Abstract

The use of small, natural chemical reporters in conjunction with catalyst-free bioorthogonal reactions will greatly streamline protein labeling in a cellular environment with minimum perturbation to their function. Here we report the discovery of a 2-cyanobenzothiazole (CBT)-reactive peptide tag, CX10R7, from a cysteine-encoded peptide phage library using the phage-assisted interrogation of reactivity method. Fusion of CX10R7 with a protein of interest allows site-specific labeling of the protein with CBT both in vitro and on the surface of E. coli cells. Mutagenesis studies indicated that the reactivity and specificity of CX10R7 are attributed to the sequence environment, in which the residues surrounding cysteine help to stabilize the ligation product.
Bioorthogonal reactions typically require the use of a non-natural chemical reporter, which is introduced into a biomolecule of interest via natural or engineered biosynthetic pathways.1 Since the overall bioorthogonal labeling efficiency is the product of the bioorthogonal reaction efficiency multiplying the incorporation efficiency, it varies greatly depending on the specific enzymes and non-natural chemical reporters used in the first step. To circumvent this limitation, sequence-specific bioorthogonal reactions involving only natural chemical reporters, e.g., natural peptides with unique chemical reactivity, would offer a streamlined approach toward biomolecular labeling. Indeed, peptide tags comprised of 20 natural amino acids have been reported, e.g., a tetracysteine tag (CCPGCC) that reacts selectively with the biarsenical fluorescent dyes such as FlAsH2 and ReAsH,3 a tetraserine tag (SSPGSS) that reacts selectively with rhodamine-derived bisboronic acid,4 a hexahistidine tag that chelates with nickel nitriloacetate,5 and a tetraaspartate tag that forms a stable Zn-coordination complex.6 Separately, a large number of peptide tags that undergo specific enzyme-mediated bioconjugation reactions have also been reported.7 Despite these advances, short peptide tags that show enzyme-free, monovalent, selective conjugation reactions with biocompatible chemical reagents are still rare.8
In our search of reactivity-based peptide tags, we turned our attention to the cysteine-containing sequences because cysteine is relatively uncommon in proteins (2.3% occurrence in human10); the sulfhydryl group exhibits exceptionally high nucleophilicity, which has been exploited in the development of many useful cysteine-specific bioconjugation reactions;11 and the cysteine reactivity can be further tuned through its sequence environment for selective conjugation with designed chemical probes such as dimaleimide14 and perfluoroaromatics.15 Specifically, we were intrigued by the selective condensation reaction between N-terminal cysteine and 2-cyanobenzothiazole (CBT)16 as well as extensive medicinal chemistry literature showing selective interactions of the enzyme active-site cysteine with the nitrile-containing ligands.17 Because N-terminal cysteine does not occur naturally, a TEV protease is needed to generate the requisite 1,2-aminothiol prior to CBT ligation (Scheme 1a). Alternatively, the 1,2-aminothiol moiety can be genetically encoded into proteins in the thiaprolyl form, which is unmasked using methoxyamine prior to CBT ligation (Scheme 1b).18 Inspired by these early reports, we envisioned that a suitable peptide tag encoding an internal cysteine could undergo CBT ligation to form a stabilized thioimidate adduct, affording efficient one-step ligation while obviating the need for protease treatment or protection/deprotection scheme (Scheme 1c). Here we report the discovery of such a cysteine tag, CX10R7, using phage-assisted interrogation of the reactivity method we reported recently.19 The mutagenesis and nitrile substrate scope studies revealed that CX10R7-CBT ligation displays a balance of reactivity and selectivity. Moreover, CX10R7 serves as a genetically encodable tag to direct specific labeling of proteins in vitro as well as on the surface of live E. coli cells.
Scheme 1. Cysteine-CBT Ligation.

