Abstract
Although the liver is the primary site of cytokine-mediated expression of acute-phase serum amyloid A (SAA) protein, extrahepatic production has also been reported. Besides its role in amyloidosis and lipid homeostasis during the acute-phase, SAA has recently been assumed to contribute to bone and cartilage destruction. However, expression of SAA in human osteogenic tissue has not been studied. Therefore, we first show that SAA1 (coding for the major SAA isoform) but not SAA2 transcripts are expressed in human trabecular and cortical bone fractions and bone marrow. Next, we show expression of (i) IL-1, IL-6, and TNF receptor transcripts; (ii) the human homolog of SAA-activating factor-1 (SAF-1, a transcription factor involved in cytokine-mediated induction of SAA genes); and (iii) SAA1/2 transcripts in non-differentiated and, to a higher extent, in osteoblast-like differentiated human mesenchymal stem cells. Third, we provide evidence that human osteoblast-like cells of tumor origin (MG-63 and SAOS-2) express SAF-1 under basal conditions. SAA1/2 transcripts are expressed under basal conditions (SAOS-2) and cytokine-mediated conditions (MG-63 and SAOS-2). RT-PCR, Western blot analysis, and immunofluorescence technique confirmed cytokine-mediated expression of SAA on RNA and protein level in osteosarcoma cell lines while SAA4, a protein of unknown function, is constitutively expressed in all osteogenic tissues investigated.
Keywords: SAA, SAF-1, cancer, inflammation, FPRL-1/ALX, SR-BI
Serum amyloid A (SAA) is a generic term for a family of cytokine-induced acute-phase proteins synthesized primarily by the liver that are coded for by different genes with a high allelic variation and a high degree of homology in mammals [Uhlar et al., 1994]. In humans, SAA1 and SAA2 genes encode for non-glycosylated acute-phase SAA (104 amino acids) proteins SAA1 (the most abundant isoform) and SAA2. The SAA4 gene encodes for constitutively expressed glycosylated SAA4 protein (112 amino acids). While no function has been attributed to SAA4, a panel of different activities has been reported for SAA [Malle et al., 1993; Uhlar and Whitehead, 1999]. SAA, an important clinical marker for inflammation [Malle and De Beer, 1996] and precursor protein of secondary reactive amyloidosis [Husebekk et al., 1985], contributes to cellular cholesterol homeostasis, modulates intracellular calcium levels, and promotes signaling cascades [Badolato et al., 1995; Artl et al., 2000; Baranova et al., 2005]. In addition, several functions of SAA, described in the context of inflammation, are compatible with the mechanisms of tumor cell invasion and metastasis. Both the capacity to induce chemotaxis, cell adhesion and migration [Badolato et al., 1994] and the ability to act as an extracellular matrix adhesion protein [Hershkoviz et al., 1997], suggested that SAA might play a role in the local inflammation of the malignant tissue. Recently, SAA transcripts have been observed in cancerous tissues of non-hepatic origin [Gutfeld et al., 2006; Kovacevic et al., 2006]. Increased levels of SAA mRNA have been verified in lymphoma and cancerous regions of human renal carcinoma [Nishie et al., 2001]. Furthermore, SAA levels are increased in a broad spectrum of neoplastic diseases [Rosenthal and Sullivan, 1979] and a number of studies proposed a direct correlation between SAA concentrations and the stage of tumor [Weinstein et al., 1984]. Animal experiments revealed that SAA levels correlated with the tumor burden [McLean et al., 2004]. This led to the assumption that SAA might be considered a marker for tumor progression and even a biomarker for specific cancer types [Howard et al., 2003]. A proteomic signature approach of plasma proteins suggested SAA as one of the discriminatory peaks between osteosarcoma and benign osteochondroma [Li et al., 2006]. SAA is also produced by inflamed synovial tissue [O’Hara et al., 2004], where, by promoting synoviocyte hyperplasia and angiogenesis via the formyl peptide receptor like 1 (FPRL-1), found to be identical with the lipoxin A4 receptor (ALX), SAA may induce destruction of bone and cartilage [Lee et al., 2006]. Cytokine-mediated induction of SAA1 transcripts have been reported in human chondrocytes and SAA protein has been shown to induce transcription of matrix metalloproteinases (MMPs) [Migita et al., 1998; Vallon et al., 2001], proteins that in turn promote tumor invasion, metastasis, and angiogenesis.
Studies on SAA and bone biology were performed primarily in diseased human synovium and cartilage and rabbit chondrocytes [Vallon et al., 2001]. As no investigations so far assessed the biosynthesis of SAA1/2 and SAA4 in human osteogenic specimens, the current study aimed at investigating the expression of SAA transcripts in bone material and differentiated stem cells with an osteoblast-like phenotype. Finally, expression of SAA was studied in two human osteosarcoma cell lines. MG-63 cells are only weakly positive for alkaline phosphatase (a biomarker for bone formation) and exhibit a premature fibroblast-like state. In contrast, SAOS-2 cells stain intensely positive for alkaline phosphatase, appear rounded and display an epithelial phenotype, and represent a more differentiated osteoblast cell type than MG-63 [Sevetson et al., 2004]. We also were interested whether the human homologue of SAA-activating factor-1 (SAF-1), a Cys2His2-type zinc finger transcription factor, known to be involved in cytokine-induced expression of SAA transcripts in hepatic tissue [Ray et al., 2002] and MMPs in chondrocytes [Ray et al., 2005], is expressed in osteoblast-like cells of non-tumor and tumor origin.
MATERIALS AND METHODS
Bone Tissue and Cells
The bone material was of femur origin (either from biopsies or bone segments removed from patients with osteoarthritis in the process of positioning prostheses), obtained from the Department of Trauma Surgery, Medical University of Graz. Material was frozen in liquid nitrogen followed by storage at −70°C and subsequently pulverized using a freezer/mill SPEX 6700 (SPEX CertiPrep, Inc., Stanmore, UK).
Mononuclear cell fractions were derived from bone marrow from three different patients suffering from arthritis of hip joint (one female, 63.9-year-old; one male, 74-year-old) or arthritis of knee joint (one female, 71.8-year-old), who gave consent after full information and approval by the hospital ethical committee (No. 12-091). Mononuclear cells were isolated from bone marrow aspirates using methods slightly modified from those described previously [Haynesworth et al., 1992]. Briefly, isolation was performed in Percoll gradient (d = 1.073 g/ml, 900g, 30 min), and human mesenchymal stem cells (hMSCs) were selected on the basis of adherence. The cells were maintained in DMEM-F12 (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), l-glutamine (Sigma) at 100 U/ml and 100 μg/ml penicillin/streptomycin (P/S), respectively. To induce osteogenic differentiation [Jaiswal et al., 1997], hMSCs (passage 4) were seeded in six-well plates, at a density of 1 × 105cells/well, and propagated in DMEM-LG (Invitrogen) supplied with 10% FBS, 100 U/ml l-glutamine, and 100 μg/ml P/S, 0.1 mM l-ascorbic acid 2-phosphate, 10 mM β-glycerophosphate, and 0.1 μM dexamethasone (all from Sigma) for 3 weeks prior to cytokine treatment. The differentiation of hMSC to osteogenic lineage was verified by Alizarin Red S (Sigma) staining as described [Kulterer et al., 2007].
