Abstract
Sulfoglycolipids are present on the surface of a variety of cells. The sulfatide SM4s is increased in lung, renal, and colon cancer and is associated with an adverse prognosis, possibly due to a low immunoreactivity of the tumor. As macrophages significantly contribute to the inflammatory infiltrate in malignancies, we postulated that SM4s may modulate macrophage function. We have investigated the effect of SM4s on the uptake of apoptotic tumor cells, macrophage cytokine profile, and receptor expression. Using flow cytometry and microscopic analyses, we found that coating apoptotic murine carcinoma cells from the colon and kidney with SM4s promoted their phagocytosis by murine macrophages up to 3-fold ex vivo and in vivo. This increased capacity was specifically inhibited by preincubation of macrophages with oxidized or acetylated low density lipoprotein and maleylated albumin, indicating involvement of scavenger receptors in this interaction. The uptake of SM4s-coated apoptotic cells significantly enhanced macrophage production of TGF-β1, expression of P-selectin, and secretion of IL-6. These data suggest that SM4s within tumors may promote apoptotic cell removal and alter the phenotype of tumor-associated macrophages.
Sulfatides are anionic and amphiphilic glycosphingolipids (GSLs)3 found in the extracellular lipid leaflet of the cell plasma membrane. GSLs are thought to participate in cell-cell and cell-matrix contacts. Sulfated galactosylceramide (Galactosylsulfatide, SM4s; Sulfatide) and sulfated galactosylalkylacylglycerol (Seminolipid, SM4g) are two major sulfoglycolipids found in mammals. SM4g is restricted to the testis where it is required for proper spermatogenesis (1). It constitutes >90% of glycolipids in spermatozoa and probably plays an important role in sperm/egg recognition (2–4). SM4s is abundantly present in the brain (being the major glycolipid of the myelin sheath) and is also found in high concentrations in glandular epithelial tissue (e.g., colon), in kidney, and in islets of Langerhans (5). Other sulfated GSLs that can be isolated in smaller quantities from the kidney include complex sulfatides such as lactosylsulfatide (SM3) (2, 3). Sulfation of glycolipid terminal galactosyl residues in 3-position is catalyzed by the enzyme cerebrosulfotransferase. SM4s concentration has been reported to be markedly increased in some cancer tissue (e.g., human renal cell carcinoma, RCC, adenocarcinoma of colon and lung) (6–9). The expression of SM4s in these tumors has been positively correlated with the occurrence of lymph node metastases and advanced disease (9). Sulfatides have been shown to bind a variety of cell surface and extracellular proteins (10–15). They may also play an important role as adhesion molecules, acting as ligands to P- and L-selectin (16–19). P-selectin expressed on tumor cells has been proposed as a “receptor in charge” of enhanced metastatic phenotype in patients with increased cancer-associated SM4s (20). Nevertheless, precise mechanisms that give insight into the pathophysiological functions and relevance of elevated SM4s in cancer tissue still remain enigmatic.
Apoptosis is a cell death modus found in both malignancy and inflammation. Apoptotic cells are rapidly ingested by phagocytes, principally macrophages, and this mechanism has been shown to inhibit macrophage proinflammatory activities (21). There is evidence that the interaction between a number of receptors expressed by professional phagocytes and their ligands exposed on apoptotic cells regulates recognition and removal of apoptotic “waste” material (22, 23). Phosphatidylserine (PS) expression on the surface of apoptotic cells is assumed to be the key “eat me” signal and the promoter of the apoptotic cell clearance (24–27). Macrophage receptors known to take part in the uptake of apoptotic particles include scavenger receptors (SRs), phosphatidylserine receptor (PSR), CD14, and the vitronectin receptor (αVβ3 integrin) (28–31). Class A and B SRs were originally reported to be involved in receptor-mediated endocytosis of selected polyanionic ligands, including modified lipoproteins (32–37). Sulfatides can bind to SRs (38), which may indicate that these molecules could function as phagocytic ligands on dying cells.
Several functional phenotypes of macrophages have been described (39). Apoptotic cell uptake has been shown to promote alternative activation of macrophages characterized by high secretion of IL-10, synthesis of arginase-1, IL-1 receptor antagonist, CCL22, up-regulation of SRs, mannose receptor, and galactose receptor. This phenotype (known as M2) is thought to promote angiogenesis and tumor growth. This is in contrast to the macrophage M1 phenotype, defined by high release of IL-12, IL-23, TNF-α, IL-1, CXCL1, reactive oxygen, and nitrogen intermediates, which is associated with a Th1 response and resistance to a tumor. Moreover, ingestion of apoptotic cells is known to induce TGF-β1 secretion by macrophages, resulting in an anti-inflammatory effect (40). Thus, the host immune response to a tumor may be influenced by clearance of apoptotic cells by macrophages (41–44).
The aim of our study was to evaluate the role of SM4s in apoptotic cell uptake and macrophage function. We hypothesized that SM4s enhances clearance of late apoptotic bodies by tissue macrophages and modulates macrophage cytokine signature. Our results showed enhanced uptake of SM4s-painted apoptotic cells by murine peritoneal macrophages.
The phenomenon was principally dependent on the sulfate group of SM4s and its recognition by macrophage SRs. SM4s-dependent increase in uptake of apoptotic cells resulted in enhanced TGF-β1 generation, IL-6 secretion, and P-selectin expression by macrophages.
Materials and Methods
Reagents
RPMI 1640 medium, Dulbecco’s PBS, HEPES solution, DMSO, propidium iodide (PI), phosphatidic acid (l-α-phosphatidic acid sodium salt), cholesterol sulfate (cholesterol 3-sulfate sodium salt), brewer thioglycollate medium, O-phospho-l-serine (soluble PS), LPS from Salmonella minnesota, and BSA were from Sigma-Aldrich. DMEM, trypsin, penicillin streptomycin, l-glutamine, FITC-conjugated annexin V, vibrant DiO solution, AlexaFluor 546 phalloidin, AlexaFluor 488 donkey anti-mouse IgG (H+L) mAb, and fluorescent-labeled galactosylceramide (BODIPY-FL C12-galactocerebroside) were from Invitrogen Life Technologies. FBS was from Biochrom. Accutase was from PAA Laboratories. Staurosporine was from Alomone Laboratories. SM4s (sulfatides from bovine brain) and galactosylceramide (cerebrosides phrenosin; galactosyl ceramide with mostly 2-hydroxy fatty acid side chains) were from Matreya. Ceramide-1-phosphate was from Avanti Polar Lipids. R-PE- and FITC-conjugated rat anti-mouse CD11b (M1/70) mAbs, purified rat anti-mouse CD16/CD32 mAb (2.4G2), FITC-conjugated rat anti-mouse CD62P mAb (RB40.34), mouse IL-6, TNF, GM-CSF, IL-12 (p70), IL-4, IL-10, and IFN-γ BD OptEIA ELISA sets, BD GolgiStop, Cytofix/Cytoperm solution and perm/wash buffer were from BD Biosciences. Anti-TGF-β R-PE Ab was purchased from IQ Products. Mouse TGF-β1 ELISA DuoSet was from R&D Systems. Mouse MIP-1α ELISA kit was from RayBiotech. Anti-sulfatide (anti-O4; oligodendrocyte marker) mAb (clone 81) was obtained from Chemicon. DRAQ5 was from Alexis. AffiniPure goat anti-mouse IgG was from Jackson ImmunoResearch Laboratories. HRP streptavidin, 3-amino-9-ethylcarbazole substrate kit for peroxidase, and hematoxylin were from Vector Laboratories. Goat-anti-mouse Ab gold particle conjugate (6 nm) and Aurion R-GENT SE-EM were purchased from Aurion ImmunoGold. SYTOX green nucleic acid stain was from Cambrex Bio Science.
Detection of SM4s in human renal and colon tissue
Presence of SM4s in human tissue was analyzed by immunohistochemistry (IHC) and thin-layer chromatography (TLC). Tissue samples from RCC patients (n = 4), colorectal adenocarcinoma patients (n = 4), and samples from noncancerous tissue parts of same patients were analyzed. For IHC, frozen samples were cut to 5-µm sections. Anti-O4 mAb followed by biotin-SP-conjugated AffiniPure goat anti-mouse IgG as secondary Ab, HRP streptavidin, and 3-amino-9-ethylcarbazole substrate for peroxidase were used as described (45). The sections were counterstained with hematoxylin. For TLC, the extraction and purification of glycosphingolipids from normal and tumor tissue was performed as described by Sandhoff et al. (46). In brief, the total lipid fraction was extracted from lyophilized tissue using solutions of chloroform/methanol/water, treated with 0.1 M KOH/methanol to saponify acylglycerolipids, desalted with RP-18 columns (Waters Associates) and separated into neutral and acidic glycolipid fractions using DEAE-Sephadex A25 column chromatography (Pharmacia Fine Chemicals). The acidic fraction was then desalted again and subjected to TLC analysis using HPTLC-Si60 plates (Merck). Samples were normalized to 1 mg of total protein. For separation of sulfated glycolipds by TLC, chloroform/methanol/water (60/35/8, v/v/v) was used. Bands were visualized by spraying the plates with orcinol/sulfuric acid solution and heating at 110°C for 10 min. Bands were identified in comparison with standard GSLs. Standard gangliosides and SM4s were obtained from Matreya. Standard complex sulfatides SM2, SM3, and SB1a were purified as described (46).
Mice
BALB/c mice (female, 10–12 wk old) were purchased from Charles River Laboratories. All mice were housed in a specific pathogen-free facility at the German Cancer Research Center. All mice experiments were approved by federal authorities (Regierungspraesidium Karlsruhe).
Cells
The C26 (derived from BALB/c Colon26 murine colon adenocarcinoma) and Renca (renal cell carcinoma of BALB/c origin) cell lines were provided by the tumor-bank of the German Cancer Research Center (Heidelberg, Germany) and cultured in DMEM medium with high Glucose-4500 mg/L (C26 cells) or RPMI 1640 medium (Renca cells). Media were supplemented with 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 10% (v/v) heat-inactivated FBS, and 1% (v/v) HEPES buffer (C26 cells). Thioglycollate-elicited murine peritoneal macrophages were isolated by peritoneal lavage 3 days after intraperitoneal injection of 1 ml of 4% (v/v) Brewer thioglycollate medium. Aspirated peritoneal cells were plated in RPMI 1640 medium supplemented with 2 mM l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 10% (v/v) heat-inactivated FBS, and 1% (v/v) HEPES buffer. Macrophages were washed 2 h after plating, fed with fresh medium, left in culture overnight, washed, and used in different assays (47–51). All cells were maintained in a humidified atmosphere of 5% CO2 at 37°C.
