Abstract
Alternative precursor-mRNA splicing is a key mechanism for regulating gene expression in mammals and is controlled by specialized RNA-binding proteins. The misregulation of splicing is implicated in multiple neurological disorders. We describe recent mouse genetic studies of alternative splicing that reveal its critical role in both neuronal development and the function of mature neurons. We discuss the challenges in understanding the extensive genetic programmes controlled by proteins that regulate splicing, both during development and in the adult brain.
The precursor-mRNA (pre-mRNA) splicing reaction is a key step in the regulation of eukaryotic gene expression. Nearly all mammalian multi-exon genes produce multiple mRNA isoforms through alterations in the choice of splice sites to produce proteins of different structures and functions, or to alter mRNA localization, translation or decay. In keeping with its cellular and functional complexity, the mammalian nervous system makes extensive use of splicing regulation to generate specialized protein isoforms that affect all aspects of neuronal development and function1–4. Splicing defects are being increasingly implicated in neurological and neurodegenerative diseases, which underscores the need to better understand these regulatory processes.
Alternative splicing patterns (BOX 1) are regulated by specialized pre-mRNA binding proteins that alter spliceosome assembly at specific splice sites5–8 (BOX 2). These proteins are structurally diverse and can exert different effects on a target transcript depending on their binding position, their modification by signalling pathways and their interactions with cofactors. Some regulators exhibit tissue-specific expression, whereas others are more ubiquitous, but they all regulate large overlapping programmes of neuronal alternative splicing events. Although each regulatory protein can affect many different RNA targets, each transcript is usually targeted by multiple regulators (FIG. 1). These compounded levels of complexity have challenged the characterization of the biological function of splicing regulatory proteins and studies of their mechanisms of action. The biological roles of splicing regulators and the cellular programmes they control are currently being elucidated through mouse genetics.
Box 1. Patterns of alternative splicing.
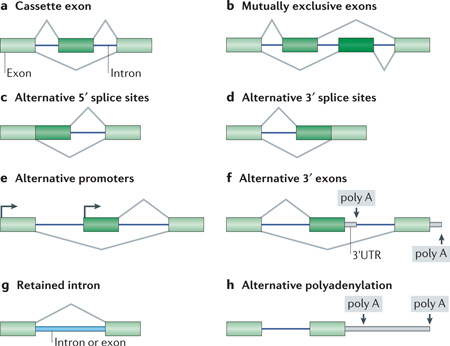
The diversity of mRNA isoforms is generated from many different patterns of alternative splicing. Genes are segmented into introns and exons. During the precursor-mRNA splicing process, introns are excised from the precursor mRNA and exons are ligated together to form the mRNA. Special sequences at the intron ends define where the cleavage and ligation reactions occur. The 5′ splice site or donor site is at the 5′ end of the intron. The 3′ splice site or acceptor site is at the 3′ end of the intron. Splicing catalysis by the spliceosome takes place in two cleavage and ligation steps. The 3′ splice site has an associated branchpoint sequence, which is joined to the 5′ splice site after the first cleavage step. This is followed by cleavage at the 3′ splice site and ligation of the two exons. In the figure, the light green boxes indicate exons and the dark green boxes indicate alternative exons. The v-shaped lines show the different ways in which the exons can be joined in a final mRNA. The most common change in splicing pattern is a cassette exon (skipped exon; see part a of the figure), the inclusion or skipping of which will insert or delete a sequence from the final mRNA. Mutually exclusive exons (see part b of the figure) are a pair of consecutive cassette exons where only one of the exons is included in the mRNA. Alternative 5′ splice sites (see part c of the figure) are consecutive ‘donor sites’ that change the length of an exon at its 3′ end. Conversely, alternative 3′ splice sites (see part d of the figure) are consecutive acceptor sites that change the 5′ end of the exon. Alternative promoters (see part e of the figure) and alternative 3′ exons (see part f of the figure) create different first exons and different last exons on the mRNA, respectively. Retained introns (see part g of the figure) can be excised as a typical intron or remain in the final mRNA. Alternative polyadenylation (see part h of the figure) in the last exon allows for the generation of three prime untranslated regions (3’ UTRs) of varying lengths. A single gene can have multiple positions and patterns of alternative splicing to create a family of many different mRNAs and proteins through the inclusion or skipping of various alternatively spliced RNA segments. Figure adapted from REF. 1, Nature Publishing Group.
Box 2. Regulation of an alternative exon by RNA-binding proteins.
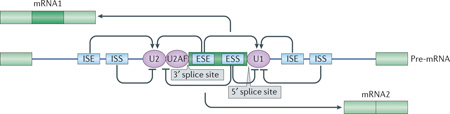
Trans-acting RNA-binding proteins (RBPs) interact with cis-sequence elements in the precursor mRNA to facilitate or inhibit the assembly of the spliceosomal machinery at nearby splice sites. The 5′ splice site is initially bound by U1 small nuclear ribonucleoprotein (snRNP, see the figure). The U2 snRNP recognizes the branchpoint and is recruited by the U2AF proteins that are bound to the polypyrimidine tract between the branchpoint and the 3′ splice site. Binding of U1 and U2 allows recognition of an exon in a process called exon definition. These snRNPs are subsequently brought into interaction across an intron to allow further spliceosome assembly and the pairing of splice sites within the catalytic centre of the spliceosome. An alternative splicing event frequently involves multiple competing weak splice sites that are subject to dynamic regulation by neighbouring cis elements. These cis elements include intronic and exonic splicing enhancers and intronic and exonic splicing silencers that recruit activator or repressor RBPs, respectively. These RBPs, through multiple modes of action that are not yet understood, collectively influence splice site recognition or splice site pairing within the spliceosome. The levels and activity of these trans-acting RBPs control the choice of splice sites for many different transcripts.
Activator RBPs binding to enhancer elements are shown as arrows, and repressors binding to silencer elements are shown as inhibitory arrows. Constitutive flanking exons are shown in light green and the alternative exon is shown in dark green.
ESE, exonic splicing enhancer; ESS, exonic splicing silencer; ISE, intronic splicing enhancer; ISS, intronic splicing silencer.
Figure 1. Splicing regulatory networks.
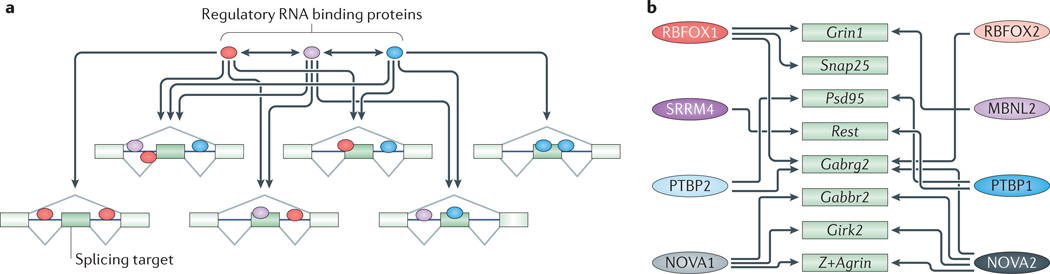
a | Splicing regulators control large target exon sets that often overlap with those regulated by other RNA-binding proteins (RBPs). Therefore, splicing of a given alternative exon can be affected by multiple RBPs. RBPs also affect their own splicing and homeostatic expression as well as that of other RBPs. The high degree of cross-regulation (indicated by arrows) between splicing regulators and their target sets creates complex splicing networks where the perturbation of a single RBP can lead to pleiotropic effects. Conversely, the splicing outcome of an exon can result from the combinatorial control of many RBPs. b | This figure shows some of the splicing target transcripts discussed in this Review (green boxes) that are cross-regulated (indicated by arrows) by multiple RBPs (coloured ovals on the left and right). Girk2, inwardly rectifying potassium channel Kir3.2; Gabrg2, GABAA receptor subunit gamma 2; Gabbr2, GABAB receptor 2; MBNL, muscleblind-like; NOVA, neuro-oncological ventral antigen; Psd95, postsynaptic density protein 95; PTBP, polypyrimidine tract binding protein; Rest, repressor element 1-silencing transcription factor; RBFOX, RNA-binding protein fox 1 homologue; Snap25, synaptosomal-associated protein 25; SRRM4, serine/arginine repetitive matrix protein 4.
The neurogenetics of splicing is still in its infancy and can be confounded by several factors. How an alteration in splicing pattern changes the function of an encoded gene product is not usually known. Nevertheless, the mutation of an individual regulator often leads to a phenotype that either exhibits a high degree of pleiotrophy and/or is lethal owing to its extensive target set. In other cases, splicing regulators often have paralogues, the partially redundant function of which reduces the phenotypic impact of single gene mutations. Although splicing regulators can be widely expressed throughout the brain, different neuronal cell types often express different combinations of regulators and show a distinct susceptibility to their mutation. Despite this complexity, it is clear that splicing regulatory programmes dramatically affect all aspects of neuronal development and biology, from neurogenesis to mature synaptic function.
In this Review, we survey recent genetic studies of individual splicing regulators and the diverse roles they have in the mammalian nervous system. These studies have been aided by powerful new methods that allow the analysis of changes in splicing across the whole transcriptomes of mutant mice and the genome-wide mapping of regulatory protein binding. These methods have been extensively reviewed elsewhere9–12, and our main focus is the phenotypic analyses that have begun to define the biological roles of these complex splicing regulatory programmes.
Alternative splicing and neurogenesis
Alternative splicing patterns change dramatically as cells progress along the neuronal lineage. These new splicing events are directed by changes in the expression of particular RNA-binding proteins, such as the polypyrimidine tract binding protein 1 (PTBP1) and serine/arginine repetitive matrix protein 4 (SRRM4; also known as nSR100), that affect neuronal fate and early neuronal differentiation.
PTBP1 in neurogenesis
The members of the PTBP family of splicing regulators are similar in structure and RNA-binding properties, but distinct in their cell type expression. PTBP1 (also known as PTB) is broadly expressed, but is largely absent from neurons, muscle cells and certain other mature cells. By contrast, PTBP2 (nPTB or brPTB) is found in neurons, myoblasts and spermatocytes. A third paralogue, PTBP3 (ROD1), expressed in haematopoietic cells and liver cells, is not known to affect splicing in neurons. Each PTBP contains four RNA recognition motif domains that together bind to extended CU-rich sequences13,14.
PTBP1 is abundant in neural stem cells and progenitors, but on mitotic exit its expression is sharply reduced by the induction of the neuronal microRNA miR-124 (REF. 15). Reduced PTBP1 expression also enhances miR-124 repression of the REST (repressor element 1-silencing transcription factor) complex16, a well-known transcriptional repressor of neuronal gene expression17,18. Strikingly, the simple depletion of PTBP1 from cultured fibroblasts is sufficient to induce their trans-differentiation into neurons16. How the many PTBP1 splicing targets contribute to maintaining pluripotency or preventing differentiation is not yet clear. One target exon repressed by PTBP1 is in the transcription factor pre-B cell leukaemia homeobox 1 (Pbx1) gene. Precocious expression of the neuronal PBX1 isoform leads to the early induction of neurogenic genes19. Another notable PTBP1 target is exon 10 of Ptbp2. The repression of exon 10 leads to nonsense-mediated mRNA decay (NMD) of the Ptbp2 transcript and prevents its expression in PTBP1+ cells15,20,21. Induction of the PTBP2 protein has a critical role in neuronal differentiation.