Human osteosarcoma cell lines (MG-63 and SAOS-2) were maintained in MEMα (Invitrogen), supplemented with 5% (MG-63) or 10% (SAOS-2, v/v) FBS and 100 U/ml l-glutamine and 100 μg/ml P/S at 37°C under 5% CO2.
Human hepatoma cells (HUH-7) and human macrophages (THP-1 cells) were grown in RPMI-1640 medium (Bio Whitaker, Austria) containing 10% (v/v) FBS as described [Malle et al., 1999; Kovacevic et al., 2006].
Cell Culture Experiments
Both, hMSCs (non-differentiated or differentiated) and osteosarcoma cells were seeded in 6 cm plates (Costar, Austria) and, upon reaching 80% confluency, incubated (at indicated time points) in medium containing interleukin-1α (IL-1α), IL-1β, IL-6, or tumor necrosis factor-α (TNFα, at indicated concentrations; R&D Systems, Inc., Minneapolis). To follow expression of FPRL-1/ALX, osteosarcoma cells were seeded in six-well plates and, upon reaching 80% confluency, stimulated for 24 h by adding dexamethasone (1 or 2 μM; Sigma) in the absence or presence of 10 ng/ml IL-1β or IL-6 to the cell culture medium.
RNA Isolation and Northern Blot Analysis
RNA from human bone material was isolated using TRIreagent (Sigma), according to the manufacturer’s instructions. Total RNA was isolated from hMSCs, osteosarcoma cells, HUH-7, and THP-1 using RNeasy kit (QIAGEN, Vienna, Austria). Northern blots were performed exactly as described [Kovacevic et al., 2006]. The SAA1/2 cDNA probe (a kind gift of Dr. P. Woo, London) was radiolabeled with [32P] dCTP by Prime-a-Gene Labeling system (Promega, Mannheim, Germany) and used as a probe. The hybridized blots were washed and exposed to X-ray film for 2–7 days.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Prior to reverse transcription, RNA was digested with DNase (Invitrogen), following manufacturer’s instructions. Reverse transcription was done with Superscript II RNase H− Reverse Transcriptase (Invitrogen) [Kovacevic et al., 2006]. PCR was performed with Taq polymerase (Solis Biodyne, Estonia) using 2 μl cDNA from reverse transcription reaction as template. The final concentrations in the PCR mix (50 μl reaction volume) were: 100 pM of each primer, 250 ng of each dNTP, 0.5 U Taq-polymerase. PCR was performed in a Mastercycler (Eppendorf) as follows: Taq-polymerase activation 15 min at 95°C; 30–40 cycles of 1 min at 95°C, 1 min at annealing temperature, 1 min at 72°C; followed by a final 7 min elongation at 72°C. In the case of SAA1 and SAA2, cycles were each of 10 s at 95°C, 5 s at annealing temperature and 10 s at 72°C. Specific primers, with annealing temperature for each primer pair and number of cycles for each PCR are listed in Table I (Supplement). Control reactions using RNA as template were performed in all experiments.
Immunofluorescence and Confocal Laser Scanning Microscopy
MG-63 and SAOS-2 cells were cultured on Lab-Tek chamber slides (Nunc) in the absence or presence of 10 ng/ml IL-1β up to 24 h. Then, the cells were washed with PBS, air dried (2 h, 22°C), and acetone-fixed (5 min). Fixed chamber slides were incubated for 30 min with either polyclonal anti-SAA1/2 rabbit antiserum [Malle et al., 1995], sequence-specific anti-human SAA1/2 peptide (positions 1–17 and 89–104) rabbit antiserum (dilutions of 1:100) [Malle et al., 1995] or sequence-specific anti-human SAA4 peptide (positions 1–17 and 94–112) rabbit antiserum [Hrzenjak et al., 2001], followed by 30 min incubation with cyanine-3-labeled goat anti-rabbit IgG (dilution 1:300, Jackson Dianova) [Kovacevic et al., 2006]. PBS was used for washing sections between different incubation steps. Sections were mounted with Moviol (Calbiochem–Novabiochem, La Jolla) and analyzed on a confocal laser-scanning microscope (Leica SP2, Leica Lasertechnik GmbH, Heidelberg, Germany) using the 543 nm laser line for excitation of cyanine-3. Control experiments encompassed immunofluorescence (i) without primary antibody, (ii) with polyclonal non-immune antiserum as primary antibody, (iii) without secondary antibody, or (iv) with pre-absorbtion of the corresponding primary antibody with human SAA1/2 [Malle et al., 1995] or SAA4 [Hrzenjak et al., 2001].
Western Blot Analysis
To separate nucleus- and cytoplasm-protein fractions, the cells were lysed in RIPA buffer (50 mM Tris–HCl, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 μg/ml of aprotinin, leupeptin, and pepstatin, 1 mM Na3VO4), kept 15 min on ice, scraped with a cell scraper and centrifuged at 600g (10 min, 4°C). Supernatant with cytoplasm-protein fractions was stored at −20°C. To obtain nucleus-protein fraction, the cell-pellet was resuspended in RIPA buffer with 270 mM NaCl, incubated 5 min on ice and centrifuged (13,000g, 10 min, 4°C).
Protein lysates (20–30 μg, as assessed by the Lowry method) were separated on 10–20% SDS–PAGE. Proteins were transferred to PVDF membranes as described [Malle et al., 1999]. The membranes were incubated with (i) either sequence-specific anti-human SAA1/2 peptide (positions 1–17, 79–94, and 89–104) rabbit antiserum (dilutions of 1:100–1:500) [Malle et al., 1995], (ii) sequence-specific anti-human SAA4 peptide (positions 1–17 and 94–112) rabbit antiserum (dilution of 1:300) [Hrzenjak et al., 2001], or (iii) rabbit anti-SAF-1 antiserum (dilution 1:1,000) [Ray et al., 2002] as primary antibodies. Peroxidase-conjugated goat anti-rabbit IgGs (Pierce Chemical Co.; 1:100,000) were used as secondary antibodies. Immunoreactive bands were visualized with the SuperSignal HRP substrate (Pierce Chemical Co.) and exposure to X-ray films (Kodak).
RESULTS
Expression of SAA Transcripts in Human Bone
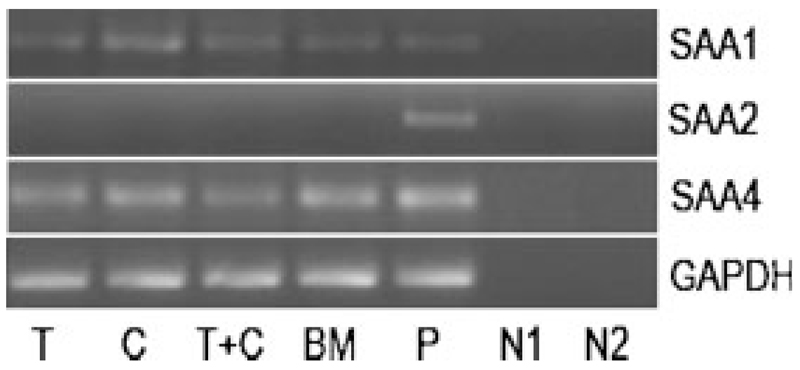
First, we investigated whether SAA transcripts are expressed in different bone RNA preparations obtained from three patients with osteoarthritis (samples were taken at least two times from each patient). SAA1 transcripts were detected in individual trabecular and cortical bone fractions, in a mixed trabecular/cortical bone fraction, as well as in RNA isolated from bone marrow (Fig. 1). Unlike SAA1, SAA2 was not detected in any bone RNA preparation. SAF-1, the transcription factor involved in cytokine-mediated induction of SAA could not be verified in all biological specimens investigated (data not shown). Subsequently, no expression of SAF-2, a splice variant of SAF-1 and negative regulator of inflammation [Ray et al., 2002], was found. However, SAA4 was constitutively expressed in trabecular and cortical fraction, and bone marrow (Fig. 1).
