Abstract
The brain development transcription factor OTX2 is overexpressed and/or genomically amplified in most medulloblastomas, but the mechanistic basis for its contributions in this setting are not understood. In this study we identified OTX2 as a transcriptional repressor and a gatekeeper of myogenic and neuronal differentiation in medulloblastoma cells. OTX2 binds to the MyoD1 core enhancer through its homeobox domain and the remarkable repressor activity exhibited by the homeobox domain renders OTX2 transcriptionally repressive. RNAi-mediated attenuation of OTX2 expression triggered myogenic and neuronal differentiation in vitro and prolonged the survival in an orthotopic medulloblastoma mouse model. Conversely, inducing myogenic conversion of medulloblastoma cells led to the loss of OTX2 expression. In medullomyoblastoma (MMB), a medulloblastoma subtype containing muscle elements, myogenic cells share cytogenetic signatures with the primitive tumor cells and OTX2 expression was lost in the differentiated myogenic cells. Thus, OTX2 functions via its homeobox domain as a suppressor of differentiation and the loss of OTX2 expression is linked to the myogenesis in medullomyoblastoma. Together, our findings illustrate the origin of muscle cells in MMBs and the oncogenic mechanism of OTX2 as a repressor of diverse differentiating potential.
Keywords: OTX2, medulloblastoma, medullomyoblastoma, MyoD, myogenic differentiation
Introduction
Medulloblastoma, the most common type of pediatric brain malignancy, is an aggressive primitive neuroectodermal tumor arising from the cerebellum. It presents a significant cause of cancer-related death in children and current treatment of radio-chemotherapy could impair children’s development and cause long-term adverse effects (1). In addition to the general classic medulloblastoma designation, current WHO classification recognizes four variants: desmoplastic/nodular medulloblastoma, medulloblastoma with extensive nodularity anaplastic medulloblastoma and large cell medulloblastoma (2). The most common classic medulloblastoma usually arises in the vermis of the cerebellum, lacks distinctive features and is composed of densely packed small undifferentiated cells. A secondary description termed Medullomyoblastoma exits for the medulloblastoma with variously differentiated myogenic cells mixed with the tumor cells (2). It is featured with rhabdomyoblastic elements immuno-reactive to desmin, myoglobin and fast myosin (3).
In some medulloblastomas, driver mutations have been identified. For example, Sonic Hedgehog (SHH) pathway mutations are mainly associated with the nodular/desmoplastic and anaplastic medulloblastomas, which presumably arise from the granule neuron precursor cells (GNPCs) (2). Activating mutations in the WNT pathway contribute to about 7–15 % of medulloblastoma and mainly present as the classic medulloblastoma (2). Recently, this group of medulloblastoma has been determined of originating outside the cerebellum from the cells of the dorsal brainstem (4). Among the majority of medulloblastomas that are non-SHH and non-WNT types, the driver genetic alterations and tumor origin are largely undefined, while the overexpression and/or amplification of MYC and OTX2 represent one of the main oncogenic features (2, 5).
Orthodenticle Homeobox 2 (OTX2), a member of a highly conserved family of the bicoid-like homeobox transcription factors, controls brain morphogenesis (6). During embryogenesis, OTX2 is required for specification and regionalization of the developing brain, and is expressed in restricted areas of the forebrain and midbrain and throughout the posterior cerebellum, particularly, within the external granular layer and the emerging internal granular layer (6, 7). The abundant expression of OTX2 in developing brain is silenced in adult rodent brain except for the pineal gland and ventral tegmental area (VTA) neurons (8, 9). OTX2 controls neuron subtype identity by antagonizing molecular and functional features of the dorsal–lateral ventral tegmental area (10). In the ventral midbrain and dorsal thalamus, OTX2 controls the identity of neuronal progenitors and is required to suppress the progenitors from generating various neuronal cell types during the brain development (11, 12). These findings indicate the pivotal repressor role of OTX2 in controlling the fate and differentiation of various progenitors in early developing brain.
Although OTX2 expression in children and adults is still found in the retina, OTX2 is mainly silenced in the central nervous system and the other organs in human (13). Genomic copy gains and amplification have been identified in various frequencies ranging from 1 % to 21% by different methods and studies (5, 14, 15). Several laboratories have reported the over-expression of OTX2 in over 60% of medulloblastoma (13, 16, 17). The sustained expression of OTX2 in the tumorigenic stem cells and progenitors could possibly achieve the similar function of maintaining the primitive status of those medulloblastoma-generating cells. This function could be highly relevant, since medulloblastoma is regarded as primitive neuroectodermal tumor (PNET), a type of embryogenic tumor, and is believed to originate from aberrant neural stem/progenitor cells.
The significance of OTX2 as a medulloblastoma oncogene is underscored by the recent whole-exon sequencing and copy number analysis of medulloblastoma genomes (15). The genomic amplification frequency of OTX2 gives statistical support to the notion that this alteration is selected as a driver mutation, and beside MYC, OTX2 is likely only one of the two amplified oncogenes in medulloblastoma. Overexpression of OTX2 in the mouse hindbrain resulted in the accumulation of proliferative clusters of cells in the cerebellar white matter and dorsal brainstem of postnatal mice (18). Additionally, knockdown by OTX2 siRNA or suppression of OTX2 promoter by 9-cis retinoic acid slowed medulloblastoma tumor growth in mice (5, 19), suggesting that OTX2 expression sustains tumor growth. Recently, Bunt et al reported that induced over-expression of OTX2 in MB cells leads to G1 arrest in cell cycle and senescence-like phenotype (20), while, however, an inducible knockdown of OTX2 activated cell cycle regulators and neuronal genes (21). Beside genomic amplification as a means of OTX2 activation, the majority of medulloblastomas overexpress OTX2 through an unknown mechanism. Despite its strong presence as a potential oncogene, the basic underlying molecular mechanism of OTX2 in medulloblastoma tumorigenesis remains to be defined.
During the course of the present study we determined that OTX2 is involved in differentiation and hypothesized that OTX2 was additionally involved in the closely related MMB, in part due to the large and unusual regions of differentiated muscle cells within this tumor. MMB makes up 3–5% of all medulloblastoma cases and the features of this tumor may provide important clues to the pathogenesis of this embryonal tumors (22, 23). MMB’s distinguishing feature is the presence of cells showing muscle differentiation among the primitive neuroectodermal tumor cells that characterize medulloblastomas (3). These myogenic elements in MMB can be found in various differentiation phases, ranging from myoblastic single cells to well developed striated muscle structure (24). The origin of the muscle components in MMB has been an interesting question since its first description. Different hypotheses include the possibilities that tumor myoblasts arise from recruitment and migration of mesenchymal or multipotent endothelial cells, or that MMB is a variant of malignant teratomas (3). More recently it has been speculated that MMB myoblasts are a result of the primitive neuroectodermal cells undergone myogenic differentiation (3, 25).
In this study, we aimed to investigate the molecular mechanism of OTX2 in medulloblastoma and shed light as to why MMB contains muscle cells and how OTX2 contributes to this process. Starting with the genes that are transcriptionally regulated by OTX2 in medulloblastoma, we built a molecular model of OTX2 that supports its function as a gatekeeper of myogenic and neuronal differentiation in medulloblastoma. Turning off OTX2 expression can cause medulloblastoma cells to progress towards myogenic and neuronal differentiation. Thus, inducing differentiation by interfering with OTX2-mediated transcriptional repression could offer therapeutic benefit, as shown by the survival extension in the orthotopic medulloblastoma mouse model.