To probe the sequence environment in an unbiased manner, we designed a randomized 11-mer peptide phage library, X5CX5 (X = any natural amino acid), in which a cysteine was placed in the middle, and performed a series of reaction-based panning with biotin-PEG4-CBT (1) (Figure S1). Representative clones that survived were subsequently sequenced (Tables S1–S3). Gratifyingly, two selections converged to a single peptide sequence encoding two cysteines, VTNQECCSIPM, hereafter referred to as CX10R7. Strikingly, several other selected clones also encoded more than one cysteine. Aside from the convergence, the selected clones encode predominantly polar and charged residues surrounding the central cysteine, presumably stabilizing the cysteine-CBT adducts. It is noteworthy that no significant enrichment was observed in all three selections, despite the sequence convergence (Tables S4–S6).
To verify the reactivity of the selected peptides, the sequences that appeared at later rounds were individually appended to the C-terminus of ubiquitin (Ub), a small protein that does not contain cysteine. The resulting Ub-peptide fusion proteins were expressed in E. coli and purified by affinity chromatography, and their identities were confirmed by mass spectrometry (Table S7). As a negative control, we appended to the Ub C-terminus a pentapeptide (CG4) encoding a cysteine flanked by two glycines at each side, on the basis that CG4 offers a sterically unhindered cysteine for sequence-independent ligation with CBT. Consistent with the literature reports,16,18 Ub-CG4 did not form a stable adduct with 6-amino-CBT because Cys is located internally and not at the N-terminus (entry 1, Table 1). In contrast, the selected peptide sequences exhibited varying degrees of reactivity toward 6-amino-CBT, with CX10R7 giving the highest yield (Table S8, Figure S2). The increased reactivity of CX10R7 is not a result of additive effect due to the presence of two cysteines, as other di- and tricysteine-containing sequences showed significantly lower yields (Table S8). Interestingly, LC-MS analysis revealed that the adduct masses correspond to a mixture of one and two molecules of 6-amino-CBT added to the Ub, consistent with thioimidate formation without subsequent loss of NH3 to form the thiazoline ring (Figure S2, Table S9). Since the thioimidate is generally labile on its own,20 we suspect that the surrounding polar residues might stabilize the thioimidate through some noncovalent interactions such as hydrogen bonding. It should be noted that a BLAST search of the bacterial and human proteomes did not reveal protein sequences that are similar to CX10R7 (Table S10).
Table 1. Reactivity Study of Ubiquitin-CX10R7 Mutantsa.


A mixture of 5 μM Ub-peptide fusion protein, 500 μM 6-amino-CBT, and 1 mM TCEP was incubated in NH4HCO3 buffer/acetonitrile (1:1), pH 8.5, at 37 °C for 1 h.
Conversion was determined by LC-MS based on the ion counts and calculated using the following equation: % conversion = Iproducts/(IUb-peptide + Iproducts), where Iproducts and IUb-peptide represent the ion counts of the CBT adducts (both mono- and di-CBT adducts) and Ub-peptide, respectively. Data shown are averages of two independent measurements ± standard deviation. See Table S9 and Figure S3 for details.
To probe the effect of sequence environment on the CBT ligation efficiency, we prepared a series of Ub-CX10R7 mutants (Table S7) and determined their ligation yields with 6-amino-CBT (Table 1 and Figure S3). When only one cysteine was present, the yield dropped by more than 4-fold (compare entries 3 and 4 to entry 2 in Table 1). Removal of both cysteines abolished the product formation, indicating that the CBT reactivity is specific toward cysteine (entry 5). Since the 7-mer retains the majority of the reactivity (entry 6), we replaced the surrounding polar residues (Asn3, Gln4, Glu5, and Ser8) that may serve as potential H-bond donors or acceptors with alanine and found these mutations reduced the reactivity by 3–10-fold (entries 7–10). The Q4A mutant showed the lowest yield, indicating that Gln4 may play a major role in stabilizing the thioimidate product. To probe which Cys adduct is stabilized by Gln4, double alanine mutants were prepared and their reactivities examined. Both mutants produced minimal adducts (entries 11 and 12), suggesting Gln4 is important for both cysteines (compare entries 11 to 3; 12 to 4). Since the amide side chain in Gln4 can serve as either an H-bond donor or acceptor, we generated the Q4E mutant in which the carboxylate side chain can only serve as an H-bond acceptor at pH 8.5 and found that this mutation completely abolished the CX10R7 reactivity (entry 13), indicating that Gln4 likely serves as an H-bond donor in stabilizing the thioimidate product and that the presence of the negatively charged carboxylate also destabilizes the adduct (compare entry 13 to 8). Since deamination from the thioimidate to the thiazoline was not observed, we further tested whether this could occur when a proximal −NH2 group, reminiscent of the N-terminal cysteine, is present. Thus, a lysine scan of CX10R7 was carried out by replacing three proximal residues on either side of cysteine with lysine. Upon incubation with 6-amino-CBT, all lysine mutants showed reduced reactivity (entries 14–19). Pairwise comparisons between the lysine mutants and the alanine mutants (compare entries 14–17 to entries 7–10) indicate that the lysine mutants afforded higher yields, consistent with the notion that the polar side chains help to stabilize the thioimidate adduct.