Fig. 1.
RT-PCR of SAA transcripts in human bone: RNA was isolated from different bone preparations, reverse transcribed, and cDNAs of SAA1, SAA2, and SAA4 amplified using specific forward and reverse oligonucleotide primers (Supplement, Table I). The PCR products were separated on 1.5% agarose gels. T, trabecular bone; C, cortical bone; BM, bone marrow; P, positive control, HUH-7 cells; N1, negative control—RNA template: negative controls were done for all samples; N2, negative control—water template. To ensure that equal amount of cDNA was used as a template RT-PCR for GAPDH was made as a control. One representative experiment out of three is shown.
Expression of SAA Transcripts in Human Mesenchymal Stem Cells
Human bone RNA preparations (e.g., bone marrow) may contain RNA from a variety of cells expressing SAA transcripts, for example, adipocytes and fibroblasts [Meek et al., 1994]; the trabecular fraction may contain osteoblasts, osteoclasts, and osteocytes, while the cortical fraction may additionally contain chondrocytes. To investigate expression of cytokine receptor and SAA transcripts in osteogenic material, mononuclear cells were isolated; hMSCs, selected on the basis of adherence, were differentiated into an osteoblast-like-lineage. As osteocalcin may not be considered a straightforward or reliable marker for human osteoblastic cell differentiation [Jaiswal et al., 1997], calcium deposition was investigated. Using Alizarin Red S, pronounced calcium deposition in the extracellular matrix of hMSCs, kept in osteogenic medium, is indicative for differentiation of cells to osteogenic lineage (Supplement, Fig. I).
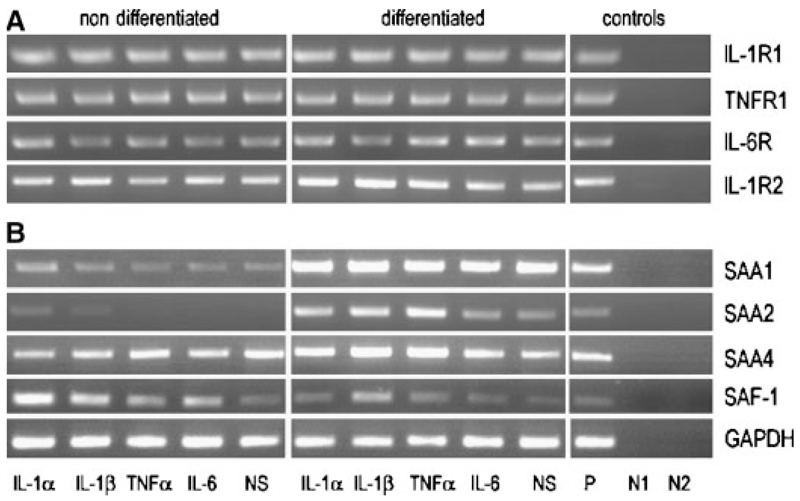
To check for the presence of cytokine receptors on hMSCs, RT-PCR analysis was performed. Both, non-differentiated and differentiated hMSCs expressed IL-1, IL-6, and TNF receptors (Fig. 2A), responsible for binding and transducing the actions of corresponding cytokines. Additionally, hMSCs expressed IL-1 receptor type 2, a decoy receptor that binds IL-1 but is unable to start a signaling cascade.
Fig. 2.
RT-PCR of cytokine receptors, SAA and SAF-1 transcripts in non-differentiated and differentiated hMSCs: hMSCs were kept in either expansion or osteogenic medium and stimulated with different cytokines (10 ng/ml) for 24 h. A: Human cytokine receptor transcripts as well as (B) SAA and SAF transcripts were amplified using specific oligonucleotide primers (Supplement, Table I). The PCR products were separated on 1.5% agarose gels. NS, non-stimulated; P, positive control: RNA was isolated from HUH-7 cells for SAF-1, SAA, cytokine receptors except IL-1R2, where RNA from THP-1 cells was used; N1, negative control— RNA template: negative controls were done for all samples; N2, negative control—water template. To ensure that equal amount of cDNA was used as a template RT-PCR for GAPDH was made as a control. One representative experiment out of three is shown.
Next, expression of SAA transcripts was studied. SAA1 was present under non-stimulated and cytokine-mediated conditions in non-differentiated hMSCs and abundantly present in differentiated hMSCs (Fig. 2B). In contrast to differentiated hMSCs, where expression of SAA2 paralleled that of SAA1, SAA2 was only present in non-differentiated cells in response to IL-1 (Fig. 2B). As shown with human bone RNA preparations (Fig. 1), SAA4 was constitutively expressed in non-differentiated hMSCs and differentiated primary osteoblasts. Figure 2B shows that SAF-1, the transcription factor involved in cytokine-mediated induction of SAA transcripts, was present in non-differentiated and differentiated hMSCs under non-treated and cytokine-treated conditions; SAF-2 transcripts were not identified.
Expression of SAA Transcripts and SAA Protein in Human Osteosarcoma Cell Lines
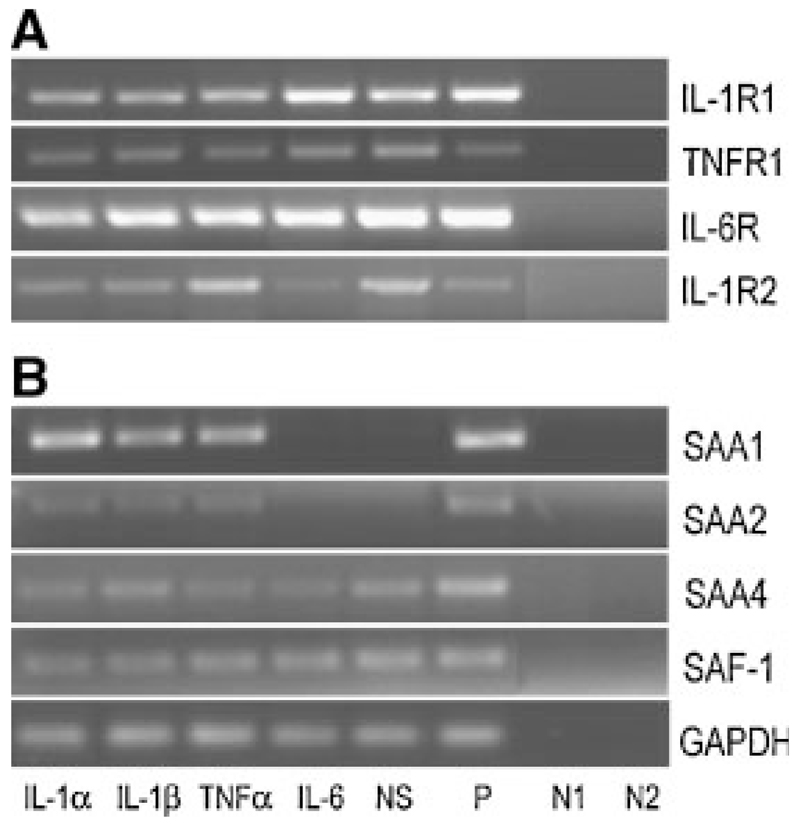
Due to limited access to hMSCs, the next series of experiments were performed with human osteoblast-like cell lines of tumor origin. First, we checked for the presence of cytokine receptors in osteosarcoma cells lines. Both, MG-63 (Fig. 3A) and SAOS-2 cell lines (Supplement, Fig. IIA) express all cytokine receptors (IL-1R1, TNFR, IL-6R, and IL-1R2) under non-treated and cytokine-treated conditions.
