Abstract
Performing sequential reactions for the orthogonal derivatization of peptides in solution often requires intermediate handling and purification steps. To solve these problems, we have exploited the distinct adsorption kinetics of peptides towards particulate reversed-phase (RP) C18 silica material, enabling consecutive reactions to be performed without intermediate elution. To illustrate this approach, sequential CuAAC/click reactions were used to modify an analog of the bicyclic peptide Sunflower Trypsin Inhibitor 1 (SFTI-1), a potent scaffold for trypsin and chymotrypsin-like enzyme inhibitors. The SFTI-1 scaffold was synthesized containing both β-azido alanine and propargyl glycine residues. Despite the mutual reactivity of these groups, site isolation on RP silica enabled consecutive click reactions and associated washing steps to be performed while the peptide remained immobilized. Importantly, this approach eliminated side products that could form between two peptides or within a single peptide. These studies suggest a broad utility for RP silica in solving both peptide handling problems and in improving synthetic workflows.
Keywords: peptides, reversed-phase silica, CuAAC/click chemistry
Graphical abstract
Introduction
Sunflower Trypsin Inhibitor 1 (SFTI-1) is a 14 amino acid bicyclic peptide that operates as a Bowman-Birk type inhibitor of trypsin and trypsin-like serine protease enzymes (Figure 1).1 The hyper-exposed P1 lysine residue (following Schechter-Berger nomenclature2) interacts with the S1 cavity of the target enzyme, accounting for up to 50% of the inhibitor-enzyme contacts and affording its specificity and sub-nanomolar Ki for trypsin (1.1×10-10). In addition to its head-to-tail cyclization and disulfide bridge, the structure features a dense network of intramolecular hydrogen bonds which contribute to its compact rigidity.3,4 Together, these properties promise its utility as a bioscaffold for inhibitor engineering and pharmaceutical development.
Figure 1.
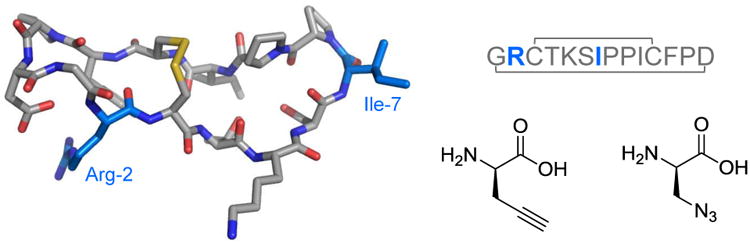
SFTI-1 wild-type structure and sequence organization. The P2′ (Ile-7) and P4 (Arg-2) sites for library substitution to propargyl glycine and β-azido alanine are highlighted in blue. PDB accession code: 1SFI.
For all protease substrates and inhibitors, specificity arises from residues at the scissile bond as well as their proximal interactions. These regions of the SFTI-1 inhibitor are named the binding loop (Thr4-Ile10) and the secondary loop (Gly1-Arg2, Phe12-Asp14).1 A typical combinatorial chemistry exercise would call for the diversification of at least two residues from these distal regions to generate new inhibitors.5 This strategy would call for a number of split syntheses and tedious purifications to build a peptide library of even a moderate size. Inspired by previous work on peptide and protein modifications using solid supports, namely peptide sequencing by membrane-supported Edman degradation,6 hydrophobic column-supported chemical and enzymatic digestions,7 protein bioconjugation reactions,8,9 and sortase-mediated ligations,10 we have developed an RP silica-supported, click-chemistry-based approach to rapidly construct a library of inhibitors from a single parent peptide. This approach leverages the polyvalent immobilization of the peptide to facilitate washing of small molecule reagents and buffers and enable selective elution of the dual-modified product. The copper-catalyzed azide-alkyne cycloaddition (CuAAC) “click” reaction was chosen for its selectivity in azide and alkyne conjugation, ease of setup, high yields, short reaction times, and broad compatibility with peptides and other biomolecules.11,12,13 To validate the synthetic approach, the SFTI-1 P4 arginine of the secondary loop and the P2′ isoleucine of the binding loop were substituted to propargyl glycine and β-azido alanine, respectively.
