Abstract
Stem cells offer a promising tool in tissue engineering strategies, as their differentiated derivatives can be used to reconstruct most biological tissues. These approaches rely on controlling the biophysical cues that tune the ultimate fate of cells. In this context, significant effort has gone to parse out the role of conflicting matrix-elicited signals (e.g., topography and elasticity) in regulation of macroscopic characteristics of cells (e.g., shape and polarity). A critical hurdle, however, lies in our inability to recapitulate the nanoscale spatiotemporal pattern of these signals. The study presented in this manuscript took an initial step to overcome this challenge by developing a carbon nanotube (CNT)-based substrate for nanoresolution control of focal adhesion formation and cell alignment. The utility of this system was studied using human umbilical vascular endothelial cells (HUVECs) and human embryonic stem cells (hESCs) at a single cell level. Our results demonstrated the ability to control cell orientation by merely controlling the alignment of focal adhesions at a nanoscale size. Our long-term vision is to use these nanoengineered substrates to mimic cell orientation in earlier development and explore the role of polarity in asymmetric division and lineage specification of dividing cells.
Keywords: Human embryonic stem cells, carbon nanotubes, cellular alignment, micropatterning
1. Introduction
Stem cells provide a potential therapeutic avenue for the treatment of a variety of diseases, as their differentiated derivatives can be used to reconstruct most biological tissues. Stem cell-based therapy approaches, however, rely on our ability to control the environmental cues that contribute to the ultimate fate of stem cell.1 As an example, the topography of the matrix on which stem cells are cultured regulates cell orientation and consequently, the migratory behavior and lineage specification of cells.2–4 This has motivated scientists to manipulate the matrix-induced biophysical cues (e.g., surface topographical features) to tune the cellular behavior according to each specific stem cell application.5
One of the stem cell's characteristics of interest to control in tissue engineering applications is polarity. The interplay between cell polarity and division axis described by early cell biologists is still used as a paradigm in developmental studies.6–8 This interplay ultimately regulates the self-renewal versus differentiation of stem cells. Mechanical signals exerted from the matrix play an important role in regulation of cell polarity and spindle orientation during cell division.9–11 These mechanical signaling pathways are hypothesized with tension forces to being transmitted from focal adhesions through the cytoskeleton and ultimately to the cell nucleus.3,6,7 These forces are mediated by transmembrane adhesion proteins and can therefore affect the expression of signaling proteins.12–14 Integrin-linked kinase activation has been reported to increase cytoplasmic levels of β-catenin, which positively affects the Wnt-β catenin signaling pathway. However, the role of individual physical forces in control of cell polarity and orientation remains controversial.10,15–17
Major technological advances in materials design have enabled the study of stem cell functions in response to substrate topographical and mechanical cues.18 For instance, stem cells have been shown to differentiate into a neuronal lineage with significant neurite extensions when confined in nanogrooves18 or along with direction of electronic spun fibers.19 Strikingly, it was shown that the degree of cell spreading and cell shape regulates cell apoptosis, proliferation, and differentiation. Another major discovery that emerged using biomaterial niches was the relationship between substrate mechanical stresses and stem cell lineage specification.13,20 Although considerable progress has been made in identifying the biophysical regulators of stem cells, we face limitations in recapitulating the cooperative effects of these signals. The main challenge remains in controlling the nanoscale organization of diverse biophysical signals presented to the cells. A major step forward is the biofabrication of novel platforms that allow for independent tuning, control and integration of morphological electrical cues in nanoscale.
The presented study takes a step toward this goal by developing a CNT-based substrate as a novel platform that allows for control over the size and direction of focal adhesions and the modulation of cell polarity, shape and cytoskeletal organization. CNTs have recently received significant interest for several unique characteristics including high mechanical modulus, electrical conductivity and inertness.21,22 In a number of recent studies, CNTs have been used as a filler for developing substrates to enhance nanoroughness23 among other characteristics.23–25 Interestingly, it has been demonstrated that CNTs can lead to an increase in the number of focal adhesions and expression of vinculin by cells.26,27 In our previous report, we demonstrated higher rates of cell proliferation and osteogenic differentiation on PLGA substrates containing CNTs, as compared to blank PLGA.23 Here, we take a step further to modulate cell polarity and shape by conjugating binding domain proteins (Geltrex or fibronectin) to CNTs. Our hypothesis is that by tuning the size and direction of focal adhesions, one can control the directionality and spreading of cells at a single cell level. To this end, we demonstrate the utility of the aligned CNT substrates using human umbilical endothelial cells (HUVECs) and human embryonic stem cells (hESCs) cultured at single cell level. The absolute control over the mechanical and structural features of CNTs presents a unique and promising approach to fabricating novel platforms with independently tailored properties to direct cardiac stem cell differentiation.