Generation and labeling of late apoptotic cells
C26 and Renca cells were grown to late-log phase. To induce apoptosis, the cells were treated with staurosporine (1 µM for 4 h and 24 h) or cycloheximide (10 µg/ml for 16 h) (52, 53). Early and late apoptosis were verified by both light microscopy and flow cytometry for FITC-labeled annexin V/PI. The presence of nuclear condensation and/or fragmentation along with both annexin V and PI positivity was considered as late apoptosis and the absence of membrane permeability (negative PI test) together with Annexin V-positivity was recognized as early apoptosis (54). Apoptotic C26 and Renca cells were suspended at a density of 1 × 106/ml in serum-free RPMI 1640 medium. For sulfatide concentration gradient experiments and biochemical control experiments, C26 cells were incubated with increasing concentrations of SM4s (1, 2, 5, 10, 25, and 50 µM), appropriate controls (galactosylceramide, cholesterol sulfate, ceramide-1-phosphate, or SM4g) or vehicle (DMSO) for 1 h at 37°C. For the analysis of Renca apoptotic cell uptake, 10 µM SM4s was applied for 1 h at 37°C. For the experiment with viable C26 cells, the cells were washed with PBS, detached using trypsin solution (5 mg/ml porcine trypsin and 2 mg/ml EDTA in PBS), resuspended in serum-containing medium, washed again, and incubated in serum-free medium with SM4s or DMSO, as described above. Before incubation with macrophages, the cells were washed three times in serum-free medium. For flow cytometry and confocal microscopy phagocytosis assays, DiO cell-labeling solution was added at 5 µl per ml of cell suspension and mixed by gentle pipetting. For video microscopy assay, apoptotic cells were labeled with Sytox Green at a concentration of 0.5 µM. After incubation at 37°C for 15 min, the suspensions were washed three times with warm medium (37°C). In all experiments, apoptotic cells were resuspended in RPMI 1640 serum-containing medium before coincubation with macrophages at a ratio of 1:5 or 1:10 (macrophages:apoptotic cells).
Detection of SM4s, SM4g, galactosylceramide, ceramide-1-phosphate, and cholesterol sulfate in cells
Apoptotic C26 or Renca cells, previously incubated with different concentrations of SM4s, controls, or vehicle, were collected and incorporation of applied compounds was evaluated using the following techniques: binding of SM4s to the cells was analyzed by immunocytochemistry, confocal microscopy, immune electron microscopy and TLC. Coupling of SM4g, ceramide-1-phosphate, and cholesterol sulfate to apoptotic cells was proven by TLC. Binding of galactosylceramide to the C26 plasma membrane was assessed by flow cytometry. For immunocytochemistry, confocal, and immune electron microscopy experiments, the cells were spun down (Shandon Cytospin 3) and stained using mouse anti-O4 mAb. AlexaFluor 488 donkey anti-mouse IgG (H+L) and gold-coupled goat-anti-mouse Abs with silver enhancement reagent (Aurion R-GENT SE-EM) were used for SM4s visualization by confocal and electron microscopy, respectively. The percentage of SM4s-positive cells was determined from confocal photomicrographs in three randomly chosen fields (magnification 400 ×). Immunocytochemistry of apoptotic cells was done as described above. For immune electron microscopy, the cells were grown on cover slips in 24-well plates, fixed in 8% (v/v) paraformaldehyde solution, incubated with indicated Abs, and embedded in araldite. Ultrathin sections were cut with Leica UCT ultramicrotome to 70–80 nm and stained with uranyl acetate (Leica UCT). The samples were viewed using a Zeiss 900 electron microscope and a SlowScan CCD camera (Zeiss). Analysis was performed using Olympus Soft Imaging software (Olympus Soft Imaging Solutions). Isolation of glycolipids from SM4s-, SM4g-, cholesterol sulfate-, and ceramide-1-phosphate-treated and untreated viable and apoptotic cells for TLC was performed essentially as described (46). Glycolipids were isolated in a single extraction step with chloroform/methanol/water 10/10/1 (v/v/v) using an ultrasound water bath at 50°C for 30 min. Aliquots of the acidic glycolipid fraction corresponding to 0.1 mg total protein were spotted on silica high performance thin layer chromatography plates. SM4s and SM4g were developed in a saturated chromatography chamber with chloroform/methanol/0.2% Ca++ in water 60/35/8 (v/v/v) and visualized with orcinol/H2SO4 spray-reagent as described. Ceramide-1-phosphate and cholesterol sulfate containing extracts were developed in chloroform/methanol/water 65/25/4 (v/v/v) and lipids were visualized with 10% CuSO4 in 8% H3PO4 at 160°C for 10 min. For assessment of galactosylceramide binding by flow cytometry, apoptotic C26 cells were incubated with fluorescent-labeled (BODIPY-FL) galacto-sylceramide and analyzed by FACS as described.
Lipoprotein isolation and modification: preparation of anti-SR-A mAb, SM4g extraction
Native low density lipoprotein (nLDL, d = 1.035 to 1.065 g/ml) was isolated by ultracentrifugation as described (55). Modification of nLDL was performed with copper (OxLDL) or sodium acetate/acetic anhydride (AcLDL) as described (55). Maleyl-BSA (MBSA) was prepared as stated by Butler et al. (56). Protein and cholesterol content as well as electrophoretic mobility of nLDL and modified LDL preparations were measured. Endotoxin levels were determined using the gel-clot method (Charles River Endosafe) and were detected to be <0.2 ng/mg for all (lipo)protein preparations used. Anti-SR-A mAb was prepared according to Fraser et al. (57). SM4g extraction from rat testes was done as indicated by Fujimoto et al. (1).
Phagocytosis assays by flow cytometry
For the ex vivo assay, murine peritoneal macrophages were washed with serum-free RPMI 1640 medium and seeded (1 × 106 per well) in 24-well tissue culture plates before coincubation with 5 × 106 apoptotic or viable cells at 37°C. In experiments with different apoptotic cell ratios, 5 × 106 and 10 × 106 of late apoptotic C26 cells were incubated with the indicated number of macrophages. After 60 min, noningested C26 or Renca cells were washed off, macrophages were detached using Accutase and were resuspended in cold PBS for further analysis. In receptor-blocking experiments, before incubation with apoptotic C26 cells, macrophages were incubated with OxLDL, AcLDL, nLDL, BSA, MBSA, soluble PS (all 100 µg/ml), anti-SR-A mAb (2F8; 1 µg/1 × 106 cells/100 µl), or vehicle for 30 min at the standard cell culture conditions (58–60). For the in vivo assay, apoptotic C26 cells were resuspended at a concentration of 5 × 106 per ml in serum-containing RPMI 1640 (37°C) and injected i.p. in a volume of 3 ml. Macrophages were harvested 1 h after injection, as described above. In both assays, macrophage Fc receptors were blocked with specific mAb, washed, incubated with anti-mouse CD11b PE according to the manufacturer’s protocol, and analyzed by flow cytometry (FACSCalibur, BD Biosciences). CD11b positive cells were considered as macrophages, gated, and analyzed for the intensity of DiO fluorescence as a parameter of phagocytosis (47–51). The FACS phagocytosis index was calculated by dividing mean relative DiO fluorescence intensities of macrophages and apoptotic bodies from the same experiment.
Confocal, electron, and video microscopy phagocytosis assays
Macrophages were incubated on cover slips in 6-well culture plates overnight. For confocal and video microscopy experiments, DiO or SYTOX green-labeled apoptotic C26 cells were added, respectively. In the confocal microscopy assay, macrophages were washed and subjected to fluorescent staining to actin (Alexa 546 phalloidin) and nucleus (DRAQ5) after 1 h of coincubation. Image acquisition and analysis was done using a Leica TCS-SL microscope and Leica confocal software (v. 2.61). For the video microscopy experiment, cover slips with macrophages and apoptotic C26 cells were positioned in an imaging chamber with standard medium and cell culture conditions (serum-containing RPMI 1640, 37°C, 5% CO2) for 2 h. Movies were acquired on a Zeiss Axiovert 200M microscope by AxioCam camera using AxioVision Rel4.5 software (Zeiss). For the electron microscopy experiment, macrophages were fixed in Karnovsky solution and embedded in araldite (61). Samples were prepared and analyzed as described above. In total, 80 macrophages were analyzed.
Cytokines
Macrophages, plated at a density of 2 × 106 cells per well in 6-well culture plates, were washed twice with PBS and cocultured with apoptotic C26 cells (10 × 106 per well, corresponding to a 1:5 ratio of macrophages: apoptotic cells), apoptotic C26 cells preincubated with SM4s (10 µM), vehicle, or LPS (1 µg/ml). In addition, macrophages were preincubated with SM4s for 1 h and then cocultured with C26 apoptotic cells. Supernatants were harvested at 2 and 12 h after start of coculture, cleared from remaining particles by centrifugation, and frozen at –80°C until being analyzed by ELISA for active TGF-β1, IL-6, TNF-α, GM-CSF, IL-12 (p70), IL-4, IL-10, IFN-γ, and MIP-1α. Immunofluorescent staining of intracellular TGF-β1 for flow cytometry analysis was performed according to standard manufacturers’ protocols (BD Biosciences and IQ Products).
Statistical analyses
All data are reported as mean ± SEM. Statistical analyses were performed using GraphPad Prism v2.01, CellQuest v3.3, BD CellQuest Pro v.5.2.1, and FCAP Array v1.0.1. softwares.
Results
Human RCC and colorectal adenocarcinoma contain sulfatides
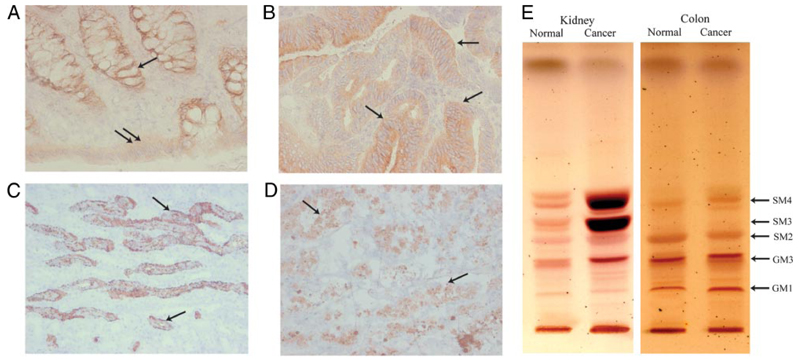
Frozen samples from noncancerous and cancerous human kidney and colon tissue were examined for the presence of sulfatides. IHC revealed abundant sulfatide in the following specimen ratios: noncancerous colon 4:4 (Fig. 1A); colon cancer 2:4 (Fig. 1B); noncancerous kidney 4:4 (Fig. 1C) and RCC 4:4 (Fig. 1D). Epithelial cells showed positivity for sulfatides. The invasive colon cancer cells and RCC cells demonstrated the strongest reactivity to sulfatide Abs (Fig. 1, B and D). TLC of sulfatides isolated form the tissue confirmed immunohistochemical observations that SM4 (kidney and colon) and SM3 (kidney) were increased in cancerous in comparison to noncancerous tissue from the same patient (Fig. 1E).
FIGURE 1. Sulfatide can be detected in both normal and cancerous human kidney and colon tissue.
A–D, Frozen samples from human renal and colon tissue were analyzed by IHC. A, Light micrograph showing sulfatide staining in basal crypt epithelium (arrow) and surface epithelium (double arrow) of normal colon (Leica, ×400). B, Colon cancer tissue showing strong intraepithelial (arrows) positivity for sulfatide (Leica, ×200). C, Light micrograph illustrating cortex of normal kidney. Collecting duct epithelium (arrows) shows presence of sulfatide. (Leica, ×200). D, Sulfatide expression (arrows) in renal cell cancer tissue, with positive apoptotic cells (Leica, ×200). E, TLC of acidic glycosphingolipid species. Normal and cancerous tissue of human kidney and colon were analyzed.