Germline knockout of Ptbp1 in mice leads to early embryonic mortality22,23. Mice with pan-neuronal loss of Ptbp1 have grossly normal brain morphology at an early age, but show a progressive loss of ependymal cells from the lateral ventricles with hydrocephaly and die by 10 weeks after birth24. PTBP1 loss may induce the precocious differentiation of radial glial cells into neurons, thereby depleting the radial glial cell pool that later gives rise to ependymal cells25 (FIG. 2). The loss of ependymal cells is restricted to the dorsal telencephalon, indicating variable roles for PTBP1 across brain regions.
Figure 2. Splicing regulators in cortical development and function.
Alternative splicing controls multiple aspects of early neuronal development. Defects in neurogenesis are seen in mouse mutants of a variety of regulatory RNA-binding proteins, including Ptbp1−/−, Ptbp2−/− and Srrm4−/−. Loss of polypyrimidine tract binding protein 1 (PTBP1) can cause precocious neurogenesis, deplete the neural stem cell pool and lead to fewer ependymal cells arising from radial glia later in development. PTBP2 loss may alter neural stem cell positioning and proliferation. Depletion of serine/arginine repetitive matrix protein 4 (SRRM4) inhibits neurogenesis of upper-layer neurons and causes the accumulation of progenitors or lower-layer neurons, resulting in abnormal cortical lamination (see inset). Defects in cortical lamination are also seen in mice lacking neuro-oncological ventral antigen 2 (NOVA2), where mis-splicing of the Reelin component disabled 1 (Dab1) leads to failure of many layer II/III and IV neurons to migrate properly (see inset). C P, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; WM, white matter; VZ, ventricular zone. Figure (left panel) adapted with permission from REF. 199, Elsevier, and from REF. 198: Poduri, A., Evrony, G. D., Cai, X. & Walsh, C. A. Somatic mutation, genomic variation, and neurological disease. Science 341, 1237758–1237758 (2013). Reprinted with permission from AAAS.
An interplay between PTBP1 and SRRM4
SRRM4 is similar to the serine/arginine (SR)-rich splicing factor (SRSF) family and SR-related proteins containing serine/arginine repeats26–28, but is unique for its brain-specific expression29. Despite lacking a canonical RNA-binding domain, SRRM4 frequently binds UGC-rich sequences located between the polypyrimidine tract and the 3′ splice site of target exons30. The most enriched motifs surrounding SRRM4-dependent exons are typical PTBP binding elements, suggesting that SRRM4 can antagonize PTBP activity, and the regulatory programmes for these two proteins significantly overlap. SRRM4 promotes splicing of the REST4 isoform, which lacks four of the nine zinc fingers found in the full-length protein31 and has reduced transcriptional repression activity32–34. Conversely, REST inhibits Srrm4 expression in non-neuronal cells31. Through their action on transcription factor pre-mRNA, splicing regulators can indirectly control the transcription of neuronal genes.
Knockdown of Srrm4 in the developing mouse cortex inhibits neuronal differentiation and leads to the accumulation of Pax6+ progenitor cells in the ventricular zone and the depletion of differentiated cells from the cortical plate31. Interestingly, the germline deletion of Srrm4 results in fewer Pax6+ cells in the ventricular zone and fewer postmitotic NeuN+ neurons35 (FIG. 2). The different observations of Pax6+ cells may result from different effects of acute versus prolonged loss of SRRM4, or may indicate that SRRM4 has different roles in early versus late neurogenesis. Indeed, Srrm4−/− mice have fewer late-born, upper-layer neurons and more early-born, lower-layer neurons, suggesting either depletion of the neural stem/progenitor cell pool or alterations in neuronal subtype specification (FIG. 2).
The seemingly mild neurogenic phenotypes of the Ptbp1 and Srrm4 knockout mice compared with the more dramatic results in tissue culture suggest that the in vivo programme of neuronal induction contains multiple fail-safe mechanisms, with PTBP1 and SRRM4 serving opposing roles in reinforcing the robustness of the regulatory network. The interplay of these factors and their interactions with miR-124 and the REST complex constitute an important genetic programme underlying neuronal cell fate commitment.
Regulation of neuronal migration
The alternative splicing of components of the Reelin signalling pathway is important for proper neuronal migration in multiple brain regions. A loss of splicing regulators, such as neuro-oncological ventral antigen 2 (NOVA2) and RNA-binding protein fox-1 homologue 2 (RBFOX2) leads to defects in cortical and cerebellar lamination.
NOVA2 ensures proper migration of late-born cortical neurons
The first NOVA protein was identified as an autoantigen in patients with paraneoplastic opsoclonus-myoclonus ataxia, a human neurological syndrome characterized by motor and cognitive deficits36. The two paralogues NOVA1 and NOVA2 each contain three K homology (KH)-type RNA-binding domains and bind clusters of YCAY elements. NOVA1 is mainly expressed in the hindbrain and ventral spinal cord, whereas NOVA2 is predominant in the forebrain and dorsal spinal cord, with some overlapping expression in portions of the midbrain and hindbrain37. Genetic knockout of Nova1, Nova2, or both has demonstrated important roles for the two proteins in multiple aspects of brain development38,39.
Proper cortical lamination requires NOVA2. In Nova2-null mice, neurons of cortical layers II/III and IV are mislocalized to lower layers without altering the layer-specific molecular markers39 (FIG. 2). Progenitor cell proliferation and radial glia morphology are largely unaffected, suggesting a defect in neuronal migration rather than subtype specification. This contrasts with lamination defects in Srrm4-null mice, where increased numbers of lower-layer neurons were attributed to the premature commitment of progenitors to neurogenesis or alterations in subtype specification.
The defective migration of Nova2−/− upper-layer neurons was attributed to the mis-splicing of disabled 1 (Dab1), a component of the Reelin signalling pathway that controls cortical neuronal migration and lamination40–42. In wild-type neurons, NOVA2 represses both exon 7b and 7c of the Dab1 transcript, and the resulting DAB1 protein isoform DAB1Δ7bc is subject to ubiquitylation upon Reelin activation43–45. In Nova2−/− neurons, the abnormal inclusion of exons 7b and 7c produces a more stable isoform that may antagonize the activity of DAB1Δ7bc46,47. The introduction of a Dab1Δ7bc transgene rescues the migration defect for a subset of the layer II–IV Nova2−/− neurons. This rescue with a single spliced isoform provides an important method of validating the source for particular aspects of a pleiotropic phenotype. Notably, Dab1 is mis-spliced in the Nova2-null cortex only between E14 and E18. This restricted regulatory window and the limited population of affected cells reflect the complicated landscape of alternative splicing during neuronal development, where overlapping splicing regulatory programmes come into play at specific times and in specific neuronal populations.
RBFOX2 is required for proper Purkinje cell radial migration
Purkinje cell migration in the cerebellum is controlled by RBFOX2, which is a member of the highly conserved RBFOX family of RNA-binding proteins: RBFOX1 (also known as A2BP1), RBFOX2 (also known as RBM9) and RBFOX3 (also known as NeuN). The RBFOX proteins all bind the RNA sequence element (U)GCAUG via a single RNA recognition motif domain. RBFOX binding upstream of, or within, an alternative exon typically inhibits exon inclusion, whereas downstream binding usually promotes splicing48. The mechanistic basis for this pattern is not known, but multiple other splicing regulators, such as NOVA49,50, exhibit the same positional dependence. All three Rbfox genes are broadly expressed in the brain, with individual neuronal cell types expressing different combinations at different developmental times51–55. For example, Purkinje cells express RBFOX2 early in development, with later onset of RBFOX1 and no expression of RBFOX3. By contrast, cerebellar granule cells switch from expressing RBFOX2 during proliferation and migration to RBFOX1 and RBFOX3 with maturation55. It is not known how the different RBFOX proteins differ in activity; however, their complex expression patterns imply that they may serve overlapping but distinct roles.
Pan-neuronal Rbfox2 knockout mice show increased mortality with frequent hydrocephaly at 1 month of age55. The cerebellum is severely affected, with a substantially reduced size and a loss of foliation. Purkinje cells normally migrate outwards from the ventricular zone to be arrayed in a single layer between the external and internal granule layers by embryonic day 18 (REFS 56,57). The Rbfox2−/− Purkinje cells show a substantial delay in migration and increased cell death, resulting in a disorganized Purkinje cell layer (FIG. 3). Rbfox2−/− brains show altered splicing in transcripts known to control cell migration. In particular, low-density lipoprotein receptor-related protein 8 (LRP8), which normally binds Reelin to control cortical and Purkinje neuron migration, produces higher amounts of a dominant-negative isoform in Rbfox2−/− brains58. Consistently, Lrp8-null mice show ectopic Purkinje cells similar to the Rbfox2-null mice59,60. It will be interesting to test the roles of LRP8 and other components of the Reelin signalling pathway in Rbfox2−/− rescue experiments, similar to those in Nova2-null mice.
Figure 3. Splicing regulators in cerebellar development and function.
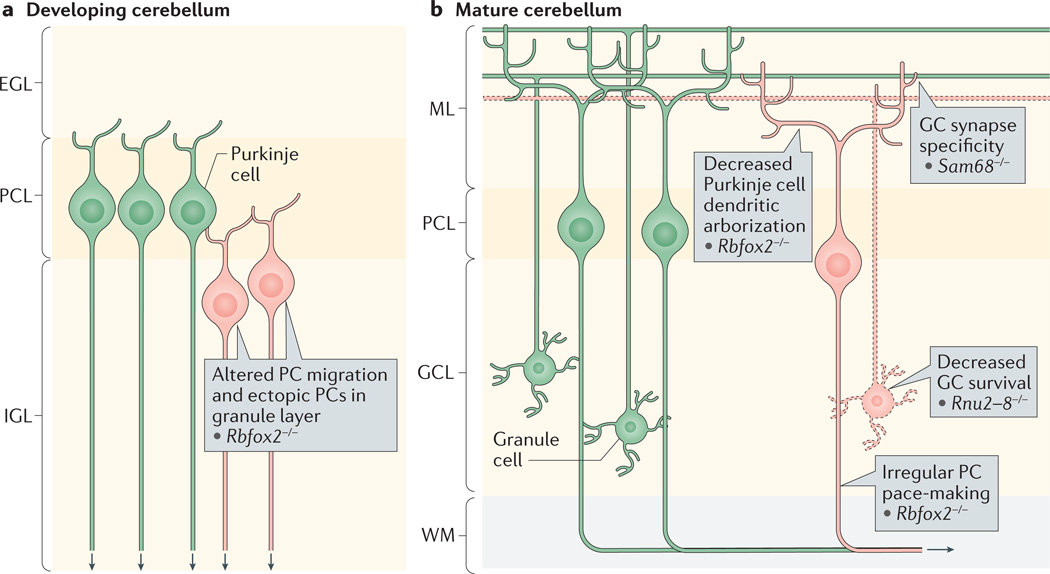
a | RNA-binding protein fox1 homologue 2 (RBFOX2) is required for both Purkinje cell (PC) migration and mature function. The cerebellum in Rbfox2−/− mouse mutants exhibits a disorganized PC layer (PCL) with ectopic PCs found in the internal granule layer (IGL), as well as reduced PC dendritic arborization later in development. b | In mature PCs, RBFOX2 controls the splicing and expression of the sodium channel gene Scn8a, which is needed for proper PC pace-making. Splicing regulation is also required for granule neuron survival and proper synaptic specificity. Loss of the U2 small nuclear RNA, a core spliceosomal component that is partially encoded by Rnu2–8, leads to increased intron retention and progressive granule neuron death. In granule neurons, SRC-associated in mitosis 68 kDa protein (SAM68) affects trans-synaptic interactions through alternative splicing of neurexin. EGL, external granule layer; GC, granule cell; GCL; granule cell layer; ML, molecular layer; WM, white matter.