2. Materials and Method
2.1. Substrate preparation
To fabricate CNT substrates, coverslips were rinsed in sterilized water for 5 min, dried with air blow, and cleaned with an ozone cleaner (BioForce Nanoscience) for 3 min. Coverslips were then coated with a 5-nm Ti layer and 10-nm Au layer through sputter coating (Denton Desk II Turbo Sputter Coater and Denton Desk II Sputter Coater respectively). Next, 100 μg of carboxyl CNTs (Nano Lab, purity> 95%) were dispersed in 0.5 mL of 0.2 mol/L 2-(N-Morpholino)ethanesulfonic acid (pH 5.6) (MES, Acros) buffer under sonication for 1 h at room temperature. About 0.25 mL of 0.8 mol/L 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC, Acros) and 0.25 mL of 0.4 mol/L N-Hydroxysuccinimide (NHS) (Acros) in MES solution were added to the activated carboxyl groups.28,29 The mixture was washed with PBS and centrifuged in a MW 100 KDa cutoff centrifugal filter (Millipore) to remove EDC and NHS at 5000 g three times for 30 min each. Then, 5 μg of a model protein was added at 4°C and incubated overnight. Finally, the mixture was washed and centrifuged in a filter to remove unconjugated protein at 5000 g six times for 30 min each29 and CNT-protein solution was collected and stored at −20°C until use. These studies used bovine serum albumin (BSA) (Sigma) and fluorescent BSA (fBSA) (Sigma) as model proteins for characterization of adhesion to CNTs. Fibronectin (Sigma) and Geltrex were used to provide cell attachment for human umbilical vein endothelial cells (HUVECs) and hESCs, respectively.
In order to obtain the aligned CNT pattern on the surface, substrates were rinsed in CNT-ligand solution for 12 h and aligned on the sample through spin coating (Chemat Technology). The solution was spun at 800 rpm for 10 s and 5000 rpm for 1 min. After incubation in ambient conditions for 30 min, CNT substrates were washed gently with PBS and ethanol to remove any remaining particles. Poly (ethylene) glycol (PEG) thiol (Sigma) was placed in ethanol solution and coated onto the surface for 10 min to prevent nonspecific binding.
2.2. Substrate characterization
To perform transmission electron microscopy (TEM), several droplets of homogenized carboxyl CNTs and protein-conjugated CNT solution were coated onto 200 mesh copper grids and dried at room temperature for 10 min. TEM images of carboxyl CNTs and Geltrex-conjugated CNTs were visualized by a Hitachi H-8000 TEM system. To perform scanning electron microscopy (SEM) characterization, substrates with carboxyl CNTs were imaged using a Zeiss Ultra Plus SEM system.
We used immunofluorescence microscopy to confirm the attachment of a model protein (fBSA) to the CNTs. Immunofluorescent images were captured with fBSA-conjugated CNTs on substrates and imaged by a Nikon Eclipse 80i microscope.
UV-Vis Spectrum characterization was performed with 5 μg/mL BSA solution and 5 μg/mL carboxyl CNT homogenized solution prepared through a sonication step. Briefly, 5 μg/mL of BSA-conjugated CNT solution was prepared with 1 h incubation of 5 μg/mL BSA and carboxyl CNTs at 37°C. The unconjugated BSA was then removed with a 100 K molecular weight cutoff centrifugal filter (Millipore). The UV-vis spectrum was recorded using a Beckman Coulter DU 730 UV-Vis spectrometer.
2.3. Cell culture
HUVECs (Lonza) at passages 8–12 were used for this study. Cells were seeded on CNT-aligned substrates at a density of 5000 cells/cm2 in Ham's F12K medium (Invitrogen) supplemented with 2 mmol/L glutamine (Invitrogen), 1.5 g/L sodium bicarbonate (Sigma), 0.1 mg/mL heparin (Fisher), 0.03–0.05 mg/mL endothelial cell growth supplement (BD Biosciences), 10% FBS (Invitrogen) and 1% penicillin-streptomycin (Invitrogen).
hESC line H9 was purchased from WiCell with cell lines grown on a mouse embryonic fibroblast (MEF) (CF-1, Millipore) feeder layer in DMEM/F12 (Invitrogen) with 20% KnockOut Serum (Invitrogen), 0.1 mM MEM NonEssential Amino Acids (Invitrogen), 3.5 mM L-Glutamine final concentration (Invitrogen), 100 μm 2-Mercaptoethanol (Sigma) and supplemented with 10 ng/mL human recombinant bFGF (PeproTech) as shown in WiCell protocols. Medium was changed daily and cells were passaged on every fifth day of culture with collagen-ase type IV (Invitrogen). MEF were grown in DMEM (Invitrogen) with 10% FBS (Invitrogen) and 100U/100 μg/mL of penicillin/streptomycin (Invitrogen).
hESC monolayer culture with single cell passaging was established as previously described.30 Briefly, hESCs were switched from feeder-dependent culture to feeder-free conditions and were grown as colonies for 2–3 passages on growth factor reduced Geltrex (Invitrogen) coated 6-well plates (BD Biosciences) in MEF conditioned medium supplemented with 10 ng/mL bFGF. Cells were passaged mechanically on day five. Conditioned medium was prepared as described: MEF cells were mitotically inactivated using 10 μg/mL Mitomycin C (Sigma) for 2.5 h and then plated onto gelatinized flasks at a density of 60 000 cells/cm2. The following day, cells were washed with PBS and medium was changed to hESC medium without bFGF addition at 0.5 mL/cm2 and collected every 24 h for 7 days. Prior to cell culture, the conditioned medium was filtered and supplemented with 10 ng/mL bFGF. On the following passage, hESC colonies were treated with TrypLE Select (Invitrogen) for 1 min, gently dissociated to single cells and plated to new wells at approximately 80–100 000 cells/cm2. Cells were grown under the same culture conditions and single cell passaged upon reaching confluence. After stabilization of the cell culture cells were routinely passaged as single cells on each fourth day with a seeding density of 50 000 cells/cm2.