Apoptosis and painting of C26 and Renca cells with SM4s
To induce a homogeneous and defined stage of apoptosis, staurosporine (a protein kinase C inhibitor) and cycloheximide (a protein translation inhibitor) were used at concentrations of 1 µM and 10 µg/ml, respectively, for different time periods. Late apoptosis in C26 and Renca cells was verified by Annexin V/PI flow cytometry assay and confirmed by quantitative ultrastructural analysis of apoptotic cells by electron microscopy. Consistent results were obtained at time points of 24 h (staurosporine) and 16 h (cycloheximide) after the start of induction, as demonstrated by both PS exposure on the outer layer of plasma membrane and complete membrane permeability in nearly all collected cells (Fig. 2A) along with nuclear condensation and/or fragmentation (Fig. 2B). Early apoptosis of C26 cells was achieved by staurosporine induction (10 µM) for 4 h and confirmed by exposure of PS on the cell surface without increase in membrane permeability.
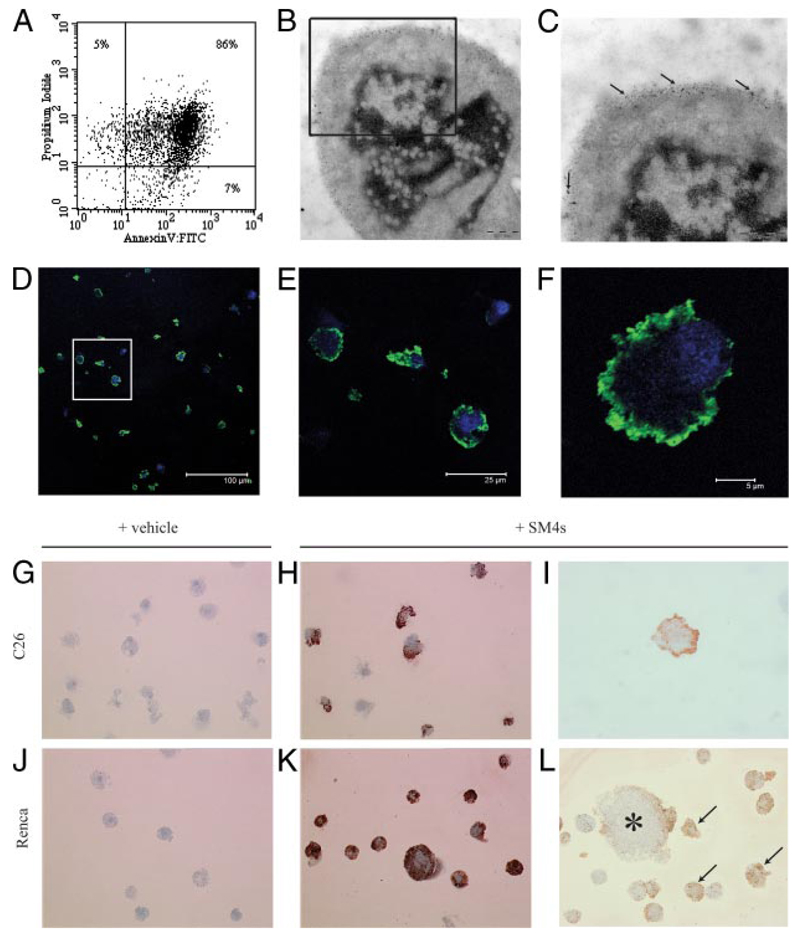
FIGURE 2. C26 and Renca cells demonstrate late apoptotic phenotype after induction with staurosporine and successful SM4s painting after exposure to SM4s.
A, C26 cells were incubated with staurosporine (1 µM) for 24 h. PS exposure and membrane permeability were detected by flow cytometry using FITC-conjugated annexin V and PI, respectively. Annexin V/PI double-positive cells were considered as late apoptotic. B, Electron photomicrograph of apoptotic C26 cell with evident nuclear condensation and fragmentation (Zeiss, ×30000). B and C, Electron photomicrographs of apoptotic C26 cells preincubated with 10 µM SM4s. Gold particle-conjugated goat anti-mouse Ab (particle size of 6 nm) was used for visualization of the sulfatide (arrows). D–F, Apoptotic C26 cells were prepared as described above, spun down, stained with DRAQ5 (nucleus), and immunostained with anti-O4 and AlexaFluor 488 donkey anti-mouse IgG (H+L) mAbs (SM4s). F, Confocal image of a single cell shows predominant membrane localization of the sulfatide (×5000). Data are presented as merge of green and blue channels. G–L, Apoptotic cells subjected to immunocytochemistry analysis with anti-O4 mAb. Photomicrographs of apoptotic cells treated with sulfatide or vehicle (DMSO). G and J, C26 and Renca control cells (Leica, ×400). H and I, C26 cells treated with 10 µM SM4s (Leica, ×400 and ×1000, respectively). K, Late apoptotic Renca cells painted with sulfatide (Leica, ×400). L, Binding of SM4s to plasma membrane of late apoptotic (arrows) and early apoptotic (star) Renca cells (Leica, ×400).
To determine presence of SM4s on the apoptotic C26 cells after incubation with different concentrations of SM4s, immunocytochemistry, confocal imaging, and immune electron microscopy of control and SM4s-painted apoptotic cells were performed (Fig. 2, B–L). SM4s was not detected in control apoptotic C26 or Renca cells (Fig. 2, G and J). A strong saturated SM4s label was achieved in apoptotic particles at a concentration of 10 µM (Fig. 2, D–F, H, and K). At this concentration, >90% of cells showed painting by sulfatide, as analyzed by confocal microscopy (Fig. 2E). In addition, TLC analysis confirmed the successful binding of SM4s to late apoptotic C26 cells (Fig. 3A). SM4s also effectively bound to late apoptotic Renca cells and early apoptotic C26 cells (Fig. 2L; Fig. 3, A and B). Binding of sulfatide to viable C26 cells was evident, but distinctly less efficient than in apoptotic cells (Fig. 3B). Immune electron microscopy and confocal imaging analyses of apoptotic C26 cells incubated with SM4s revealed pronounced plasma membrane localization of the sulfatide in apoptotic cells (Fig. 2, B and C).
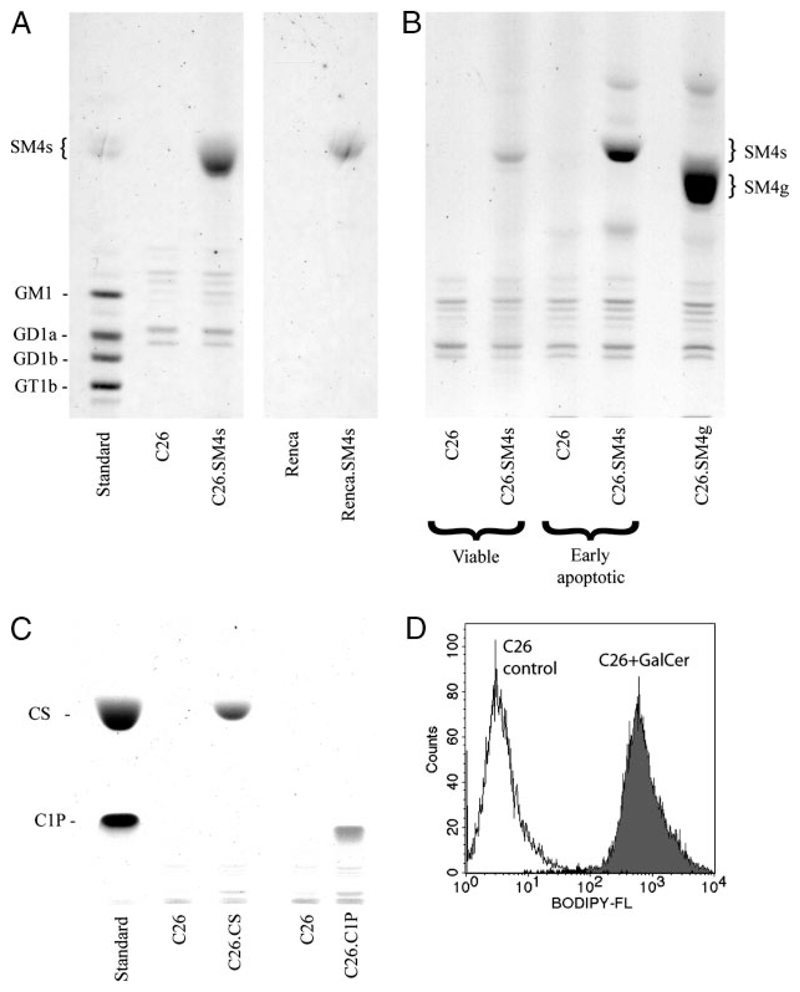
FIGURE 3. TLC of the cells treated with SM4s or controls.
Cells were incubated with indicated concentrations of SM4s, SM4g, cholesterol sulfate (CS), or ceramide-1-phosphate (C1P) and subjected to thin layer chromatography. A, Chromatograms of lipids from late apoptotic C26 (left) and Renca (right) cells treated with SM4s or vehicle. B, Binding of sulfatide to early apoptotic and viable C26 cells and binding of seminolipid to late apoptotic cells. C, Chromatogram of lipids from late apoptotic C26 cells treated with CS and C1P. D, Late apoptotic C26 cells were incubated with fluorescent galactosylceramide (BODIPY-FL C12-galactocerebroside) for 1 h at 37°C and analyzed by flow cytometry. Histogram shows binding of galactosylceramide (GalCer) to the apoptotic cell.
Painting of late apoptotic C26 cells with galactosylceramide, SM4g, ceramide-1-phosphate, and cholesterol sulfate
TLC of late apoptotic C26 cells incubated with SM4g, ceramide-1-phosphate, and cholesterol sulfate showed effective binding of these compounds to the cells (Fig. 3B, C). Strong coupling of galactosylceramide with apoptotic C26 plasma membrane was confirmed by flow cytometry (Fig. 3D).