Multiple components of the Reelin pathway and other signalling pathways affecting neuronal migration are expressed as alternatively spliced isoforms61,62. The regulators of these splicing events are largely unknown, and it is likely that other splicing regulator mutants will show migration defects in particular brain structures.
Synaptogenesis and cell survival
After commitment to differentiation and migration to their proper location, neurons undergo a long period of maturation that includes the formation and maturation of synapses. Alternative splicing defines the gene products involved in these processes, and particular splicing regulator mutations have been shown to dramatically affect these developmental steps.
PTBP2 is required for proper neuronal maturation
The downregulation of PTBP1 in neural stem/progenitor cells as they exit mitosis induces the expression of PTBP2, which is required for neuronal development and survival. The role of PTBP2 was revealed in different mutant mice carrying either germline null alleles or pan-neuronal conditional alleles63,64. These mice are paralysed and unresponsive to touch at birth and show perinatal lethality with respiratory failure, possibly owing to a loss of innervation to the diaphragm. The brains of these mice appear grossly normal. In one analysis, small ectopic clusters of S and M phase cells were found in reverse orientation from their normal positions63 (FIG. 2). These cellular abnormalities were possibly mediated by an observed change in the splicing of Numb, a known regulator of asymmetrical neural stem cell division65,66. Early differentiation defects were not reported in another germline Ptbp2-null allele, but these mice were not subjected to the same analyses64. This null mutant exhibited a loss of some early developing white matter tracts. Overall, the early lethality of the Ptbp2-null alleles and of the pan-neuronal knockout limited analyses of their phenotypes.
The role of PTBP2 in later development was revealed by its depletion from excitatory neurons of the dorsal telencephalon using an Emx1-Cre line64. At birth, the Emx1– Ptbp2−/− brain appears similar to the wild-type brain in morphology, size, neuronal fate commitment and cortical lamination. However, Emx1–Ptbp2−/− brains begin to show cortical atrophy as early as P5, and by P15 the cortex shows massive cell death and is almost completely degenerated. Similarly, cultured Ptbp2−/− embryonic cortical neurons initially appear normal in plating efficiency and neurite outgrowth, but exhibit progressive cell death beginning in the second week. The cell death is possibly due to a failure of synapse formation or another aspect of maturation, with a resulting lack of activity-dependent survival signals (FIG. 2). Because synaptogenesis occurs later in the forebrain than elsewhere in the central nervous system, similar defects in synaptogenesis with neuronal death may also occur in the lower brain of Ptbp2-null mice and lead to the perinatal lethality.
All of the Ptbp2 mutant mouse models show a precocious expression of many adult mRNA isoforms encoding proteins that affect a variety of cellular functions, including the regulation of transcription, synaptic transmission, synapse organization and endocytosis63,64. Crosslinking immunoprecipitation (CLIP) analysis indicates that many of these transcripts are direct PTBP2 targets63. In early development, the embryonic splicing pattern of many neuronal genes is maintained during the switch from PTBP1 to PTBP2. Later in maturation, coincident with marked PTBP2 downregulation and synaptogenesis, the adult isoforms become more prevalent63,64,67. Thus the high expression of PTBP2 during early development extends the expression of the embryonic splicing programme until late in neuronal maturation. How the premature induction of the adult isoforms contributes to neuronal cell death and other phenotypes of Ptbp2−/− neurons is unclear.
One synaptic target of PTBP1 and PTBP2 is exon 18 of postsynaptic density protein 95 (Psd95; also known as Dlg4), which encodes the major scaffold protein of excitatory synapses. The Psd95 transcript is expressed in many non-neuronal cells, where skipping exon 18 leads to NMD of the transcript, preventing its productive translation. Overexpression of either PTBP1 or PTBP2 in mature neurons inhibits exon 18 inclusion, PSD95 protein expression and excitatory synapse formation67. PTBP1 restricts PSD95 protein expression to neurons, whereas PTBP2 controls the temporal induction of PSD95 late in neuronal maturation. The sequential expression of PTBP1 and PTBP2 thus serves to precisely time the production of this key synaptic protein.
SRRM4 in synaptogenesis and development
Two mutant alleles of Srrm4 have been analysed and shown to exhibit different phenotypes. The Bronx waltzer (bv) mouse arose from a forward genetic screen68 and produces an unstable truncated or translationally defective protein. Bv/bv mutants show deafness, head tossing and circling69. A second conditional Srrm4 allele produces a frame-shifted product lacking the critical RS-rich domain and splicing activity when crossed to a germline Cre strain (Srrm4Δ7–8)35. The phenotypes of homozygous Srrm4Δ7–8 mice are more severe than those of the bv/bv mice, with defects in multiple neurodevelopmental processes. Although bv/bv mice live to adulthood, only 15% of Srrm4Δ7–8 mice survive beyond birth and adult survivors show severe tremors with some balance defects similar to the bv/bv mice.
The bv/bv mice have defects in the differentiation and/or survival of inner hair cells and vestibular hair cells of the cochlea70,71. Both inner hair cells and vestibular hair cells, normally densely innervated by spiral ganglion neurons, progressively degenerate and are completely lost by the first postnatal week. By contrast, the outer hair cells, which normally require only sparse innervation, are unaffected72. The bv/bv inner ear shows aberrant splicing in genes enriched in neurotransmission and secretion. This is similar to the Srrm4Δ7–8 brain, which shows aberrant splicing in genes implicated in vesicle trafficking and recycling. Although it remains unclear how these splicing defects lead to decreased cell survival, one possibility is that synaptogenesis and synaptic transmission, which are required for the survival of inner hair cells and vestibular hair cells, are impaired. As seen in the Ptbp2−/− forebrain, the bv/bv mouse provides evidence that correct alternative splicing is necessary for proper synaptogenesis and cell survival.
The phenotypes of Srrm4Δ7–8 mice have similarities to those of the Ptbp2−/− mice. Srrm4Δ7–8 mice show no gross morphological phenotype during embryonic development, but most die from respiratory failure within a few hours of birth. This appears to result from insufficient phrenic innervation to the diaphragm35, as secondary branching of motor neuron axons in this region is reduced by twofold. Although the molecular events underlying the phenotypic defects are not yet defined, one-third of in vivo SRRM4 targets overlap with PTBP2 targets. Comparisons of these systems will provide interesting insights into how developmental alternative splicing programmes control synaptogenesis.
NOVA control of motor neuron development and survival
Defects in muscle innervation and neuromuscular junction (NMJ) development and function are also seen in Nova1 and Nova2 double-knockout (dKO) mice. Nova1−/− mice appear normal at birth, but die in the second postnatal week with motor neuron apoptosis, profound motor failure and action-induced tremors37,38. Nova1/2 dKO mice are born paralysed and die from respiratory failure indicative of NMJ defects73. Indeed, the dKO mice, but not the single-knockout mice, show fewer acetylcholine receptor (AChR) clusters and a loss of apposition between AChR clusters and phrenic nerve terminals, suggesting redundancy of the NOVA proteins for controlling NMJ development38,73. Although NOVA1 and NOVA2 are detected in the ventral and dorsal spinal cord, respectively37, the loss of NOVA1 might upregulate NOVA2.
The impaired NMJ synaptogenesis in Nova1/2 dKO mice is due to aberrant agrin (Agrn) splicing. The AGRN protein promotes the clustering of AChRs within the postsynaptic membrane of the innervated muscle. Neuronal-specific Agrn isoforms containing the Z exons (32 and 33) are the most potent in promoting AChR aggregation74–77. Targeted deletion of the Z exons leads to paralysis and perinatal lethality similar to the phenotype of Nova1/2 dKO mice78,79. Although exon 32 is only slightly affected in single Nova knockouts, it is almost completely skipped in dKO mice. Strikingly, restoring Z+ Agrn expression in dKO motor neurons via a transgene rescues AchR clustering, nerve terminal apposition, NMJ morphology and muscle responses to stimuli73. The paralysis phenotype and early mortality are unchanged by the transgene, indicating the contributions of additional NOVA targets, possibly in other brain regions. These studies demonstrate how the activity of a particular alternatively spliced isoform can play a crucial part in ensuring proper synaptic development.
Regulation of synaptic function
Alternative splicing contributes to many aspects of synaptic function, including synapse specificity through the action of the KH domain-containing, RNA-binding, signal transduction-associated (KHDRBS) family, regulation of inhibitory synapse function by NOVA2, and splicing of many ion channels and synaptic components by muscleblind-like 2 (MBNL2), the neuronal ELAV-like (nELAVL) proteins, RBFOX1 and RBFOX2, and sodium channel modifier 1 (SCNM1).
KHDRBSs control alternative splicing of neurexin
The KHDRBS family has three members: SRC-associated in mitosis 68 kDa protein (SAM68; also known as KHDRBS1), SLM1 (also known as KHDRBS2) and SLM2 (also known as KHDRBS3)80. Each contains a single KH domain for RNA binding and dimerization. SAM68 is found in both the nucleus and the cytosol of many cell types81 and affects a variety of cellular processes, including splicing in the nucleus82,83. SLM1 and SLM2 are more restricted to the nervous system and have distinct expression patterns. For example, SLM1 is found in the dentate gyrus, some cortical neurons and Purkinje cells, whereas SLM2 is in found in the CA1 and CA3 neurons of the hippocampus, most cortical neurons, and sparsely in the granule and molecular layers of the cerebellum84. The mutually exclusive expression pattern of SLM1 and SLM2 is enforced by their cross-regulation via alternative splicing coupled with NMD, as seen with other paralogous pairs of RNA-binding proteins15,20,21,49,53,85,86. Specifically, SLM2 depletion shifts the splicing of Slm1 away from an NMD-targeted isoform and towards a productive transcript87.
One target of the KHDRBS proteins is neurexin (Nrxn), encoding presynaptic cell surface proteins that promote synaptogenesis through trans-synaptic signalling88. Pre-mRNAs from the three Nrxn genes undergo extensive alternative splicing to produce more than 3,000 protein isoforms88,89, presumably to define synapse specificity90,91. Regulation of Nrxn exon 20 generates AS4+ or AS4– isoforms that show differential binding to cell-type-specific postsynaptic partners88,92. NRXNβ AS4– isoforms preferentially bind neurolignin 1 (B) concentrated at glutamatergic synapses, whereas NRXNβ AS4+ isoforms preferentially bind NLGN2(A) at GABAergic and glycin-ergic synapses93 (FIG. 4a). Most interestingly, the inclusion of exon 20 is negatively regulated by KHDRBS proteins.