2.4. Immunochemistry
HUVEC and ESC single cells were allowed 2–4 h to attach onto the CNT-based substrates and were fixed with formalin (Sigma), permeabilized in 0.1% Triton X-100 (Sigma) at 4°C, and blocked in 4% goat serum solution at room temperature for 30 min. HUVECs were stained primarily for PECAM-1 at a 1:500 dilution ratio (Cell Signaling). ESC single cells were stained primarily with Oct4 at a 1:500 dilution (Santa Cruz Technology) and vinculin at a 1:100 dilution (Sigma). Secondary antibodies were purchased from Invitrogen and used at a 1:100 dilution. Rhodamine phalloidin at a 1:100 dilution (Invitrogen) was used to visualize cytoskeleton structure of HUVECs and human embryonic single cells and DAPI (Sigma) were used to visualize nuclei.
3. Results and Discussion
3.1. Preparation and characterization of aligned CNT substrate
Figure 1 demonstrates a schematic representation of steps involved in developing aligned CNT substrates. We experimentally observed that, CNT–ligand conjugates adhered to Au-coated coverslips due to electrostatic interactions. When centrifugal force was applied through spin coating, CNTs–ligand conjugates would be organized in an aligned fashion over the substrate. The concentration of conjugates, rinse time and spin coating procedure were adjusted to achieve a specific density of CNT–ligand conjugates on the surface to facilitate cell adhesion. To prevent nonspecific binding, the area without CNT–ligand conjugates was rendered nonadhesive to protein through a PEG-thiol (PEG-SH) solution. This was done to guide cell adhesion solely to the CNT–ligand conjugated areas.
Fig. 1.
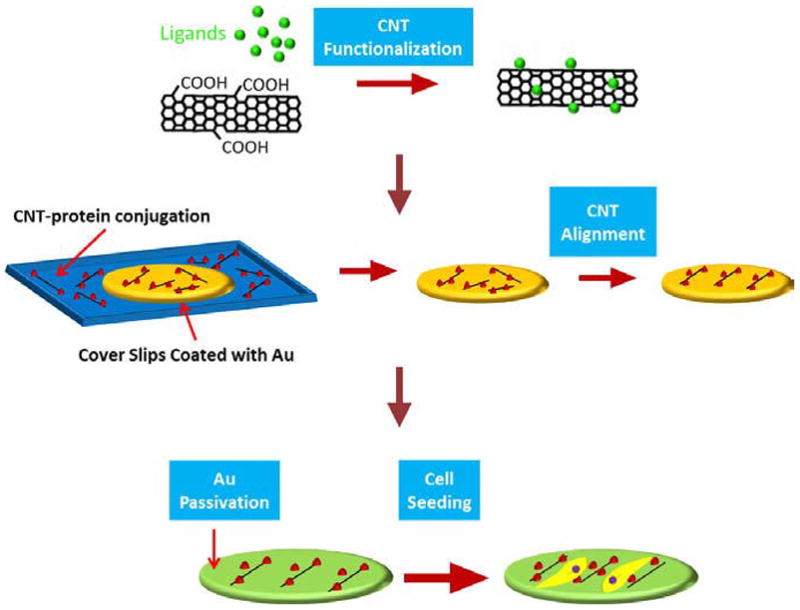
Schematic diagram showing experimental procedure for CNT substrate. Carboxyl CNTs are functionalized with protein and CNT–protein conjugates are aligned on Au surface by spin coating. Then Au surfaces without CNTs are rendered nonadhesive by thiolated polyethylene glycol (PEG-SH) and cells are cultured on the substrates.