Effect of SM4s on the uptake of late apoptotic cells by macrophages
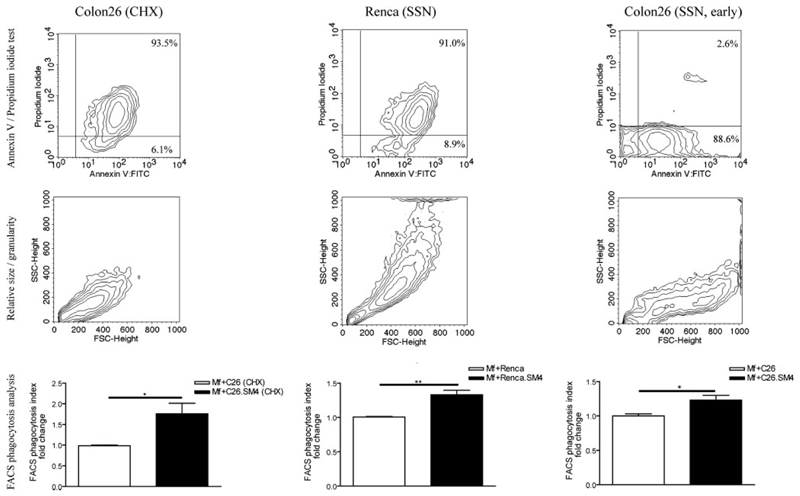
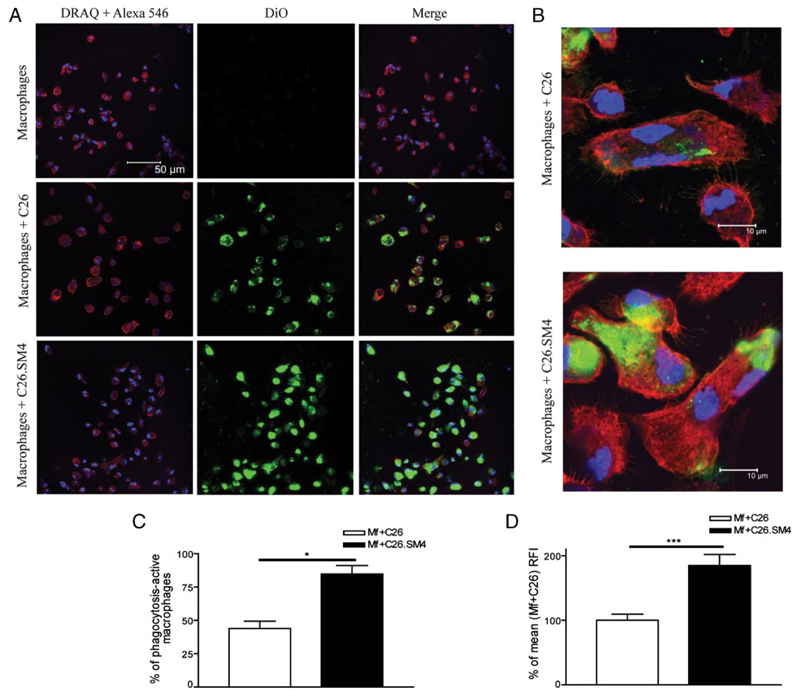
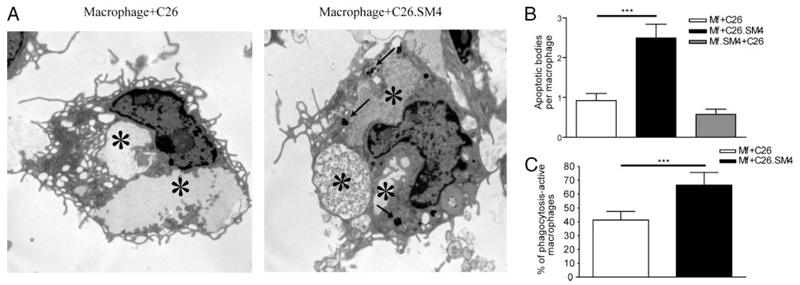
As analyzed by flow cytometry, incubation of fluorescently labeled apoptotic cells with macrophages resulted in formation of a double-positive (CD11b+/DiO+) population that represented macrophages with tethered and/or ingested apoptotic cells (Fig. 4). Results obtained from the ex vivo experiments showed that painting of apoptotic cell membranes with SM4s significantly increased the FACS phagocytosis index (Fig. 4, A–C). The increased uptake was evident and significant for apoptotic cells previously exposed to SM4s at concentrations of 10 µM or higher. The effect was not significantly enhanced with higher (1:10) concentration of apoptotic cells (3.258 ± 0.647 vs 3.660 ± 0.619, p = 0.1236; data not shown). The experiments with late apoptotic C26 cells induced by cycloheximide and late apoptotic Renca cells also showed a significant difference in the uptake between SM4s-free and SM4s-positive cells (1.01 ± 0.01 vs 1.33 ± 0.06, p = 0.0012; and 0.99 ± 0.02 vs 1.76 ± 0.25, p = 0.0201, respectively) (Fig. 5). We could observe a similar effect in the case of early apoptotic C26 cell uptake, as measured by fold increase in FACS phagocytosis index (1.00 ± 0.03 vs 1.23 ± 0.07, p = 0.0266) (Fig. 5). The in vivo experiments confirmed a significant increase in phagocytosis of apoptotic C26 cells painted with SM4s, by flow cytometry (Fig. 4, D–F). The in vitro video microscopy assay showed that most of the uptake process was completed at 1 h after the start of coincubation.4 Confocal microscopy was used to confirm these results and to distinguish between tethering and internalisation of apoptotic cells. Both percent of DiO positive macrophages (43.93 ± 5.58 vs 84.75 ± 6.37; p = 0.0286) and percent change of mean DiO fluorescence intensity per macrophage (100 ± 9.39 vs 184.8 ± 17.08; p = 0.0002) were again found to be strongly increased in the population ingesting SM4s-positive apoptotic cells (Fig. 6, A–D). Electron microscopy revealed that the number of intraphagosomal apoptotic particles per macrophage was increased up to 3-fold in macrophages coincubated with SM4s-positive apoptotic cells. We also detected a significant difference in the percent of apoptotic particle-containing macrophages between the two groups (41.32 ± 6.18% vs 66.59 ± 9.09%, p < 0.0001) (Fig. 7A–C).
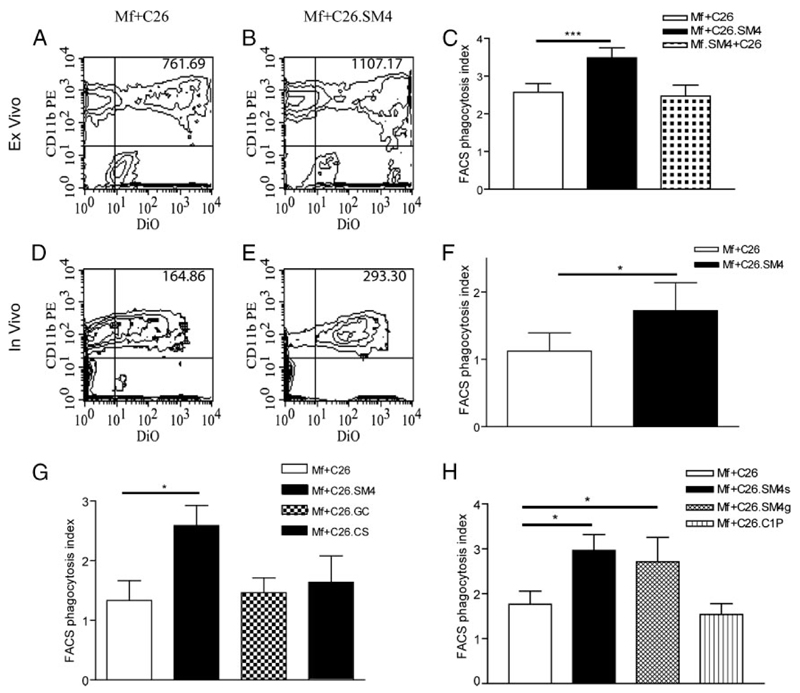
FIGURE 4. Effect of SM4s on phagocytosis of apoptotic C26 cells by macrophages.
A and B, For ex vivo phagocytosis assay, murine peritoneal macrophages were coincubated with DiO-labeled SM4s- positive or SM4s-free apoptotic C26 at 37°C for 1 h, washed, stained with anti-mouse CD11b-PE Ab, and analyzed by flow cytometry. Alternatively, macrophages were directly exposed to SM4s before coincubation with SM4s-free apoptotic C26 cells. Double-positive population represented macrophages that took part in tethering and/or uptake of apoptotic C26. C, FACS phagocytosis index was calculated by dividing mean relative DiO fluorescence intensities of macrophages (upper quadrants of density plots) and apoptotic C26 (lower right quadrant). D–F, FACS analysis of in vivo uptake experiments. Number of macrophages per mouse was counted to ascertain constant ratio macrophages:apoptotic cells (1:2). G and H, Macrophages were cocultured with apoptotic cells preincubated with 10 µM SM4s (SM4), galactosylceramide (GC), cholesterol sulfate (CS), seminolipid (SM4g), ceramide-1-phosphate (C1P), or vehicle at 37°C for 1 h. Phagocytosis was analyzed by flow cytometry. Results represent mean ± SEM of three experiments in duplicates. Statistical differences were evaluated by paired t test (ex vivo data) or Mann-Whitney U test (in vivo data). *, p < 0,05; and ***, p < 0,001.
FIGURE 5. Comparative analysis of uptake of apoptotic cells from different cell lines, distinct apoptotic inducers, and different apoptotic stages.
Late apoptotic C26 cells induced by cycloheximide (left), late apoptotic Renca cells induced by staurosporine (middle), and early apoptotic C26 cells (right) were subjected to flow cytometric analysis for PS exposure on the cell surface and membrane permeability (upper lane) and cell size and granularity (middle lane). The cells were used in FACS phagocytosis assay as described (bottom lane). Data in the graphs are presented as mean ± SEM of three experiments in duplicates. Statistical differences were evaluated by paired t test. *, p < 0,05 and **, p < 0,01.
FIGURE 6. Confocal microscopy and SM4s enhancement of uptake of apoptotic cells by macrophages.
A, DiO-labeled apoptotic cells were cocultured with macrophages at 37°C for 1 h. The macrophages were washed from noningested apoptotic cells, stained with Alexa-546 phalloidin (actin) and DRAQ5 (nucleus), and visualized with confocal microscopy. Data are presented in red and blue channel (Alexa-546 and DRAQ5, respectively; left), green channel (DiO; middle), and merge (right). B, Higher magnification confocal images of macrophages with intracellular localization of apoptotic material (×2500). C, Percent of phagocytosis-active macrophages was calculated by dividing numbers of DiO-positive and negative macrophages, multiplied by 100. D, Single macrophages were analyzed for the intensity of green fluorescence, as a parameter of intensity of phagocytosis. Mean green fluorescence intensity of macrophages from macrophages + C26 samples was taken as “standard” (100%). Fluorescence intensity of single macrophages was calculated as percentage of standard 20 macrophages per condition per experiment were analyzed. All data are presented as mean ± SEM of three experiments in duplicates. Statistical differences were evaluated by Mann-Whitney U test. *, p < 0.05 and ***, p < 0.001.
FIGURE 7. Increased number of apoptotic particles in the phagosomal compartment in macrophages that ingested SM4s-painted apoptotic C26.
Murine peritoneal macrophages were grown on cover slips placed in 6-well plates and cocultured with apoptotic cells at 37°C for 1 h and subjected to ultrastructural analysis by electron microscopy. A, Quantification of apoptotic particles within phagosomes (arrows) between the two groups. Dense apoptotic material in the phagolysosomes (stars) of apoptotic cell-SM4s phagocytosing macrophages in comparison to the control. B and C, Number of apoptotic particle-containing phagosomes per macrophage and percent of macrophages that contain apoptotic material were evaluated. Results are presented as mean ± SEM from total of 80 macrophages from three experiments. Statistical differences were evaluated by Mann-Whitney U test. ***, p < 0.001.
