmogorov-Smirnov test | Weekly | Acceptable (sensitivity 1.00; specificity 0.88) | No comparisons | |
| Martínez-Beneito et al18 | Markov model (hidden) V.1 | Weekly | Outstanding (AUWROC 0.97–0.98) | Markov model (switching)>Markov model (hidden)>regression (Serfling)>CUSUM>regression (simple) | |
| Regression (Serfling) | Outstanding (AUWROC 0.93) | Markov model (switching)>Markov model (hidden)>regression (Serfling)>CUSUM>regression (simple) | |||
| Markov model (hidden) V.2 | Outstanding (AUWROC 0.93–0.95) | Markov model (switching)>Markov model (hidden)>regression (Serfling)>CUSUM>regression (simple) | |||
| Regression (simple) | Poor (AUWROC 0.57) | Markov model (switching)>Markov model (hidden)>regression (Serfling)>CUSUM>regression (simple) | |||
| SPC (CUSUM) | Poor (AUWROC 0.65–0.70) | Markov model (switching)>Markov model (hidden)>regression (Serfling)>CUSUM>regression (simple) | |||
| Cowling et al19 | Time series, dynamic linear model | Different parameter combinations tested. W represents the assumed smoothness of the underlying system. Range: 0.025, 0.050, 0.075 or 0.100 | Weekly | Hong Kong: acceptable (VUTROC 0.77, sensitivity 1.00, timeliness 1.40 weeks), with fixed specificity=0.95 USA: excellent (VUTROC 0.81, sensitivity 1.00, timeliness 0.75 weeks), with fixed specificity=0.95 |
Hong Kong data: time series (dynamic linear model)>regression (simple)>CUSUM US data: time series (dynamic linear model)>CUSUM>regression (simple) |
| Regression (simple) | Different parameter combinations tested. m represents the number of prior weeks used to calculate the running mean and variance. Range: 3, 5, 7 or 9 | Hong Kong: acceptable (VUTROC 0.75, sensitivity 1.00, timeliness 1.72 weeks), with fixed specificity=0.95 USA: excellent (VUTROC 0.81, sensitivity 0.90, timeliness 1.45 weeks), with fixed specificity=0.95 |
Hong Kong data: time series (dynamic linear model)>regression (simple)>CUSUMUS data: time series (dynamic linear model)>CUSUM>regression (simple) | ||
| SPC (CUSUM) | Different parameter combinations tested. d represents the number of weeks t separating the baseline and the index day of the outbreak. Range: 2 or 3. k represents the minimum standardised difference. Range: 1 or 2 | Hong Kong: poor (VUTROC 0.56, sensitivity 0.86, timeliness 2.00 weeks), with fixed specificity=0.95 USA: excellent (VUTROC 0.90, sensitivity 0.82, timeliness 1.51 weeks), with fixed specificity=0.95 |
Hong Kong data: time series (dynamic linear model)>regression (simple)>CUSUMUS data: time series (dynamic linear model)>CUSUM>regression (simple) | ||
| Outbreak prediction | |||||
| Timpka et al20 | Shewhart type | Daily and weekly | Pandemic outbreak: poor (AUC 0.84; PPV 0.58) on a daily basis and poor (at most acceptable) (AUC 0.78; PPV 0.79) on a weekly basis Seasonal outbreaks: outstanding (AUC 0.89; PPV 0.93) on a daily basis and excellent (AUC 0.83; PPV 1.00) on a weekly basis |
No comparisons | |
| Yuan et al21 | Multiple linear regression | Monthly | NA. Limits not defined for the adjusted metrics of residuals used (APE) | No comparisons | |
| Jiang et al22 | Bayesian network | Daily | Outstanding (r=0.97, prediction on day 13; r=0.94, prediction on day 22) | No comparisons | |
| Burkom et al23 | Regression (log-linear, non-adaptive) | Non-adaptive | Daily | NA. Limits not defined for the adjusted metrics of residuals used (MAD, MedAPE) | Ten series of case count data: Holt-Winters>regression (log-linear, adaptive)>regression (log-linear, non-adaptive) |
| Regression (log-linear, adaptive) | Adaptive | Ten series of case count data: Holt-Winters>regression (log-linear, adaptive)>regression (log-linear, non-adaptive) | |||
| Holt-Winters (generalised exponential smoothing) | Ten series of case count data: Holt-Winters>regression (log-linear, adaptive)>regression (log-linear, non-adaptive) | ||||
| Viboud et al24 | Method of analogues (non-parametric time-series forecasting method) | Weekly | From poor (r=0.66, for 10-week-ahead prediction) to excellent (r=0.81, for 1-week-ahead prediction) | Method of analogues>autoregressive model (linear)>Stone's naive method | |
| Autoregressive model (linear) | From poor (r=–0.07, for 10-week-ahead prediction) to acceptable (r=0.73, for 1-week-ahead prediction) | Method of analogues>autoregressive model (linear)>Stone's naive method | |||
| The naive method | Poor (r=–0.09, for 10-week-ahead prediction; r=0.65, for 1-week-ahead prediction) | Method of analogues>autoregressive model (linear)>Stone’s naive method | |||
APE, absolute percentage error; AUC, area under the curve; AUWROC, area under the weighted receiver operating characteristic curve; CUSUM, cumulative sum; MAD, median absolute residual; MedAPE, median absolute percentage error; NA, not applicable; PPV, positive predictive value; SPC, statistical process control; VUTROC, volume under the time-ROC surface.