Fig. 3.
RT-PCR of cytokine receptors, SAA and SAF-1 in human osteosarcoma cells: MG-63 cells were stimulated with different cytokines (10 ng/ml) for 24 h. A: Cytokine receptor transcripts as well as (B) SAA and SAF-1 transcripts were amplified using specific oligonucleotide primers (Supplement, Table I). The PCR products were separated on 1.5% agarose gels. NS, non-stimulated; P, positive control: RNA was isolated from HUH-7 cells for SAF-1, SAA, cytokine receptors except IL-1R2, where RNA from THP-1 cells was used; N1, negative control—RNA template: negative controls were done for all samples; N2, negative control—water template. To ensure that equal amount of cDNA was used as a template RT-PCR for GAPDH was made as a control. One representative experiment out of three is shown.
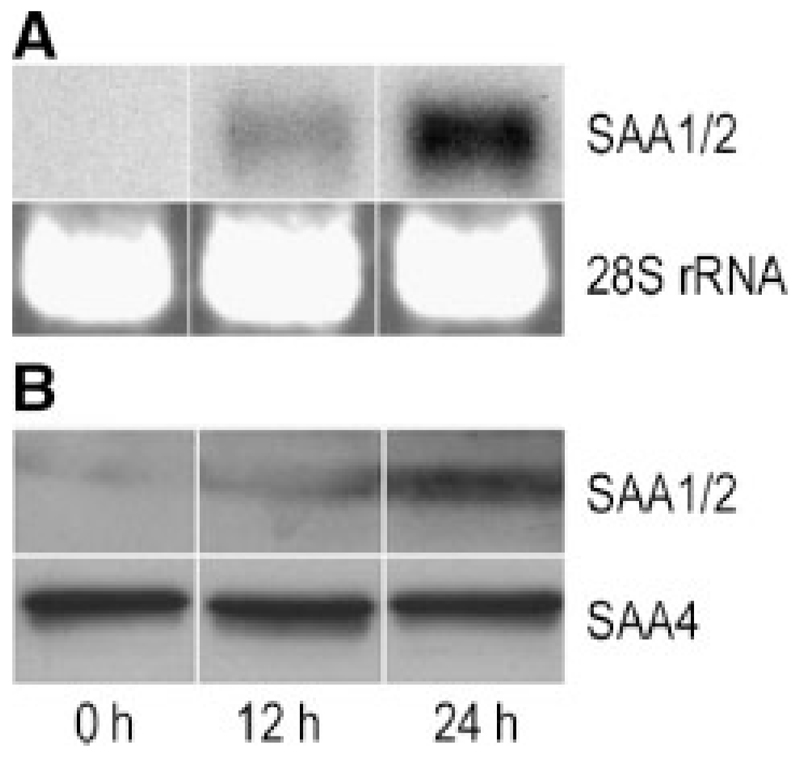
We then studied time-dependent expression of SAA on RNA level in osteosarcoma cells. Northern blot experiments revealed time-dependent SAA expression in MG-63 cells in response to IL-1β (Fig. 4A). Next, expression of SAA was followed on a protein level. Because the results of Western blot analysis (Fig. 4B) and immunofluorescence technique (Supplement, Fig. III) were consistent with that of RT-PCR and Northern blot, we had evidence that pronounced SAA protein expression in MG-63 (Fig. 4) occurred after a 24-h stimulation period. The modest increase in staining for SAA1/2 in SAOS-2 cells after 24 h (Supplement, Fig. III) is in line with high basal expression of SAA1/2 transcripts in these cells under non-stimulated conditions (Supplement, Fig. II). Most importantly, the use of sequence-specific antibodies (raised against the N- and C-terminal portion of SAA) confirms that the SAA protein is intact and not cleaved/degraded.
Fig. 4.
Time-dependent expression of SAA in osteosarcoma cells: MG-63 cells were stimulated with 10 ng/ml IL-1β for 12 and 24 h. A: RNA was isolated and Northern blot experiments were performed using SAA1/2 cDNA as a probe. The 28S rRNA was used as gel loading control. B: Cells were lysed and total cellular protein was subjected to SDS–PAGE and transferred to membranes. Western blot experiments were performed using sequence-specific anti-human SAA1/2 or SAA4 peptide antisera (see Materials and Methods section) as primary antibodies. One representative experiment out of three is shown.
Because of the high sequence similarity between SAA1 and SAA2 [Uhlar et al., 1994], the cDNA used in Northern blot experiments cannot differentiate between both SAA isoforms. Therefore, an RT-PCR using specific primer pairs (Supplement, Table I) was done. Neither SAA1 nor SAA2 were present in non-stimulated MG-63 cells (Fig. 3B). However, IL-1α, IL-1β and TNFα induced expression of both SAA transcripts with SAA1 being preferentially expressed (Fig. 3B). Although abundant IL-6 receptor expression was found on MG-63 cells (Fig. 3A), IL-6 did not induce SAA1 and SAA2 transcripts (Fig. 3B). These observations are in line with findings that the IL-6 receptor is non-functional in MG-63 cells as no significant phosphorylation of the signal-transducing molecule glycoprotein 130 did occur in the presence of IL-6 [Vermes et al., 2002]. However, in SAOS-2 cells, both SAA transcripts were detected in non-stimulated and cytokine-stimulated cells with SAA1 being also preferentially expressed (Supplement, Fig. IIB).
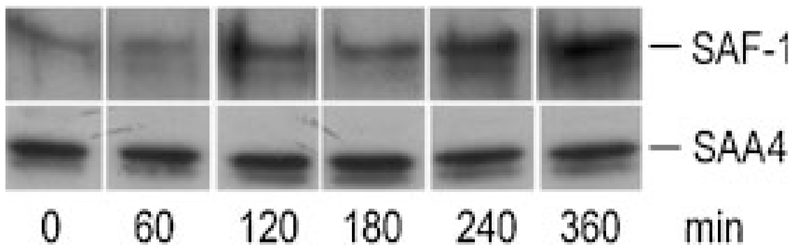
Next, expression of SAF-1/2 was investigated in both osteosarcoma cells. SAF-1 was present to a similar extent in non-stimulated and cytokine-stimulated MG-63 cells (Fig. 3B). In SAOS-2 cells, pronounced SAF-1 expression was observed in non-stimulated cells, while the presence of cytokines led to a decrease of SAF-1 (Supplement, Fig. IIB). SAF-2 was undetectable in both cell lines cultured in the absence or presence of cytokines. As immunofluorescence technique was not successful to confirm that activation of SAF-1 precedes expression of SAA, MG-63 cells were stimulated with IL-1β up to 6 h. Then, the cells were lysed, and Western blot analyses of cell lysates (cytoplasm- and nucleus-protein fraction) were performed. In the cytoplasm of MG-63 cells only a faint band corresponding to immunoreactive SAF-1 was detected (data not shown). However, in the nuclear fraction of MG-63 cells, pronounced staining for immunoreactive SAF-1 protein became apparent after 2 h of stimulation and increased up to 6 h (Fig. 5).