Materials and Methods
Cell Lines and Cell Culture
Cos 1, Hela, mouse neuroblastoma cell line Neuro2a and mouse myoblast cell line C2C12 were acquired from ATCC, Manassas, VA. The following Medulloblastoma cell lines were used in the study were obtained from the Duke University Brain Tumor Center or indicated in the references: D283Med (D283)(26), D341Med (D341), D425Med (D425), D487Med (D487), D556Med (D556), DAOY, MCD1, UW228-2 (UW228) and Mhh-Med-1 (Mhh1)(27). All cells were maintained in DMEM media supplemented with 10 % fetal bovine serum and antibiotics. All cells were kept in frozen stocks upon reception and were not additionally authenticated.
Plasmids and Constructs
OTX2-L or OTX2 was subcloned into pCDNA3.1 and transfected into Cos1 cells by Lipofectamine 2000 (Invitrogen, Calsbad, CA). OTX2 was fused with 3 copies of FLAG sequence on the N-terminus and subcloned in pCMV-TAG-2B (Stratagene, La Jolla, CA). This construct was transfected in D425 cells by electroporation at 200 volts/ 25 ms with a GenePulser (Bio-Rad, Calsbad, CA) and selected by 1 mg/ml of Geneticin. Single clones were selected by limiting dilution. Homeobox triple mutant OTX2-3M and OTX2-HD-3M (R89G, P133T and P134A) were generated, which lost the DNA-binding affinity of the homeobox domain as described before (28). OTX2-HD contains the HD domain (aa 35–95). OTX2-ΔHD was created by deleting the aa 35–95 of human OTX2 cDNA.
Human MyoD core enhancer sequence was cloned into pGL3-P vector containing SV40 minimal promoter and luciferase to create the pMyoD-CER construct (29). The promoter region of human synapsin I (−408 to +47), with or without the Re1 sequence as previously described (30), was cloned in pGL3 luciferase construct (Promega, Madison, WI). Plasmids were transfected into Hela or Neuro2a cells by Lipofectamine 2000, or into D425 cells by electroporation as described above. pRL-CMV from Promega encoding Renilla luciferase was co-transfected as the transfection control. After 36 h, a luciferase assay was performed with the dual-luciferase reporter assay system from Promega using the FB12 luminometer (Zylux, Oak Ridge, TN). At least three independent experiments were performed. Luciferase activity was normalized to Renilla luciferase activity.
siRNA Knockdowns
The following siRNA duplexes were ordered from (Integrated DNA Technologies, Coralville, IA): OTX2_562: rGrGrArUrArUrGrCrUrGrGrCrUrCrArArCrUrUrCrCrUrACT/rArGrUrArGrGrArArGrUrUrGr ArGrCrCrArGrCrArUrArUrCrCrUrU, OTX2_758: rCrCrArCrUrGrArUrUrGrCrUrUrGrGrArUrUrArUrArArGGA/rUrCrCrUrUrArUrArArUrCrCr ArArGrCrArArUrCrArGrUrGrGrUrU, REST_502: rGrCrArGrArArUrCrUrGrArArGrArArCrArGrUrUrUrGrUGC/rGrCrArCrArArArCrUrGrUrUr CrUrUrCrArGrArUrUrCrUrGrCrUrU. Randomized siRNA control was purchased from Ambion, Austin, TX. In each experiment, 10 µg siRNA were tranfected into 2 million cells by electroporation as described above. Inducible OTX2 Knockdown by shRNA was achieved with the V2THS_87164 lentivirus construct of Open Biosystems, Huntsville, AL. OTX2 shRNAmir in pTRIPZ were transfected along with SPAX2 and pMD.G in 293T cells by Lipofectamine 2000 (Invitrogen). Virus was harvested after 48 hour and infected D425 cells by incubating with 8 µg/ml polybrene (Sigma, St. Louis, MO). Control cells were infected with lentivirus carrying the same construct but lacking the OTX2-shRNAmir sequence. Cells were selected by puromycin and transcription of shRNAmir were induced by adding 0.1 µg/ml doxycycline and assessed by the expression of RFP.
Isolation of RNA and RT-PCR
Total RNA was isolated by SV Total RNA Isolation Kit (Promega). cDNA was made by cDNA Synthesis System (Invitrogen). PCR was performed using Platinum Taq polymerase (Invitrogen) for 27 cycles using gene specific custom primer.
Antibodies and Western blotting
Cells were lysed in lysis buffer as described before and immunostaining was performed according to standard procedure (31). The following antibodies were used in this study: mouse anti-OTX2 against the human OTX2 aa1-289 (MAB1979) and anti-β-TubIII clone Tuj1 (R&D Systems, Minneapolis, MN); rabbit anti-OTX2 against the human full length OTX2 (AB9566) and anti-synapsin I (Millipore, Billerica, MA); mouse anti-MyoG clone F5D, mouse anti-Gal4, rabbit anti-MYH H-300 and anti-GAPDH (Santa Cruz Biotech., Santa Cruz, CA); rabbit anti-REST (Millipore), rabbit and mouse anti-FLAG (Sigma, St. Louis, MO);mouse anti-desmin antibody clone D33 (Dako, Carpinteria, CA), rabbit anti-desmin antibody clone Y66 (Millipore) and mouse anti-p21 (Cell Signaling Tech., Danvers, MA). Mouse anti-REST antibody was kindly provided by D.J. Anderson. Signals were visualized by the SuperSignal chemiluminescent system (Pierce, Rockford, IL).
Chromatin Immunoprecipitation (ChIP)
The OTX2 ChIP assay was carried out with the ChIP assay kit according to the manufacturer’s instructions (Upstate). Briefly, D425 cells were first crosslinked with 1% formalin for 15 min, lysed by SDS buffer and sonicated for 1 min at 30% power output by a VC505 sonicater (Sonic & Materials Inc., Newtown, CT). Rabbit anti-OTX2 antibody or a control rabbit antibody was used for immunoprecipitation. PCR reactions were carried out with primers targeting the indicated regions.
Immunofluorescence Staining
Paraffin-embeded sections of confirmed cases of medullomyoblastomas, MMB1-3, were obtained from the Department of Pathology of the Children’s Hospital Boston, and MMB4 was obtained from the Department of Pathology of Johns Hopkins Hospital. Mouse brain tumor sections as well as human normal brain sample were embedded in paraffin. Slides were deparrafinized using a standard procedure and treated with antigen retrieval citra solution (BioGenex, San Ramon, CA). The Immunofluorescence Staining staining using anti-OTX2 or anti-desmin antibodies followed the procedure described before (19).
Co-Culturing of C2C12 and D425-GFP
D425 cells were transfected with pCDNA3.1-GFP and a polyclonal population (D425-GFP) was selected with 0.2 mg/ml zeocin. Equal proportions of D425-GFP and C2C12 cells were mixed and grown on chamber slides with medium containing 10 % FBS for 2 days and later with differentiation mediam containing 2 % horse serum for 4–6 days. The cells were then fixed and stained as described above.
Fluorescent In Situ Hybridization (FISH)
Human BAC clone RP11-1085N6 from Invitrogen was used for detecting OTX2 locus and was directly labeled with SpectrumGreen-dUTP. Pre-labeled SpectrumOrange c-MYC (8q24.12-q24.13) probe was purchased from Abbott Moleculare-Vysis, Des Plaines, IL. FISH on paraffin sections followed the procedure as described before(32). FISH signals were assessed by an Axioplan2 Imaging microscope by Zeiss with the Isis FISH Imaging System V5.4 by MetaSystem GmbH, Germany. The combination of immunofluerescent staining and FISH was carried out according to Peters et al with modifications (33). Briefly, the paraffin sections were deparaffinized in Xylene and rehydrated by an ethanol series. Then sections were incubated overnight at 70°C in citra antigen retrieval buffer (BioGenex), rinsed in PBS, and incubated in 0.5 mg/ml pepsin at 37°C for 3 min. Sections were stained by mouse anti-desmin antibody and DyLight649 (Cy5) anti-mouse secondary antibody from Vector Lab with the procedure described above. Subsequently, the sections were fixed for 10 min in ice cold Clarke’s fixative (3 parts 100% ethanol, 1 part glacial acetic acid), then washed in 95% ethanol, dehydrated in 100% ethanol and dried in the dark. To proceed with FISH, sections were rehydrated in water and processed as described above. Fluorescent signals were analyzed and photographed with a fluorescent microscope equipped with DAPI, FITC, TRITC/Orange and Cy5 filters.