To identify the specificity of SM4s-induced enhanced binding and uptake of apoptotic particles by macrophages, the effect of molecules with similar structure and/or charge to SM4s was analyzed. Apoptotic cells were incubated with the same concentration (10 µM) of SM4s, galactosylceramide (GalCer “bone” of SM4s), or cholesterol sulfate (sulfate group) and examined in flow cytometry assays as described. Cholesterol sulfate-painted apoptotic C26 cells demonstrated a slight but not significant increase in the uptake by macrophages. Galactosylceramide did not change the apoptotic cell uptake pattern, compared with the control. A significantly enhanced phagocytosis effect ( p = 0.0440) was evident only in the SM4s samples (Fig. 4G). An additional set of experiments included seminolipid (SM4g; sulfate group and alkylacylglycerol instead of ceramide structure) and ceramide-1-phosphate (ceramide structure and negative charge) next to SM4s. Enhanced uptake was evident in SM4s ( p = 0.0136) and SM4g ( p = 0.0318) samples, while C1P did not lead to an increase in the FACS phagocytosis index (Fig. 4H).
The increased uptake phenomenon is mainly mediated by macrophage scavenger receptors
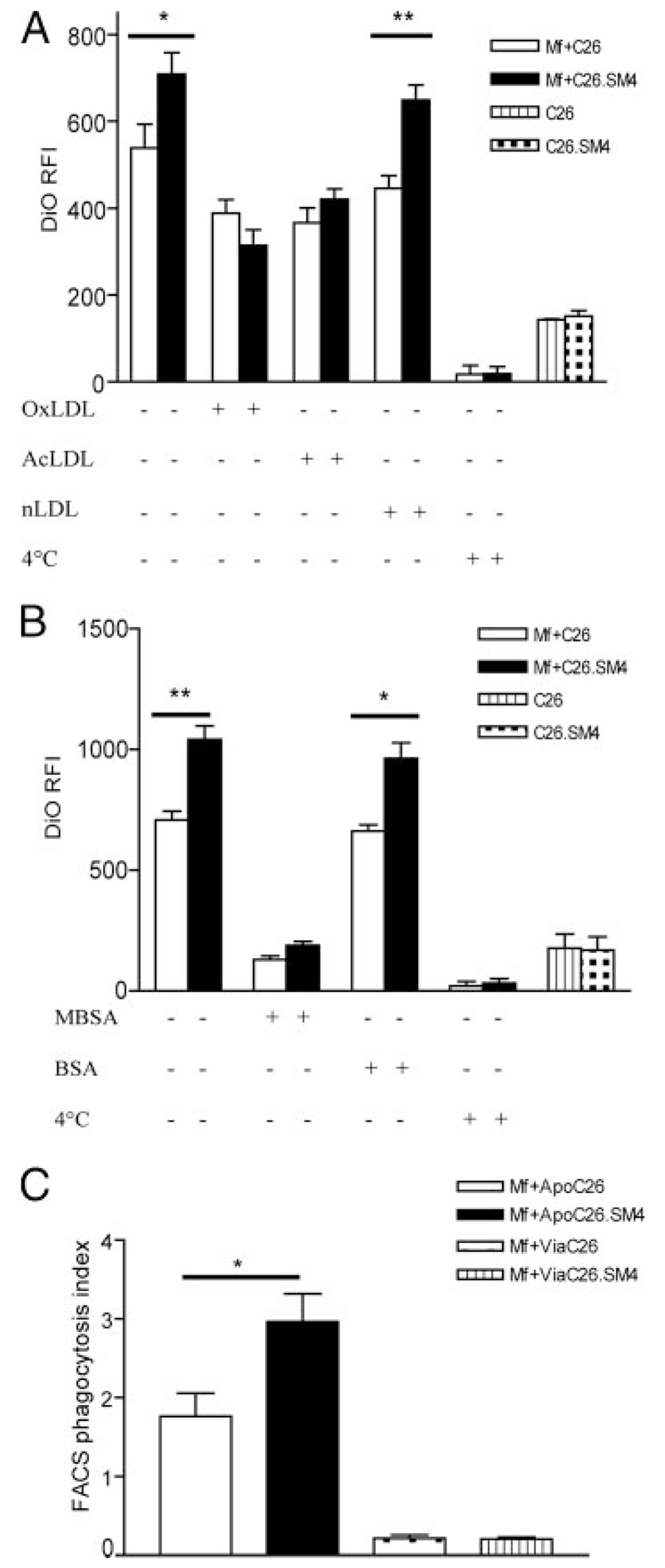
In the next step, it was investigated whether blocking of SRs is paralleled by impaired uptake of SM4s-painted apoptotic cells. Macrophages were incubated with OxLDL, AcLDL, nLDL, soluble PS, or vehicle (PBS) before incubation with apoptotic cells. In addition, a set of experiments with MBSA as a nonspecific SR blocker and anti-SR-A mAb (2F8) were included (Fig. 8, A and B). Inhibition of increased uptake (up to 100%) was achieved with SR blockers (OxLDL, AcLDL, and MBSA), with an insignificantly stronger inhibition by OxLDL in comparison to AcLDL. Blockade with anti-SR-A mAb gave inconsistent results (data not shown). The uptake pattern in the presence of nLDL was similar to that when uptake was studied in the absence of lipoproteins. Exposure of macrophages to soluble PS had only a small, statistically insignificant influence on clearance of either control or SM4s painted apoptotic cells (data not shown). Viable nonpainted (control) and viable SM4s-painted C26 cells were incubated with macrophages, as described earlier, to elaborate if SM4s alone would suffice for the effect. Macrophages incubated with viable cells showed only minimal uptake, with no evident difference between control and SM4s samples (Fig. 8C).
FIGURE 8. Increased uptake of SM4s-painted apoptotic cells is dominantly mediated by scavenger receptor SR-A.
A, Before incubation with DiO-labeled apoptotic C26 cells, macrophages were incubated with a general scavenger receptor blocker (OxLDL), a SR-A blocker (AcLDL), control (nLDL), or vehicle at 37°C and subjected to flow cytometric analysis. B, Effect of maleylated BSA (MBSA) as a SR-blocker and BSA as a control on the uptake of apoptotic cells. C, Apoptotic (ApoC26) vs viable (ViaC26) cell uptake by macrophages. Data are presented as mean ± SEM of three experiments in duplicates. Statistical differences were evaluated by paired t test. *, p < 0.05 and **, p < 0.01.
Increased recognition of SM4s-painted apoptotic particles enhances generation of TGF-β1, P-selectin surface expression, and IL-6 secretion by peritoneal macrophages
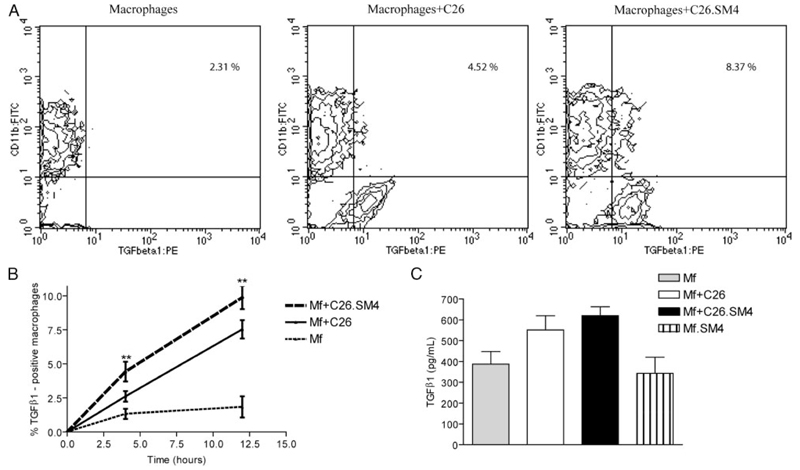
Flow cytometry immunostaining of macrophages incubated with late apoptotic C26 cells for 4 h and 12 h showed a significant increase in the percent of TGF-β1-positive macrophages in the sulfatide group, compared with the control, for both time points (average increase of 1.69-fold, p = 0.0069; and 1.30-fold, p = 0.0027, for 4 h and 12 h, respectively; Fig. 9, A and B). Analysis of macrophage supernatants for active TGF-β1 12 h after induction showed a slight increase in TGF-β1 concentration by macrophages from the sulfatide group (on average 1.13-fold, p = 0.1924; Fig. 9C).
FIGURE 9. Biosynthesis of TGF-β1 by macrophages after phagocytosis of SM4s-painted late apoptotic C26 cells.
A, Macrophages were grown in 6-well plates, incubated with control and SM4s-painted apoptotic C26 cells for 4 h and 12 h, and subjected to immunostaining for intracellular TGF-β1. Representative contour plots with percent of TGF-β1-positive macrophages 4 h after start of coincubation. B, Statistical analysis with time course of intracellular total TGF-β1 for 4 h and 12 h time points. C, Macrophages were grown in 6-well plates and cocultured with apoptotic C26 cells, SM4s-positive C26 cells, SM4s (10 µM), or vehicle at 37°C for 12 h. Supernatants were collected and subjected to ELISA. Data represent mean ± SEM of three experiments in duplicates. Statistical differences were evaluated by paired t test. **, p < 0.01.
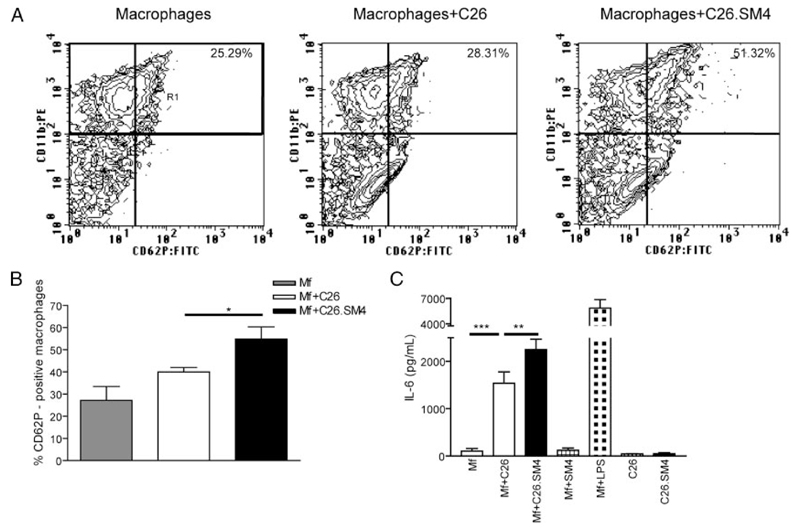
P-selectin was constitutively expressed by resident nonstimulated peritoneal macrophages. However, expression of P-selectin was enhanced within 2 h after incubation with C26 apoptotic cells with a significant increase both in percent of CD62P-positive macrophages (39.93 ± 2.07 vs 54.83 ± 5.48; p = 0.0448; Fig. 10, A and B) and intensity of expression (169.8 ± 18.31 vs 239.4 ± 21.91; p = 0.0003) in macrophages that had recognized SM4s-containig apoptotic C26 cells.
FIGURE 10. Phagocytosis of SM4s-positive apoptotic C26 cells increases IL-6 secretion and enhances P-selectin expression by murine peritoneal macrophages.
After coculture with apoptotic cells, macrophages were incubated with anti-mouse CD11b:PE and anti-mouse CD62P:FITC mAbs. A and B, FACS analysis of macrophage P-selectin expression. Macrophage population (CD11b+) was gated (R1) and percent of P-selectin-positive (CD62P+) macrophages was evaluated by flow cytometry. C, Macrophages were grown in 6-well plates and cocultured with apoptotic C26 cells, SM4s-positive C26 cells, SM4s (10 µM), LPS (1 µg/ml), or vehicle at 37°C for 2 h. IL-6 levels from the supernatants collected at 12 h after the start of coculture were analyzed by ELISA. Data represent mean ± SEM of three experiments in duplicates. Statistical differences were evaluated by paired t test. *, p < 0.05; **, p < 0.01, and ***, p < 0.001.