In addition to peptide modification, click chemistry has been used for the intermolecular cyclodimerization of peptides containing azide and alkyne groups on-resin, without the occurrence of intramolecular cycloaddition.14 In another example, a head-to-tail peptide cyclization was achieved.15 This approach has also been used for the generation of side chain tethered peptides for helical peptide stabilization.16 The CuAAC reaction is a flexible method that is compatible with numerous product profiles and can be tailored to solid support chemistries as well.
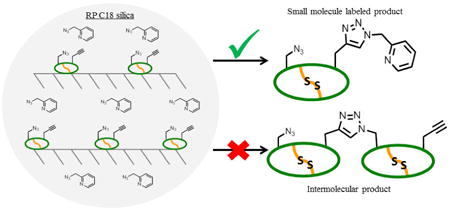
Silica and reversed-phase silica-supported approaches have been used previously in a variety of applications, including methane couplings with covalently-supported tantalum catalysts, asymmetric hydrogenation reactions with supported iridium catalysts, asymmetric Aldol reactions in water with supported prolinamide organocatalysts, and palladium-catalyzed Heck reactions and enantio selective allylic substitution reactions.17,18,19,20 These approaches all rely on covalent attachment of the catalyst to the solid support. For our system, RP C18 silica was chosen to non-covalently immobilize peptide reactants, inspired by previous work using solution-solid mixed phase derivitization for proteomics.21,22 The silica surface offers the unique conditions of site isolation of peptides, thereby disfavoring undesirable intermolecular reactions while favoring products labeled with the small molecule. In addition, the silica surface provides a unique reaction environment by co-localizing the peptide and reagents through adsorption. Importantly, the peptide is a macromolecule that makes multivalent hydrophobic interactions with the RP C18 material, while the small molecule reactants are in rapid equilibrium and move on and off the silica more evenly (Figure 2). As a result, peptides are effectively immobilized while small molecules can be washed away using aqueous / organic mixtures that are below the critical elution window for the polyvalent peptide. This can be illustrated by the isocratic elution of small molecules from an RP-HPLC column while macromolecules such as peptides require gradient elution.23
Figure 2.
Gradient-eluting versus isocratic-eluting binding behavior. Binding of SFTI-1 peptide to the RP C18 silica surface is multivalent, leading to a higher population of bound peptide. This localization and site isolation prevents intermolecular peptide reactions. In contrast, small molecule reactants bind in a monovalent manner and exhibit more equivalent silica and solution occupancies. This allows the peptide to react exclusively with the small molecule via CuAAC.
Results and Discussion
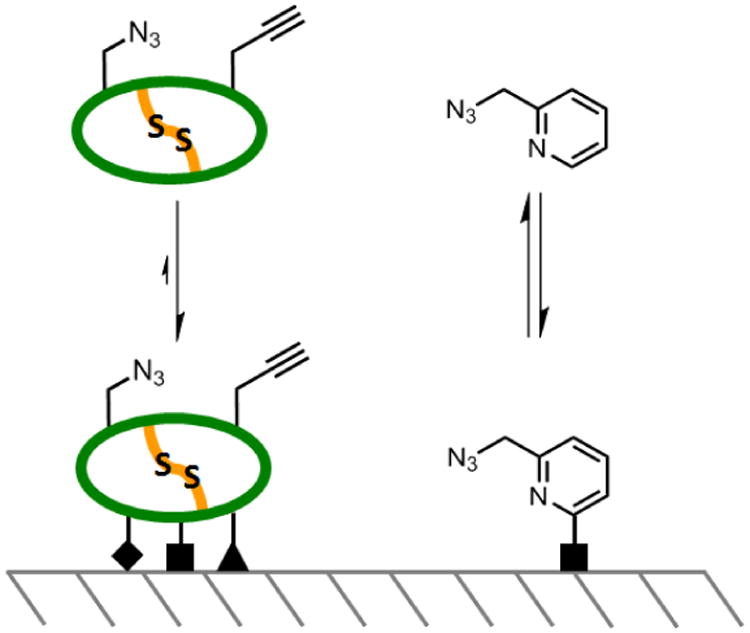
The P4/P2′ doubly-substituted SFTI-1 peptide was chain assembled by the Boc/Bzl-protection strategy for solid phase peptide synthesis (SPPS), cleaved from the resin support by anhydrous HF,24 head-to-tail cyclized by Native Chemical Ligation,25,26,27 and oxidized in solution to yield the disulfide peptide product (Supporting Information). From this parent peptide, a library of 16 doubly-modified peptides was generated by using a non-covalent, silica-supported, sequential-click reaction approach. To illustrate the approach, the first click reaction of picolyl azide (molecule X1, Figure 4) to P4/P2′ SFTI-1 peptide is described in detail. RP-C18 silica (20 mg) was swelled overnight in 1:1 H2O:EtOH (500 μL). Following centrifugation and removal of the supernatant, the reaction was set up as detailed in Figure 3. The peptide and small molecule were combined in the reaction tube containing the silica, while the ligand and copper were mixed separately. Following the mixing of these components, sodium ascorbate was added to start the reaction.13
Figure 4.