Regarding prediction algorithms, a French study predicted national-level influenza outbreaks over 18 seasons,24 observing excellent performance for a non-parametric time-series method in 1-week-ahead predictions and poor performance in 10-week-ahead predictions. A study using county-level data from the USA22 reported outstanding predictive performance for a Bayesian network algorithm. However, the predictions in that study were made on days 13 and 22 of one single ongoing outbreak. Another study using telenursing data from a Swedish county to predict influenza outbreaks over three seasons, including the H1N1 pandemic in 2009, showed outstanding performance for seasonal influenza outbreaks on a daily basis and excellent performance on a weekly basis.20 However, the performance for the pandemic was poor on a daily and on a weekly basis (see online supplementary material file).
bmjopen-2015-010683supp.pdf (276.1KB, pdf)
An explanation of the apparent diversity of evaluation methods and findings is that the methodological perspectives and experiences from algorithm evaluations were reported in two distinct narrative formats. These narrative formats can be interpreted to represent biodefence informatics and health policy research, respectively (table 3).
Table 3.
Summary of narrative characteristics
| Narrative | Storyline | Intended audience* | Learning period dilemma | Theoretical proofs | Population descriptions | End point measures |
|---|---|---|---|---|---|---|
| Biodefence informatics17 18 22 23 |
System verification | Engineers and modellers | Irregular attention | Included in argument | Summary | Various statistical |
| Health policy research19–21 24 |
System validation | Policymakers | Binding attention | Excluded | Extensive | Standard epidemiological |
*In addition to researchers.
The biodefence informatics narrative
Assessments informing construction of technically and mathematically sound algorithms for outbreak detection and prediction were reported from mathematical modelling and health informatics contexts. Research in these fields was described in a biodefence informatics narrative. The setting for this narrative is formative evaluation and justification of algorithms for detection and prediction of atypical outbreaks of infectious diseases and bioterror attacks. In other words, these studies can be said to answer the system verification question: ‘Did we build the system right?’25 The narrative is set in a context where algorithms need to be modified and assured for detection and prediction of microbiological agents with unusual or unknown characteristics, for example, novel influenza virus strains or anthrax.26 The number of studies presented in the biodefence informatics narrative grew rapidly after the terrorist attacks in 2001.27 Reporting of influenza algorithm performance in this narrative is characterised by presentation of statistical or technical advancements, for example, making use of increments instead of rates or introduction of methods based on Markov models.18 As empirical data for logical reasons are scarce in biodefence settings, limited attention is in this narrative paid to the learning period dilemma. This dilemma represents a generic methodological challenge in algorithm development, that is, the statistical associations between indicative observations and the events to be predicted are determined in one time interval (the learning period) and used to predict the occurrence of corresponding events in a later interval (the evaluation period).28 When trying to detect or predict a novel infectious agent, the learning period dilemma primarily shows unavailability of learning data for calibration of model-based algorithms. For instance, for prediction algorithms based on the reproductive number,29 series of learning data of sufficient length for empirical determination of the serial interval cannot be made available during early outbreak stages, implying that the method cannot be used as supposed.30 Moreover, the microbiological features of the pathogen and the environmental conditions in effect during the learning period can change after the algorithm has been defined, requiring adjustments of algorithm components and parameters to be made for preserving the predictive performance. Algorithm performance can in the biodefence informatics be narrative verified by combining prospective evaluations with formal proofs and analyses of simulated and retrospective data. Although it is commonly emphasised that the evaluation results are preliminary with regard to population outcomes,22 the evaluation results are still included in the narrative.