Analysis of macrophage supernatants showed an increase in the release of IL-6 by macrophages incubated with SM4s-positive apoptotic C26 cells, compared with the nonpainted apoptotic particles (1538.0 ± 240.8 pg/ml vs 2249.0 ± 222.1 pg/ml, p = 0.0026). The effect was apoptotic cell dependent, because release of IL-6 by macrophages incubated with SM4s alone was undetectable (Fig. 10C). Analysis of levels of GM-CSF, IL-10, IL-4, IFN-γ, MIP-1α, IL-12 p70, and TNF-α in the supernatants of the macrophages incubated with C26 or sulfatide-coupled SM4s did not demonstrate significant differences (data not shown).
Discussion
Sulfatides can be found in human kidney and colon tissue. Enhanced expression of SM4s in RCC and adenocarcinoma of the colon and lung has been reported (6–9). Concordant with these observations, we have shown in this study expression of sulfatides in both cancerous and noncancerous tissue and an increase of SM4s in tumors of kidney and colon. Nevertheless, the function of sulfatides in cancer remains unknown up to date. As macrophage SRs contribute to apoptotic cell uptake and as they have been reported to bind sulfatides, we hypothesized that SM4s might play a potent role as a phagocytic ligand and contribute to macrophage clearance of cancer apoptotic cells rich in sulfatides.
Apoptotic cell clearance by phagocytes requires surface presentation of a variety of ligands by the dying cell. Far less is known about the nature of ligands involved in phagocytosis of apoptotic cells than we do of the multiple receptors they may interact with. Exposure of PS on the cell surface has been suggested as a major signal in macrophage recognition of apoptotic particles (26). Macrophage SR-B (CD36), SR-B1, PSR, vitronectin receptor, CD14, and LRP/CD91 are among a number of macrophage receptors detected to bind PS (28, 29, 62–65). Oxidation of PS has been recently identified as a key factor for its recognition by macrophage CD36 (66). Next to PS exposure as a “general” apoptotic recognition pattern, other ligands (such as annexin I and II, lipoxins, calreticulin, sialoglycoprotein CD43) have been reported to promote phagocytosis, mainly in a cell- and tissue-dependent manner (67–70). Moreover, many of the apoptotic ligands do not bind directly to the corresponding macrophage receptors, but interact with bridging molecules (i.e., MFG-E8; β-2-glycoprotein 1) to facilitate binding and uptake of apoptotic particles (65, 71). We propose in this study that SM4s can act as a phagocytic ligand when expressed in sulfatide-rich apoptotic cancer cells. Our data indicate that the presence of SM4s on apoptotic cell membrane facilitates their uptake by macrophages. In ex vivo assays, both the number of phagocytosed late apoptotic C26 cells per macrophage and the percent of phagocytosis-involved macrophages were elevated, as shown by flow cytometry and confocal and electron microscopy. Moreover, the increased uptake was evident in the case of late apoptotic Renca cells and early apoptotic C26 cells as well as apoptotic cells induced by cycloheximide, indicating that the enhanced uptake phenomenon does neither depend on cell line nor on apoptotic inducer. We could not observe a significant difference in apoptotic cell clearance with different macrophage:apoptotic cell ratios (1:5 vs 1:10). Based on the dynamics of uptake as observed by video microscopy (supplemental movies), we suggest that macrophages have limited phagocytosis capacity and that, in our experiments, maximal capacity has already been reached with the lower (1:5) ratio. Ultrastructural analyses revealed a strong increase in the number of intraphagosomal apoptotic particles in macrophages ingesting apoptotic cells coupled to SM4s compared with the control group, corroborating that the effect of SM4s was not limited to tethering only. Results from our in vivo experiments, based on the capacity of thioglycollate-elicited murine peritoneal macrophages to ingest apoptotic cancer cells within the peritoneal cavity, were concordant with the ex vivo findings. SM4s alone did not promote ingestion. The sulfated glycolipid has to be present on the surface of apoptotic cells to enhance the interaction between the particle and the macrophage that results in greater levels of internalisation. Binding of SM4s to viable cells was strongly diminished, in comparison to early and late apoptotic cells. The ligand for SM4s on the surface of apoptotic cells was not identified. Because external exposure of PS represents a major difference between a viable and apoptotic cell, it can be assumed that PS may serve as a SM4s-binding molecule on the apoptotic cell surface. Consistent with this, SM4s painting of viable cells did not lead to enhanced binding to macrophages. We suggest that SM4s constitutes a bridging molecule between the apoptotic particle and macrophage SRs. In addition to their proposed role in the removal of apoptotic cells, SRs have been implicated in the clearance of aged platelets (72), which also express sulfatides (73).
We have identified the sulfate group of SM4s as a component critical for its recognition on apoptotic cancer cells by macrophages. Clearance of Galactosylceramide-painted apoptotic cells was not enhanced, while SM4g showed an effect similar to SM4s. These data are in accordance with those for the recognition of SM4 by SR-A, which similarly identified the sulfate moiety as essential (38). In this study, it was found that the ceramide moiety could be replaced with another bearing two long chains and that the galactose moiety could be substituted or deleted without loss of binding activity (38). However, our data have shown that negative charge (ceramide-1-phosphate) or sulfate group (cholesterol sulfate) alone are not sufficient per se for enhanced phagocytosis. Rather, the positioning of the galactosyl residue between the sulfate group and a lipid anchor would appear to be critical, suggesting the requirement for specific charge density. SR-A and CD36 clearly played a major role in the recognition of apoptotic cells, independent of SM4s, which is in agreement with published data (29, 66).
Macrophages constitute a dominant population of the local mononuclear cell infiltrate at cancer sites. They are known to take part in a variety of functions crucial for tissue remodelling and immune responses to cancer (74–76). Increased clearance of cancer apoptotic cells by macrophages via recognition of SM4s may have several implications: first, apoptotic cancer cells ingested by macrophages are not an effective immunogenic substrate as macrophages exhibit poor Ag presentation capacity (77). This mechanism may dampen the host anti-tumor Th1 response, contributing to a tumor immune escape. Another important aspect of apoptotic cell uptake by macrophages toward Th2-like immune response is TGF-β1 biosynthesis (21, 40, 78, 79). We have shown in this study an increase in intracellular TGF-β1 in macrophages that ingested sulfatide-painted apoptotic cells. However, we could not observe a significant increase in active TGF-β1 content in macrophage supernatants for the same time points. This may be partially explained by strong binding and autocrine regulation of macrophages by endogenously produced TGF-β1, which would prevent accumulation of the active cytokine form in supernatants in our closed in vitro system (80). We observed an increased macrophage P-selectin expression after incubation with SM4s-painted apoptotic cells, which may be caused either by a primary effect of the uptake or by a potential soluble factor secreted by macrophages secondary to the uptake. This may contribute to enhanced formation of macrophage-to-macrophage and macrophage-to-cancer cell aggregates (81). Increased interaction between apoptotic tumor cells and tumor-associated macrophages (TAM) could lead to an amplification of mitogenic and angiogenic activity of the macrophages. It is known that cancer cells can attract TAM and sustain their survival (82). TAM may respond to microenvironmental factors in tumors by producing mitogens and humoral factors that stimulate tumor angiogenesis (83). IL-6 secretion by macrophages after uptake of SM4s-painted apoptotic cells may induce and foster tumor proliferation. It has been reported that colon cancer growth can be inhibited and apoptosis can be stimulated by a neutralizing Ab against IL-6 receptor and gp130, a soluble IL-6 receptor fragment (84). IL-6 trans-signaling would thus support tumor progression (84). IL-6 has also been reported as an inducer of tumor angiogenic potential (85, 86). Clinical studies have identified IL-6 as a prognostic factor in some cancer patients. These investigations have found a highly significant correlation between increased IL-6 in patient sera with enhanced tumor growth at later cancer stages and reduced survival (87).
The results presented in this study are based on a macrophage-apoptotic cell phagocytosis model that used murine peritoneal macrophages and murine colon and renal cancer cells. We suggest that tumor-associated macrophages may well exhibit similar phagocytic characteristics, but this will require verification in vivo. Our data indicate that sulfated glycolipids such as SM4s in combination with an apoptotic ‘eat me’ signal can increase the extent of apoptotic cell elimination and significantly alter the phenotype of phagocytosing macrophages.
Supplementary Material
Acknowledgments
H.-J.G., Z.V.P., and R.S. designed research. S.K., E.K., T.P.S., R.J., and E.T. helped in performing experiments. F.A., N.P., and E.M. contributed key reagents and oversaw study design. Z.V.P. performed experiments and analyzed data. Z.V.P., N.P., and H.-J.G. wrote the manuscript. We thank Stefan Porubsky for critical discussion of the data; Herbert Wiegandt and Elisabeth Gröne for suggestions; and Benita von Tümpling-Radosta, Claudia Schmidt, Ulrike Rothermel, and Mahnaz Bonrouhi for expert technical assistance.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This work was supported by a Deutsche Forschungsgemeinschaft Grant Gr 728-6 and SFB 405, B10 (to H.-J.G.) and Fonds zur Förderung der Wissenschaftlichen Forschung Grant P19074-B05 (to E.M.).