We then tested the reactivity of other aryl nitriles toward Ub-CX10R7. As expected, the reactivity of nitriles correlates with their electrophilicity21 (Table 2). To evaluate selectivity of the reactive aryl nitriles, we also determined their ligation yield with Ub-CG4, a negative control for sequence-independent reactivity. Simple nitrile-containing heterocycles that are less electrophilic than 6-amino-CBT, including 2- and 4-cyanopyridine, 8-cyanoquinoline, 2-cyanopyrrole, and 2-cyanothiophene, showed no reactivity (entries 2–4, 6, and 7, Table 2). A more electrophilic nitrile, 2-cyanopyrimidine, showed increased reactivity (entry 5); however, up to four molecules can be added to Ub-CX10R7 based on LC-MS analysis (Figure S4a), suggesting selectivity was lost by reacting with other nucleophiles, probably the two lysines on the surface of Ub. 2-Cyanothiazole (half of CBT) reacted with Ub-CX10R7 at a much lower yield (entry 8), indicating that the benzene ring in the CBT structure is important. Among three CBT bioisosteres, 7-methylthiazolo[5,4-b]pyridine-2-carbonitrile (2) showed higher reactivity but suffered poor selectivity, 2-cyanobenzimidazole (3) showed no reactivity, and 2-cyanobenzoxazole (4) showed modest yield along with poor selectivity (entries 9–11).
Table 2. Reactivity and Selectivity of Aryl Nitrilesa.


A mixture of 5 μM Ub-peptide fusion protein, 500 μM aryl nitrile, and 1 mM TCEP was incubated in NH4HCO3 buffer/acetonitrile (1:1), pH 8.5, at 37 °C for 1 h.
Conversion was determined by LC-MS as described previously. Data shown are averages of two independent measurements ± standard deviation. See Table S11 and Figure S4 for details. ND, not determined.
The initial aryl nitrile scope studies suggest that the CBT core is crucial for the ligation and that its moderate electrophilicity ensures exquisite selectivity toward Ub-CX10R7. To fine-tune the CBT reactivity and selectivity, we placed various electron-donating and -withdrawing groups on the benzene ring. Compound 5, containing two electron-donating methoxy groups, thus decreasing the electrophilicity of the nitrile group, showed essentially no reactivity (entry 12, Table 2). Substitution of the electron-withdrawing bromo and nitro groups resulted in higher reactivity toward Ub-CX10R7, but selectivity was lost (entries 13 and 14). This brief survey of the various aryl nitriles and substituted CBTs indicates that 6-amino-CBT displays a delicate balance between reactivity and selectivity. Further testing of the various reaction parameters for the ligation reaction between Ub-CX10R7 and biotin-PEG4-CBT (Figure S5) revealed that the optimal reaction conditions involved the use of PBS buffer, pH 7.4, at 37 °C for 1 h. The second-order rate constant, k2, for this ligation was measured to be 17 M–1 s–1 (Figure S6), about twice as fast as the condensation between cysteine and CBT.16 The adducts appear to be stable at room temperature after removal of the excess biotin-PEG4-CBT (Figure S7). Notably, replacing biotin in the biotin-PEG4-CBT with a fluorophore (Figure S8) abolished the reaction, indicating that the remote functional groups present on the CBT structure may participate in secondary interactions with the CX10R7.