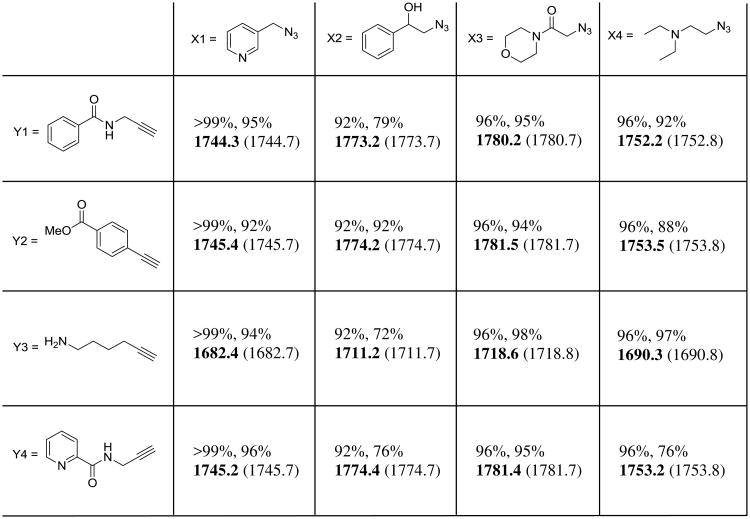
P4/P2′ SFTI-1 peptide library results. Percent conversion by HPLC presented as (X1%, Y1%). Mass observed by ESI-MS in bold, expected mass in brackets. Most reactions exhibited a high conversion rate (>90%) across two sequential steps.
Figure 3.
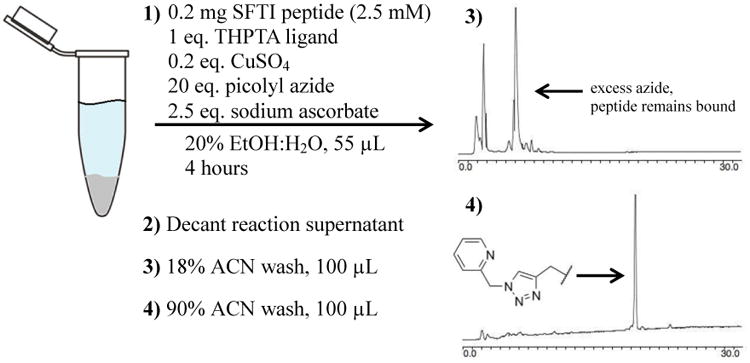
SFTI-1 sequential click reaction setup. The elution requirement of the peptide of greater than 18% ACN in H2O affords a facile wash protocol (3) to prepare the silica-bound single-clicked peptide (4) for the second click reaction in situ.