The health policy research narrative
For evaluation study results to qualify as input to recommendations regarding infectious disease control practice, they should conform to general criteria established for health policy evidence. The analyses must be unbiased and not open for manipulation, for example, the data sources and analytic models should be described and fixed before data are accessed for analyses.31 In the corresponding research paradigm, the use of prospective study designs is regarded as the cornerstone in the research process.32 Correspondingly, the studies reported in the health policy research narrative answer the validation question: ‘Have we built the right system for detection and prediction of influenza seasons and outbreaks?’ Although the studies reported in this narrative mainly used data on clinical diagnoses and from laboratory tests, the two most recent studies also employed syndromic data: one study used data from telenursing call centres20 and the other study used data from an internet search engine.21 In the health policy research narrative, the foundation in real-world validation of alerts and predictions was shown, for instance, by pointing out that usually only a small number of annual infectious disease cycles of data are available for evaluations of new algorithms, leading to a constant lack of evidence-based information on which to base policy.19 It was also shown by that space was provided for discussions regarding whether algorithms would yield worse performances when outbreak conditions change, for example, that pandemic incidences are higher than those recorded during interpandemic periods.20 24 Moreover, evaluations presented in the health policy research narrative highlight the quantitative strength of the research evidence. For instance, in the study reporting excellent predictive performance of a non-parametric time-series method,24 the evaluation period lasted 938 weeks and covered an entire nation. In comparison, a prospective study reported in the biodefence informatics narrative accounted for an evaluation of a Bayesian network model22 that lasted 26 weeks and covered one US county.
Discussion
In a metanarrative review of studies evaluating the prospective performance of influenza outbreak detection and prediction algorithms, we found that methodological perspectives and experiences have, over time, been reported in two narratives, representing biodefence informatics and health policy discourse, respectively. Differences between the narratives are found in elements ranging from the evaluation settings and end point measures used to the structure of the argument. The biodefence informatics narrative, having an emphasis on verification of technically and mathematically sound algorithms, originates from the need to rapidly respond to evolving outbreaks of influenza pandemics and agents disseminated in bioterror attacks. Only more recently, studies presented in the biodefence informatics narrative have been directed to common public health problems, such as seasonal influenza and air pollution.33 Although evidence-based practices have been promoted by public health agencies during the period the assessed studies were published,34 only four prospective evaluations of influenza detection and prediction algorithms were reported as a health policy research narrative. However, despite being scarce for influenza, algorithm evaluations emphasising real-world validation of algorithm performance are relatively common for several other infectious diseases, for example, dengue fever.35 One reason for not choosing to report evaluations of influenza detection and prediction algorithms in the health policy narrative may be that the urgent quest for knowledge in association with atypical influenza outbreaks has led to an acceptance of evaluation accounts with limited empirical grounding. These accounts agree with mathematical and engineering research practices in biodefence informatics and are thus accepted as scientific evidence within those domains. This implies that awareness of the narrative format in which evidence is reported is essential when interpreting algorithm evaluations.