To evaluate whether CX10R7 can serve as a general peptide tag for selective protein labeling, we fused CX10R7 to the N-terminus of superfolder green fluorescent protein to obtain CX10R7-sfGFP (Figures 1a and S9). After incubation with biotin-PEG4-CBT in PBS buffer, pH 7.4, at 37 °C for 1 h, CX10R7-sfGFP gave the desired ligation products in 65% yield (Figure 1b). As a control, wild-type sfGFP without the CX10R7 peptide tag did not yield any ligation product, despite the presence of two free cysteines (Cys48, Cys70)22 on its surface (Figure 1b). Furthermore, streptavidin blot confirmed that the biotin conjugate was stable and detectable after denaturing electrophoresis and protein blotting (Figures 1c and S10).
Figure 1.
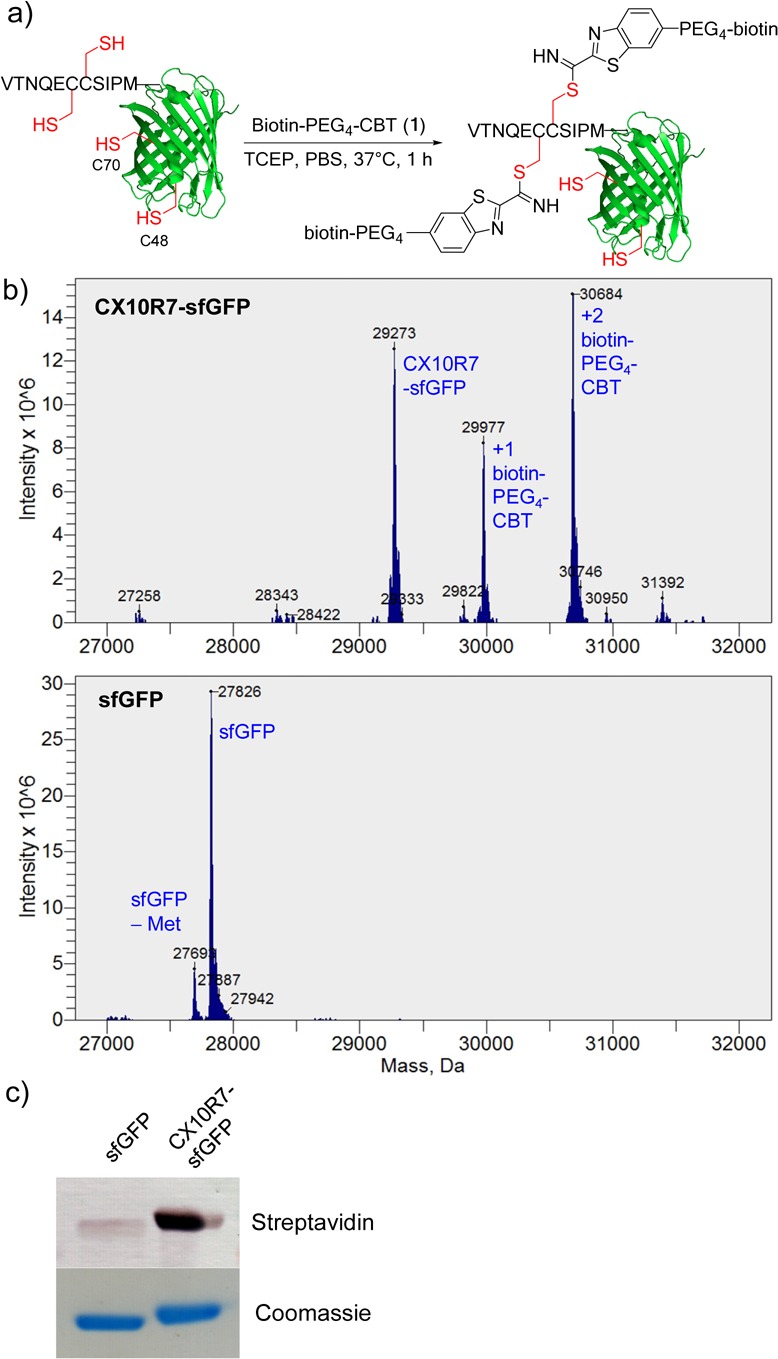
Selective labeling of the CX10R7-tagged sfGFP by biotin-PEG4-CBT. (a) Reaction scheme. The reaction was set up by incubating 5 μM of CX10R7-sfGFP (or sfGFP), 500 μM of biotin-PEG4-CBT, and 1 mM of TCEP in PBS, pH 7.4, at 37 °C for 1 h. (b) Deconvoluted masses of the product mixtures. See Table S12 for mass calculations. (c) Streptavidin blot to probe selective biotinylation of CX10R7-sfGFP by biotin-PEG4-CBT. Equal protein loading was verified by Coomassie blue staining.