Fig. 5.
Western blot of SAF-1 and SAA4 in osteosarcoma cells: MG-63 cells were stimulated with 10 ng/ml IL-1β up to 6 h in six-well plates. The cells were lysed and the nuclear fraction was isolated as described in Materials and Methods. The protein was subjected to SDS–PAGE and transferred to membranes. Western blot experiments were performed using polyclonal anti-SAF-1 antiserum or sequence-specific anti-human SAA4 peptide antisera as primary antibodies. One representative experiment out of two is shown.
Finally, we checked for the presence of SAA4 on RNA and protein level in human osteosarcoma cell lines. Both, RT-PCR (Fig. 3B, Supplement, Fig. IIB), Western blotting experiments (Figs. 4B and 5), and immunofluorescence staining (Supplement, Fig. III) confirmed that SAA4 is constitutively expressed in MG-63 and SAOS-2 cells and that its expression is not cytokine-responsive.
Expression of Candidate Receptors for SAA in Human Osteosarcoma Cell Lines
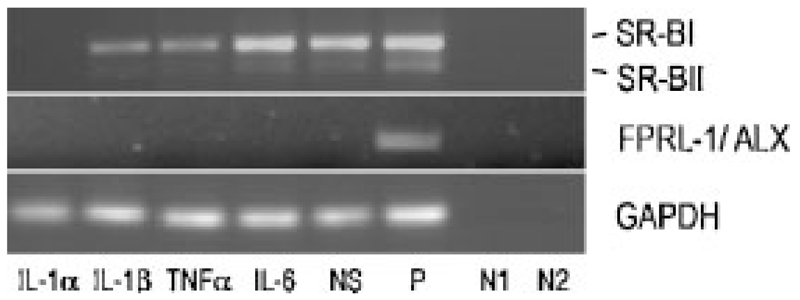
Binding of SAA to FPRL-1/ALX may promote expression of MMPs that in turn could degrade a variety of proteins including SAA [Stix et al., 2001]. As slight expression of MMPs was detected on MG-63 and SAOS-2 cells [Pautke et al., 2004] and SAA is expressed in response to cytokines (Fig. 4B, Supplement, Fig. III), expression of FPRL-1/ALX was followed by RT-PCR. Neither in non-stimulated nor cytokine-stimulated MG-63 (Fig. 6) and SAOS-2 cells (Supplement, Fig. IV), FPRL-1/ALX transcripts could be identified. Even the gluco-corticoid dexamethasone, recently reported to promote induction of FPRL-1/ALX (Giner et al., 2007), failed to induce FPRL-1/ALX transcripts in MG-63 and SAOS-2 cells (Supplement, Fig. V).
Fig. 6.
RT-PCR of SR-BI/II and FPRL-1/ALX in human MG63 cells: Cells were stimulated with different cytokines (10 ng/ml) for 24 h. Human SR-BI/II and FPRL-1/ALX transcripts were amplified using specific oligonucleotide primers (Supplement, Table I). The PCR products were separated on 1% agarose gels. NS, non-stimulated; P, positive control: RNA was isolated from differentiated THP-1 cells for SR-BI/II, for FPRL-1/ALX, HUH-7 genomic DNA was used; N1, negative control—RNA template: negative controls were done for all samples; N2, negative control—water template. To ensure that equal amount of cDNA was used as a template RT-PCR for GAPDH was made as a control. One representative experiment out of three is shown.
SAA has been recently reported to bind to SR-BI [Baranova et al., 2005; Marsche et al., 2007], a class B scavenger receptor that mediates both cellular cholesterol efflux and cellular lipid uptake, respectively. Indeed, SR-BI and its splice variant SR-BII are expressed in non-treated MG-63 (Fig. 6) and SAOS-2 cells (Supplement, Fig. V). Addition of cytokines revealed that IL-6 had no effect on SR-BI/II expression, while TNFα and IL-1 down-regulated expression of SR-BI/II transcripts (Fig. 6). Similar findings have been observed in human monocytes/macrophages, where both lipopolysaccharide and cytokines decreased SR-BI expression [Buechler et al., 1999].
DISCUSSION
High SAA levels have been found in serum of patients with different neoplastic diseases, for example, renal and colorectal cancers [Kimura et al., 2001; Glojnaric et al., 2001]. SAA has also been shown to be a prognostic marker in some cancers [Biran et al., 1986] and was used to monitor disease and therapeutic response in prostate cancer [Kaneti et al., 1984]. Recent reports even proposed SAA as a specific biomarker for specific types of cancer [Cho et al., 2004; Khan et al., 2004]. High levels of SAA were found in prostate cancer with bone metastasis [Le et al., 2005] and in patients with osteosarcoma [Li et al., 2006]. We here show expression of SAA transcripts in osteogenic tissue, in particular in human bone material, non-differentiated hMSCs and osteoblast-like differentiated hMSCs.
Cytokines are important local mediators of bone remodeling. Released by various cell types in bone microenvironment, including osteoblasts, IL-1 and TNFα are potent stimulators of bone resorption in cell culture, organ culture systems and in vivo [Kwan Tat et al., 2004]. IL-1 directly stimulates osteoclast function through the IL-1 type 1 receptor expressed by osteoclasts. TNFα may induce resorption, either indirectly, by affecting the production of other osteoclast differentiation factors or directly, by stimulating the differentiation of osteoclast progenitors into osteoclasts in the presence of M-CSF. IL-6 also stimulates osteoclast activity and bone resorption, but only when osteoclasts are co-cultured with osteoblasts. We here show cytokine-mediated expression of SAA on RNA and protein level by osteoblast-like cancer cells. The high expression of SAA in osteoblast-like human sarcoma cells under cytokine-mediated conditions provides clear evidence of extrahepatic SAA synthesis in cancerous tissue of bone origin. One of the important findings in the present study is the detection of SAF-1 as a regulator of A-SAA expression in hMSCs and osteosarcoma cells. SAF-1 expression level is increased in hMSCs in response to cytokines under both non-differentiated and differentiating conditions but levels of SAA1/2 increased at a significantly higher level under differentiating conditions (Fig. 2). This result suggests that differentiated cells may contain nuclear factor(s) that synergize SAF-1 activity. Indeed SAF-1 activity is seen to be facilitated by members of the AP-1 family [Ray et al., 2005] and it will be interesting to see whether AP-1 activity is induced in the differentiated mesenchymal stem cells. Unlike hMSCs, SAF-1 is constitutively present in MG-63 osteosarcoma cells and its level remains unchanged during cytokine exposure (Fig. 3). However, cytokine treatment increased nuclear translocation of SAF-1 (Fig. 5), which correlates with a significant increase of SAA1/2 level in MG-63 cells (Figs. 3 and 4). SAF-1 protein contains two nuclear localization signals that efficiently translocate this protein to the nucleus [Ray et al., 2004a]. Also noticeable was that between the two isoforms, expression of SAA1 is higher to SAA2, which indicates for a differential regulation of SAAs by SAF-1. The observed differences in SAF-1 and SAA1/2 expression in MG-63 and SAOS-2 could be due to alterations in cyclin-dependent kinase inhibitors [Park et al., 2002], molecules that are regulated by SAF-1 expression [Ray et al., 2004b].