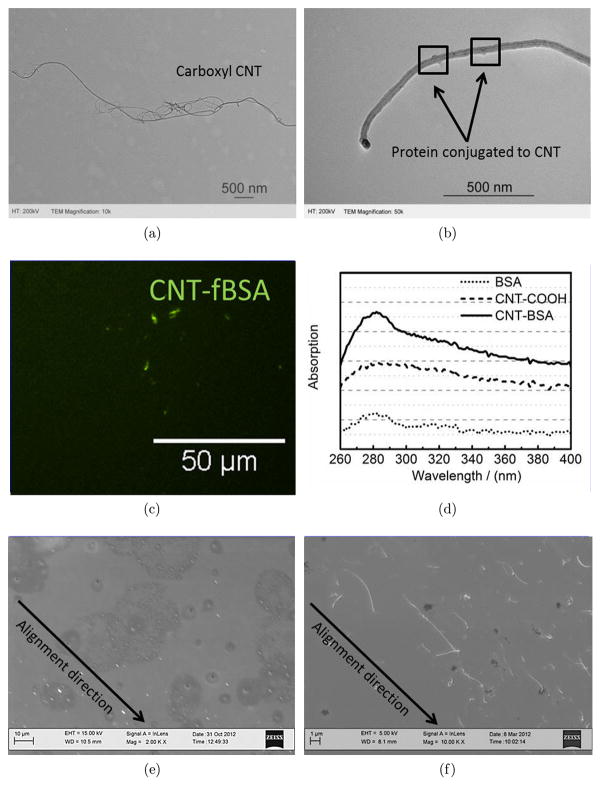
Multiwall carboxyl CNTs were commercially purchased from Nanolab and characterized through TEM. As shown in Fig. 2(a), the length of the carboxyl CNTs was approximately 5 μm with a diameter of approximately 20 nm shown in Fig. 2(b). These specifications are larger than the single wall CNTs and consequently the density of conjugated protein per CNT will be higher. We hypothesized that the higher concentration of protein adhered to CNTs will facilitate cell adhesion.31,32 The appropriate functionalization and protein conjugation to CNTs is a challenging step in preparation of aligned CNT substrates. There are two major types of functionalization approaches including covalent and noncovalent reaction processes.33 To achieve covalent binding, reactive groups34,35 are formulated to allow for further attachment of polymer,36 protein,37 and DNA.38 In contrast, noncovalent binding is based on π- π stacking between CNTs and aromatic groups from the linkers.39 However, there are reports suggesting that noncovalent binding of biological molecules may not remain stable in serum containing situations.40 Thus, based on our goal to provide cell attachment sites on CNTs, we functionalized CNTs through covalent reactions.
Fig. 2.
Characterization of CNT–ligand conjugates and aligned CNT substrate. (a) TEM of carboxylated CNTs. (b) TEM image of Geltrex-conjugated CNTs. (c) Fluorescent image of CNT-fBSA conjugation on Au substrate. (d) UV-vis spectrum of CNT, BSA and CNT-BSA conjugates. CNT-protein conjugates and its dispersion confirmed on Au surfaces which allow cell attachment for HUVEC and hESCs. (e) Low-magnification SEM image of aligned CNT substrate. (f) High-magnification SEM image of aligned CNT substrate.
Our results demonstrated highly specific protein conjugation to CNTs for cell attachment as shown in Fig. 2. The magnification TEM images show Geltrex protein conjugated to CNTs with protrusions from the sidewall of these CNTs to the space. The sizes of these protrusions were found to be approximately 2 nm, which correlate with the calculated size of the proteins comprised in Geltrex according to the equation ,41 in which MW is the molecular weight of the proteins, ρ is the density of solvent, and NA is Avogadro's number. fBSA was also used as a sample protein for the demonstration of protein conjugation to CNTs as shown in the fluorescent stained image Fig. 2. UV-vis absorption was also used to test the conjugation spectrometrically. As shown in Fig. 2(d), CNT–BSA conjugates exhibited an enhanced absorption at 280 nm, which is the typical protein adsorption wavelength. This type of UV absorption is similar to other studies which analyzed protein attachment to materials.42 These results together demonstrate the successful conjugation of proteins to multiwall CNTs through the covalent binding approach. In the following experiments, we used these CNT–protein conjugates to provide unidirectional binding sites to cells with the ultimate objective of dictating the degree of polarization and cell shape on 2D substrates.
Recent studies have shown that aligned topographies are highly effective in directing cell differentiation and adopting specific cell behavior.12,43–48 Various methods including dip pen nanolithography,49 UV lithography50 and electro-spun nanofibers50,51 have been utilized to align cells in microscale. In this study, we used aligned CNTs to guide the direction of focal adhesions and polarization of cells. The utility of this system was tested using HUVECs and ESCs cultured at a single cell level. As observed in low-magnification SEM images [see Fig. 2(e)], the rod-shaped tubes representing carboxyl CNTs were well aligned in the direction of centrifugal force. In the high-magnification SEM image of Fig. 2(f), carboxyl CNTs are clearly observed to be aligned throughout the gold-coated passivated substrates. The CNT alignment pattern was consistent with the patterns produced by Namgung et al., who used spin coating to homogenously distribute single wall CNTs on a poly-l-lysine substrate.52,53 However, our design parameters differed with this study in that (i) we covalently attached proteins to multiwall CNTs to ensure higher stability and (ii) we used multiwall CNTs to provide larger surface areas for protein and cell adhesion.