Beadarray Expression Profiling
Total RNA was isolated and subjected to reverse transcription and amplification using recommended procedures from Illumina, San Diego, CA. In the Johns Hopkins Illumina core laboratory led by Dr. Christopher Cheadle, samples were loaded onto a HumanRef-8 Beadarray slide, which contains 8 identical arrays with probes for 22,000 transcripts as annotated by the NCI cDNA annotation. Data were analyzed by the BeadStudio Gene Expression Module program of Illumina.
Measurement of Cell Growth
Cells were plated in 96 well flat-bottomed plates and mixed with 10% of WST-1 solution (Alexis, Lausen, Switzerland). After incubation for 1–2 h at 37 °C, the plates were measured by a Victor3 plate reader at an absorbance of 450 nm (PerkinElmer, Waltham, MA). Wells filled with WST-1 and media were used as blank controls.
Animal Experiments
Female athymic nude mice (NCr-nu/nu) of 4–5 week old were purchased from the Frederick National Laboratory for Cancer Research of NCI, Maryland. For the implantation procedure, mice were anesthetized via intraperitoneal injection of ketamine/xylazine. On a stereotactic frame, 1 mio D425 cells transfected with empty vector or with doxycycline-inducible shRNAmir construct targeting on OTX2 were injected in a prepared burr hole 2mm lateral to the sagittal suture and 1 mm anterior to the coronal suture, at a depth of 3 mm below the dura at a rate of 1 µl/minute. Eight days after the implantation, mice were gavaged with 50 mg/kg doxycycline daily for the first three days and once every five days for the rest of the period.
Results
OTX2 Knockdown Led to Induction of Myogenic Genes in Medulloblastoma Cells
D425, a medulloblastoma cell line with genomic amplification of OTX2 locus (5), expresses the short isoform of OTX2 and is among a panel of OTX2-positive medulloblastoma cell lines (Supplemental Figure S1A & B) (19). OTX2 knockdown by siRNA in D425 cells, led to extensive induction of myogenic pathways. In Fig. 1A, we compared transcripts in D425 cells transfected with a control siRNA to those transfected by OTX2 siRNA (OTX2_562) at 36, 48 and 72 hrs, on the llumina gene expression bead array HumanRef-8 containing probes for 22,000 transcripts. The regulated genes were defined as 2 fold change with a minimal expression of 50 in the array counts. In this single experiment, the significance as well as the false positive counts were determined based on the consistency in the time course of 24, 48 and 72 h. It revealed 277 transcripts with >2 fold induction and 137 transcripts suppressed by more than 50%, with possible replicates corrected. The largest and clearly obvious functional cluster of genes that were activated was a group related to myogenic differentiation. These genes included MyoD1 (MyoD), MyoG, MYH3, MYL4 p21, GADD45γ,MEF2C and p57, all of which were markedly induced after 36 h of OTX2 knockdown. Apoptotic gene caspase 9 was also induced and a moderate sub-G1 fraction indicating the apoptotic cells was observed by flow cytometry (Supplemental Figure S1C). Fifty of the 277 transcripts which were increased at least two-fold were closely related to skeletal, cardiac and smooth muscle function or growth (Supplemental Table S1).
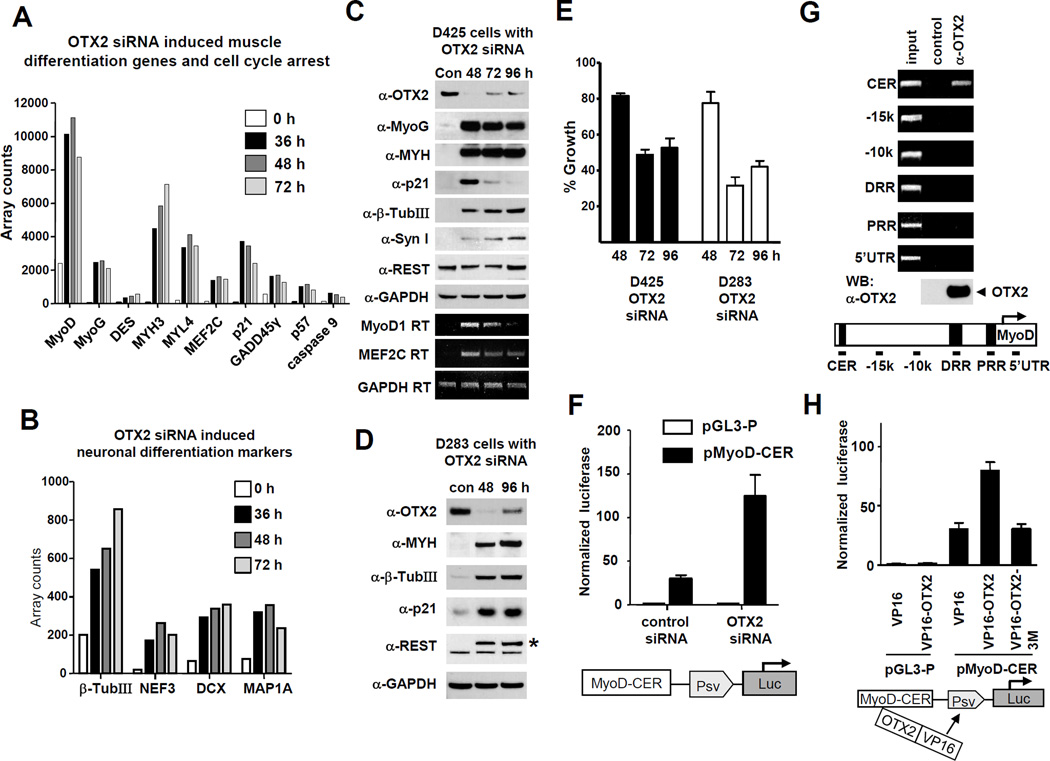
Figure 1. OTX2 knockdown activates myogenic pathway in medulloblastoma cells and OTX2’s association with the core enhancer of MyoD locus.
(A) OTX2 knockdown activated myogenic pathway genes and caspase 9. D425 cells were transfected with OTX2 siRNA (OTX2_562) or with randomized control siRNA. Total RNA was isolated from the cells after 36, 48 or 72 h of transfection. RNA of the control (Con) was made after 48 h of the transfection with control siRNA. Gene expression was analyzed by Illumina Beadarray technology and select sets of genes involved in muscle differentiation were displayed in the graph. MyoD (MyoD1): myogenic determination/myogenic factor 3; MyoG: myogenin/myogenic factor 4; MYH3: myosin heavy polypeptide 3, skeletal muscle; MYL4: myosin light polypeptide 4; p21: CDKN1A; GADD45γ: growth arrest and DNA-damage-inducible gamma; MEF2C: MADS box transcription enhancer factor 2; p57: CDKN1C.
(B) Neuronal differentiation markers are activated by OTX2 siRNA in D425 cells. As described in (A), select sets of genes involved in neuronal differentiation were displayed. β-TubIII: β-tubulin III; NEF3: neurofilament 3; DCX: doublecortex (doublecortin) transcript variant 3.