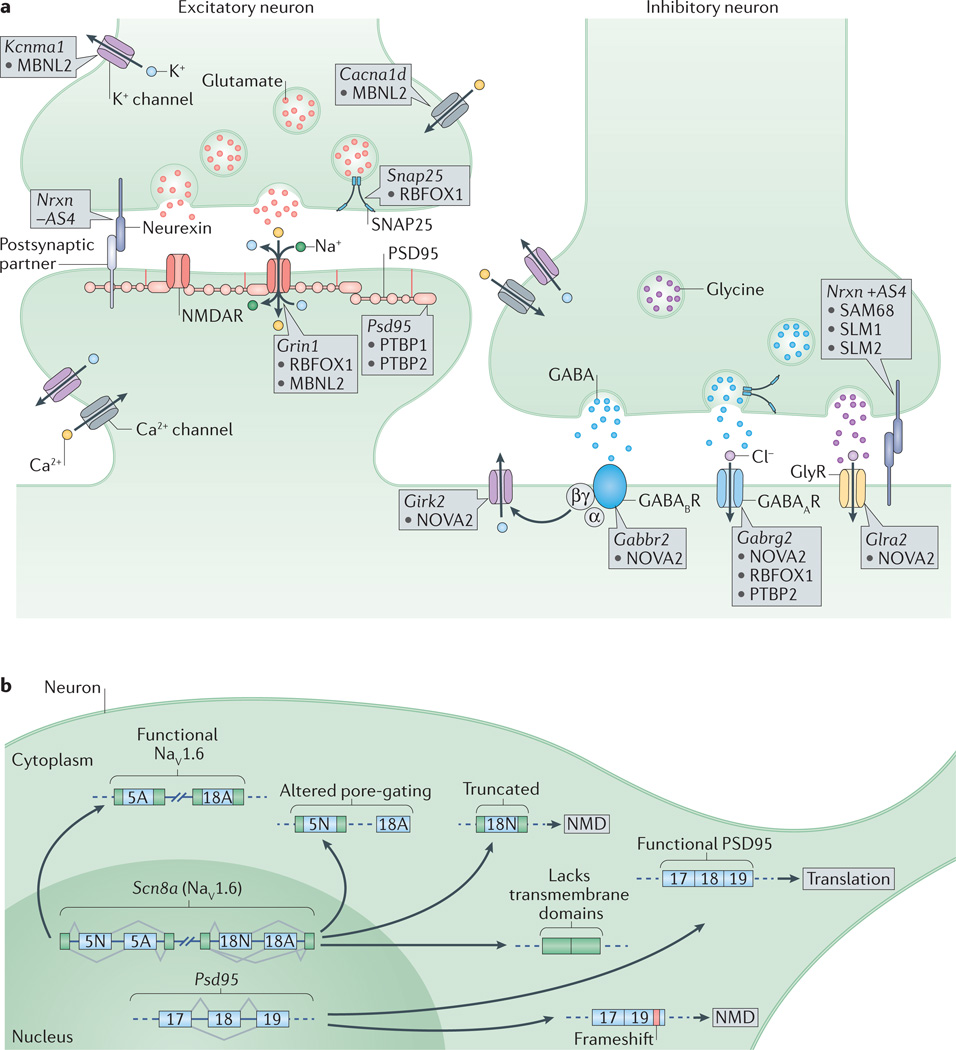
Figure 4. Alternative splicing regulation of synaptogenesis and synaptic function.
a |At the presynaptic terminal, alternative splicing of synaptosomal-associated protein 25 (Snap25) by RNA-binding protein fox 1 homologue 1 (RBFOX1) and of the calcium-activated potassium channel subunit alpha 1 (Kcnma1) by muscleblind-like 2 (MBNL2) are important to control neurotransmitter release. Differential splicing of the presynaptic neurexins (Nrxns) at AS4 by KHDRBS proteins (SAM68, SLM1 and SLM2) controls targeting to postsynaptic partners. At excitatory synapses, alternative splicing of the transcript encoding the NMDA receptor subunit GluN1, Grin1, is regulated by RBFOX1 and MBNL2, whereas the polypyrimidine tract binding proteins (PTBPs) control productive splicing of the scaffold protein, postsynaptic density protein 95 (Psd95). Splicing of the transcripts encoding L-type voltage-gated calcium channels, such as the pore-forming subunit Cav1.3 (encoded by Cacna1d), by MBNL2 may allow the voltage sensitivity, conductance, or other properties to be tuned as synapses differentiate. At inhibitory synapses, neuro-oncological ventral antigen 2 (NOVA2) mediates alternative splicing of the transcripts encoding many postsynaptic components such as the metabotropic GABAB receptor (Gabbr2), the inwardly rectifying potassium channel Kir3.2 (Girk2) and the glycine receptor alpha 2 (Glra2). Splicing of the GABAA receptor subunit transcript (Gabrg2) is controlled by multiple splicing regulators including NOVA2, RBFOX1 and PTBP2. b | Alternative splicing controls the expression and function of many synaptic components. Expression of PSD95 is repressed by PTBP-controlled exclusion of exon 18 until late in neuronal maturation when it is required for synaptogenesis. The gene encoding the voltage-gated sodium channel Nav1.6, Scn8a, has multiple alternative exons (such as 5N, 5A, 18N and 18A as shown in the figure) that can change its gating properties, determine its localization or alter its overall function. GABAAR, GABAA receptor; GABABR, GABAB receptor; GlyR, glycine receptor; NMD, nonsense-mediated decay; NMDAR, NMDA receptor; SAM68, SRC-associated in mitosis 68 kDa protein. Figure adapted from REF. 1, Nature Publishing Group.
Although exon 20 is similarly regulated by the three KHDRBS proteins in cell culture87,94, Khdrbs mutant mice show region-specific differential Nrxn splicing defects as a result of cell-type-specific expression of the individual KHDRBS proteins. Control of exon 20 by KHDBRS also appears to be modulated by synaptic activity. In cerebellar granule neurons, both KCl depolarization and kainic acid treatment increase SAM68 phosphorylation at Ser20 via CamKIV activation, leading to increased repression of Nrxn1 exon 20 and changes in trans-synaptic interactions94 (FIGS 3b,4a). Thus the KHDRBS proteins probably play a key part in modulating synaptic specificity and the plasticity of neural circuits during development and in adults.
Germline Sam68-null mice have deficits of bone metabolism, sexual organ development, motor coordination and motor learning95,96; recent work indicates possible alterations in long-term depression97. As SAM68 is almost ubiquitously expressed throughout the cerebellum, it is a question as to which neuronal subtypes contribute to the motor phenotypes and whether Nrxn1 is one of the relevant targets.
In contrast to Sam68-null mice, Slm1 and Slm2 single-null mice are viable with no apparent behavioural or anatomical defects84,87,98. Slm1/2 dKO mice also show no apparent behavioural abnormalities, although possible morphological defects have not been characterized87. Sam68/Slm1 dKO mice were similar to Sam68 single-null mice, except for additional defects in cerebellar foliation and scattered ectopic Purkinje cells in the molecular layer. SAM68 and SLM1 are both expressed in Purkinje cells and probably have redundant roles in their development. It will be interesting to assess the Slm2/Sam68 dKO or the triple Khdrbs knockout mice. The interplay between the specific effects of particular KHDRBS paralogues and their partial redundancy typifies the complexity of analysing splicing regulator mutants and indicates how a given splicing pattern may be controlled differently across different neuronal subtypes.
NOVA2 and synaptic plasticity
Many NOVA2 target genes, including the GABAB receptor 2 (Gabbr2), glycine receptor α2 (Glra2), gephyrin (Gphn) and the inward rectifier potassium channel Kir3.2 (Girk2), are involved in inhibitory synapse function99 (FIG. 4a). Mis-splicing of GABAB receptors and GIRK channels probably leads to deficient long-term potentiation of the slow inhibitory current seen in the Nova2-null hippocampus100. By contrast, long-term potentiation of excitatory post-synaptic currents is unchanged, as are basal excitatory and inhibitory synaptic transmission. This high degree of phenotypic specificity highlights the variable sensitivity of different forms of synaptic transmission and plasticity to splicing alteration. How the alternative splicing of ion channels and neurotransmitter receptors changes precise physiological functions and how this regulation defines circuit function will be a rich area of investigation going forward.
MBNL2 and neurological symptoms of myotonic dystrophy
The MBNL family of RNA-binding zinc finger proteins has three members in mice and humans. MBNL proteins have been studied extensively in relation to the neuromuscular disorder myotonic dystrophy. In myotonic dystrophy, CTG or CCTG repeat expansions in expressed RNAs sequester MBNL proteins from their normal binding sites, altering MBNL-dependent splicing patterns101–105. Although all three MBNLs are expressed in the brain, only Mbnl2-null mice exhibit obvious central nervous system phenotypes. Germline deletion of Mbnl2 results in abnormal sleep patterns, memory loss and learning deficits. Mbnl2-null mice are also more susceptible to pentylenetetrazole-induced seizures. Muscle function is, however, unperturbed, probably as a result of abundant MBNL1 expression106. Hundreds of exons are mis-spliced in the Mbnl2−/− brain and overlap significantly with those known to be mis-spliced in myotonic dystrophy. MBNL2 overall promotes adult-like splicing patterns and its loss leads to continued expression of the fetal isoforms of ion channel-encoding genes such as the calcium-activated potassium channel subunit alpha 1 gene (Kcnma1; which encodes BK (also known as Slo1)), the voltage-gated calcium channel subunit alpha 1D gene (Cacna1d; which encodes Cav1.3) and the NMDA receptor subunit gene (Grin1; which encodes GluN1) (FIG. 4a). NMDAR-mediated responses and pattern-induced long-term potentiation are impaired in Mbnl2-null mice. The observed alterations in synaptic plasticity and perturbations in neuronal excitability may be a result of the continued expression of fetal ion channel isoforms.
It is notable that about half of the MBNL2 CLIP tags are found in three prime untranslated region (3′ UTR) sequences, indicating the non-splicing functions of MBNL2 (REF. 106). Although Mbnl2-null brains did not show major changes in transcript levels, recent studies of Mbnl1/2 dKOs have highlighted MBNL activity in controlling alternative polyadenylation events105,107,108. Binding in 3′ UTRs is commonly observed for other splicing regulators and points to the need to distinguish phenotypes driven by splicing changes from those arising from altered mRNA stability, localization and/or translation (FIG. 5).
Figure 5. Regulatory outcomes of RNA-binding proteins.
RNA-binding proteins (RBPs) regulate transcript splicing, stability, translation and localization, and many RBPs studied as splicing regulators have extended functions in the cytoplasm, affecting every subsequent step of the mRNA life cycle. a | Nuclear roles of RBPs. Step 1: RBPs control alternative splicing of precursor mRNA (pre-mRNA) to generate multiple isoforms that differ in functional activity, interactions with cofactors or post-translational modifications. Step 2: regulated intron retention targets transcript isoforms for degradation by nuclear surveillance mechanisms200,201, or by the introduction of a premature termination codon leading to nonsense-mediated decay (NMD, shown in red; see below). Step 3: by shifting the reading frame or by including a ‘poison exon’ containing a premature translation termination codon, alternative splicing produces transcript isoforms that are degraded by NMD202. Alternative splicing coupled with NMD can control the overall abundance of gene transcripts203. b | Cytoplasmic roles of RBPs. Step 4: RBPs compete with AU-binding proteins for binding at AU-rich elements in the 3′ untranslated region (UTR) to stabilize their target transcripts110,111. Other RBPs regulate transcript stability in the cytoplasm by either competing with microRNAs for their binding sites or facilitating microRNA binding131,204,205. RBP binding in both the 5′ UTRs and 3′ UTRs also affects translational efficiency206. Step 5: RBPs regulate the transport and differential localization of mRNA, which are crucial for spatial and temporal control of translation in response to activity-dependent signalling162,207,208. The establishment of neuronal polarity and consolidation of synaptic strength through local translation of mRNAs in response to synaptic activity are some well-known examples. AAA, poly-A tail; Gppp, 5′ cap; Pol II, RNA polymerase II.