3.2. Adhesion of HUVEC and hESC single cell on aligned CNT substrate
We used two cell types, HUVECs and hESCs, to demonstrate the utility of aligned CNT substrates. Cell studies were performed on both randomly assembled CNT and aligned CNT substrates. As generally accepted, cell adhesion is fundamental for cell behavior and function. The ability of different cell types to attach to a specified surface varies with corresponding integrins expressed by the cell. In general, stem cells are more sensitive to attachment sites provided on a surface than differentiated cells. hESCs are yet more complex to culture at single cell level with very few proteins able to sustain hESC culture. We propose that by promoting cell adhesion and alignment on CNT platforms, this would provide a chance to conduct hESC culture at single cell level and thus direct hESC differentiation at single cell levels. Note that under these ligand density conditions only a very small amount of cells were able to attach on both random and aligned CNT substrates. This was to promote cell adhesion to CNTs and be used as a pilot study to determine the potential of aligned CNT substrates.
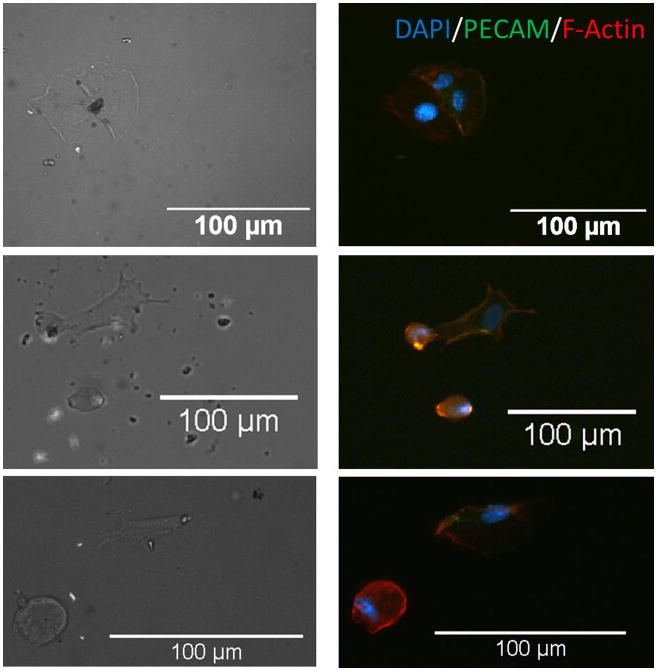
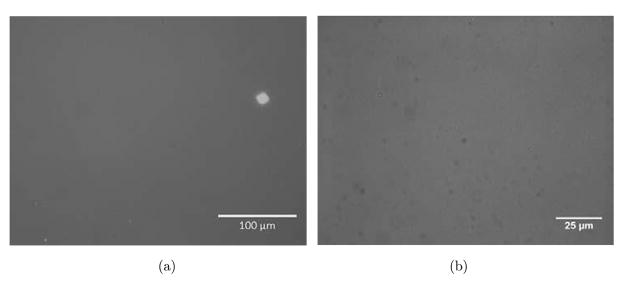
Our results demonstrated HUVECs to be randomly distributed and adopt an irregular shape on randomly assembled CNT substrates (see Fig. 3). HUVECs were able to organize cytoskeleton in random orientations and were confined in a small adhesive area. PECAM-1 staining showed a significant lack of cell–cell contact junction indicating that cells possessed limited mobility on randomly assembly CNT substrate.54 Nonetheless, HUVECs were able to sense and recognize the binding sites on CNT–fibronectin conjugates randomly distributed on the substrates (see Fig. 4). In contrast, immunofluorescent images of HUVECs on aligned CNT substrates showed that cells adopted an aligned shape in direction of CNTs. The HUVEC cells that were able to attach to CNT patterns were all organized in an aligned fashion. PECAM-1 staining shows HUVECs to possess distinct cell–cell contact junctions along the aligned CNT direction. Cyto-skeletal organization showed highly prominent stressed fibres aligned with the CNT as well. It is important to note that HUVECs were able to migrate along the aligned CNT substrate (results not shown). We also conducted simultaneous cellular attachment studies on PEG-SH blocked substrates, but found no HUVEC or single hESC attachment to substrates (see Fig. 5). These results demonstrate the potential of aligned CNT substrates to be used as novel platforms for the study of cell migration and orientation processes associated with angiogenesis, currently a direction of great interest for future studies.
Fig. 3.
Immunofluorescent images of HUVECs on CNT substrates. DAPI (blue), PECAM (green), F-actin (red) are shown. HUVECs are randomly distributed on CNT randomly assembled substrate, either confined by CNTs or spread over CNTs.
Fig. 4.