This study has methodological strengths and limitations that need to be taken into account when interpreting the results. A strength is that it was based on a metanarrative review. This is a relatively new method of systematic analyses of published literature, designed for topics that have been conceptualised differently and studied by different groups of researchers.36 We found that in a historical perspective, researchers from different paradigms have evaluated algorithms for influenza outbreak detection and prediction with different means and purposes. Some researchers have conceptualised algorithm evaluations as an engineering discipline, others as a subarea of epidemiology. The intention was not to conclude recommendations for algorithm use. Instead, the aim was to summarise different perspectives on algorithm development and reporting in overarching narratives, highlighting what different researchers might learn from one another's approaches. Regarding the limitations of the review, it must be taken into consideration that the ambition was to base the narrative analysis on evaluations with relevance for operational readiness and real-world application. There is a possibility that we failed to identify some relevant evaluations due to the absence of specific indexing terms for infection disease detection and prediction methods and that we excluded studies that were not indexed in research databases. However, we believe that the probability that we missed relevant evaluations for these reasons is low. We initially identified 1084 studies out of which 116 had relevant abstracts. Following examination of the corresponding articles, the majority had to be excluded from the final review because they did not fulfil the inclusion criteria at the detailed level (figure 1). One overall interpretation of this finding is that more research activity had been associated with developing detection and prediction algorithms than evaluating them and carefully reporting the results. For instance, a large number of interesting studies had to be excluded because non-prospective data were used for the evaluations, for example, the models were developed from learning data and evaluated against out-of-sample verification data from the same set using a leave-one-season-out approach.37 38 Regarding prediction algorithms, numerous potentially interesting studies were excluded because they did not report standard evaluation metrics. One example is a prospective Japanese study of predictions conducted during the pandemic outbreak in 2009, which reported only descriptive results.39 We found no prospective algorithm evaluations that applied an integrated outbreak detection and prediction. An Australian study applied an algorithm including detection and prediction functions,40 but this study used simulated data for the evaluation. Nonetheless, the eligibility criteria applied in this review accepted syndromic definitions of influenza as the gold standard, that is, specified sets of symptoms not requiring laboratory confirmation for diagnosis.41 If laboratory-confirmed diagnosis of influenza would have been included in the criteria, almost no studies would have qualified for inclusion in the review.
In summary, two narratives for reporting influenza detection and prediction algorithm evaluations have been identified. In the biodefence informatics narrative, technical and mathematical verification of algorithms is described, while the health policy narrative is employed to allow conclusions to be drawn about public health policy. A main dissimilarity between the narratives is the attention paid to the learning period dilemma. This dilemma represents a generic methodological challenge in the development of biosurveillance algorithms; the statistical models used to detect or predict an influenza-related event must be determined in a preceding time interval (the learning period). This means that there is always a shortage of time when algorithms for novel infectious diseases are to be validated in real-world settings. We offer two suggestions for future research and development based on these results. First, a sequence of evaluation research phases interconnected by a translation process should be defined, starting from theoretical research on construction of new algorithms in the biodefence informatics setting and proceeding stepwise to prospective field trials performed as health policy research. In the latter setting, the evaluation study design should be registered in an international trial database, such as ClinicalTrials.gov, before the start of prospective data collection. Second, standardised and transparent reporting criteria should be formulated for all types of algorithm evaluation research. The recent development of consensus statements for evaluations of prognostic models in clinical epidemiology42 can here be used as a reference.
Acknowledgments
Elin A. Gursky, ScD, MSc, provided valuable comments on drafts of this manuscript.
Footnotes
Contributors: AS and TT designed the study, analysed the information contained in the manuscript, wrote the manuscript and provided final approval of the version to be published. AS searched for relevant papers and documented data about the manuscripts. TT revised the results of the search and the documentation and is the guarantor of the content.