Abbreviations used in this paper: GSL, glycosphingolipid; PS, phosphatidylserine; SR, scavenger receptors; PSR, phosphatidylserine receptor; PI, propidium iodide; IHC, immunohistochemistry; TLC, thin-layer chromatography; nLDL, native low density lipoprotein; OxLDL, copper low density lipoprotein; AcLDL, sodium acetate/acetic anhydride low density lipoprotein; MBSA, Maleyl-BSA; TAM, tumor-associated macrophages.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Fujimoto H, Tadano-Aritomi K, Tokumasu A, Ito K, Hikita T, Suzuki K, Ishizuka I. Requirement of seminolipid in spermatogenesis revealed by UDP-galactose: Ceramide galactosyltransferase-deficient mice. J Biol Chem. 2000;275:22623–22626. doi: 10.1074/jbc.C000200200. [DOI] [PubMed] [Google Scholar]
- 2.Ishizuka I. Chemistry and functional distribution of sulfoglycolipids. Prog Lipid Res. 1997;36:245–319. doi: 10.1016/s0163-7827(97)00011-8. [DOI] [PubMed] [Google Scholar]
- 3.Tadano-Aritomi K, Ishizuka I. Structure and function of sulfoglycolipids in the kidney and testis. Trends Glycoscience Glycotechnology. 2003;15:15–27. [Google Scholar]
- 4.Honke K, Zhang Y, Cheng X, Kotani N, Taniguchi N. Biological roles of sulfoglycolipids and pathophysiology of their deficiency. Glycoconj J. 2004;21:59–62. doi: 10.1023/B:GLYC.0000043749.06556.3d. [DOI] [PubMed] [Google Scholar]
- 5.Tadano-Aritomi K, Hikita T, Fujimoto H, Suzuki K, Motegi K, Ishizuka I. Kidney lipids in galactosylceramide synthase-deficient mice: absence of galactosylsulfatide and compensatory increase in more polar sulfoglycolipids. J Lipid Res. 2000;41:1237–1243. [PubMed] [Google Scholar]
- 6.Yoda Y, Gasa S, Makita A, Fujioka Y, Kikuchi Y, Hashimoto M. Glycolipids in human lung carcinoma of histologically different types. J Natl Cancer Inst. 1979;63:1153–1160. [PubMed] [Google Scholar]
- 7.Kobayashi T, Honke K, Kamio K, Sakakibara N, Gasa S, Miyao N, Tsukamoto T, Ishizuka I, Miyazaki T, Makita A. Sulfolipids and glycolipid sulfotransferase activities in human renal cell carcinoma cells. Br J Cancer. 1993;67:76–80. doi: 10.1038/bjc.1993.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakakibara N, Gasa S, Kamio K, Makita A, Koyanagi T. Association of elevated sulfatides and sulfotransferase activities with human renal cell carcinoma. Cancer Res. 1989;49:335–339. [PubMed] [Google Scholar]
- 9.Morichika H, Hamanaka Y, Tai T, Ishizuka I. Sulfatides as a predictive factor of lymph node metastasis in patients with colorectal adenocarcinoma. Cancer. 1996;78:43–47. doi: 10.1002/(SICI)1097-0142(19960701)78:1<43::AID-CNCR8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Sandhoff R, Grieshaber H, Djafarzadeh R, Sijmonsma TP, Proudfoot AE, Handel TM, Wiegandt H, Nelson PJ, Grone HJ. Chemokines bind to sulfatides as revealed by surface plasmon resonance. Biochim Biophys Acta. 2005;1687:52–63. doi: 10.1016/j.bbalip.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Roberts DD. Interactions of thrombospondin with sulfated glycolipids and proteoglycans of human melanoma cells. Cancer Res. 1988;48:6785–6793. [PubMed] [Google Scholar]
- 12.Miura R, Aspberg A, Ethell IM, Hagihara K, Schnaar RL, Ruoslahti E, Yamaguchi Y. The proteoglycan lectin domain binds sulfated cell surface glycolipids and promotes cell adhesion. J Biol Chem. 1999;274:11431–11438. doi: 10.1074/jbc.274.16.11431. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Honke K, Miyazaki T, Matsumoto K, Nakamura T, Ishizuka I, Makita A. Hepatocyte growth factor specifically binds to sulfoglycolipids. J Biol Chem. 1994;269:9817–9821. [PubMed] [Google Scholar]
- 14.Kobayashi T, Honke K, Kuramitsu Y, Hosokawa M, Miyazaki T, Murata J, Saiki I, Ishizuka I, Makita A. Cell-surface sulfoglycolipids are involved in the attachment of renal-cancer cells to laminin. Int J Cancer. 1994;56:281–285. doi: 10.1002/ijc.2910560223. [DOI] [PubMed] [Google Scholar]
- 15.Jennemann R, Schulze M, Bauer BL, Kurtz C, Wiegandt H. Serum immunoglobulins in Heymann’s experimental nephritis modulate binding of properdin and factor-H to sulpho-glycosphingolipids II3SO3(-)-Gg3Cer and III3SO3(-)-,II3SO3(-)-Gg3Cer. J Biochem. 1994;116:450–456. doi: 10.1093/oxfordjournals.jbchem.a124545. [DOI] [PubMed] [Google Scholar]
- 16.Merten M, Beythien C, Gutensohn K, Kuhnl P, Meinertz T, Thiagarajan P. Sulfatides activate platelets through P-selectin and enhance platelet and platelet-leukocyte aggregation. Arterioscler Thromb Vasc Biol. 2005;25:258–263. doi: 10.1161/01.ATV.0000149675.83552.83. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa D, Shikata K, Honke K, Sato S, Matsuda M, Nagase R, Tone A, Okada S, Usui H, Wada J, et al. Cerebroside sulfotransferase deficiency ameliorates L-selectin-dependent monocyte infiltration in the kidney after ureteral obstruction. J Biol Chem. 2004;279:2085–2090. doi: 10.1074/jbc.M305809200. [DOI] [PubMed] [Google Scholar]
- 18.Kajihara J, Guoji Y, Kato K, Suzuki Y. Sulfatide, a specific sugar ligand for L-selectin, blocks CC14-induced liver inflammation in rats. Biosci Biotechnol Biochem. 1995;59:155–157. doi: 10.1271/bbb.59.155. [DOI] [PubMed] [Google Scholar]
- 19.Mulligan MS, Warner RL, Lowe JB, Smith PL, Suzuki Y, Miyasaka M, Yamaguchi S, Ohta Y, Tsukada Y, Kiso M, et al. In vitro and in vivo selectin-blocking activities of sulfated lipids and sulfated sialyl compounds. Int Immunol. 1998;10:569–575. doi: 10.1093/intimm/10.5.569. [DOI] [PubMed] [Google Scholar]
- 20.Garcia J, Callewaert N, Borsig L. P-selectin mediates metastatic progression through binding to sulfatides on tumor cells. Glycobiology. 2006;17:185–196. doi: 10.1093/glycob/cwl059. [DOI] [PubMed] [Google Scholar]
- 21.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-β, PGE2, and PAF. J Clin Invest. 1998;101:890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savill J. Phagocyte clearance of cells dying by apoptosis and the regulation of glomerular inflammation. Adv Nephrol Necker Hosp. 2001;31:21–28. [PubMed] [Google Scholar]
- 23.Henson PM, Bratton DL, Fadok VA. Apoptotic cell removal. Curr Biol. 2001;11:R795–R805. doi: 10.1016/s0960-9822(01)00474-2. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann PR, deCathelineau AM, Ogden CA, Leverrier Y, Bratton DL, Daleke DL, Ridley AJ, Fadok VA, Henson PM. Phosphatidylserine (PS) induces PS receptor-mediated macropinocytosis and promotes clearance of apoptotic cells. J Cell Biol. 2001;155:649–659. doi: 10.1083/jcb.200108080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlegel RA, Williamson P. Phosphatidylserine, a death knell. Cell Death Differ. 2001;8:551–563. doi: 10.1038/sj.cdd.4400817. [DOI] [PubMed] [Google Scholar]
- 26.Fadok VA, Bratton DL, Henson PM. Phagocyte receptors for apoptotic cells: recognition, uptake, and consequences. J Clin Invest. 2001;108:957–962. doi: 10.1172/JCI14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 28.Fadok VA, Savill JS, Haslett C, Bratton DL, Doherty DE, Campbell PA, Henson PM. Different populations of macrophages use either the vitronectin receptor or the phosphatidylserine receptor to recognize and remove apoptotic cells. J Immunol. 1992;149:4029–4035. [PubMed] [Google Scholar]
- 29.Platt N, Suzuki H, Kurihara Y, Kodama T, Gordon S. Role for the class A macrophage scavenger receptor in the phagocytosis of apoptotic thymocytes in vitro. Proc Natl Acad Sci USA. 1996;93:12456–12460. doi: 10.1073/pnas.93.22.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platt N, da Silva RP, Gordon S. Recognizing death: the phagocytosis of apoptotic cells. Trends Cell Biol. 1998;8:365–372. doi: 10.1016/s0962-8924(98)01329-4. [DOI] [PubMed] [Google Scholar]
- 31.Savill J, Dransfield I, Hogg N, Haslett C. Vitronectin receptor-mediated phagocytosis of cells undergoing apoptosis. Nature. 1990;343:170–173. doi: 10.1038/343170a0. [DOI] [PubMed] [Google Scholar]
- 32.de Villiers WJ, Smith JD, Miyata M, Dansky HM, Darley E, Gordon S. Macrophage phenotype in mice deficient in both macrophage-colony-stimulating factor (op) and apolipoprotein E. Arterioscler Thromb Vasc Biol. 1998;18:631–640. doi: 10.1161/01.atv.18.4.631. [DOI] [PubMed] [Google Scholar]
- 33.Boullier A, Gillotte KL, Horkko S, Green SR, Friedman P, Dennis EA, Witztum JL, Steinberg D, Quehenberger O. The binding of oxidized low density lipoprotein to mouse CD36 is mediated in part by oxidized phospholipids that are associated with both the lipid and protein moieties of the lipoprotein. J Biol Chem. 2000;275:9163–9169. doi: 10.1074/jbc.275.13.9163. [DOI] [PubMed] [Google Scholar]
- 34.Febbraio M, Podrez EA, Smith JD, Hajjar DP, Hazen SL, Hoff HF, Sharma K, Silverstein RL. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J Clin Invest. 2000;105:1049–1056. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramprasad MP, Fischer W, Witztum JL, Sambrano GR, Quehenberger O, Steinberg D. The 94- to 97-kDa mouse macrophage membrane protein that recognizes oxidized low density lipoprotein and phosphatidylserine-rich liposomes is identical to macrosialin, the mouse homologue of human CD68. Proc Natl Acad Sci USA. 1995;92:9580–9584. doi: 10.1073/pnas.92.21.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huszar D, Varban ML, Rinninger F, Feeley R, Arai T, Fairchild-Huntress V, Donovan MJ, Tall AR. Increased LDL cholesterol and atherosclerosis in LDL receptor-deficient mice with attenuated expression of scavenger receptor B1. Arterioscler Thromb Vasc Biol. 2000;20:1068–1073. doi: 10.1161/01.atv.20.4.1068. [DOI] [PubMed] [Google Scholar]
- 37.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 38.Yoshiizumi K, Nakajima F, Dobashi R, Nishimura N, Ikeda S. Studies on scavenger receptor inhibitors. Part 1: synthesis and structure-activity relationships of novel derivatives of sulfatides. Bioorg Med Chem. 2002;10:2445–2460. doi: 10.1016/s0968-0896(02)00120-7. [DOI] [PubMed] [Google Scholar]
- 39.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 40.Huynh ML, Fadok VA, Henson PM. Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-β1 secretion and the resolution of inflammation. J Clin Invest. 2002;109:41–50. doi: 10.1172/JCI11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 42.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 43.Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, Mantovani A, Sozzani S. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol. 2005;174:6561. doi: 10.4049/jimmunol.174.11.6561. [DOI] [PubMed] [Google Scholar]