After 4 hours, the silica was rinsed with 100 μL 18% acetonitrile (ACN) in H2O containing 0.05% trifluroacetic acid (TFA). The elution point of the peptide from RP-C18 silica is 40% ACN, leaving the peptide immobilized. Therefore, this wash selectively removes only the excess reactants, as evidenced by the RP-HPLC trace of Figure 3-3. To fully elute a sample of peptide for analysis, an aliquot of silica was rinsed with 90% ACN in H2O with 0.05% TFA, revealing the singly-modified peptide shown in the RP-HPLC trace of Figure 3-4. Note that no intra- or intermolecular peptide triazole products were observed, likely due to conformational restriction and site isolation on the silica, respectively. Following three additional wash cycles with 100 μL 18% acetonitrile (ACN) in H2O with 0.05% TFA, the second click reaction was set up in the same manner as the first for the functionalization of the peptide azide.
Using this approach, reactants can be removed without eluting the peptide (Figure 3-3 and 3-4), and a second reaction to the immobilized singly-modified peptide can be set up in minutes. Percent completion by RP-HPLC peak area of each reaction step and corresponding ESI-MS data for all 16 reactions are shown in Figure 4. Final eluted peptide products (prior to RP-HPLC purification) from representative reaction series X1Y1-X4Y1 are shown in Figure 5. As a representative example, RP-HPLC traces showing the entire workflow for reaction X1Y1 as well as RP-HPLC traces of the eluted products for all 16 reactions are presented in the Supporting Information (Figures S1 and S2a-S2d).
Figure 5.
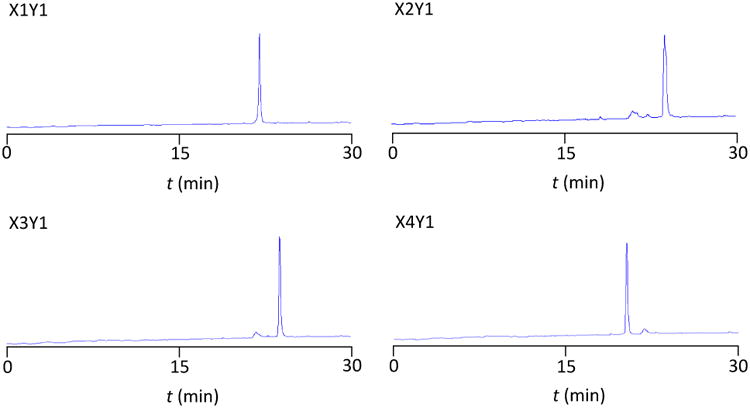
Final eluted peptide products for representative reaction series X1Y1-X4Y1. A dual-clicked peptide product can be obtained without an intermediate purification step between CuAAC reactions. RP-HPLC traces monitored at 214 nm, on a 30 min gradient from 100% H2O with 0.05% TFA (Buffer A) to 90% ACN, 10% H2O with 0.05% TFA (Buffer B).
Conclusion
We have developed an efficient, peptide-economical method to perform subsequent click reactions on a peptide to build a library of diverse compounds. The application of reversed-phase C18 silica contributes a number of properties which make this possible. Monovalent-binding, isocratic-eluting small molecules move off the silica at low ratios of ACN to H2O while multivalent-binding, gradient-eluting peptides remain bound for subsequent reactions. These contrasting modes of non-covalent adsorption of the peptide and reactants to silica affords a distinct reactivity profile favoring small molecule CuAAC peptide labeling reactions over non-productive intermolecular peptide side reactions. In addition, the silica allows us to set up and effectively handle small reactions (0.2 mg peptide), a problem universal to solution methodology.
Beyond generating a small, chemically diverse library of protease inhibitors in a short period, we have established that the application of reversed-phase silica to mainstay bioorganic solution reactions allows for highly selective product profiles and greatly enhanced peptide handling and recovery. We envision the expansion of this methodology to peptide labeling, bioconjugation, Native Chemical Ligation and other prevalent reactions. The application of these techniques and their implicit handling advantages to derivitization-ready macrocyclic peptide scaffolds, which are enjoying renewed interest as therapeutic candidates, as well as macromolecular natural products as a whole would be of high utility.28,29
Supplementary Material
Acknowledgments
The present work has been funded by the National Institutes of Health R01 AI113867. The authors thank Anthony P. Silvestri and the laboratory of Valery V. Fokin for supplying the small molecule azides X1-X4 and alkynes Y1-Y4 used in these studies.