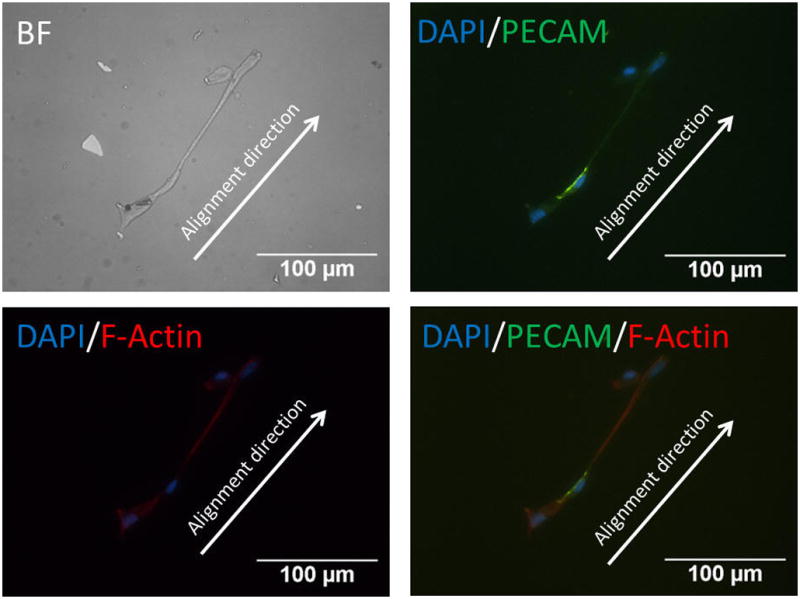
Immunofluorescent images of HUVECs on CNT substrates. DAPI (blue), PECAM (green), F-actin (red) are stained. HUVECs are able to bind to aligned CNT–ligand conjugates and show a spread, aligned phenotype along CNTs aligning direction.
Fig. 5.
Bright field microscopic images of HUVEC and ESC controls on substrates. (a) HUVECs did not attach to substrate in absence of conjugated CNTs (b) ESCs also did not attach to substrates without conjugated CNTs.
Another innovative aspect of this study is the culture of hESCs as single cells on a 2D synthesized substrate. Traditionally, maintaining hESC cultures in vitro requires MEF feeder layers and manual passaging of individual colonies. This method does not lend itself to large-scale production of hESCs for high throughput experiments. Enzymatic dissociation of hESCs has recently been reported as a method for higher scale passaging of cells in regards to future clinical possibilities.55–57 However, there are many limitations associated with enzymatic passaging of hESCs. Among these limitations are the challenges to maintaining hESCs within a certain size56,58,59 resulting in a heterogeneous population of stem cells. Secondly, there is the possibility of genetic and epigenetic changes in hESCs60–62 when cultured in colonies. Overcoming these obstacles is sparamount in harnessing hESC research to decipher single cell level interactions. With potential applications of hESCs on the horizon, a significant challenge is the establishment of controllable and scalable culture methods that allow for control over the ECM signals defining the ultimate fate of cells. The recent work by Denning et al., Braam et al. and Burridge et al. address this challenge by culturing hPSCs in a monolayer under feeder and serum-free conditions at a single cell level.63–65 hESCs cultured under this condition demonstrated stable cell self-renewal and the expression of pluripotency markers.65,66 Despite the versatility of the single cell culture systems, the behavior of hESCs at single cell level remains poorly characterized. Toward optimizing the hPSC single cell culture conditions, additional studies are warranted to fully characterize the pluripotency potential and early stage differentiation processes of cells on monolayer single cell cultures opposed to cells within colonies.
In this work, hESC adhesion experiments also demonstrated the utility of aligned CNT substrates to be a versatile platform to control cell polarity. We studied adhesion of H9 cells at single cell culture levels to demonstrate the ability to selectively bind to a CNT–Geltrex conjugate and retain initial pluripotency. Although there were a large number of initial cells seeded, we were only able to obtain a small amount of cell attachment on the substrate. Single cells were able to bind to CNT substrate with a small percentage of cells being able to fully spread. To test whether CNT substrates affect initial hESC properties such as pluripotency, cytoskeletal formation or spreading tendencies of single ESCs, markers for Oct4 and F-actin were labeled with antibodies. As shown in Fig. 6, cells retained pluripotency by expressing markers of Oct4 and flattened cell morphology. As shown in phase contrast in Fig. 6, hESCs exhibited a more irregular outline showing more prominent filapodia extensions to surrounding CNT–ligand conjugates. Focal adhesions were shown to be expressed in normal pattern and intact indicating a normal cell phenotype on the CNTs as shown in Fig. 6(a). This demonstrated the functionally of the CNT substrate and the ability to parse the substrate–hESC interaction in future studies.
Fig. 6.
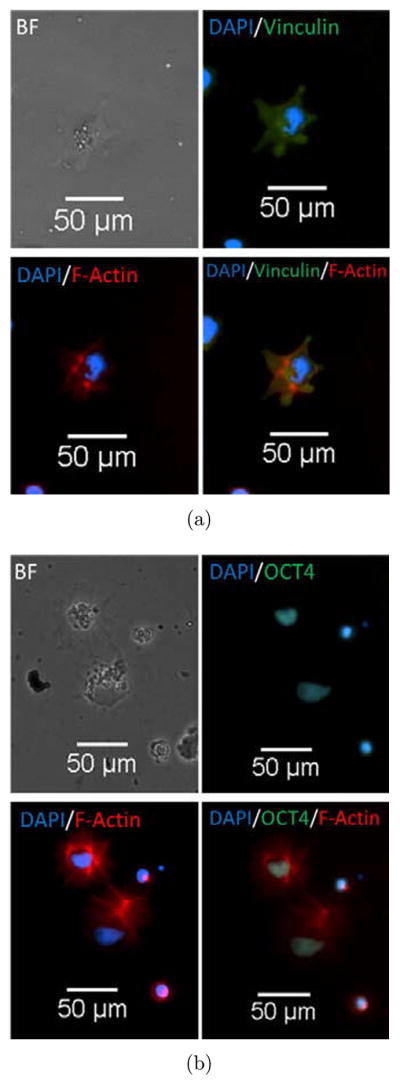
(a) Top left showing brightfield image of single H9 cell line, top right showing vinculin in green and DAPI in blue, bottom left showing actin in red and DAPI in blue, bottom right showing merged image. This demonstrates that H9 cells are able to bind to CNT substrate with normal focal adhesion structure and spreading tendencies. (b) Top left showing brightfield image of single H9 cell line, top right showing Oct4 in green and DAPI in blue, bottom left showing actin in red and DAPI in blue, bottom right showing merged image. H9 cells are able to attach and spread on CNT substrate while retaining initial pluripotency.