(C) D425 cells were transfected with OTX2_562 siRNA or a random sequence control siRNA (Con). Western blot (white background) or RT-PCR (black background) was performed at 48, 72 and 96 h for the indicated targets or GAPDH normalization control. Asterix (*) indicates a non-specific band. MYH: skeletal and cardiac myosin heavy chain isoforms.
(D) In a similar experiment as (C), medulloblastoma cell line D283 was transfected with OTX2_562 siRNA or a random sequence control siRNA (con).
(E) OTX2 siRNA reduces growth in D425 cells and D283 cells. D425 and D283 cells treated by control siRNA, OTX2 siRNAs (OTX2_562) and incubated for 48, 72 or 96 h. Viable cells were quantified by WST-1 reagent and graphed as percentage of the control siRNA cells.
(F) Knockdown of OTX2 by siRNA activates the reporter construct of MyoD core enhancer (CER). D425 cells were first transfected with OTX2_562 siRNA or control siRNA for 36 hours. Subsquently, a luciferase construct (pMyoD-CER) containing the 258 bp MyoD CER and a SV40 minimal promoter was transfected and luciferase activity was determined after 24h.
(G) Chromatin immunoprecipitation (ChIP) of OTX2 with MyoD CER. ChIP was performed with D425 cells using control rabbit antibody or OTX2 antibody. Bound DNA fragments were detected by PCR with primers encompassing the indicated regions of MyoD locus.
(H) OTX2 homeobox domain mediates the interaction with MyoD CER. OTX2 or OTX2 homeobox triple mutant OTX2-3M (R89G, P133T and P134A) was cloned in fusion with VP16 transactivation domain and co-transfected with vector pGL3-P or pMyoD-CER in D425 cells. Luciferase activity was measured and normalized.
Myogenic differentiation is governed by a group of basic-helix-loop-helix (b-HLH) DNA-binding proteins, in which MyoD is regarded as the master regulator capable of inducing MyoG and initiating the myogenic pathway (34). Myosin proteins such as MYH3 and MYL4 observed in our data are found either in embryonic or mature muscle cells. MyoD engages the cell cycle machinery to activate cell cycle withdrawal prior to myogenic differentiation. p21/CDKN1A, p57/CDKN1C and GADD45γ are downstream targets of MyoD pathway that are responsible for cell cycle arrest. The induction of the apoptotic initiator caspase 9 indicates apoptosis in the cells with OTX2 knockdown.
Neuronal markers such as the neuron-specific β-tubulin III (β-TubIII/TUBB4), neurofilament 3 (NEF3), NCAM1 and doublecortex (DCX) were also induced (Fig. 1B & Supplemental Table S1), reflecting the activation of neuronal differentiation subsequent to OTX2 knockdown.
At the protein level, myogenic differentiation factors MyoG, myosin heavy chain (MYH), downstream target p21, as well as neuronal markers β-TubIII and synapsin I, were induced after 48 h of OTX2 knockdown by OTX2_562 siRNA, as shown by western blotting (Figure 1C). The inductions of MyoD and MEF2C were confirmed by RT-PCR (Figure 1C). Use of OTX2_758 siRNA targeting a different region of the OTX2 mRNA confirmed these results (Supplemental Figure S2A). A similar induction of differentiation following OTX2 knockdown was also observed in the other OTX2-positive medulloblastoma cell lines, D283, D458 and D341 (Figure 1D & S2B, C). Re1-silencing transcription factor (REST/NRSF) is a major suppressor of neurogenesis, which is turned off after neural stem/progenitor cells undergo neural differentiation (35, 36). We did not observe any significant change in REST protein with both OTX2_562 and OTX2_758 siRNA. The knockdown of OTX2 led to a dramatic decrease of cell growth (Figure 1E), consistent with a previous report (16).
Association of OTX2 with MyoD Core Enhancer
The expression of MyoD is tightly controlled by three regulatory units. The 258 bp core enhancer region (CER) located −20 kb relative to the MyoD transcriptional start site regulates the initial expression of MyoD in muscle precursors and determines the tissue-specific expression (29). The distal regulatory region (DDR) at around −5 kb, is required for the maintenance of MyoD expression in muscle cells (37). The sequence surrounding the core promoter is termed as the proximal regulatory region (PRR). MyoD core enhancer contains four E-box motives, which could provide binding sites for the E-box proteins during heterodimerization with b-HLH myogenic factors (29). The regulation of tissue-specific expression by the 258 bp CER of MyoD can be conveyed by a construct combining CER with a heterologous promoter (29). In this study, we used a CER/SV40 minimal promoter luciferase construct (pMyoD-CER) in D425 cells, where it showed significant activity that reveals myogenic potential (Figure 1F, control siRNA). OTX2 knockdown activated pMyoD-CER substantially further (Figure 1F, OTX2 siRNA), indicating that the MyoD CER is likely involved in the activation of MyoD by OTX2 knockdown.
In chromatin immunoprecipitation (ChIP) with an anti-OTX2 antibody to determine the potential in vivo association of OTX2 with the MyoD CER, DRR, PRR and/or various positions in the MyoD locus, OTX2 showed affinity only with CER (Figure 1G). While OTX2 displayed no intrinsic trans-activity when fused with Gal4-binding domain (Gal4-BD) in D425 cells (Figure 1H), we fused OTX2 with the VP16 trans-activation domain to test its trans-activation ability with the pMyoD-CER construct, since this can serve as an indirect readout of the association of OTX2 with the MyoD CER (38). VP16-OTX2 was capable of activating pMyoD-CER significantly above VP16 vector alone, while the VP16-OTX2-3M construct containing triple DNA-binding mutations in the homeobox domain (HD) lost this trans-activity (Figure 1H). This result indicates that attachment of OTX2 protein with MyoD CER is mediated by the DNA-binding property of the OTX2 HD. It is well established that the homeoprotein Msx1 represses MyoD by directly binding to CER, presumably in cooperation with histone H1b (39). In a mammalian two-hybrid assay, OTX2 showed no binding with the MyoD or H1b protein (Supplemental Figure S3A), suggesting OTX2 does not affect MyoD function via direct protein-protein interaction.
OTX2 HD Functions as a Transcriptional Repressor
We employed a one-hybrid system that uses the binding properties of the Gal4 DNA-binding domain to recruit a Gal4-OTX2 fusion protein onto a Gal4-binding luciferase reporter. The full length OTX2 fusion protein, as well as the OTX2-3M mutant and OTX2 HD alone (OTX2-HD), appeared transcriptionally inert in the Gal4 one-hybrid system in D425 (Figure 2A). Among a series of Gal4-OTX2 deletion mutants, we observed that all constructs containing the HD remained transcriptionally inactive (data not shown). In contrast, the deletion of the homeobox domain rendered OTX2 potently trans-active, suggesting that HD was crucial for OTX2’s ability to suppress transcription.