During inflammation, SAA seems to exhibit Janus-faced effects. SAA has been reported to exert cytokine-like properties [Patel et al., 1998] or even to exert a feedback mechanism of IL-1-induced inflammatory events [Shainkin-Kestenbaum et al., 1991]. However, the role of SAA in bone-derived cells or in cells contributing to bone remodeling remains unknown. A possible function of apolipoprotein SAA during inflammation is its role in lipid metabolism. Indeed, SR-BI is abundantly expressed in MG-63 (Fig. 6) and SAOS-2 cells (Supplement, Fig. IV) and thus may contribute to proliferation and cellular growth of osteosarcoma as shown in other cancer cell lines [Wadsack et al., 2003]. However, under basal conditions, SAA is not expressed; the fact that cytokine-mediated expression of A-SAA is paralleled by down regulation of SR-BI transcripts suggests that SAA may probably not favor SR-BI-mediated bidirectional cholesterol flux under inflammatory conditions in osteosarcoma cells.
The findings that FPRL-1/ALX is a functional receptor for amyloidogenic proteins and peptide agonists including A-SAA may have important implications in several disease states. Phagocytic leukocytes are the source of enzymes that can fragment and degrade SAA. The usage of FPRL-1/ALX by SAA to chemoattract leukocytes may serve to recruit these cells to participate in degradation of SAA, a process assumed to occur at sites of amyloidogenic deposits. Indeed, binding of SAA to FPRL-1/ALX is paralleled by expression of MMPs. MMPs are endopeptidases with specific and selective activities against many components of the extracellular matrix, that may also degrade SAA as shown with THP-1 cells [Stix et al., 2001] and chondrocytes [Zerega et al., 2004]. However, an autologous degradation of SAA via the FPRL-1/ALX/MMP axis in human osteosarcoma may be excluded as both MG-63 and SAOS-2 cells are lacking FPRL-1/ALX transcripts. Our findings are in line with recent observations that the pentapeptide WKYMVm, a specific ligand for FPRL-1/ALX, failed to induce chemotactic response in wild-type HOS osteosarcoma cell lines while over expression of FPRL-1 in these cells mediated the chemotactic response [Li et al., 2001].
Summarizing, we could show that both benign and malign bone-derived tissues express SAA transcripts and that the constitutively expressed SAA4 is also present. In particular human osteosarcoma cell lines represent a suitable cellular model to follow cytokine-mediated induction of SAA on RNA and protein level. Our findings that SAA is synthesized by osteosarcoma in vitro could support a recent study [Li et al., 2006] that reports high SAA1 levels in plasma of osteosarcoma patients; whether SAA1 is a direct tumor product under in vivo conditions needs to be verified. The present study further demonstrates that purified hMSCs express SAA transcripts and that culture-expanded differentiated hMSCs express higher levels of SAA transcripts. The transcriptional regulation of genes in non-differentiated and differentiated hMSCs incubated with SAA is currently under investigation.
Supplementary Material
This article contains supplementary material, which may be viewed at the Journal of Cellular Biochemistry website at http://www.interscience.wiley.com/jpages/0730-2312/suppmat/index.html.
ACKNOWLEDGMENTS
This work was supported by grants from the Kamillo Eisner Foundation (Switzerland) and the Austrian Science Fund (FWF P14186-B05, P17013-B05, P19074-B05) to AH and EM.
Footnotes
Grant sponsor: Kamillo Eisner Foundation; Grant sponsor: Austrian Science Fund; Grant numbers: FWF P14186-B05, P17013-B05, P19074-B05.
References
- Artl A, Marsche G, Lestavel S, Sattler W, Malle E. Role of serum amyloid A during metabolism of acute-phase HDL by macrophages. Arterioscler Thromb Vasc Biol. 2000;20:763–772. doi: 10.1161/01.atv.20.3.763. [DOI] [PubMed] [Google Scholar]
- Badolato R, Wang JM, Murphy WJ, Lloyd AR, Michiel DF, Bausserman LL, Kelvin DJ, Oppenheim JJ. Serum amyloid A is a chemoattractant: Induction of migration, adhesion, and tissue infiltration of monocytes and polymorphonuclear leukocytes. J Exp Med. 1994;180:203–209. doi: 10.1084/jem.180.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badolato R, Johnston JA, Wang JM, McVicar D, Xu LL, Oppenheim JJ, Kelvin DJ. Serum amyloid A induces calcium mobilization and chemotaxis of human monocytes by activating a pertussis toxin-sensitive signaling pathway. J Immunol. 1995;155:4004–4010. [PubMed] [Google Scholar]
- Baranova IN, Vishnyakova TG, Bocharov AV, Kurlander R, Chen Z, Kimelman ML, Remaley AT, Csako G, Thomas F, Eggerman TL, Patterson AP. Serum amyloid A binding to CLA-1 (CD36 and LIMPII analogous-1) mediates serum amyloid A protein-induced activation of ERK1/2 and p38 mitogen-activated protein kinases. J Biol Chem. 2005;280:8031–8040. doi: 10.1074/jbc.M405009200. [DOI] [PubMed] [Google Scholar]
- Biran H, Friedman N, Neumann L, Pras M, Shainkin-Kestenbaum R. Serum amyloid A (SAA) variations in patients with cancer: Correlation with disease activity, stage, primary site, and prognosis. J Clin Pathol. 1986;39:794–797. doi: 10.1136/jcp.39.7.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechler C, Ritter M, Quoc CD, Agildere A, Schmitz G. Lipopolysaccharide inhibits the expression of the scavenger receptor Cla-1 in human monocytes and macrophages. Biochem Biophys Res Commun. 1999;262:251–254. doi: 10.1006/bbrc.1999.1193. [DOI] [PubMed] [Google Scholar]
- Cho WC, Yip TT, Yip C, Yip V, Thulasiraman V, Ngan RK, Yip TT, Lau WH, Au JS, Law SC, Cheng WW, et al. Identification of serum amyloid A protein as a potentially useful biomarker to monitor relapse of nasopharyngeal cancer by serum proteomic profiling. Clin Cancer Res. 2004;10:43–52. doi: 10.1158/1078-0432.ccr-0413-3. [DOI] [PubMed] [Google Scholar]
- Giner RM, Mancini L, Kamal AM, Perretti M. Uneven modulation of the annexin 1 system in osteoblast-like cells by dexamethasone. Biochem Biophys Res Commun. 2007;354:414–419. doi: 10.1016/j.bbrc.2006.12.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glojnaric I, Casl MT, Simic D, Lukac J. Serum amyloid A protein (SAA) in colorectal carcinoma. Clin Chem Lab Med. 2001;39:129–133. doi: 10.1515/CCLM.2001.022. [DOI] [PubMed] [Google Scholar]