- 45.Porubsky S, Schmid H, Bonrouhi M, Kretzler M, Malle E, Nelson PJ, Grone HJ. Influence of native and hypochlorite-modified low-density lipoprotein on gene expression in human proximal tubular epithelium. Am J Pathol. 2004;164:2175–2187. doi: 10.1016/S0002-9440(10)63775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandhoff R, Hepbildikler ST, Jennemann R, Geyer R, Gieselmann V, Proia RL, Wiegandt H, Grone HJ. Kidney sulfatides in mouse models of inherited glycosphingolipid disorders: determination by nano-electrospray ionization tandem mass spectrometry. J Biol Chem. 2002;277:20386–20398. doi: 10.1074/jbc.M110641200. [DOI] [PubMed] [Google Scholar]
- 47.Nauta AJ, Castellano G, Xu W, Woltman AM, Borrias MC, Daha MR, van Kooten C, Roos A. Opsonization with C1q and mannose-binding lectin targets apoptotic cells to dendritic cells. J Immunol. 2004;173:3044–3050. doi: 10.4049/jimmunol.173.5.3044. [DOI] [PubMed] [Google Scholar]
- 48.Welch JS, Ricote M, Akiyama TE, Gonzalez FJ, Glass CK. PPARγ and PPARδ negatively regulate specific subsets of lipopolysaccharide and IFN-γ target genes in macrophages. Proc Natl Acad Sci USA. 2003;100:6712–6717. doi: 10.1073/pnas.1031789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou W, Patel H, Li K, Peng Q, Villiers MB, Sacks SH. Macrophages from C3-deficient mice have impaired potency to stimulate alloreactive T cells. Blood. 2006;107:2461–2469. doi: 10.1182/blood-2005-08-3144. [DOI] [PubMed] [Google Scholar]
- 50.Davies JQ, Gordon S. Isolation and culture of murine macrophages. Methods Mol Biol. 2005;290:91–103. doi: 10.1385/1-59259-838-2:091. [DOI] [PubMed] [Google Scholar]
- 51.Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417:182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- 52.Nozawa K, Casiano CA, Hamel JC, Molinaro C, Fritzler MJ, Chan EK. Fragmentation of Golgi complex and Golgi autoantigens during apoptosis and necrosis. Arthritis Res. 2002;4:R3. doi: 10.1186/ar422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ossina NK, Cannas A, Powers VC, Fitzpatrick PA, Knight JD, Gilbert JR, Shekhtman EM, Tomei LD, Umansky SR, Kiefer MC. Interferon-γ modulates a p53-independent apoptotic pathway and apoptosis-related gene expression. J Biol Chem. 1997;272:16351–16357. doi: 10.1074/jbc.272.26.16351. [DOI] [PubMed] [Google Scholar]
- 54.Kelly KJ, Sandoval RM, Dunn KW, Molitoris BA, Dagher PC. A novel method to determine specificity and sensitivity of the TUNEL reaction in the quantitation of apoptosis. Am J Physiol. 2003;284:C1309–C1318. doi: 10.1152/ajpcell.00353.2002. [DOI] [PubMed] [Google Scholar]
- 55.Malle E, Hazell L, Stocker R, Sattler W, Esterbauer H, Waeg G. Immunologic detection and measurement of hypochlorite-modified LDL with specific monoclonal antibodies. Arterioscler Thromb Vasc Biol. 1995;15:982–989. doi: 10.1161/01.atv.15.7.982. [DOI] [PubMed] [Google Scholar]
- 56.Butler PJ, Harris JI, Hartley BS, Lebeman R. The use of maleic anhydride for the reversible blocking of amino groups in polypeptide chains. Biochem J. 1969;112:679–689. doi: 10.1042/bj1120679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraser I, Hughes D, Gordon S. Divalent cation-independent macrophage adhesion inhibited by monoclonal antibody to murine scavenger receptor. Nature. 1993;364:343–346. doi: 10.1038/364343a0. [DOI] [PubMed] [Google Scholar]
- 58.Gurnani K, Kennedy J, Sad S, Sprott GD, Krishnan L. Phosphatidylserine receptor-mediated recognition of archaeosome adjuvant promotes endocytosis and MHC class I cross-presentation of the entrapped antigen by phagosome-to-cytosol transport and classical processing. J Immunol. 2004;173:566–578. doi: 10.4049/jimmunol.173.1.566. [DOI] [PubMed] [Google Scholar]
- 59.Wintergerst ES, Jelk J, Rahner C, Asmis R. Apoptosis induced by oxidized low density lipoprotein in human monocyte-derived macrophages involves CD36 and activation of caspase-3. Eur J Biochem. 2000;267:6050–6059. doi: 10.1046/j.1432-1327.2000.01682.x. [DOI] [PubMed] [Google Scholar]
- 60.Terpstra V, Kondratenko N, Steinberg D. Macrophages lacking scavenger receptor A show a decrease in binding and uptake of acetylated low-density lipoprotein and of apoptotic thymocytes, but not of oxidatively damaged red blood cells. Proc Natl Acad Sci USA. 1997;94:8127–8131. doi: 10.1073/pnas.94.15.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eitner F, Cui Y, Hudkins KL, Schmidt A, Birkebak T, Agy MB, Hu SL, Morton WR, Anderson DM, Alpers CE. Thrombotic microangiopathy in the HIV-2-infected macaque. Am J Pathol. 1999;155:649–661. doi: 10.1016/S0002-9440(10)65161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arur S, Uche UE, Rezaul K, Fong M, Scranton V, Cowan AE, Mohler W, Han DK. Annexin I is an endogenous ligand that mediates apoptotic cell engulfment. Dev Cell. 2003;4:587–598. doi: 10.1016/s1534-5807(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 63.Devitt A, Pierce S, Oldreive C, Shingler WH, Gregory CD. CD14-dependent clearance of apoptotic cells by human macrophages: the role of phosphatidylserine. Cell Death Differ. 2003;10:371–382. doi: 10.1038/sj.cdd.4401168. [DOI] [PubMed] [Google Scholar]
- 64.Tait JF, Smith C. Phosphatidylserine receptors: role of CD36 in binding of anionic phospholipid vesicles to monocytic cells. J Biol Chem. 1999;274:3048–3054. doi: 10.1074/jbc.274.5.3048. [DOI] [PubMed] [Google Scholar]
- 65.Gregory CD, Brown SB. Apoptosis: eating sensibly. Nat Cell Biol. 2005;7:1161–1163. doi: 10.1038/ncb1205-1061. [DOI] [PubMed] [Google Scholar]
- 66.Greenberg ME, Sun M, Zhang R, Febbraio M, Silverstein R, Hazen SL. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J Exp Med. 2006;203:2613–2625. doi: 10.1084/jem.20060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eda S, Yamanaka M, Beppu M. Carbohydrate-mediated phagocytic recognition of early apoptotic cells undergoing transient capping of CD43 glycoprotein. J Biol Chem. 2004;279:5967–5974. doi: 10.1074/jbc.M310805200. [DOI] [PubMed] [Google Scholar]
- 68.Fan X, Krahling S, Smith D, Williamson P, Schlegel RA. Macrophage surface expression of annexins I and II in the phagocytosis of apoptotic lymphocytes. Mol Biol Cell. 2004;15:2863–2872. doi: 10.1091/mbc.E03-09-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitchell S, Thomas G, Harvey K, Cottell D, Reville K, Berlasconi G, Petasis NA, Erwig L, Rees AJ, Savill J, et al. Lipoxins, aspirin-triggered epi-lipoxins, lipoxin stable analogues, and the resolution of inflammation: stimulation of macrophage phagocytosis of apoptotic neutrophils in vivo. J Am Soc Nephrol. 2002;13:2497–2507. doi: 10.1097/01.asn.0000032417.73640.72. [DOI] [PubMed] [Google Scholar]
- 70.Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, Henson PM. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123:321–334. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 71.Balasubramanian K, Maiti SN, Schroit AJ. Recruitment of β-2-glycoprotein 1 to cell surfaces in extrinsic and intrinsic apoptosis. Apoptosis. 2005;10:439–446. doi: 10.1007/s10495-005-0817-3. [DOI] [PubMed] [Google Scholar]
- 72.Brown SB, Clarke MC, Magowan L, Sanderson H, Savill J. Constitutive death of platelets leading to scavenger receptor-mediated phagocytosis. A caspase-independent cell clearance program. J Biol Chem. 2000;275:5987–5996. doi: 10.1074/jbc.275.8.5987. [DOI] [PubMed] [Google Scholar]
- 73.Ida M, Satoh A, Matsumoto I, Kojima-Aikawa K. Human annexin V binds to sulfatide: contribution to regulation of blood coagulation. J Biochem. 2004;135:583–588. doi: 10.1093/jb/mvh071. [DOI] [PubMed] [Google Scholar]
- 74.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 75.Valkovic T, Dobrila F, Melato M, Sasso F, Rizzardi C, Jonjic N. Correlation between vascular endothelial growth factor, angiogenesis, and tumor-associated macrophages in invasive ductal breast carcinoma. Virchows Arch. 2002;440:583–588. doi: 10.1007/s004280100458. [DOI] [PubMed] [Google Scholar]
- 76.Visscher DW, Tabaczka P, Long D, Crissman JD. Clinicopathologic analysis of macrophage infiltrates in breast carcinoma. Pathol Res Pract. 1995;191:1133–1139. doi: 10.1016/S0344-0338(11)80658-8. [DOI] [PubMed] [Google Scholar]
- 77.Oh YK, Harding CV, Swanson JA. The efficiency of antigen delivery from macrophage phagosomes into cytoplasm for MHC class I-restricted antigen presentation. Vaccine. 1997;15:511–518. doi: 10.1016/s0264-410x(97)00221-1. [DOI] [PubMed] [Google Scholar]
- 78.Freire-de-Lima CG, Xiao YQ, Gardai SJ, Bratton DL, Schiemann WP, Henson PM. Apoptotic cells, through transforming growth factor-β coordinately induce anti-inflammatory and suppress pro-inflammatory eicosanoid and NO synthesis in murine macrophages. J Biol Chem. 2006;281:38376–38384. doi: 10.1074/jbc.M605146200. [DOI] [PubMed] [Google Scholar]
- 79.McDonald PP, Fadok VA, Bratton D, Henson PM. Transcriptional and translational regulation of inflammatory mediator production by endogenous TGF-β in macrophages that have ingested apoptotic cells. J Immunol. 1999;163:6164–6172. [PubMed] [Google Scholar]
- 80.Comalada M, Cardo M, Xaus J, Valledor AF, Lloberas J, Ventura F, Celada A. Decorin reverses the repressive effect of autocrine-produced TGF-β on mouse macrophage activation. J Immunol. 2003;170:4450–4456. doi: 10.4049/jimmunol.170.9.4450. [DOI] [PubMed] [Google Scholar]
- 81.Tchernychev B, Furie B, Furie BC. Peritoneal macrophages express both P-selectin and PSGL-1. J Cell Biol. 2003;163:1145–1155. doi: 10.1083/jcb.200310079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196:254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 83.Chen JJ, Lin YC, Yao PL, Yuan A, Chen HY, Shun CT, Tsai MF, Chen CH, Yang PC. Tumor-associated macrophages: the double-edged sword in cancer progression. J Clin Oncol. 2005;23:953–964. doi: 10.1200/JCO.2005.12.172. [DOI] [PubMed] [Google Scholar]
- 84.Rose-John S, Scheller J, Elson G, Jones SA. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. J Leukocyte Biol. 2006;80:227–236. doi: 10.1189/jlb.1105674. [DOI] [PubMed] [Google Scholar]
- 85.Aoki Y, Jaffe ES, Chang Y, Jones K, Teruya-Feldstein J, Moore PS, Tosato G. Angiogenesis and hematopoiesis induced by Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93:4034–4043. [PubMed] [Google Scholar]
- 86.Legrand-Poels S, Schoonbroodt S, Piette J. Regulation of interleukin-6 gene expression by pro-inflammatory cytokines in a colon cancer cell line. Biochem J. 2000;349:765–773. doi: 10.1042/bj3490765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salgado R, Junius S, Benoy I, Van Dam P, Vermeulen P, Van Marck E, Huget P, Dirix LY. Circulating interleukin-6 predicts survival in patients with metastatic breast cancer. Int J Cancer. 2003;103:642–646. doi: 10.1002/ijc.10833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.