4. Conclusions
In summary, we have developed a substrate patterned with aligned CNT–ligand conjugates to be used for the tuning of focal adhesions, cell directionality and spreading. We tested the utility of this system using HUVECs and hESC single cells. Our results demonstrated that CNTs conjugated with ligands were able to guide the spatial and directional formation of focal adhesions in cells. Furthermore, we found that through aligning focal adhesions, the spreading and polarity of cells can be controlled. Our long-term vision is to use these nanoengineered substrates for the control of asymmetric/symmetric division in stem cells and thereby tuning germ layer specification of hESCs in vitro.
Acknowledgments
We gratefully acknowledge support from the National Institute of Health (Grant NIH P20 GM103641) and National Science Foundation (Grant EPS-0903795).
References
- 1.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song W, et al. Langmuir. 2011;27:6155. doi: 10.1021/la200487w. [DOI] [PubMed] [Google Scholar]
- 3.Chen CS, et al. Biochem Biophys Res Commun. 2003;307:355. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 4.McBeath R, et al. Dev Cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 5.Zhu BS, et al. Tissue Eng. 2005;11:825. doi: 10.1089/ten.2005.11.825. [DOI] [PubMed] [Google Scholar]
- 6.Zernicka-Goetz M. Semin Cell Dev Biol. 2004;15:563. doi: 10.1016/j.semcdb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 7.De Smet I, Beeckman T. Nat Rev Mol Cell Biol. 2011;12:177. doi: 10.1038/nrm3064. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita YM. Curr Opin Cell Biol. 2010;22:605. doi: 10.1016/j.ceb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li D, et al. J Cell Biol. 2010;191:631. doi: 10.1083/jcb.201006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mammoto T, Ingber DE. Development. 2010;137:1407. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daga RR, Nurse P. J Cell Sci. 2008;121:1973. doi: 10.1242/jcs.011825. [DOI] [PubMed] [Google Scholar]
- 12.Jones ENB, Mallapragada SK. J Biomater Sci Polym Ed. 2007;18:999. doi: 10.1163/156856207781494449. [DOI] [PubMed] [Google Scholar]
- 13.Engler AJ, et al. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 14.Forte G, et al. Stem Cells. 2008;26:2093. doi: 10.1634/stemcells.2008-0061. [DOI] [PubMed] [Google Scholar]
- 15.Thery M, Bornens M. Curr Opin Cell Biol. 2006;18:648. doi: 10.1016/j.ceb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Torres-Padilla ME, et al. Nature. 2007;445:214. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piotrowska-Nitsche K, et al. Development. 2005;132:479. doi: 10.1242/dev.01602. [DOI] [PubMed] [Google Scholar]
- 18.Lee MR, et al. Biomaterials. 2010;31:4360. doi: 10.1016/j.biomaterials.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Xie JW, et al. Biomaterials. 2009;30:354. doi: 10.1016/j.biomaterials.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engler AJ, et al. J Musculoskelet Neuronal Interact. 2007;7:335. [PubMed] [Google Scholar]
- 21.Dai HJ, Wong EW, Lieber CM. Science. 1996;272:523. [Google Scholar]
- 22.Shokrieh MM, Rafiee R. Mech Compos Mater. 2010;46:155. [Google Scholar]
- 23.Cheng Q, Rutledge K, Jabbarzadeh E. Ann Biomed Eng. 2013;41:904. doi: 10.1007/s10439-012-0728-8. [DOI] [PubMed] [Google Scholar]
- 24.Ishii M, Hamilton B, Poolton N. J Appl Phys. 2008;104:103535. [Google Scholar]
- 25.Borka D, et al. Nucl Instr Methods Phys Res B. 2012;279:169. [Google Scholar]
- 26.Ryoo SR, et al. ACS Nano. 2010;4:6587. doi: 10.1021/nn1018279. [DOI] [PubMed] [Google Scholar]
- 27.Zhang XK, et al. Carbon. 2008;46:453. [Google Scholar]
- 28.McCarron PA, et al. J Biomed Mater Res A. 2008;87A:873. doi: 10.1002/jbm.a.31835. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, et al. Nat Protoc. 2009;4:1372. doi: 10.1038/nprot.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saha K, et al. Proc Natl Acad Sci USA. 2001;108:18714. [Google Scholar]