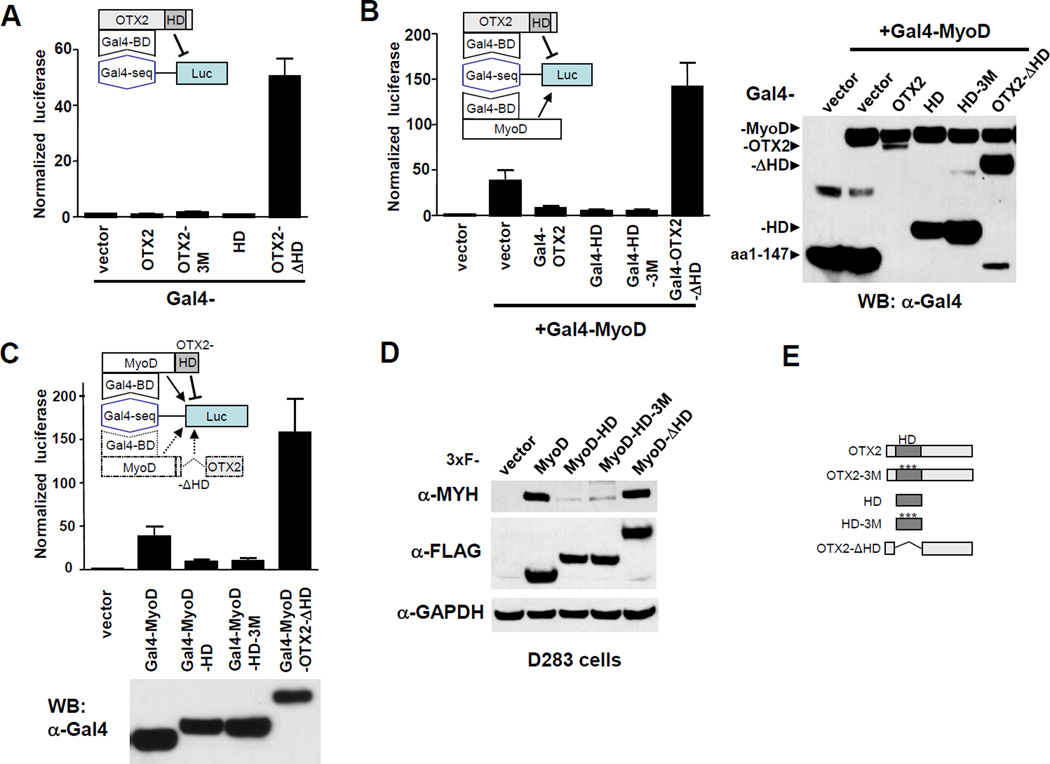
Figure 2. OTX2 functions as a transcriptional repressor of MyoD.
(A) OTX2 homeobox domain (HD) mediates transcriptional repression. OTX2, OTX2-3M, OTX2-HD (HD) or OTX2 deleted of HD (OTX2-ΔHD) were cloned in fusion with Gal4 DNA binding domain (BD, aa1-147) and co-transfected in D425 cells along with a luciferase reporter construct containing 4x Gal4 binding sites (Gal4-seq).
(B) Gal4-OTX2 represses the trans-activity of Gal4-MyoD in D425 cells. Gal4-MyoD was transfected along with empty vector or with indicated Gal4-OTX2 constructs. On the right panel, anti-Gal4 western blot showed the expression levels of transfected constructs.
(C) Fusion protein of MyoD and OTX2-HD (HD) showed repressed transcription activity. OTX2-HD (HD), OTX2-HD-3M (HD-3M) or OTX2-ΔHD were cloned in fusion with Gal4-MyoD and tested for their trans-activities in D425 cells.
(D) Fusion of OTX2-HD decreases the myogenic potential of MyoD protein in D283 medulloblastoma cells. 3xFLAG-tagged MyoD, MyoD-OTX2-HD (MyoD-HD), MyoD-OTX2-HD-3M (MyoD-HD-3M) or MyoD-OTX2-ΔHD (MyoD-ΔHD) was transiently transfected in D283 cells for 2 days. Cell lysates were blotted for myogenic marker MYH.
(E) A summary of the OTX2 mutants used in the study.
Figure 2B shows the result of co-expressing Gal4-MyoD and various Gal4-OTX2 constructs in the one-hybrid trans-activation assay. This experiment reveals that Gal4-OTX2, Gal4-OTX2-HD (Gal4-HD) and Gal4-OTX2-HD-3M (Gal4-HD-3M) were capable of suppressing the trans-activity of Gal4-MyoD, while Gal4-OTX2-ΔHD (Gal4-ΔHD) in combination further elevated the trans-activating potential. The Gal4 DNA binding domain (aa 1–147) is reported to homodimerize and be recruited by the Gal4 binding sites located in the luciferase reporter (40). Since OTX2 and MyoD can be brought into physical proximity with this system as a heterodimer, the repression of MyoD’s transcriptional activity by OTX2 can be observed. The similar repression of MyoD trans-activity was displayed when Gal4-MyoD was fused with OTX2-HD or OTX2-HD-3M and conversely, fusion of Gal4-MyoD with OTX2-ΔHD was able to further enhance the trans-activity of Gal4-MyoD (Figure 2C). These experiments all support the hypothesis that OTX2 suppresses MyoD transcription and that this suppression is mediated by its homeobox domain.
It has been previously described that the trans-activity of Gal4-MyoD is a good indicator of the in vivo myogenic potential of MyoD regarding its induction of myogenic pathway (41). We transiently over-expressed 3xFLAG-tagged MyoD constructs in D283 cells rather than in D425 cells since the latter did not transfect well with full length MyoD. In D283, MyoD-OTX2-HD (MyoD-HD) and MyoD-OTX2-HD-3M (MyoD-HD-3M) fusion proteins showed only a marginal induction of the myogenic marker MYH, while the wildtype MyoD and MyoD-OTX2-ΔHD (MyoD-ΔHD) displayed high levels of MYH induction and reflected similar myogenic potential (Figure 2D). Similar results were obtained with pMyo-CER in D425 cells (Supplemental Figure S3B). Therefore, OTX2, in particular its HD, can suppress MyoD’s myogenic potential of inducing endogenous myogenic gene expression in medulloblastoma cells and fibroblasts.