Abbreviations
- RP
reversed-phase
- SFTI-1
sunflower trypsin inhibitor 1
- CuAAC
copper-catalyzed azide alkyne cycloaddition
- RP-HPLC
reversed-phase high-performance liquid chromatography
- ESI-MS
electrospray ionization mass spectrometry
- SPPS
solid-phase peptide synthesis
- TFA
trifluoroacetic acid
- ACN
acetonitrile
Footnotes
Supporting Information: Materials, peptide synthesis details, reaction setup, and HPLC traces showing the entire workflow for reaction X1Y1 as well as HPLC traces of the eluted products for all 16 reactions. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions: The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes: The authors declare no competing financial interest.
References
- 1.Lesner A, Łȩgowska A, Wysocka M, Rolka K. Sunflower Trypsin Inhibitor 1 as a Molecular Scaffold for Drug Discovery. Curr Pharm Des. 2011;17:4308–4317. doi: 10.2174/138161211798999393. [DOI] [PubMed] [Google Scholar]
- 2.Schechter I, Berger A. On the Active Site in Proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–62. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 3.Luckett S, Garcia RS, Barker JJ, Konarev AV, Shewry PR, Clarke AR, Brady RL. High-Resolution Structure of a Potent, Cyclic Proteinase Inhibitor from Sunflower Seeds. J Mol Bio. 1999;290:525–533. doi: 10.1006/jmbi.1999.2891. [DOI] [PubMed] [Google Scholar]
- 4.Korsinczky MLJ, Schirra HJ, Rosengren KJ, West J, Condie BA, Otvos L, Anderson MA, Craik DJ. Solution Structures by 1H NMR of the Novel Cyclic Trypsin Inhibitor SFTI-1 from Sunflower Seeds and an Acyclic Permutant. J Mol Bio. 2001;311:579–591. doi: 10.1006/jmbi.2001.4887. [DOI] [PubMed] [Google Scholar]
- 5.Combinatorial Chemistry. Nat Biotechnol. 2000;18:IT50–IT52. [PubMed] [Google Scholar]
- 6.Edman P. Method for Determination of the Amino Acid Sequence in Peptides. Acta Chem Scand. 1950;4:283–293. [Google Scholar]
- 7.Ward M. In Situ Chemical and Enzymatic Digestions of Proteins Immobilized on Miniature Hydrophobic Columns. Protein Protocols Handbook. 1996;IV:399–403. [Google Scholar]
- 8.Houen G, Jakobsen MH, Svaerke C, Koch C, Barkholt V. Conjugation to preadsorbed preactivated proteins and efficient generation of anti peptide antibodies. J Immunol Methods. 1997;206:125–34. doi: 10.1016/s0022-1759(97)00097-5. [DOI] [PubMed] [Google Scholar]
- 9.Houen G, Olsen DT, Hansen PR, Petersen KB, Barkholt V. Preparation of bioconjugates by solid-phase conjugation to ion exchange matrix-adsorbed carrier proteins. Bioconj Chem. 2003;14:75–79. doi: 10.1021/bc025622j. [DOI] [PubMed] [Google Scholar]
- 10.Policarpo RL, Kang H, Liao X, Rabideau AE, Simon MD, Pentelute BL. Flow-Based Enzymatic Ligation by Sortase A. Angew Chem Int Ed. 2014;53:9203–9208. doi: 10.1002/anie.201403582. [DOI] [PubMed] [Google Scholar]
- 11.Tornøe CW, Christensen C, Meldal M. Peptidotriazoles on Solid Phase: [1,2,3]-Triazoles by Regiospecific Copper(i)-Catalyzed 1,3-Dipolar Cycloadditions of Terminal Alkynes to Azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 12.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective Ligation of Azides and Terminal Alkynes. Angew Chem Eng Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Hong V, Presolski SI, Ma C, Finn MG. Analysis and Optimization of Copper-Catalyzed Azide-Alkyne Cycloaddition for Bioconjugation. Angew Chem Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sreenivas P, Kuzelka J, Wang Q, Finn MG. Head-to-Tail Peptide Cyclodimerization by Copper-Catalyzed Azide–Alkyne Cycloaddition. Angew Chem Int Ed. 2005;44:2215–2220. doi: 10.1002/anie.200461656. [DOI] [PubMed] [Google Scholar]