ELAVL proteins regulate neuronal excitation
The ELAVL (also known as Hu) family of proteins consists of four highly homologous members109–111 that recognize U- and AU-rich elements112–118 (FIG. 5b). ELAVL1 (also known as HuR or HuA) is widely expressed in non-neuronal tissues, whereas ELAVL2 (also known as HuB), ELAVL3 (also known as HuC) and ELAVL4 (also known as HuD) show neuronal-specific expression and are called neuronal ELAVLs (nELAVLs)51,119,120. Like NOVA, nELAVLs are target antigens in patients with paraneo-plastic neurological disorders121,122. The ELAVLs have primarily been studied as regulators of mRNA stability and translation efficiency through their binding to 3′ UTRs110,111,123 (FIG. 5b), but recent genome-wide profiling analyses have revealed intronic binding of nELAVLs and hundreds of splicing changes in Elavl3/4 dKO brains. These splicing targets are enriched for proteins involved in microtubule assembly and disassembly at synapses and axons. Interestingly, the biological processes affected at the level of splicing are different from those affected at the level of transcript abundance, suggesting that the regulatory programmes of the nuclear and cytoplasmic nELAVL proteins are distinct118.
Depletion of nElavl in the brain leads to multiple neurological defects124,125. Although Elavl3-null mice are born grossly normal and fertile, most of the adult animals show poor motor coordination. The specificity of the motor defect may be because ELAVL3 is the only nELAVL protein in Purkinje cells118. These mice also show spontaneous cortical hypersynchrony and non-convulsive electrographic seizures. These phenotypes are attributed to aberrant glutamate levels, based on binding of nELAVL to the 3′ UTRs of genes affecting glutamate synthesis. The multiple splicing regulator mutants that show seizure phenotypes may reflect the large number of synaptic and membrane proteins regulated at the level of splicing, with hyperexcitability being a common consequence of their perturbation.
RBFOX1 control of neuronal excitability
Another splicing regulator whose mutation leads to a hyperexcitability phenotype is RBFOX1. Human mutations in RBFOX1 have been identified in patients with epilepsy126–128 and autism spectrum disorder129–131. Pan-neuronal deletion of Rbfox1 (Rbfox1loxp/loxp; Nestin-Cre) leads to increased susceptibility to spontaneous seizures and seizures induced by kainic acid, as well as hyperexcitation in the dentate gyrus53. Relatively few splicing and expression changes were detected in the Rbfox1−/− whole brain, presumably as a result of the redundancy of RBFOX2 and RBFOX3 function53,132. However, these splicing changes affect transcripts encoding ion channels, neurotransmitter receptors, structural proteins and Ca2+ signalling molecules, many of which are associated with seizure disorders in humans or mice, such as the GABAA receptor subunit Gabrg2, Grin1, the voltage-gated sodium channel Scn8a (which encodes NaV1.6), and synaptosomal-associated protein 25 (Snap25)133–137 (FIGS 1b,4). Changes in the isoform ratios for these proteins may increase action potential firing or disrupt the excitation/inhibition balance in neuronal circuits. For example, the gene for the SNARE protein SNAP25 uses a pair of mutually exclusive exons (5a and 5b) to produce two alternative isoforms that show differences in the kinetics of synaptic vesicle release138,139. The Rbfox1−/− brains show decreased 5b and increased 5a inclusion. The Rbfox1−/− seizure phenotype is thus consistent with studies showing that mice carrying genetic substitution of exon 5b with exon 5a also exhibit seizures140.
The Rbfox1−/− and Elavl3−/− mice present interesting animal models for the study of human epileptogenesis and mechanisms controlling neuronal excitability. It will be interesting to compare their molecular targets and mutant physiology to understand whether they affect a common regulatory programme or perhaps drive different splicing changes that have similar physiological outcomes.
RBFOX proteins control Purkinje cell pace-making
In addition to affecting Purkinje cell migration, RBFOX2, in conjunction with RBFOX1, regulates mature Purkinje cell function. Rbfox2−/− Purkinje cells eventually form a proper layer, but show decreased dendritic arborization in the molecular layer, as well as irregular and less frequent spontaneous action potentials. This pace-making defect becomes more severe in Rbfox1 heterozygous, Rbfox2-null brains (Rbfox1+/−; Rbfox2−/−)55. Specific depletion of Rbfox1 and Rbfox2 from mature Purkinje cells after the completion of migration and development, using the L7(Pcp1)-Cre strain (L7-DKO), results in a similar pace-making phenotype and motor defects55 (FIG. 3). The early and late phenotypes of Rbfox mutation demonstrate roles for these proteins in both cerebellar development and mature function.
The pace-making defect in the L7-DKO mice is highly reminiscent of mice lacking the voltage-gated sodium channel subunit Scn8a (REFS 141–143). Nav1.6 functions, in part, to maintain a resurgent sodium current that enables regular, spontaneous firing. Several alternatively spliced exons in Scn8a contribute to the resurgent sodium current144,145. Mutually exclusive exons 5A and 5N alter voltage-dependent gating and/or interactions with a blocking subunit145. Another pair of mutually exclusive exons, 18A and 18N, determine whether a functional 18A+ Scn8a mRNA is produced. Inclusion of 18N introduces a premature termination codon and leads to nonsense-mediated decay, whereas skipping both 18A and 18N produces an isoform that lacks large portions of the third and fourth transmembrane domains146,147 (FIG. 4b). Although the single Rbfox1−/− and Rbfox2−/− brains show modest changes in Scn8a splicing, the Rbfox1+/−; Rbfox2−/− brains show dramatic splicing changes at both exons 5 and 18, including a twofold decrease in exon18A that decreases the amount of functional Nav1.6 (REF. 55). Thus the Purkinje cell pace-making defect in Rbfox mutant mice could largely arise from the loss of Nav1.6. These results further confirm the partial redundancy of the Rbfox family members.
SCNM1 enhances non-consensus splicing of Scn8a
Forward genetic studies have identified an Scn8a splicing mutation that causes severe neurological defects. This mutation, medJ, does not alter the coding sequence of Scn8a but instead has a four base pair deletion in the 5′ splice site of intron 3, which causes skipping of both exons 2 and 3 in a majority of Scn8a transcripts (FIG. 4b), and produces a severely truncated, non-functional Nav1.6 protein148. The medJ mice show hindlimb paralysis, muscle atrophy and the degeneration of Purkinje cells149. Normally, Nav1.6 replaces fetal Nav1.2 at the nodes of Ranvier during the first few weeks of postnatal development150. In medJ/C3H mice, the replacement of fetal Nav1.2 is delayed and the amount of Nav1.6 at the nodes of Ranvier reaches only 10–20% of that seen in wild-type mice. Nerve conduction velocity in medJ mutants is decreased by half151, probably as a consequence of insufficient Nav1.6 expression.
The severity of the medJ phenotype was found to be affected by genetic background and to correlate with the amount of correctly spliced transcript152. In the C3H background, medJ mice live a normal lifespan with dystonia and ataxia, with 10% of Scn8a transcripts correctly spliced. The same mutation in a C57Bl/6J background produces only 5% correctly spliced transcript and the mice show progressive paralysis and lethality by 1 month of age151. The phenotypic severity of the Scn8amedJ hypomorphic allele is determined by a single gene modifier, sodium channel modifier 1 (Scnm1). C67Bl/6J mice have a nonsense mutation in Scnm1 and targeted deletion of Scnm1 in the C3H strain confirmed that SCNM1 affects both the splicing of the Scn8amedJ transcript and the mouse phenotype152,153. Finally, a bacterial artificial chromosome transgene expressing wild-type Scnm1 can rescue the lethality and paralysis of Scn8amedJ in C57BL/6J mice154.
Although the genetic interaction between Scnm1 and Scn8a splicing has been studied in detail, the mechanism of SCNM1 function as a splicing regulator is less clear. The protein has one zinc finger domain, a basic nuclear localization signal and an acidic carboxy terminus. Its overexpression in heterologous cells can enhance the correct splicing of an Scn8a mini-gene, possibly via interactions with the spliceosomal proteins U1 small nuclear ribonucleoprotein 70 (U1-70k) and putative RNA-binding protein Luc7-like 2 (LUC7L2)153. The studies of Scnm1 indicate that splicing regulators can be an important class of phenotypic modifiers. Given the many human disease-causing mutations that affect pre-mRNA splicing, polymorphisms in splicing regulatory genes may play a large part in modifying disease severity across individuals.
Splicing regulators and neurodegeneration
Splicing misregulation is increasingly implicated in neurodegenerative disorders. Dysfunction of transactivating response DNA binding protein (TDP43; also known as TARDBP) and fused in sarcoma (FUS; also known as TLS) lead to phenotypes that are characteristic of amyotrophic lateral sclerosis (ALS) and frontal temporal lobar disease (FTLD), whereas mutation of Rnu2–8, which encodes part of a core spliceosomal component, the U2 small nuclear RNA (snRNA), leads to specific neurodegeneration in the cerebellum.
TDP43 and FUS in amyotrophic lateral sclerosis
Errors of splicing regulation are increasingly implicated in a variety of neurodegenerative disorders, including ALS and FTLD155,156. Familial and sporadic forms of ALS and some cases of FTLD have been associated with mutations in TDP43, FUS155,156, heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), hnRNPA2B1 (REF. 157) and matrin 3 (REF. 158) — all of which are regulators of splicing. Disruption of TDP43 and FUS function through protein aggregation or mislocalization is a characteristic of ALS and FTLD derived from many different mutations159–161 and neurogenetic analyses of TDP43 and FUS have been the most extensive.
TDP43 is a major component of cytoplasmic inclusions found in 95% cases of sporadic ALS and FTLD156. Widely expressed in many tissues and predominantly nuclear, TDP43 affects multiple steps of RNA metabolism, including transcription, splicing, decay, transport and translation162 (FIG. 5). One hypothesis for the pathogenic role of TDP43 is that the formation of TDP43 cytoplasmic inclusions leads to its depletion from the nucleus and a loss of splicing function155,156. Consistent with this, ALS-like phenotypes are seen in mice with partial depletion of TDP43 by RNA interference, or with targeted deletion of Tdp43 in motor neurons, whereas germline Tdp43-null mice are embryonic lethal163–165. Dominant missense mutations are sufficient to cause familial disease in humans166, and transgenic rodents expressing either wild-type or disease-associated mutants also show neurodegeneration. These phenotypes could be indicative of a toxic gain of function167–169; alternatively the mutations and overexpression could somehow both promote cytoplasmic inclusion formation with a loss of function166,170–173.
Both the ALS-associated mutations and the changes in wild-type TDP43 expression alter a large programme of alternative exons in mutant mice170,174,175. However, less than a quarter of affected exons are shared between the transgenic and RNA interference-depletion mouse models. Motor neurons may be particularly sensitive to aberrant splicing changes, or some of the common TDP43 dependent splicing events might be sufficient to cause the phenotype. These many models will allow rich comparisons in identifying potentially causative splicing changes.
Another ALS and FTLD gene, FUS156, is a widely expressed, predominantly nuclear RNA-binding protein176,177 with an RNA recognition motif that, like TDP43, has multiple roles in RNA processing, including splicing regulation162,178. Fus-null mice die soon after birth179, whereas a transgenic rat model of mutant Fus shows various ALS-like phenotypes180. Interestingly, TDP43 and FUS regulate distinct groups of alternative exons and target different sets of mRNAs in the cytoplasm178,181, suggesting different roles for the wild-type proteins. It will be interesting to compare the targets of these two proteins with the targets of other RNA-binding proteins implicated in ALS, including hnRNPA1, hnRN-PA2B1 and matrin 3, and to assess how their mutation might converge on similar disease pathologies.