- Gutfeld O, Prus D, Ackerman Z, Dishon S, Linke RP, Levin M, Urieli-Shoval S. Expression of serum amyloid A, in normal, dysplastic, and neoplastic human colonic mucosa: Implication for a role in colonic tumorigenesis. J Histochem Cytochem. 2006;54:63–73. doi: 10.1369/jhc.5A6645.2005. [DOI] [PubMed] [Google Scholar]
- Haynesworth SE, Baber MA, Caplan AI. Cell surface antigens on human marrow-derived mesenchymal cells are detected by monoclonal antibodies. Bone. 1992;13:69–80. doi: 10.1016/8756-3282(92)90363-2. [DOI] [PubMed] [Google Scholar]
- Hershkoviz R, Preciado-Patt L, Lider O, Fridkin M, Dastych J, Metcalfe DD, Mekori YA. Extracellular matrix-anchored serum amyloid A preferentially induces mast cell adhesion. Am J Physiol. 1997;273:C179–C187. doi: 10.1152/ajpcell.1997.273.1.C179. [DOI] [PubMed] [Google Scholar]
- Howard BA, Wang MZ, Campa MJ, Corro C, Fitzgerald MC, Patz EF., Jr Identification and validation of a potential lung cancer serum biomarker detected by matrix-assisted laser desorption/ionization-time of flight spectra analysis. Proteomics. 2003;3:1720–1724. doi: 10.1002/pmic.200300514. [DOI] [PubMed] [Google Scholar]
- Hrzenjak A, Artl A, Knipping G, Kostner G, Sattler W, Malle E. Silent mutations in secondary Shine–Dalgarno sequences in the cDNA of human serum amyloid A4 promotes expression of recombinant protein in Escherichia coli. Protein Eng. 2001;12:949–952. doi: 10.1093/protein/14.12.949. [DOI] [PubMed] [Google Scholar]
- Husebekk A, Skogen B, Husby G, Marhaug G. Transformation of amyloid precursor SAA to protein AA and incorporation in amyloid fibrils in vivo. Scand J Immunol. 1985;21:283–287. doi: 10.1111/j.1365-3083.1985.tb01431.x. [DOI] [PubMed] [Google Scholar]
- Jaiswal N, Haynesworth SE, Caplan AI, Bruder SP. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem. 1997;64:295–312. [PubMed] [Google Scholar]
- Kaneti J, Winikoff Y, Zimlichman S, Shainkin-Kestenbaum R. Importance of serum amyloid A (SAA) level in monitoring disease activity and response to therapy in patients with prostate cancer. Urol Res. 1984;12:239–241. doi: 10.1007/BF00256147. [DOI] [PubMed] [Google Scholar]
- Khan N, Cromer CJ, Campa M, Patz EF., Jr Clinical utility of serum amyloid A and macrophage migration inhibitory factor as serum biomarkers for the detection of nonsmall cell lung carcinoma. Cancer. 2004;101:379–384. doi: 10.1002/cncr.20377. [DOI] [PubMed] [Google Scholar]
- Kimura M, Tomita Y, Imai T, Saito T, Katagiri A, Ohara-Mikami Y, Matsudo T, Takahashi K. Significance of serum amyloid A on the prognosis in patients with renal cell carcinoma. Cancer. 2001;92:2072–2075. doi: 10.1002/1097-0142(20011015)92:8<2072::aid-cncr1547>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Kovacevic A, Hammer A, Sundl M, Pfister B, Hrzenjak A, Ray A, Ray BK, Sattler W, Malle E. Expression of serum amyloid A transcripts in human trophoblast and fetal-derived trophoblast-like choriocarcinoma cells. FEBS Lett. 2006;580:161–167. doi: 10.1016/j.febslet.2005.11.067. [DOI] [PubMed] [Google Scholar]
- Kulterer B, Friedl G, Jandrositz A, Sanchez-Cabo F, Prokesch A, Paar C, Scheideler M, Windhager R, Preisegger KH, Trajanoski Z. Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genomics. 2007;8:70. doi: 10.1186/1471-2164-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan Tat S, Padrines M, Theoleyre S, Heymann D, Fortun Y. IL-6, RANKL, TNF-alpha/IL-1: Interrelations in bone resorption pathophysiology. Cytokine Growth Factor Rev. 2004;15:49–60. doi: 10.1016/j.cytogfr.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Le L, Chi K, Tyldesley S, Flibotte S, Diamond DL, Kuzyk MA, Sadar MD. Identification of serum amyloid A as a biomarker to distinguish prostate cancer patients with bone lesions. Clin Chem. 2005;51:695–707. doi: 10.1373/clinchem.2004.041087. [DOI] [PubMed] [Google Scholar]
- Lee MS, Yoo SA, Cho CS, Suh PG, Kim WU, Ryu SH. Serum amyloid A binding to formyl peptide receptor-like 1 induces synovial hyperplasia and angiogenesis. J Immunol. 2006;177:5585–5594. doi: 10.4049/jimmunol.177.8.5585. [DOI] [PubMed] [Google Scholar]
- Li BQ, Wetzel MA, Mikovits JA, Henderson EE, Rogers TJ, Gong W, Le Y, Ruscetti FW, Wang JM. The synthetic peptide WKYMVm attenuates the function of the chemokine receptors CCR5 and CXCR4 through activation of formyl peptide receptor-like 1. Blood. 2001;97:2941–2947. doi: 10.1182/blood.v97.10.2941. [DOI] [PubMed] [Google Scholar]
- Li Y, Dang TA, Shen J, Perlaky L, Hicks J, Murray J, Meyer W, Chintagumpala M, Lau CC, Man TK. Identification of a plasma proteomic signature to distinguish pediatric osteosarcoma from benign osteochondroma. Proteomics. 2006;6:3426–3435. doi: 10.1002/pmic.200500472. [DOI] [PubMed] [Google Scholar]
- Malle E, De Beer FC. Human serum amyloid A (SAA) protein: A prominent acute-phase reactant for clinical practice. Eur J Clin Invest. 1996;26:427–435. doi: 10.1046/j.1365-2362.1996.159291.x. [DOI] [PubMed] [Google Scholar]
- Malle E, Steinmetz A, Raynes JG. Serum amyloid A (SAA): An acute phase protein and apolipoprotein. Atherosclerosis. 1993;102:131–146. doi: 10.1016/0021-9150(93)90155-n. [DOI] [PubMed] [Google Scholar]
- Malle E, Münscher G, Müller T, Vermeer H, Ibovnik A. Quantification and mapping of antigenic determinants of serum amyloid A (SAA) protein utilizing sequence-specific immunoglobulins and Eu3+ as a specific probe for time-resolved fluorometric immunoassay. J Immunol Methods. 1995;182:131–144. doi: 10.1016/0022-1759(95)00035-9. [DOI] [PubMed] [Google Scholar]
- Malle E, Leonhard B, Knipping G, Sattler W. Effects of cytokines, butyrate and dexamethasone on serum amyloid A and apolipoprotein A-I synthesis in human HUH-7 hepatoma cells. Scand J Immunol. 1999;50:183–187. doi: 10.1046/j.1365-3083.1999.00574.x. [DOI] [PubMed] [Google Scholar]