To probe whether CX10R7 can direct selective protein labeling on live cells, we inserted the sequence onto the outer loop of the bacterial membrane protein OmpX to obtain OmpX-CX10R7. A string of seven alanines was also placed in the same location to obtain OmpX-A7 as a negative control. The E. coli cells expressing either OmpX-CX10R7 or OmpX-A7 were treated with 500 μM of biotin-PEG4-CBT. The biotinylation of OmpX on cell surface was probed with streptavidin-Alexa Fluor 568 and visualized using confocal microscopy. In the Alexa Fluor channel (ex 580 nm, em 585–712 nm), only cells expressing OmpX-CX10R7 showed red fluorescence (Figure 2b). The zoomed-in single-cell image showed that the fluorescent-labeled OmpX-CX10R7 accumulated preferentially at the two poles of the E. coli cell (last row in Figure 2b), similar to the previous observation.23
Figure 2.

Selective labeling of the CX10R7-tagged membrane protein OmpX on E. coli surface with biotin-PEG4-CBT. (a) Labeling scheme. (b) Micrographs of E. coli expressing either OmpX-A7 or OmpX-CX10R7. The cells were treated with 500 μM biotin- PEG4-CBT for 1 h before PBS wash and subsequent staining with streptavidin-Alexa Fluor 568 for 45 min. The last row shows the 3× digitally zoomed-in images. Scale bar = 2 μm.
In summary, we have identified a unique cysteine-containing peptide tag, CX10R7, which reacts selectively with amino-CBT to form stable adducts without the need of the 1,2-aminothiol group. The CX10R7 tag represents a new natural chemical reporter with which the CBT ligation reaction can be performed both in vitro and on live cells. Mutagenesis and aryl nitrile scope studies indicate that both the surrounding environment and the requisite CBT structure are important to the reactivity and selectivity of this novel peptide-CBT ligation reaction. The examples shown here indicate that CX10R7 can serve as a peptide tag at either the C- or N-terminus as well as the internal loop of a target protein. With these insights, it is possible that the ligation efficiency and versatility can be further improved in the future by incorporating secondary interaction motifs into the CBT structure and/or a more focused library design.
Acknowledgments
We gratefully acknowledge the NSF (CHE-1305826) and NIH (GM 85092) for financial support. We thank Prof. Wenshe Liu at Texas A&M University for generously sharing the pBAD-sfGFP-134TAG and pETDuet-OmpX-A7 plasmids.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b00982.
Figures S1−S10, Tables S1−S13, synthetic and experimental details, and characterization data (PDF)
Author Present Address
† Z.Y.: Sichuan University College of Chemistry, China.
The authors declare no competing financial interest.