Rnu2–8 is required for cerebellar granule neuron survival
A forward genetic screen recently uncovered a novel form of neurodegeneration caused by mutation of a core component of the spliceosome. The mouse mutant NMF291 strain contains a functionally compromised allele of Rnu2-8 (REF. 182), one of multiple genes encoding the U2 snRNA183. U2 snRNA binds to the branch point during spliceosome assembly and then forms base pairs with the U6 snRNA to become a key portion of the spliceosome catalytic centre for all major class introns6,8,183,184 (BOX 2); its dysfunction might be expected to be lethal for all cell growth. Mammalian genomes contain multiple clustered copies of the U2 snRNA gene that allow production of the extremely high levels of U2 snRNA found in cells, which were previously thought to be equally expressed across tissues. The highly tissue-specific phenotype of NMF291 was therefore unanticipated.
NMF291 mice show progressive and severe degeneration of the cerebellum as a result of the loss of cerebellar granule neurons beginning at postnatal week 4 (FIG. 3) and develop tremors at 8 weeks, progressing to truncal ataxia by 12 weeks. Consistent with the phenotype, both wild-type and mutant Rnu2-8 RNA are selectively expressed in the cerebellum and increase in expression after granule neuron maturation. A transgenic mouse expressing the mutant Rnu2-8 in the wild-type background displays a similar course of granule neuron loss and ataxia. Conversely, increasing the dose of the wild-type RNA in the NMF291 mutant decreases neurodegeneration in the granule layer. These data demonstrate that not all U2 genes are the same, but that individual genes within a cluster can show temporal and cell-type-specific patterns of expression. Their mutation can thus lead to a highly specific phenotype.
The NMF291 mutation is a 5nt deletion that removes the first 2nt of the branch site recognition sequence within the U2 snRNA, as well as a 3nt linker between the branch site recognition sequence and the U2/U6 helix IA. When highly expressed, the mutant U2 snRNA decreases the overall splicing efficiency and affects alternative splicing patterns. In particular, about 3,000 annotated introns show higher levels of retention in the NMF291 cerebellum (BOX 1; FIG. 5a). These results are reminiscent of recent data on myelodysplastic syndromes, where mutations in several components of the core splicing apparatus, such as U2 small nuclear ribonucleoprotein auxiliary factor 2 (U2AF65), splicing factor 3B, subunit 1 (SF3B1) and other proteins, were found to cause very specific splicing defects in particular transcripts and to lead to a highly tissue-specific phenotype185–187. These studies open the possibility for other neurological disorders being caused by the mutation of general splicing factors.
Conclusions
Alternative splicing as a regulatory process has been studied for many years at the level of individual proteins and target transcripts. The advent of whole genome analyses10,12,188,189 brought a new appreciation of its pervasiveness and regulatory reach in metazoan organisms190,191. However, understanding the biology of splicing regulatory programmes and, in particular, the role of these programmes in the mammalian brain, needs to be addressed through a combination of genetics, neuroanatomy and physiology. The neurogenetics of splicing is still in its infancy. Of the many hundreds of potential splicing regulatory proteins, only a few are beginning to be analysed, and we have focused on those studied through mouse genetics (see Supplementary information S1 (table)). Although not yet studied in the mouse, another family of regulators that will probably have important splicing roles in the brain is the CUGBP, Elav-like family (CELF) proteins192–194. The initial targeted genetic studies described here make it clear that changes in the splice site choice have essential roles in nearly all aspects of neuronal development and function.
The analysis of splicing factor mutations is challenged by their highly pleiotropic phenotypes. Mutations are often lethal or lead to developmental abnormalities that obscure additional later functions. Several studies have overcome these obstacles using ever more precise Cre expression to ablate a regulatory gene in particular cell types at particular times. RNA sequencing and CLIP–seq now allow the relatively simple identification of mis-spliced targets potentially determining the mutant phenotype. These genome-wide analyses indicate that individual splicing regulators affect coherent sets of transcripts that can be involved in common biological pathways195,196. However, the large number of targets makes it difficult to link a phenotype to a particular splicing event. As described above, phenotypes can be connected to splicing events using transgenes expressing single spliced isoforms to rescue particular functions. This strategy will need to be applied to more refined populations of cells and circuits, perhaps through in utero electroporation or viral transduction. One challenge will be to match the expression from the rescuing gene to that of an endogenous locus in time and quantity. Developmental phenotypes may be reverted by an overexpressed transgene, but the rescue of physiological defects will probably require the precise control of isoform ratios, perhaps through genome editing of endogenous loci.
The obverse problem to the many targets of splicing regulators is that the regulators frequently occur in highly related gene families. Groups of paralogous regulators that show partially redundant functions can mute the effect of single-gene mutations. In cells where they are co-expressed, double mutation will often lead to new splicing changes that are not seen in either single mutant and reveal new phenotypes. However, regulators are rarely entirely redundant and usually show differences in their range of expression, as seen with the NOVA, nELAVL and RBFOX proteins.
In this Review, we have focused on splicing regulation. It is important to keep in mind that many RNA-binding proteins controlling splicing choices also affect the choice of poly-A site and can also be found in the cytoplasm, where they control the translation or stability of target transcripts through binding in 3′ UTRs197 (FIG. 5b). The consequences of their mutation will include the loss of these functions in addition to splicing changes. Cytoplasmic functions can be examined by measuring overall changes in expression by RNAseq, rather than splicing changes, and by identifying 3′ UTR targets in CLIP-seq data sets. The relationships between the nuclear and cytoplasmic regulatory programmes controlled by RNA-binding proteins are only beginning to be examined. It will also be important to define those effects that arise from the direct regulation of a transcript rather than as an indirect consequence of splicing factor loss. Splicing regulators extensively modulate each other’s activity, as well as controlling the activity of transcriptional regulators (FIG. 1). Gene expression changes in mutant mice may thus derive in part from extensive secondary effects downstream of the protein being examined.
Recent human genetic studies have clearly implicated splicing regulators in neurodegenerative diseases such as ALS. Other work has connected RNA-binding proteins to mis-splicing in neuropsychiatric disorders, including epilepsy, autism spectrum disorder, inherited ataxias and schizophrenia. Splicing programmes provide a highly interconnected layer of regulation that can alter protein activity without easily discernible changes in the overall expression. Perturbations of these programmes have the potential to alter neuronal connectivity and firing properties in a manner that has dramatic consequences for overall circuit function and behaviour. The mouse mutations described here provide the first glimpses of these regulatory programmes. In the RNA-binding protein knockouts so far analysed, the heterozygous mice develop largely normally, but some splicing targets are still altered by the reduction in regulatory protein dose. It will be particularly interesting to assess these heterozygous mice for behavioural defects. Future work in these genetic systems will provide potential new models for a variety of disorders.
In addition to relating splicing regulation to neurological disease, there are many questions to be addressed. Although many alternative splicing events are conserved across mammalian or vertebrate species, the effect of these splicing changes on protein activity is usually unknown. It will be important to characterize the set of protein isoforms expressed from each gene and understand their different roles in cell biology. This will be a particular challenge for physiology, but such analyses are needed to relate changes in synaptic and membrane protein structure to changes in synaptic activity and firing. Another issue is how the programmes controlled by different regulators interact (FIG. 1a). RNA-binding proteins can antagonize each other or synergize in RNA binding. The complex overlap between their regulatory programmes allows for a high degree of specificity in where and when particular splicing events occur. It will be very interesting to assess how the expression of spliced iso-forms contributes to defining specific neuronal subtypes. The future application of mouse genetics to the characterization of splicing regulatory programmes should allow some of these questions to be answered.
Supplementary Material
Acknowledgments
The authors thank researchers in the laboratory of D.B., the laboratory of S.Z. and other colleagues for many helpful discussions. Their work is supported by the US National Institutes of Health grants R01 GM49662 to D.B., K99 MH096807 to S.Z. and F31 NS093923 to C.V.
Abbreviations
- Spliceosome
A large macromolecular assembly of small nuclear ribonucleoproteins and proteins that catalyses the excision of introns from precursor mRNAs
- Pleiotropy
A condition where one gene can influence multiple phenotypic traits
- Paralogues
Related genes within a genome that often arise from gene duplication during evolution
- Polypyrimidine tract
A sequence element located between the branch point and the 3′ splice site that is bound by U2AF during normal spliceosome assemblyit is also a binding site for the polypyrimidine tract binding protein that can be found elsewhere in the precursor mRNA.
- 3′ splice site
The sequence at the 3′ end of the intron that is recognized by the U2AF protein component of the spliceosome. Also called an acceptor site.
- YCAY elements
Sequences consisting of a cytosine (C) and an adenine (A) flanked by two pyrimidines (Y)
- 5′ splice site
The sequence at the 5′ end of the intron that is recognized by the U1 small nuclear ribonucleoprotein component of the spliceosome. Also called a donor site.
- Branch point
An intronic sequence element upstream from the polypyrimidine tract of the 3′ splice site where a branched nucleotide at a specific adenosine will be formed by the first transesterification step of splicing. The branch point is recognized by the U2 small nuclear ribonucleoprotein.