- 31.Gunawan RC, et al. Langmuir. 2006;22:4250. doi: 10.1021/la0531493. [DOI] [PubMed] [Google Scholar]
- 32.Lagunas A, et al. Nanomedicine. 2012;8:432. doi: 10.1016/j.nano.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, et al. Nano Res. 2009;2:85. doi: 10.1007/s12274-009-9009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosca ID, et al. Carbon. 2005;43:3124. [Google Scholar]
- 35.Niyogi S, et al. Acc Chem Res. 2002;35:1105. doi: 10.1021/ar010155r. [DOI] [PubMed] [Google Scholar]
- 36.Zhao B, et al. J Am Chem Soc. 2005;127:8197. doi: 10.1021/ja042924i. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, et al. Nat Biotechnol. 2008;26:1285. doi: 10.1038/nbt.1501. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z, et al. Angew Chem Int Ed. 2009;48:7668. doi: 10.1002/anie.200902612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, et al. J Am Chem Soc. 2002;124:9034. doi: 10.1021/ja026104m. [DOI] [PubMed] [Google Scholar]
- 40.Moon HK, et al. Nano Res. 2008;1:351. [Google Scholar]
- 41.Fournier RL. Basic Transport Phenomena in Biomedical Engineering. 2. Taylor & Francis; New York: 2007. p. xxii.p. 450. [Google Scholar]
- 42.Chen JY, et al. J Am Chem Soc. 2008;130:16778. doi: 10.1021/ja805570f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang JX, et al. J Biomed Mater Res A. 2012;100A:632. [Google Scholar]
- 44.Guan JJ, et al. Biomaterials. 2011;32:5568. doi: 10.1016/j.biomaterials.2011.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pijnappels DA, et al. Circ Res. 2008;103:167. doi: 10.1161/CIRCRESAHA.108.176131. [DOI] [PubMed] [Google Scholar]
- 46.Wang PY, et al. Acta Biomater. 2011;7:3285. doi: 10.1016/j.actbio.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 47.Huang CY, et al. Biomaterials. 2012;33:1791. doi: 10.1016/j.biomaterials.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng CP, Swartz MA. Ann Biomed Eng. 2006;34:446. doi: 10.1007/s10439-005-9067-3. [DOI] [PubMed] [Google Scholar]
- 49.Curran JM, et al. Lab Chip. 2010;10:1662. doi: 10.1039/c004149a. [DOI] [PubMed] [Google Scholar]
- 50.You MH, et al. Biomacromolecules. 2010;11:1856. doi: 10.1021/bm100374n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dror Y, et al. Langmuir. 2003;19:7012. [Google Scholar]
- 52.Opatkiewicz J, LeMieux MC, Bao ZN. ACS Nano. 2010;4:2975. doi: 10.1021/nn101092d. [DOI] [PubMed] [Google Scholar]
- 53.Lin DW, et al. ACS Nano. 2011;5:10026. doi: 10.1021/nn203870c. [DOI] [PubMed] [Google Scholar]
- 54.Schimmenti LA, et al. J Cell Physiol. 1992;153:417. doi: 10.1002/jcp.1041530222. [DOI] [PubMed] [Google Scholar]
- 55.Xu CH, et al. Nat Biotechnol. 2001;19:971. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 56.Sjogren-Jansson E, et al. Dev Dyn. 2005;233:1304. doi: 10.1002/dvdy.20459. [DOI] [PubMed] [Google Scholar]
- 57.Cowan CA, et al. N Engl J Med. 2004;350:1353. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 58.Rosler ES, et al. Dev Dyn. 2004;229:259. doi: 10.1002/dvdy.10430. [DOI] [PubMed] [Google Scholar]
- 59.Carpenter MK, et al. Dev Dyn. 2004;229:243. doi: 10.1002/dvdy.10431. [DOI] [PubMed] [Google Scholar]
- 60.Hasegawa K, et al. Stem Cells. 2006;24:2649. doi: 10.1634/stemcells.2005-0657. [DOI] [PubMed] [Google Scholar]
- 61.Draper JS, et al. Nat Biotechnol. 2004;22:53. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 62.Herszfeld D, et al. Nat Biotechnol. 2006;24:351. doi: 10.1038/nbt1197. [DOI] [PubMed] [Google Scholar]
- 63.Denning C, et al. Int J Dev Biol. 2006;50:27. doi: 10.1387/ijdb.052107cd. [DOI] [PubMed] [Google Scholar]
- 64.Braam SR, Denning C, Mummery CL. Methods Mol Biol. 2010;584:413. doi: 10.1007/978-1-60761-369-5_22. [DOI] [PubMed] [Google Scholar]
- 65.Burridge PW, et al. PLoS One. 6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hudson JE, et al. Stem Cells Dev. 2012;21:1513. doi: 10.1089/scd.2011.0254. [DOI] [PubMed] [Google Scholar]