Supplementary Material
References
- Ramil C. P.; Lin Q. Chem. Commun. 2013, 49, 11007. 10.1039/c3cc44272a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. A.; Adams S. R.; Tsien R. Y. Science 1998, 281, 269. 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Adams S. R.; Campbell R. E.; Gross L. A.; Martin B. R.; Walkup G. K.; Yao Y.; Llopis J.; Tsien R. Y. J. Am. Chem. Soc. 2002, 124, 6063. 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Halo T. L.; Appelbaum J.; Hobert E. M.; Balkin D. M.; Schepartz A. J. Am. Chem. Soc. 2009, 131, 438. 10.1021/ja807872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis A. N.; Ebright Y. W.; Ebright R. H. J. Am. Chem. Soc. 2001, 123, 12123. 10.1021/ja017074a. [DOI] [PubMed] [Google Scholar]
- Ojida A.; Honda K.; Shinmi D.; Kiyonaka S.; Mori Y.; Hamachi I. J. Am. Chem. Soc. 2006, 128, 10452. 10.1021/ja0618604. [DOI] [PubMed] [Google Scholar]
- Rashidian M.; Dozier J. K.; Distefano M. D. Bioconjugate Chem. 2013, 24, 1277. 10.1021/bc400102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Tanaka F.; Fuller R.; Asawapornmongkol L.; Warsinke A.; Gobuty S. Bioconjugate Chem. 2007, 18, 1318. 10.1021/bc070080x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Eldridge G. M.; Weiss G. A. Bioconjugate Chem. 2011, 22, 2143. 10.1021/bc200415v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miseta A.; Csutora P. Mol. Biol. Evol. 2000, 17, 1232. 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- a Chalker J. M.; Bernardes G. J.; Lin Y. A.; Davis B. G. Chem.-Asian J. 2009, 4, 630. 10.1002/asia.200800427. [DOI] [PubMed] [Google Scholar]; b Vinogradova E. V.; Zhang C.; Spokoyny A. M.; Pentelute B. L.; Buchwald S. L. Nature 2015, 526, 687. 10.1038/nature15739. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Gianatassio R.; Lopchuk J. M.; Wang J.; Pan C.-M.; Malins L. R.; Prieto L.; Brandt T. A.; Collins M. R.; Gallego G. M.; Sach N. W.; Spangler J. E.; Zhu H.; Zhu J.; Baran P. S. Science 2016, 351, 241. 10.1126/science.aad6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Clouthier C. M.; Tsao K.; Strmiskova M.; Lachance H.; Keillor J. W. Angew. Chem., Int. Ed. 2014, 53, 13785. 10.1002/anie.201408015. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Welborn M.; Zhu T.; Yang N. J.; Santos M. S.; Van Voorhis T.; Pentelute B. L. Nat. Chem. 2016, 8, 120. 10.1038/nchem.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H.; Xiao F.; Zhan K.; Kim Y.-P.; Xie H.; Xia Z.; Rao J. Angew. Chem., Int. Ed. 2009, 48, 9658. 10.1002/anie.200903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming F. F.; Yao L.; Ravikumar P. C.; Funk L.; Shook B. C. J. Med. Chem. 2010, 53, 7902. 10.1021/jm100762r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. P.; Elliott T.; Holt M.; Muir T. W.; Chin J. W. J. Am. Chem. Soc. 2011, 133, 11418. 10.1021/ja203111c. [DOI] [PubMed] [Google Scholar]
- Lim R. K. V.; Li N.; Ramil C. P.; Lin Q. ACS Chem. Biol. 2014, 9, 2139. 10.1021/cb500443x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFaul P. A.; Morley A. D.; Crawford J. J. Bioorg. Med. Chem. Lett. 2009, 19, 1136. 10.1016/j.bmcl.2008.12.105. [DOI] [PubMed] [Google Scholar]
- Oballa R. M.; Truchon J.-F.; Bayly C. I.; Chauret N.; Day S.; Crane S.; Berthelette C. Bioorg. Med. Chem. Lett. 2007, 17, 998. 10.1016/j.bmcl.2006.11.044. [DOI] [PubMed] [Google Scholar]
- Pedelacq J. D.; Cabantous S.; Tran T.; Terwilliger T. C.; Waldo G. S. Nat. Biotechnol. 2006, 24, 79. 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Lee Y.-J.; Wu B.; Raymond J. E.; Zeng Y.; Fang X.; Wooley K. L.; Liu W. R. ACS Chem. Biol. 2013, 8, 1664. 10.1021/cb400267m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