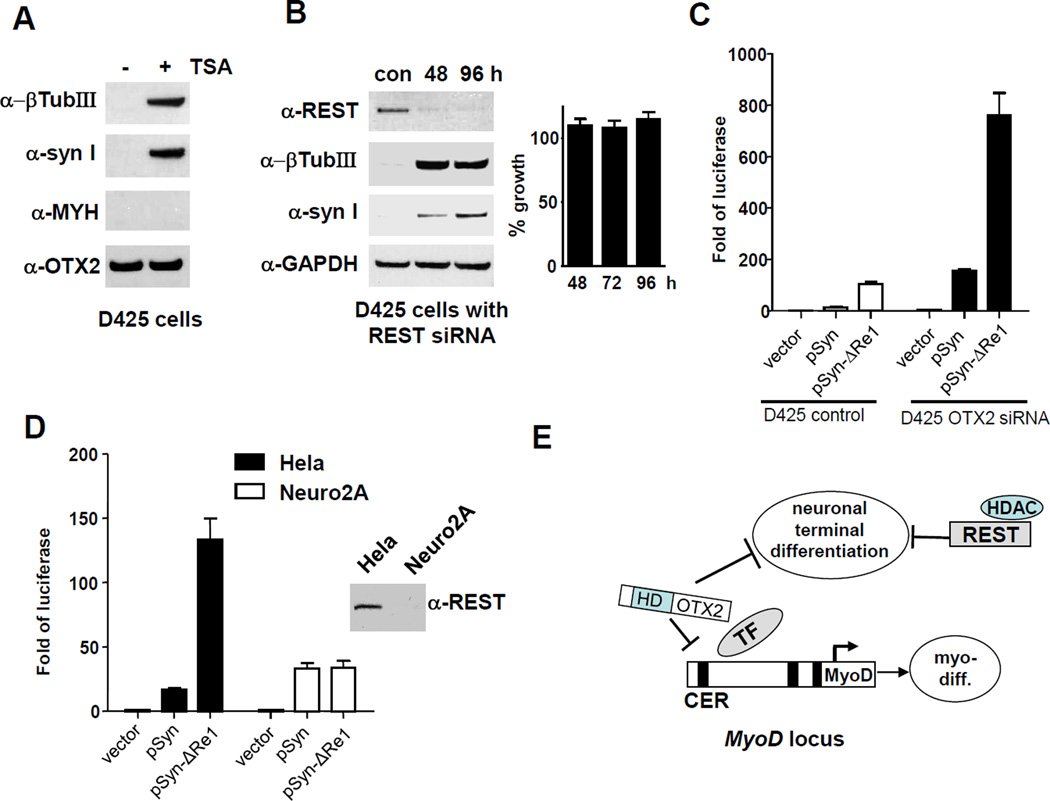
The transcription suppression mediated by OTX2 appeared to be independent of histone deacetylases (HDAC). Incubating D425 cells with trichostatin A (TSA), a HDAC inhibitor, did not activate the myogenic pathway as indicated by the absence of MYH expression, in contrast to the induction of neuronal marker β-TubIII and synapsin I by TSA (Figure 3A). Neuronal differentiation is known to be controlled by REST in association with HDAC (42). REST mainly represses the terminal neuronal differentiation genes such as SCG10, sodium channel type II, synapsin I, β-TubIII, neurofilament 3 (150 kD), synaptophysin, and glutamate receptor through its specific binding to the Re1 consensus sequence within the promoters of these genes (43). In general, REST is expressed in stem cells and non-neuronal cells, and is down-regulated for induction and maintenance of the neuronal phenotype (35). In our study, siRNA-mediated knockdown of REST induced neuronal markers directly repressed by REST, but failed to have any impact on medulloblastoma cell growth. Among the genes induced by OTX2 siRNA knockdown, some are subject to Re1-mediated REST repression, such as β-TubIII and synapsin I, while others like MAP1A and DCX are not known targets of REST (Supplemental Table S1). The promoter constructs with (pSyn) or without (pSyn-ΔRe1) the REST-binding Re1 sequence have been described before (30). In D425 control cells, we observed the expected repression of pSyn and de-repression of pSyn-ΔRe1 (Fig. 3C, left side). Knockdown of OTX2 by siRNA resulted in a marked increase of both pSyn and pSyn-ΔRe1 (760 folds) activities, which lifted pSyn activity well above the level of the de-repressed pSyn-ΔRe1 (104 folds) in D425 control cells (Figure 3C, right side). Therefore, the transcriptional repression on pSyn by OTX2 is not mediated by Re1 sequence and should be independent of REST. The specificity of the REST-mediated repression via Re1 is confirmed by the assay in Hela and Neuro2D cells. In Hela cells expressing endogenous wildtype REST, pSyn was significantly repressed in comparison to pSyn-ΔRe1 (Figure 3D, left side). In contrast, pSyn and pSyn-ΔRe1 displayed the same levels of activity in Neuro2a cells, where no functional REST is expressed (44). Our results indicate that REST is not a direct target of OTX2-mediated suppression and the induction of neuronal markers by OTX2 siRNA knockdown revealed the transcriptional suppression mediated by OTX2 unrelated to REST activity, whereas REST can function as an independent suppressor of neuronal differentiation in association with HDAC (model in Figure 3E). However, in order to suppress neuronal differentiation, both OTX2 and REST are needed as revealed by the fact that knockdown of OTX2 and REST individually was sufficient for the induction of neuronal genes in D425 cells (Figure 3 A and B).
Figure 3. OTX2 represses neuronal markers independently of REST.
(A) Trichostatin A (TSA) induces neuronal differentiation but not muscle differentiation. D425 cells were incubated with 300 nM of histone deacetylase inhibitor TSA for 48 h and subjected to western blotting analysis of neuronal and muscle differentiation markers.
(B) siRNA knockdown of REST induces neuronal differentiation marker β-tubulin III and synapsin I, but shows no effect on cell growth. D425 cells were transfected with control siRNA or REST siRNA in a time course of 48 and 96 h. Western blots revealed the knockdown of REST and the induction of β-tubulin III and synapsin I.
(C) OTX2 knockdown greatly increased the activity of synapsin promoter with and without the REST binding site: Re1 motif. Synapsin I promoter with (pSyn) or without (pSyn-ΔRe1) the 23 bp Re1 motif was cloned in pGL3 luciferase construct. D425 cells treated with control siRNA or OTX2_562 siRNA for 36 h were transfected with pGL3 vector, pSyn or pSyn-ΔRe1 for another 36 h.
(D) A control experiment of pSyn and pSyn-ΔRe1 showed the Re1 specificity. Neuro2D cells do not express functional REST and Hela cells do. pSyn-ΔRe1 activity was de-repressed compared to pSyn in Hela cells, while both construct showed similar activities in Neuro2D cells. Anti-REST western blot confirmed the REST statues in these cells.
(E) Schematic of a proposed model of OTX2 action in medulloblastoma. The homeobox domain of OTX2 acts directly on the core enhancer region (CER) of MyoD to suppress muscle differentiation. Both OTX2 and REST/HDAC are required for the repression of neuronal differentiation markers.
OTX2 Expression and Myogenic Conversion in D425 Cells are Mutually Exclusive
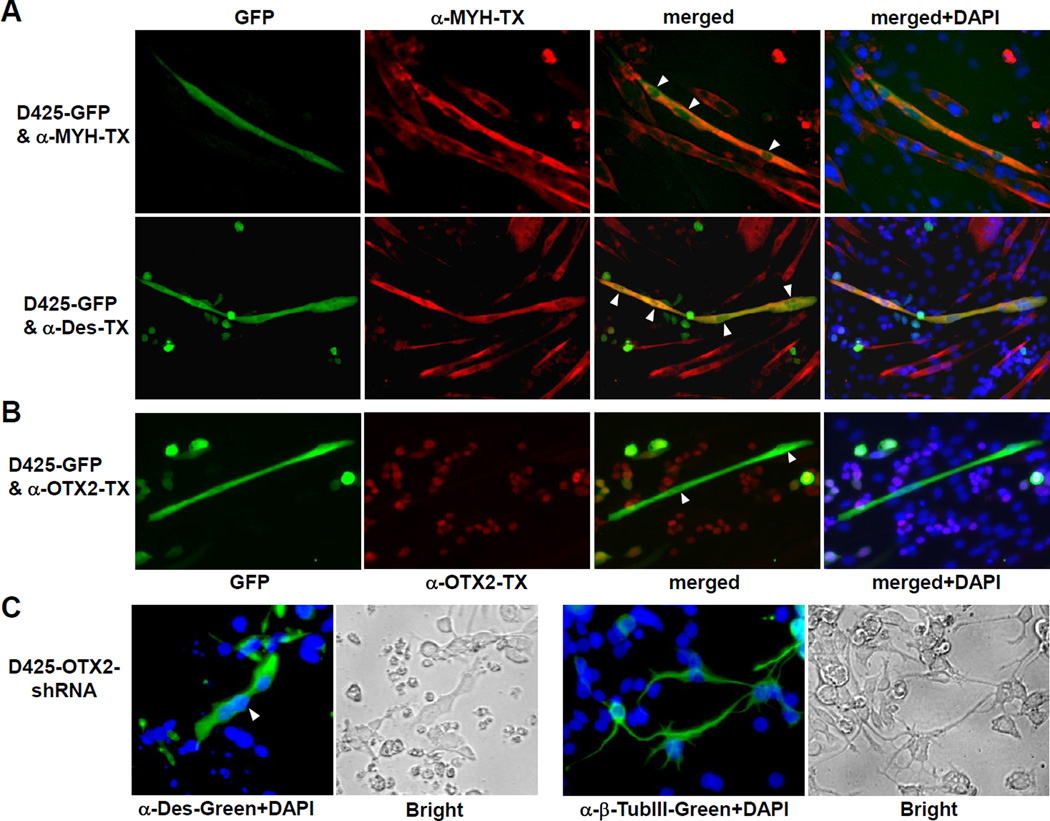
We next examined the relationship of OTX2 expression and potential myogenic differentiation of medulloblastoma cells. It has been demonstrated that the neural stem cells can undergo myogenic conversion when co-cultured with mouse C2C12 myoblasts (45). Whether medulloblastoma cells propagated in serum-containing media are able to be induced into similar myogenic conversion has been unknown before. D425 cells usually grow semi-adherently in single cell form mixed with floating clusters in suspension form. We first generated a stable polyclonal D425 population transfected with pCDNA3.1-GFP and co-cultured it with C2C12 cells in equal proportions on chamber slides. After being cultured in the low serum differentiation condition for 6 days, a small population of GFP-positive myotubes emerged among the myotubes derived from C2C12 myoblasts, along with undifferentiated D425-GFP cells remaining in round single cell form (Figure 4A). The GFP-positive multinucleated cells (green) were stained positive with MYH or desmin antibody (red) and displayed similar morphology and alignment with surrounding myotubes, which indicated the myogenic conversion of D425-GFP cells. These GFP-positive myotubes have lost the OTX2 expression, as revealed by the negative nuclear staining with OTX2 antibody (Figure 4B). In order to rule out the possible fusion of D425 and C2C12 cells, we incubated D425-GFP and C2C12-RFP cells and observed similar transformation of D425 cells (Supplemental Figure S4A). These data indicate that D425 medulloblastoma cells undergoing myogenic conversion lose OTX2 expression.