- 15.Jagasia R, Holub JM, Bollinger M, Kirshenbaum K, Finn MG. Peptide Cyclization and Cyclodimerization by Cu(I)-Mediated Azide-Alkyne Cycloaddition. J Org Chem. 2009;74:2964–74. doi: 10.1021/jo802097m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingale S, Dawson PE. On Resin Side-Chain Cyclization of Complex Peptides Using CuAAC. Org Lett. 2011;13:2822–2825. doi: 10.1021/ol200775h. [DOI] [PubMed] [Google Scholar]
- 17.Lin X, Xi Y, Zhang G, Phillips DL, Guo W. A Reaction Mechanism of Methane Coupling on a Silica-Supported Single-Site Tantalum Catalyst. Organometallics. 2014;33:2172–2181. [Google Scholar]
- 18.Liu G, Wang J, Huang T, Liang X, Zhang Y, Li H. Mesoporous Silica-Supported Iridium Catalysts for Asymmetric Hydrogenation Reactions. J Mater Chem. 2010;20:1970–1975. [Google Scholar]
- 19.Monge-Marcet A, Cattoën X, Alonso DA, Nájera C, Man MWC, Pleixats R. Recyclable Silica-Supported Prolinamide Organocatalysts for Direct Asymmetric Aldol Reaction in Water. Green Chem. 2012;14:1601–1610. [Google Scholar]
- 20.Anson MS, Mirza AR, Tonks L, Williams JMJ. Palladium Catalysed Heck and Enantioselective Allylic Substitution Reactions Using Reverse Phase Silica Supports. Tetrahedron Lett. 1999;40:7147–7150. [Google Scholar]
- 21.Nika H, Hawke DH, Angeletti RH. N-Terminal Protein Characterization by Mass Spectrometry after Cyanogen Bromide Cleavage using Combined Microscale Liquid- and Solid-Phase Derivatization. J Biomol Tech. 2014;1:19–30. doi: 10.7171/jbt.14-2501-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasslen KV, Tan LH, Manthorpe JM, Smith JC. Trimethylation Enhancement Using Diazomethane (TrEnDi): Rapid On-Column Quaternization of Peptide Amino Groups via Reaction with Diazomethane Significantly Enhances Sensitivity in Mass Spectrometry Analyses via a Fixed, Permanent Positive Charge. Anal Chem. 2014;7:3291–3299. doi: 10.1021/ac403349c. [DOI] [PubMed] [Google Scholar]
- 23.Schellinger AP, Carr PW. Isocratic and Gradient Elution Chromatography: A Comparison in Terms of Speed, Retention Reproducibility and Quantitation. J Chromatogr A. 2006;2:253–266. doi: 10.1016/j.chroma.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 24.Muttenthaler M, Albericio F, Dawson PE. Methods, Setup and Safe Handling for Anhydrous Hydrogen Fluoride Cleavage in Boc Solid-Phase Peptide Synthesis. Nat Protoc. 2015;10:1067–1083. doi: 10.1038/nprot.2015.061. [DOI] [PubMed] [Google Scholar]
- 25.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of Proteins by Native Chemical Ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 26.Dawson PE, Churchill MJ, Ghadiri MR, Kent SBH. Modulation of Reactivity in Native Chemical Ligation through the Use of Thiol Additives. J Am Chem Soc. 1997;119:4325–4329. [Google Scholar]
- 27.Hackeng TM, Griffin JH, Dawson PE. Protein Synthesis by Native Chemical Ligation: Expanded Scope by Straightforward Methodology. Proc Natl Acad Sci. 1999;96:10068–10073. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assem N, Ferreira DJ, Wolan DW, Dawson PE. Acetone-Linked Peptides: A Convergent Approach for Peptide Macrocyclization and Labeling. Angew Chem Int Ed. 2015;54:8665–8668. doi: 10.1002/anie.201502607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yudin AK. Macrocycles: Lessons From the Distant Past, Recent Developments, and Future Directions. Chem Sci. 2015;6:30–49. doi: 10.1039/c4sc03089c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.