- Marsche G, Frank S, Raynes JG, Kozarsky KF, Sattler W, Malle E. The lipidation status of acute-phase protein serum amyloid A determines cholesterol mobilization via scavenger receptor class B type I. Biochem J. 2007;402:117–124. doi: 10.1042/BJ20061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M, Wallace HL, Sharma A, Hill HC, Sabel MS, Egilmez NK. A BALB/c murine lung alveolar carcinoma used to establish a surgical spontaneous metastasis model. Clin Exp Metastasis. 2004;21:363–369. doi: 10.1023/b:clin.0000046176.33867.c5. [DOI] [PubMed] [Google Scholar]
- Meek RL, Urieli-Shoval S, Benditt EP. Expression of apolipoprotein serum amyloid A mRNA in human atherosclerotic lesions and cultured vascular cells: Implications for serum amyloid A function. Proc Natl Acad Sci USA. 1994;91:3186–3190. doi: 10.1073/pnas.91.8.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita K, Kawabe Y, Tominaga M, Origuchi T, Aoyagi T, Eguchi K. Serum amyloid A protein induces production of matrix metalloproteinases by human synovial fibroblasts. Lab Invest. 1998;78:535–539. [PubMed] [Google Scholar]
- Nishie A, Masuda K, Otsubo M, Migita T, Tsuneyoshi M, Kohno K, Shuin T, Naito S, Ono M, Kuwano M. High expression of the Cap43 gene in infiltrating macrophages of human renal cell carcinomas. Clin Cancer Res. 2001;7:2145–2151. [PubMed] [Google Scholar]
- O’Hara R, Murphy EP, Whitehead AS, FitzGerald O, Bresnihan B. Local expression of the serum amyloid A and formyl peptide receptor-like 1 genes in synovial tissue is associated with matrix metalloproteinase production in patients with inflammatory arthritis. Arthritis Rheum. 2004;50:1788–1799. doi: 10.1002/art.20301. [DOI] [PubMed] [Google Scholar]
- Park YB, Park MJ, Kimura K, Shimizu K, Lee SH, Yokota J. Alterations in the INK4a/ARF locus and their effects on the growth of human osteosarcoma cell lines. Cancer Genet Cytogenet. 2002;133:105–111. doi: 10.1016/s0165-4608(01)00575-1. [DOI] [PubMed] [Google Scholar]
- Patel H, Fellowes R, Coade S, Woo P. Human serum amyloid A has cytokine-like properties. Scand J Immunol. 1998;48:410–418. doi: 10.1046/j.1365-3083.1998.00394.x. [DOI] [PubMed] [Google Scholar]
- Pautke C, Schieker M, Tischer T, Kolk A, Neth P, Mutschler W, Milz S. Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 OS in comparison to human osteoblasts. Anticancer Res. 2004;24:3743–3748. [PubMed] [Google Scholar]
- Ray BK, Murphy R, Ray P, Ray A. SAF-2, a splice variant of SAF-1, acts as a negative regulator of transcription. J Biol Chem. 2002;277:46822–46830. doi: 10.1074/jbc.M206299200. [DOI] [PubMed] [Google Scholar]
- Ray A, Kumar D, Ray P, Ray BK. Transcriptional activity of serum amyloid A-activating factor-1 is regulated by distinct functional modules. J Biol Chem. 2004a;279:54637–54646. doi: 10.1074/jbc.M411830200. [DOI] [PubMed] [Google Scholar]
- Ray A, Shakya A, Kumar D, Ray BK. Overexpression of serum amyloid A-activating factor 1 inhibits cell proliferation by the induction of cyclin-dependent protein kinase inhibitor p21WAF-1/Cip-1/Sdi-1 expression. J Immunol. 2004b;172:5006–5015. doi: 10.4049/jimmunol.172.8.5006. [DOI] [PubMed] [Google Scholar]
- Ray A, Bal BS, Ray BK. Transcriptional induction of matrix metalloproteinase-9 in the chondrocyte and synoviocyte cells is regulated via a novel mechanism: Evidence for functional cooperation between serum amyloid A-activating factor-1 and AP-1. J Immunol. 2005;175:4039–4048. doi: 10.4049/jimmunol.175.6.4039. [DOI] [PubMed] [Google Scholar]
- Rosenthal CJ, Sullivan LM. Serum amyloid A to monitor cancer dissemination. Ann Intern Med. 1979;91:383–390. doi: 10.7326/0003-4819-91-3-383. [DOI] [PubMed] [Google Scholar]
- Sevetson B, Taylor S, Pan Y. Cbfa1/RUNX2 directs specific expression of the sclerosteosis gene (SOST) J Biol Chem. 2004;279:13849–13858. doi: 10.1074/jbc.M306249200. [DOI] [PubMed] [Google Scholar]
- Shainkin-Kestenbaum R, Berlyne G, Zimlichman S, Sorin HR, Nyska M, Danon A. Acute phase protein, serum amyloid A, inhibits IL-1- and TNF-induced fever and hypothalamic PGE2 in mice. Scand J Immunol. 1991;34:179–183. doi: 10.1111/j.1365-3083.1991.tb01535.x. [DOI] [PubMed] [Google Scholar]
- Stix B, Kahne T, Sletten K, Raynes J, Roessner A, Röcken C. Proteolysis of AA amyloid fibril proteins by matrix metalloproteinases-1, -2, and -3. Am J Pathol. 2001;159:561–570. doi: 10.1016/S0002-9440(10)61727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- Uhlar CM, Burgess CJ, Sharp PM, Whitehead AS. Evolution of the serum amyloid A (SAA) protein superfamily. Genomics. 1994;19:228–235. doi: 10.1006/geno.1994.1052. [DOI] [PubMed] [Google Scholar]
- Vallon R, Freuler F, Desta-Tsedu N, Robeva A, Dawson J, Wenner P, Engelhardt P, Boes L, Schnyder J, Tschopp C, Urfer R, et al. Serum amyloid A (apoSAA) expression is up-regulated in rheumatoid arthritis and induces transcription of matrix metalloproteinases. J Immunol. 2001;166:2801–2807. doi: 10.4049/jimmunol.166.4.2801. [DOI] [PubMed] [Google Scholar]
- Vermes C, Jacobs JJ, Zhang J, Firneisz G, Roebuck KA, Glant TT. Shedding of the interleukin-6 (IL-6) receptor (gp80) determines the ability of IL-6 to induce gp130 phosphorylation in human osteoblasts. J Biol Chem. 2002;277:16879–81687. doi: 10.1074/jbc.M200546200. [DOI] [PubMed] [Google Scholar]
- Wadsack C, Hrzenjak A, Hammer A, Hirschmugl B, Levak-Frank S, Desoye G, Sattler W, Malle E. Trophoblast-like human choriocarcinoma cells serve as a suitable in vitro model for selective cholesteryl ester uptake from high density lipoproteins. Eur J Biochem. 2003;270:451–462. doi: 10.1046/j.1432-1033.2003.03394.x. [DOI] [PubMed] [Google Scholar]
- Weinstein PS, Skinner M, Sipe JD, Lokich JJ, Zamcheck N, Cohen AS. Acute-phase proteins or tumour markers: The role of SAA, SAP, CRP and CEA as indicators of metastasis in a broad spectrum of neoplastic diseases. Scand J Immunol. 1984;19:193–198. doi: 10.1111/j.1365-3083.1984.tb00919.x. [DOI] [PubMed] [Google Scholar]
- Zerega B, Pagano A, Pianezzi A, Ulivi V, Camardella L, Cancedda R, Cancedda FD. Expression of serum amyloid A in chondrocytes and myoblasts differentiation and inflammation: Possible role in cholesterol homeostasis. Matrix Biol. 2004;23:35–46. doi: 10.1016/j.matbio.2004.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.