Funding: This study was supported by grants from the Swedish Civil Contingencies Agency (TT, grant number 2010-2788); and the Swedish Science Council (TT, grant number 2008-5252). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1.Butler D. When Google got flu wrong. Nature 2013;494:155–6. 10.1038/494155a [DOI] [PubMed] [Google Scholar]
- 2.Santos-Preciado J, Franco-Paredes C, Hernandez-Flores I et al. What have we learned from the novel influenza A (H1N1) pandemic in 2009 for strengthening pandemic influenza preparedness? Arch Med Res 2009;40:673–6. 10.1016/j.arcmed.2009.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller M, Blench M, Tolentino H et al. Use of unstructured event-based reports for global infectious disease surveillance. Emerg Infect Dis 2009;15:689–95. 10.3201/eid1505.081114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timpka T, Eriksson H, Gursky EA et al. Requirements and design of the PROSPER protocol for implementation of information infrastructures supporting pandemic response: a Nominal Group study. PLoS ONE 2011;6:e17941 10.1371/journal.pone.0017941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nsoesie EO, Brownstein JS, Ramakrishnan N et al. A systematic review of studies on forecasting the dynamics of influenza outbreaks. Influenza Other Respir Viruses 2014;8:309–16. 10.1111/irv.12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckeridge DL. Outbreak detection through automated surveillance: a review of the determinants of detection. J Biomed Inform 2007;40:370–9. 10.1016/j.jbi.2006.09.003 [DOI] [PubMed] [Google Scholar]
- 7.Louz D, Bergmans HE, Loos BP et al. Emergence of viral diseases: mathematical modeling as a tool for infection control, policy and decision making. Crit Rev Microbiol 2010;36:195–211. 10.3109/10408411003604619 [DOI] [PubMed] [Google Scholar]
- 8.Neuberger A, Paul M, Nizar A et al. Modelling in infectious diseases: between haphazard and hazard. Clin Microbiol Infect 2013;19:993–8. 10.1111/1469-0691.12309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corley CD, Pullum LL, Hartley DM et al. Disease prediction models and operational readiness. PLoS ONE 2014;9:e91989 10.1371/journal.pone.0091989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chretien JP, George D, Shaman J et al. Influenza forecasting in human populations: a scoping review. PLoS ONE 2014;9:e94130 10.1371/journal.pone.0094130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. 2nd edn New York: Springer-Verlag, 2009. [Google Scholar]
- 12.Wong G, Greenhalgh T, Westhorp G et al. RAMESES publication standards: meta-narrative reviews. BMC Med 2013;11:20 10.1186/1741-7015-11-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mays N, Pope C, Popay J. Systematically reviewing qualitative and quantitative evidence to inform management and policy-making in the health field. J Health Serv Res Policy 2005;10(Suppl 1):6–20. 10.1258/1355819054308576 [DOI] [PubMed] [Google Scholar]
- 14.Unkel S, Farrington CP, Garthwaite PH et al. Statistical methods for the prospective detection of infectious disease outbreaks: a review. J Roy Stat Soc A 2012;175:49–82. 10.1111/j.1467-985X.2011.00714.x [DOI] [Google Scholar]
- 15.Hosmer DW, Lemeshow S. Applied logistic regression. 2nd edn London: John Wiley, 2000:228–30. [Google Scholar]
- 16.Cohen J. Statistical power analysis for the behavioral sciences. 2nd edn Mahwah, NJ: Lawrence Erlbaum, 1988. [Google Scholar]
- 17.Closas P, Coma E, Méndez L. Sequential detection of influenza epidemics by the Kolmogorov-Smirnov test. BMC Med Inform Decis Mak 2012;12:112 10.1186/1472-6947-12-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Beneito MA, Conesa D, López-Quílez A et al. Bayesian markov switching models for the early detection of influenza epidemics. Stat Med 2008;27:4455–68. 10.1002/sim.3320 [DOI] [PubMed] [Google Scholar]
- 19.Cowling BJ, Wong IO, Ho LM et al. Methods for monitoring influenza surveillance data. Int J Epidemiol 2006;35:1314–21. 10.1093/ije/dyl162 [DOI] [PubMed] [Google Scholar]
- 20.Timpka T, Spreco A, Eriksson O et al. Predictive performance of telenursing complaints in influenza surveillance: a prospective cohort study in Sweden. Euro Surveill 2014;19:pii: 20966 10.2807/1560-7917.ES2014.19.46.20966 [DOI] [PubMed] [Google Scholar]