- Major class introns
Also called GT/AG introns, which are excised by the major spliceosome containing the U1 and U2 small nuclear ribonucleoproteins
- CLIP–seq
(Crosslinking immunoprecipita-tion followed by high-throughput sequencing). A method used to identify the binding sites of RNA-binding proteins across the transcriptome
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Li Q, Lee J-A, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 2.Zheng S, Black DL. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet. 2013;29:442–448. doi: 10.1016/j.tig.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Darnell RB. RNA protein interaction in neurons. Annu. Rev. Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matera AG, Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell. Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu X-D, Ares M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip. Rev. RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.König J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat. Rev. Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 11.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev. RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015;38:226–236. doi: 10.1016/j.tins.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spellman R, et al. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- 15. Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. This study demonstrates that an important splicing regulator, PTBP1, is controlled during neuronal development by the microRNA miR-124
- 16. Xue Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. This study demonstrates that PTBP1 acts as a master regulator of cell identity by driving a post-transcriptional regulatory programme that inhibits neuronal differentiation
- 17.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl Acad. Sci. USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linares AJ, et al. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. eLife. 2015;4:6778. doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spellman R, Llorian M, Smith CWJ. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. References 15, 20 and 21 provide an early example of cross-regulation by splicing regulators
- 22.Suckale J, et al. PTBP1 is required for embryonic development before gastrulation. PLoS ONE. 2011;6:e16992. doi: 10.1371/journal.pone.0016992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibayama M, et al. Polypyrimidine tract-binding protein is essential for early mouse development and embryonic stem cell proliferation. FEBS J. 2009;276:6658–6668. doi: 10.1111/j.1742-4658.2009.07380.x. [DOI] [PubMed] [Google Scholar]
- 24.Shibasaki T, et al. PTB deficiency causes the loss of adherens junctions in the dorsal telencephalon and leads to lethal hydrocephalus. Cereb. Cortex. 2013;23:1824–1835. doi: 10.1093/cercor/bhs161. [DOI] [PubMed] [Google Scholar]
- 25.Spassky N, et al. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, Fu X-D. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 27.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell. Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 2014;12:1195–1204. doi: 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calarco JA, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 30.Raj B, et al. A global regulatory mechanism for activating an exon network required for neurogenesis. Mol. Cell. 2014;56:90–103. doi: 10.1016/j.molcel.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj B, et al. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol. Cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Palm K, Metsis M, Timmusk T. Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain Res. Mol. Brain Res. 1999;72:30–39. doi: 10.1016/s0169-328x(99)00196-5. [DOI] [PubMed] [Google Scholar]
- 33.Shimojo M, Lee JH, Hersh LB. Role of zinc finger domains of the transcription factor neuron-restrictive silencer factor/repressor element-1 silencing transcription factor in DNA binding and nuclear localization. J. Biol. Chem. 2001;276:13121–13126. doi: 10.1074/jbc.M011193200. [DOI] [PubMed] [Google Scholar]
- 34.Tabuchi A, et al. REST4-mediated modulation of REST/NRSF-silencing function during BDNF gene promoter activation. Biochem. Biophys. Res. Commun. 2002;290:415–420. doi: 10.1006/bbrc.2001.6194. [DOI] [PubMed] [Google Scholar]
- 35. Quesnel-Vallières M, Irimia M, Cordes SP, Blencowe BJ. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev. 2015;29:746–759. doi: 10.1101/gad.256115.114. References 29, 30, 31 and 35 lay out a compelling description of a splicing regulatory network controlled by a neuronal splicing factor
- 36.Darnell RB, Posner JB. Paraneoplastic syndromes involving the nervous system. N. Engl. J. Med. 2003;349:1543–1554. doi: 10.1056/NEJMra023009. [DOI] [PubMed] [Google Scholar]
- 37.Yang YY, Yin GL, Darnell RB. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc. Natl Acad. Sci. USA. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen KB, et al. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 39. Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010;66:848–858. doi: 10.1016/j.neuron.2010.05.007. This paper beautifully demonstrates how NOVA2-dependent alternative splicing regulates the function of a receptor protein important for neuronal migration in the cortex and highlights the spatial specificity of splicing regulation through the use of knockout and transgenic mice
- 40.Rice DS, Curran T. Role of the reelin signaling pathway in central nervous system development. Annu. Rev. Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- 41.Ayala R, Shu T, Tsai L-H. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 42.Förster E, et al. Emerging topics in Reelin function. Eur. J. Neurosci. 2010;31:1511–1518. doi: 10.1111/j.1460-9568.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice DS, et al. Disabled-1 acts downstream of Reelin in a signaling pathway that controls laminar organization in the mammalian brain. Development. 1998;125:3719–3729. doi: 10.1242/dev.125.18.3719. [DOI] [PubMed] [Google Scholar]
- 44.Arnaud L, Ballif BA, Cooper JA. Regulation of protein tyrosine kinase signaling by substrate degradation during brain development. Mol. Cell. Biol. 2003;23:9293–9302. doi: 10.1128/MCB.23.24.9293-9302.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bock HH, Jossin Y, May P, Bergner O, Herz J. Apolipoprotein E receptors are required for reelin-induced proteasomal degradation of the neuronal adaptor protein Disabled-1. J. Biol. Chem. 2004;279:33471–33479. doi: 10.1074/jbc.M401770200. [DOI] [PubMed] [Google Scholar]
- 46.Feng L, Allen NS, Simo S, Cooper JA. Cullin 5 regulates Dab1 protein levels and neuron positioning during cortical development. Genes Dev. 2007;21:2717–2730. doi: 10.1101/gad.1604207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simo S, Jossin Y, Cooper JA. Cullin 5 regulates cortical layering by modulating the speed and duration of Dab1-dependent neuronal migration. J. Neurosci. 2010;30:5668–5676. doi: 10.1523/JNEUROSCI.0035-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell. Mol. Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 51.McKee AE, et al. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 2011;43:706–711. doi: 10.1038/ng.841. This study demonstrates the role of the RBFOX1 splicing regulator in controlling neuronal excitation, and the involvement of alternative splicing in epileptogenesis. Along with reference 55, this study exemplifies how families of paralogous splicing regulators can have either redundant or separate functions depending on the brain region
- 54.Hammock EAD, Levitt P. Developmental expression mapping of a gene implicated in multiple neurodevelopmental disorders, A2bp1 (Fox1) Dev. Neurosci. 2011;33:64–74. doi: 10.1159/000323732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gehman LT, et al. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu. Rev. Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 57.Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat. Rev. Neurosci. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- 58.Koch S, et al. A secreted soluble form of ApoE receptor 2 acts as a dominant-negative receptor and inhibits Reelin signaling. EMBO J. 2002;21:5996–6004. doi: 10.1093/emboj/cdf599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trommsdorff M, et al. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 60.Larouche M, Beffert U, Herz J, Hawkes R. The Reelin receptors Apoer2 and Vldlr coordinate the patterning of Purkinje cell topography in the developing mouse cerebellum. PLoS ONE. 2008;3:e1653. doi: 10.1371/journal.pone.0001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beffert U, et al. Modulation of synaptic plasticity and memory by reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 62.Gao Z, et al. Splice-mediated motif switching regulates disabled-1 phosphorylation and SH2 domain interactions. Mol. Cell. Biol. 2012;32:2794–2808. doi: 10.1128/MCB.00570-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Licatalosi DD, et al. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li Q, et al. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife. 2014;3:e01201. doi: 10.7554/eLife.01201. References 63 and 64 demonstrate that PTBP2 controls a transition from fetal to adult splicing programmes that is required for proper neuronal maturation and survival
- 65.Cayouette M, Raff M. Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat. Neurosci. 2002;5:1265–1269. doi: 10.1038/nn1202-1265. [DOI] [PubMed] [Google Scholar]
- 66.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell. Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zheng S, et al. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012;15:381–S1. doi: 10.1038/nn.3026. This study describes how an alternative splicing event can control the overall expression of a neuronal-specific gene, and how PTBP1 and PTBP2 control splicing programmes required for synapse formation
- 68.Nakano Y, et al. A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS Genet. 2012;8:e1002966. doi: 10.1371/journal.pgen.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deol MS, Gluecksohn-Waelsch S. The role of inner hair cells in hearing. Nature. 1979;278:250–252. doi: 10.1038/278250a0. [DOI] [PubMed] [Google Scholar]
- 70.Whitlon DS, Gabel C, Zhang X. Cochlear inner hair cells exist transiently in the fetal Bronx Waltzer (bv/bv) mouse. J. Comp. Neurol. 1996;364:515–522. doi: 10.1002/(SICI)1096-9861(19960115)364:3<515::AID-CNE9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 71.Sobkowicz HM, Inagaki M, August BK, Slapnick SM. Abortive synaptogenesis as a factor in the inner hair cell degeneration in the Bronx Waltzer (bv) mutant mouse. J. Neurocytol. 1999;28:17–38. doi: 10.1023/a:1007059616607. [DOI] [PubMed] [Google Scholar]
- 72.Cheong MA, Steel KP. Early development and degeneration of vestibular hair cells in bronx waltzer mutant mice. Hear. Res. 2002;164:179–189. doi: 10.1016/s0378-5955(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 73. Ruggiu M, et al. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc. Natl Acad. Sci. 2009;106:3513–3518. doi: 10.1073/pnas.0813112106. This reference reports a good example of using transgene rescue to understand the function of alternatively spliced isoforms and to dissect a portion of a highly pleiotropic phenotype
- 74.Nitkin RM, et al. Identification of agrin, a synaptic organizing protein from Torpedo electric organ. J. Cell Biol. 1987;105:2471–2478. doi: 10.1083/jcb.105.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reist NE, Werle MJ, McMahan UJ. Agrin released by motor neurons induces the aggregation of acetylcholine receptors at neuromuscular junctions. Neuron. 1992;8:865–868. doi: 10.1016/0896-6273(92)90200-w. [DOI] [PubMed] [Google Scholar]
- 76.Gesemann M, Denzer AJ, Ruegg MA. Acetylcholine receptor-aggregating activity of agrin isoforms and mapping of the active site. J. Cell Biol. 1995;128:625–636. doi: 10.1083/jcb.128.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 78.Gautam M, et al. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- 79.Burgess RW, Nguyen QT, Son YJ, Lichtman JW, Sanes JR. Alternatively spliced isoforms of nerve- and muscle-derived agrin: their roles at the neuromuscular junction. Neuron. 1999;23:33–44. doi: 10.1016/s0896-6273(00)80751-5. [DOI] [PubMed] [Google Scholar]
- 80.Di Fruscio M, Chen T, Richard S. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2 SLM-1 is a Src substrate during mitosis. Proc. Natl Acad. Sci. USA. 1999;96:2710–2715. doi: 10.1073/pnas.96.6.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim. Biophys. Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Matter N, Herrlich P, König H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 83.Grange J, et al. Somatodendritic localization and mRNA association of the splicing regulatory protein Sam68 in the hippocampus and cortex. J. Neurosci. Res. 2004;75:654–666. doi: 10.1002/jnr.20003. [DOI] [PubMed] [Google Scholar]
- 84.Iijima T, Iijima Y, Witte H, Scheiffele P. Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. J. Cell Biol. 2014;204:331–342. doi: 10.1083/jcb.201310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rossbach O, et al. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol. Cell. Biol. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA. 2010;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Traunmüller L, Bornmann C, Scheiffele P. Alternative splicing coupled nonsense-mediated decay generates neuronal cell type-specific expression of SLM proteins. J. Neurosci. 2014;34:16755–16761. doi: 10.1523/JNEUROSCI.3395-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baudouin S, Scheiffele P. SnapShot: neuroligin- neurexin complexes. Cell. 2010;141:908–908.e1. doi: 10.1016/j.cell.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 89.Tabuchi K, Südhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- 90.Graf ER, Zhang X, Jin S-X, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Uemura T, et al. Trans-synaptic interaction of GluRδ2 and neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 93.Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 94. Iijima T, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. doi: 10.1016/j.cell.2011.11.028. This paper and the preceding references describe how a key determinant of synaptogenesis is regulated at the level of splicing
- 95.Richard S, et al. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 2005;1:e74. doi: 10.1371/journal.pgen.0010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lukong KE, Richard S. Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav. Brain Res. 2008;189:357–363. doi: 10.1016/j.bbr.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 97.Klein ME, Castillo PE, Jordan BA. Coordination between translation and degradation regulates inducibility of mGluR-LTD. Cell Rep. 2015;10:1459–1466. doi: 10.1016/j.celrep.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ehrmann I, et al. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS Genet. 2013;9:e1003474. doi: 10.1371/journal.pgen.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 100.Huang CS, et al. Common molecular pathways mediate long-term potentiation of synaptic excitation and slow synaptic inhibition. Cell. 2005;123:105–118. doi: 10.1016/j.cell.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 101.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 102.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 103.Du H, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Poulos MG, Batra R, Charizanis K, Swanson MS. Developments in RNA splicing and disease. Cold Spring Harb. Perspect. Biol. 2011;3:a000778. doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goodwin M, et al. MBNL sequestration by toxic RNAs and RNA misprocessing in the myotonic dystrophy brain. Cell Rep. 2015;12:1159–1168. doi: 10.1016/j.celrep.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Charizanis K, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. This paper demonstrates that MBNL2 promotes adult splice isoforms of ion channels and the association of neuronal defects caused by the loss of MBNL2 with the neurological disorder myotonic dystrophy