Figure 4. Mutual exclusion of myogenic differentiation and OTX2 expression.
(A and B) Loss of OTX2 expression by myogenic conversion of D425 cells. GFP-transfected D425 cells were co-cultured with C2C12 mouse myoblasts in differentiation medium. Cells were stained with myogenic marker MYH or desmin (Des) antibody and Texas Red (TX) secondary antibody (A). Expression of OTX2 was visualized by OTX2 antibody and TX secondary antibody (B). Thirty GFP-positive cells with myotube morphology were examined and none of them showed significant OTX2 staining. White arrow heads indicate the multipile nuclei in the myogenic D245 cells, which were stained by DAPI (merge+DAPI). It is worth noting that the polyclonal D425-GFP cells expressed GFP in various levels, which remained OTX2-positive when maintained in the undifferentiated single cell form.
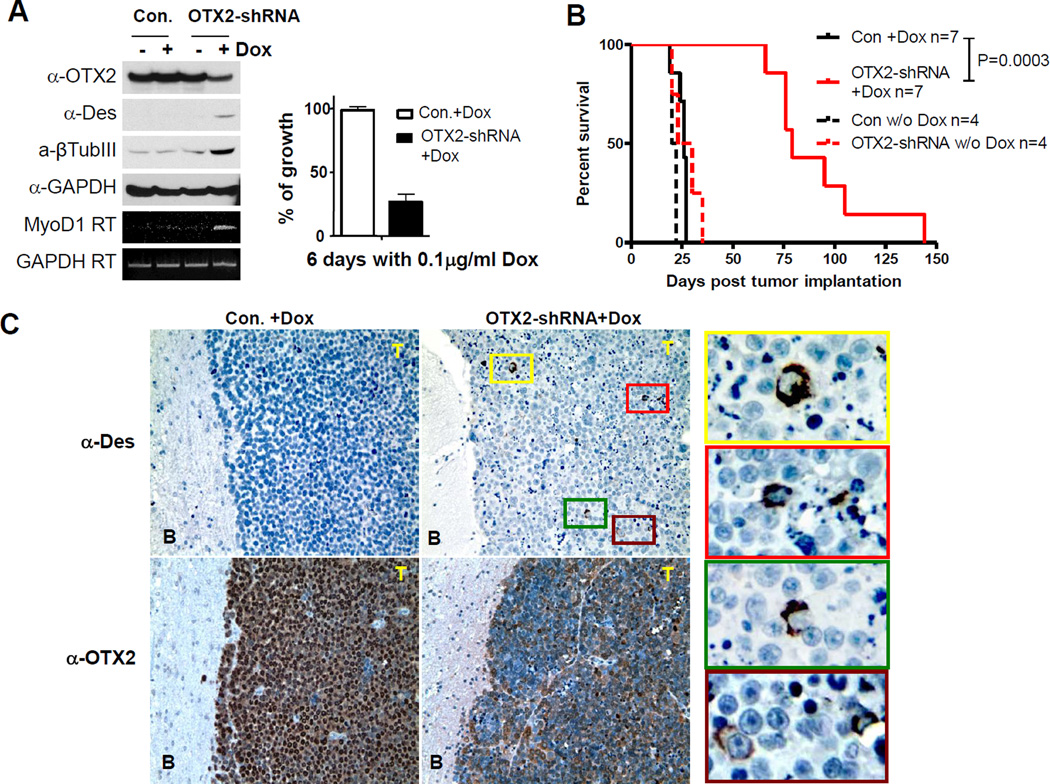
(C) OTX2 knockdown activated myogenic and neuronal differentiation in D425 cells. D425 cells infected with lentivirus of Dox-inducible OTX2-shRNA construct were incubated with 0.1 µg/ml Dox on surface coated with poly-D-lysine for 10 days. Desmin or β-tubulin III staining visualized the cells undergone myogenic or neuronal differentiation. The white arrow head indicates a desmin-positive D425 cell with three nuclei. Bright field pictures with phase contrast were shown next to the immunofluorescent staining. Control D425 cells infected with mock lentivirus did not shown positive staining of desmin and β-tubulin III (data not shown).
Next we created stable D425-OTX2-shRNA lines by infecting D425 with lentivirus carrying a doxycycline (Dox) inducible OTX2-shRNA knockdown construct. After incubating with Dox for 7–14 days, staining with muscle marker desmin and neuronal marker β-tubulin III revealed diverse populations undergone myogenic or/and neuronal differentiation, with the appropriate multinucleated myogenic or network-forming neuronal features (Figure 4C). Similar results of OTX2 knockdown in D283 cells were shown in Supplemental Figure S4B & C. The effective Dox-induced knockdown of OTX2 and the resulted inductions of MyoD and desmin in D425 cells were confirmed in Figure 5A. Thus, the knockdown of OTX2 led to myogenic and neuronal phenotypes in D425 and D283 medulloblastoma cells.
Figure 5. OTX2 knockdown induced myogenic marker in D425 xenograft tumor.
(A) D425 cells were infected with lentivirus of Dox-inducible OTX2 shRNA. Four days after incubation with Dox, induction of the indicated genes were analyzed by Western blot and RT-PCR. Control cells were infected with the control lentivirus of vector lacking OTX2-shRNA.
(B) Survival curve of the nude mice implanted intracranially with D425 cells infected with control lentiviral construct or lentiviral construct with Dox-inducible OTX2 shRNA. Solid lines indicate the groups implanted with control (Con) and OTX2-shRNA cells treated with Dox after 8 days of implantation. Dashed lines indicate the untreated (w/o DOX) mice implanted with control and OTX2-shRNA cells. The mean survival of Con+Dox, OTX2-shRNA+Dox, Con w/o Dox and OTX2-shRNA w/o Dox are 26, 79, 21 and 26.5 days, respectively.
(C) Paraffin-embedded brain samples of control D425 xenograft or OTX2 knockdown D425 xenograft were stained by desmin (Des) or OTX2 antibody. 100x objective was used and the areas with colored frames were further magnified and shown on the right side of the picture.