- 21.Yuan Q, Nsoesie EO, Lv B et al. Monitoring influenza epidemics in China with search query from Baidu. PLoS ONE 2013;8:e64323 10.1371/journal.pone.0064323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang X, Wallstrom G, Cooper GF et al. Bayesian prediction of an epidemic curve. J Biomed Inform 2009;42:90–9. 10.1016/j.jbi.2008.05.013 [DOI] [PubMed] [Google Scholar]
- 23.Burkom HS, Murphy SP, Shmueli G. Automated time series forecasting for biosurveillance. Stat Med 2007;26:4202–18. 10.1002/sim.2835 [DOI] [PubMed] [Google Scholar]
- 24.Viboud C, Boelle PY, Carrat F et al. Prediction of the spread of influenza epidemics by the method of analogues. Am J Epidemiol 2003;158:996–1006. 10.1093/aje/kwg239 [DOI] [PubMed] [Google Scholar]
- 25.Boehm BW. Software engineering economics. New York: Prentice-Hall, 1981. [Google Scholar]
- 26.Gursky EA, Bice G. Assessing a decade of public health preparedness: progress on the precipice? Biosecur Bioterror 2012;10:55–65. 10.1089/bsp.2011.0085 [DOI] [PubMed] [Google Scholar]
- 27.Paterson BJ, Durrheim DN. The remarkable adaptability of syndromic surveillance to meet public health needs. J Epidemiol Glob Health 2013;3:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altman DG, Royston P. What do we mean by validating a prognostic model? Stat Med 2000;19:453–73. [DOI] [PubMed] [Google Scholar]
- 29.White LF, Wallinga J, Finelli L et al. Estimation of the reproductive number and the serial interval in early phase of the 2009 influenza A/H1N1 pandemic in the USA. Influenza Other Respir Viruses 2009;3:267–76. 10.1111/j.1750-2659.2009.00106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klick B, Nishiura H, Leung GM et al. Optimal design of studies of influenza transmission in households II: comparison between cohort and case-ascertained studies. Epidemiol Infect 2014;142:744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly M, Morgan A, Ellis S et al. Evidence based public health: a review of the experience of The National Institute of Health and Clinical Excellence (NICE) of developing public health guidance in England. Soc Sci Med 2010;71:1056–62. 10.1016/j.socscimed.2010.06.032 [DOI] [PubMed] [Google Scholar]
- 32.Derose SF, Schuster MA, Fielding JE et al. Public health quality measurement: concepts and challenges. Annu Rev Public Health 2002;23:1–21. 10.1146/annurev.publhealth.23.092601.095644 [DOI] [PubMed] [Google Scholar]
- 33.Buehler JW, Whitney EA, Smith D et al. Situational uses of syndromic surveillance. Biosecur Bioterror 2009;7:165–77. 10.1089/bsp.2009.0013 [DOI] [PubMed] [Google Scholar]
- 34.European Centre for Disease Prevention and Control. Evidence-based methodologies for public health – how to assess the best available evidence when time is limited and there is lack of sound evidence. Stockholm: ECDC, 2011. [Google Scholar]
- 35.Hii YL, Zhu H, Ng N et al. Forecast of dengue incidence using temperature and rainfall. PLOS Negl Trop Dis 2012;6:e1908 10.1371/journal.pntd.0001908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenhalgh T, Robert G, Macfarlane F et al. Storylines of research in diffusion of innovation: a meta-narrative approach to systematic review. Soc Sci Med 2005;61:417–30. 10.1016/j.socscimed.2004.12.001 [DOI] [PubMed] [Google Scholar]
- 37.Reich NG, Cummings DA, Lauer SA et al. Triggering Interventions for Influenza: the ALERT Algorithm. Clin Infect Dis 2015;60:499–504. 10.1093/cid/ciu749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dugas AF, Jalalpour M, Gel Y et al. Influenza Forecasting with Google Flu Trends. PLoS ONE 2013;8:e56176 10.1371/journal.pone.0056176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohkusa Y, Sugawara T, Taniguchi K et al. Real-time estimation and prediction for pandemic A/H1N1(2009) in Japan. J Infect Chemother 2011;17:468–72. 10.1007/s10156-010-0200-3 [DOI] [PubMed] [Google Scholar]
- 40.Boyle JR, Sparks RS, Keijzers GB et al. Prediction and surveillance of influenza epidemics. Med J Aust 2011;194:S28–33. [DOI] [PubMed] [Google Scholar]
- 41.Last JM, ed. A dictionary of epidemiology. Oxford: Oxford University Press, 2001. [Google Scholar]
- 42.Collins GS, Reitsma JB, Altman DG et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 2015;350:g7594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2015-010683supp.pdf (276.1KB, pdf)