- 107.Batra R, et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shi Y, Manley JL. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 2015;29:889–897. doi: 10.1101/gad.261974.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Good PJ. A conserved family of elav-like genes in vertebrates. Proc. Natl Acad. Sci. USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brennan CM, Steitz JA. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.2, Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: an autoimmune RNA-binding protein with specificity for 3’ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.3, Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 114.4, Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J. Biol. Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 115.Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen C-YA, Xu N, Shyu A-B. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell. Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ince-Dunn G, et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012;75:1067–1080. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.9, Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pascale A, Amadio M, Quattrone A. Defining a neuron: neuronal ELAV proteins. Cell. Mol. Life Sci. 2008;65:128–140. doi: 10.1007/s00018-007-7017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Szabo A, et al. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 122.Sakai K, et al. A hippocampal protein associated with paraneoplastic neurologic syndrome and small cell lung carcinoma. Biochem. Biophys. Res. Commun. 1994;199:1200–1208. doi: 10.1006/bbrc.1994.1358. [DOI] [PubMed] [Google Scholar]
- 123.Srikantan S, Gorospe M. HuR function in disease. Front. Biosci. (Landmark Ed) 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Akamatsu W, et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl Acad. Sci. USA. 2005;102:4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.DeBoer EM, et al. Prenatal deletion of the RNA-binding protein HuD disrupts postnatal cortical circuit maturation and behavior. J. Neurosci. 2014;34:3674–3686. doi: 10.1523/JNEUROSCI.3703-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bhalla K, et al. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J. Hum. Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- 127.Lal D, et al. RBFOX1 and RBFOX3 mutations in rolandic epilepsy. PLoS ONE. 2013;8:e73323. doi: 10.1371/journal.pone.0073323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lal D, et al. Extending the phenotypic spectrum of RBFOX1 deletions: sporadic focal epilepsy. Epilepsia. 2015;56:e129–e133. doi: 10.1111/epi.13076. [DOI] [PubMed] [Google Scholar]
- 129.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bill BR, Lowe JK, Dybuncio CT, Fogel BL. Orchestration of neurodevelopmental programs by RBFOX1: implications for autism spectrum disorder. Int. Rev. Neurobiol. 2013;113:251–267. doi: 10.1016/B978-0-12-418700-9.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weyn-Vanhentenryck SM, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lovci MT, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mulley JC, Scheffer IE, Petrou S, Berkovic SF. Channelopathies as a genetic cause of epilepsy. Curr. Opin. Neurol. 2003;16:171–176. doi: 10.1097/01.wco.0000063767.15877.c7. [DOI] [PubMed] [Google Scholar]
- 134.Chapman AG, Woodburn VL, Woodruff GN, Meldrum BS. Anticonvulsant effect of reduced NMDA receptor expression in audiogenic DBA/2 mice. Epilepsy Res. 1996;26:25–35. doi: 10.1016/s0920-1211(96)00036-8. [DOI] [PubMed] [Google Scholar]
- 135.Zapata A, et al. Effects of NMDA-R1 antisense oligodeoxynucleotide administration: behavioral and radioligand binding studies. Brain Res. 1997;745:114–120. doi: 10.1016/s0006-8993(96)01134-1. [DOI] [PubMed] [Google Scholar]
- 136.Papale LA, et al. Heterozygous mutations of the voltage-gated sodium channel SCN8A are associated with spike-wave discharges and absence epilepsy in mice. Hum. Mol. Genet. 2009;18:1633–1641. doi: 10.1093/hmg/ddp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Corradini I, Verderio C, Sala M, Wilson MC, Matteoli M. SNAP-25 in neuropsychiatric disorders. Ann. NY Acad. Sci. 2009;1152:93–99. doi: 10.1111/j.1749-6632.2008.03995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sørensen JB, et al. Differential control of the releasable vesicle pools by SNAP-25 splice variants and SNAP-23. Cell. 2003;114:75–86. doi: 10.1016/s0092-8674(03)00477-x. [DOI] [PubMed] [Google Scholar]
- 139.Bark C, et al. Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission. J. Neurosci. 2004;24:8796–8805. doi: 10.1523/JNEUROSCI.1940-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Johansson JU, et al. An ancient duplication of exon 5 in the Snap25 gene is required for complex neuronal development/function. PLoS Genet. 2008;4:e1000278. doi: 10.1371/journal.pgen.1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Raman IM, Sprunger LK, Meisler MH, Bean BP. Altered subthreshold sodium currents and disrupted firing patterns in Purkinje neurons of Scn8a mutant mice. Neuron. 1997;19:881–891. doi: 10.1016/s0896-6273(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 142.Meisler MH, Kearney J, Escayg A, MacDonald BT, Sprunger LK. Sodium channels and neurological disease: insights from Scn8a mutations in the mouse. Neuroscientist. 2001;7:136–145. doi: 10.1177/107385840100700208. [DOI] [PubMed] [Google Scholar]
- 143.Levin SI, et al. Impaired motor function in mice with cell-specific knockout of sodium channel Scn8a (NaV1.6) in cerebellar purkinje neurons and granule cells. J. Neurophysiol. 2006;96:785–793. doi: 10.1152/jn.01193.2005. [DOI] [PubMed] [Google Scholar]
- 144.Raman IM, Bean BP. Resurgent sodium current and action potential formation in dissociated cerebellar Purkinje neurons. J. Neurosci. 1997;17:4517–4526. doi: 10.1523/JNEUROSCI.17-12-04517.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Grieco TM, Malhotra JD, Chen C, Isom LL, Raman IM. Open-channel block by the cytoplasmic tail of sodium channel β4 as a mechanism for resurgent sodium current. Neuron. 2005;45:233–244. doi: 10.1016/j.neuron.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 146.Plummer NW, McBurney MW, Meisler MH. Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J. Biol. Chem. 1997;272:24008–24015. doi: 10.1074/jbc.272.38.24008. [DOI] [PubMed] [Google Scholar]
- 147.O’Brien JE, et al. Rbfox proteins regulate alternative splicing of neuronal sodium channel SCN8A. Mol. Cell. Neurosci. 2012;49:120–126. doi: 10.1016/j.mcn.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kohrman DC, Harris JB, Meisler MH. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J. Biol. Chem. 1996;271:17576–17581. doi: 10.1074/jbc.271.29.17576. [DOI] [PubMed] [Google Scholar]
- 149.Sidman RL, Cowen JS, Eicher EM. Inherited muscle and nerve diseases in mice: a tabulation with commentary. Ann. NY Acad. Sci. 1979;317:497–505. doi: 10.1111/j.1749-6632.1979.tb56567.x. [DOI] [PubMed] [Google Scholar]
- 150.Caldwell JH, Schaller KL, Lasher RS, Peles E, Levinson SR. Sodium channel Nav1.6 is localized at nodes of ranvier, dendrites, and synapses. Proc. Natl Acad. Sci. USA. 2000;97:5616–5620. doi: 10.1073/pnas.090034797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kearney JA, et al. Molecular and pathological effects of a modifier gene on deficiency of the sodium channel Scn8a (Nav1.6) Hum. Mol. Genet. 2002;11:2765–2775. doi: 10.1093/hmg/11.22.2765. [DOI] [PubMed] [Google Scholar]
- 152.Sprunger LK, Escayg A, Tallaksen-Greene S, Albin RL, Meisler MH. Dystonia associated with mutation of the neuronal sodium channel Scn8a and identification of the modifier locus Scnm1 on mouse chromosome 3. Hum. Mol. Genet. 1999;8:471–479. doi: 10.1093/hmg/8.3.471. [DOI] [PubMed] [Google Scholar]
- 153. Howell VM, et al. A targeted deleterious allele of the splicing factor SCNM1 in the mouse. Genetics. 2008;180:1419–1427. doi: 10.1534/genetics.108.094227. References 151–153 demonstrate how a small change in the amount of a particular transcript isoform can dramatically affect neuronal function, and highlight the action of splicing regulators as phenotypic modifiers
- 154.Buchner DA, Trudeau M, Meisler MH. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science. 2003;301:967–969. doi: 10.1126/science.1086187. [DOI] [PubMed] [Google Scholar]
- 155.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ling S-C, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Johnson JO, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 160.Polymenidou M, et al. Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 2012;1462:3–15. doi: 10.1016/j.brainres.2012.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Buratti E, Baralle FE. TDP-43: gumming up neurons through protein-protein and protein-RNA interactions. Trends Biochem. Sci. 2012;37:237–247. doi: 10.1016/j.tibs.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 162.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kraemer BC, et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sephton CF, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu L-S, et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 166.Lee EB, Lee VM-Y, Trojanowski JQ. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl Acad. Sci. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang Y-J, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl Acad. Sci. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yang C, et al. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. 2014;111:E1121–E1129. doi: 10.1073/pnas.1322641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vanden Broeck L, Callaerts P, Dermaut B. TDP-43-mediated neurodegeneration: towards a loss-of-function hypothesis? Trends Mol. Med. 2014;20:66–71. doi: 10.1016/j.molmed.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 173.Budini M, Romano V, Quadri Z, Buratti E, Baralle FE. TDP-43 loss of cellular function through aggregation requires additional structural determinants beyond its C-terminal Q/N prion-like domain. Hum. Mol. Genet. 2015;24:9–20. doi: 10.1093/hmg/ddu415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Arnold ES, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl Acad. Sci. 2013;110:E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Iko Y, et al. Domain architectures and characterization of an RNA-binding protein, TLS. J. Biol. Chem. 2004;279:44834–44840. doi: 10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- 177.Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell. Sci. 1997;110:1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- 178.Rogelj B, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci. Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hicks GG, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 180.Huang C, et al. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLoS Genet. 2011;7:e1002011. doi: 10.1371/journal.pgen.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fujioka Y, et al. FUS-regulated region- and cell-type-specific transcriptome is associated with cell selectivity in ALS/FTLD. Sci. Rep. 2013;3:2388. doi: 10.1038/srep02388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Jia Y, Mu JC, Ackerman SL. Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell. 2012;148:296–308. doi: 10.1016/j.cell.2011.11.057. This paper demonstrates that mutation of a small RNA component of the spliceosome core can lead to a highly specific neurodegenerative phenotype
- 183.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 184.Black DL, et al. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 185.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 186.Lindsley RC, Ebert BL. Molecular pathophysiology of myelodysplastic syndromes. Annu. Rev. Pathol. 2013;8:21–47. doi: 10.1146/annurev-pathol-011811-132436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yoshida K, Ogawa S. Splicing factor mutations and cancer. Wiley Interdiscip. Rev. RNA. 2014;5:445–459. doi: 10.1002/wrna.1222. [DOI] [PubMed] [Google Scholar]
- 188.Blencowe BJ, Ahmad S, Lee LJ. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- 189.Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 2012;24:323–332. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 190.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 191.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhang W, et al. Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR. RNA. 2002;8:671–685. doi: 10.1017/s1355838202027036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ladd AN. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Mol. Cell. Neurosci. 2013;56:456–464. doi: 10.1016/j.mcn.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Norris AD, et al. A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol. Cell. 2014;54:946–959. doi: 10.1016/j.molcel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Calarco JA, Zhen M, Blencowe BJ. Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. RNA. 2011;17:775–791. doi: 10.1261/rna.2603911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jangi M, Sharp PA. Building robust transcriptomes with master splicing factors. Cell. 2014;159:487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lee J-A, et al. Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron. 2016;89:113–128. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758–1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 200.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 201.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 202.Popp MW-L, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yap K, Makeyev EV. Regulation of gene expression in mammalian nervous system through alternative pre?mRNA splicing coupled with RNA quality control mechanisms. Mol. Cell. Neurosci. 2013;56:420–428. doi: 10.1016/j.mcn.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 204.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 205.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 2012;69:3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Racca C, et al. The neuronal splicing factor Nova co-localizes with target RNAs in the dendrite. Front. Neural Circuits. 2010;4:5. doi: 10.3389/neuro.04.005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Klein ME, Younts TJ, Castillo PE, Jordan BA. RNA-binding protein Sam68 controls synapse number and local β-actin mRNA metabolism in dendrites. Proc. Natl Acad. Sci. 2013;110:3125–3130. doi: 10.1073/pnas.1209811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.