OTX2 Knockdown Led to Myogenic Induction and Prolonged Survival in Orthotopic Xenografts
The D425-OTX2-shRNA cells were implanted in the frontal lobe of athymic nude mouse brain and the animals were fed with Dox 8 days following the implantation. Previous report using D425 cells that were stably transfected with OTX2-shRNA showed moderate survival extension compared to the control (5). In Figure 5B, in vivo induction of OTX2-knockdown demonstrated a marked survival benefit in mice, extending the mean survival from 26 days of the control animals to 79 days. Due to the reported potential anti-tumor activity of Dox (46, 47), the control animals were equally treated with Dox. Staining of the paraffin-embedded brain slides with anti-OTX2 antibody showed reduced OTX2 expression by OTX2 knockdown (Figure 5C). Myogenic marker desmin was negative in the control D425 tumor, but induced in cells within the D425-OTX2-shRNA tumors (Figure 5C, upper pictures and the color framed magnifications on the right side).
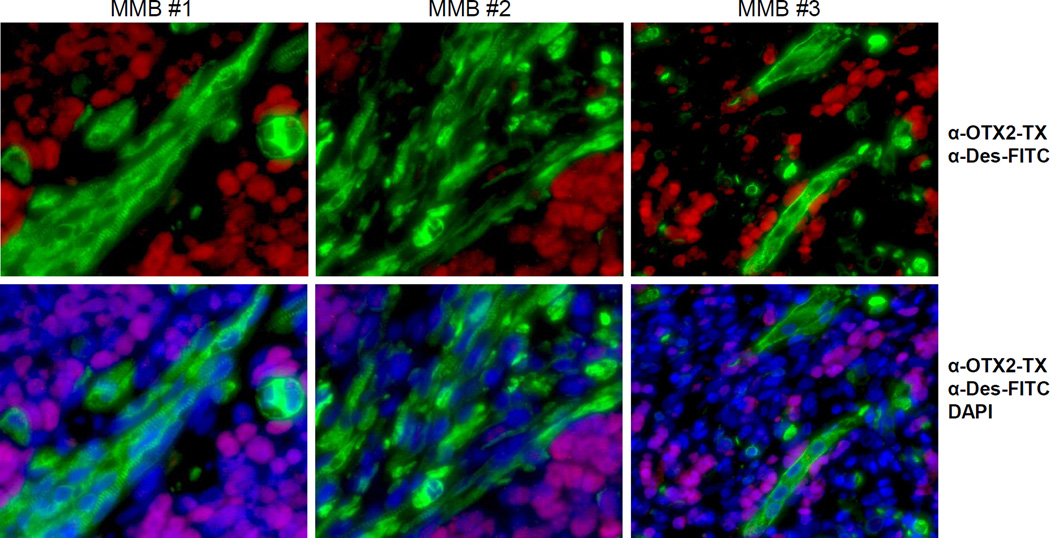
Mutually Exclusive Expression of OTX2 and Myogenic Marker in MMBs
We obtained four MMB samples (#1–4) as paraffin-embedded slides. The limited slide number of MMB #4 only allowed anti-OTX2 DAB and H&E staining, in which the myoblasts are distinguished by rhabdoid cytoplasm stained in red by eosin (Supplemental Figure S5). Immunostaining with OTX2 antibody revealed that all 4 MMBs expressed OTX2 protein (Figure 6 & S5). On immunofluorescent staining of MMB #1–3, the tumor cells with nuclear expression of OTX2 presented a distribution distinct to the desmin-positive myogenic/myoblastic cells, some of which displayed multi-nucleated structure and characteristic muscle striations. Desmin-positive cells in those MMBs demonstrated various phases of myogenic differentiation, from single cells with single nucleus to the ones undergone nuclear fusion and myotube alignment. Overall, myogenic populations in MMB #1 and MMB #2 are more pronounced than in MMB #3. Among these myogenic cells, OTX2 expression was either non-detectable or significantly reduced (Figure 6).
Figure 6. Mutually exclusive expression of OTX2 and myogenic marker desmin in MMBs.
Paraffin-embedded MMB samples #1-3 were co-stained with rabbit OTX2 antibody and mouse desmin (Des) antibody, and subsequently with anti-rabbit Texas Red (TX) and anti-mouse FITC secondary antibodies. DAPI staining was merged in the lower panels to indicate the nuclei, where OTX2 is normally located and markedly absent in the desmin-positive cells.
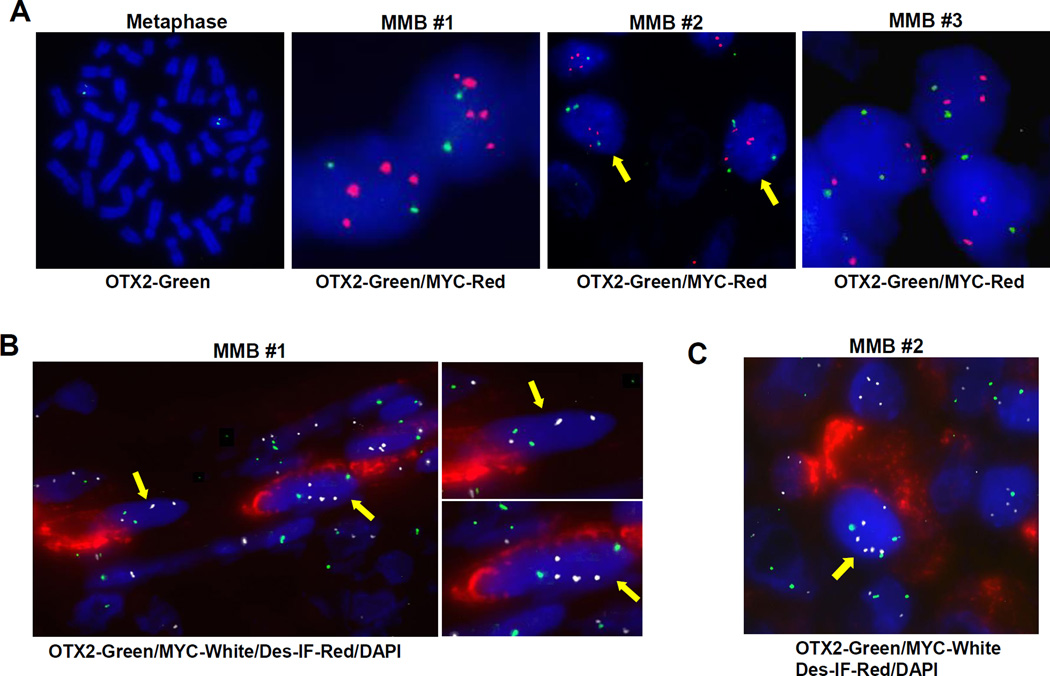
Myogenic Cells Share the Same Cytogenetic Signature with MMB Tumor Cells
The origin of the myogenic element among the primitive neuroectodermal tumor cells in MMB has been under speculation since the first description in 1933 (3). To address this issue, we investigated cytogenetic features including OTX2 and c-MYC genomic copy number in myogenic and primitive neuroectodermal tumor cells, since the often amplified MYC could serve as a marker of MB tumor cells to distinguish them from the possible normal tissue population in the primary tumor. The round-shaped neuroectodermal medulloblastoma cells can be recognized by small sizes with scant cytoplasm in relation to the pronounced nuclei. FISH with OTX2 and c-MYC probes revealed that the primitive tumor cells in MMB #1 and #3 carry two copies of OTX2, while MMB #2 has three copies of OTX2 gene and c-MYC copy numbers were increased at low levels in all three MMB samples (Figure 7A). It is worth noting that these sections often contain cells with incomplete nuclei. In order to further assess the cytogenetic features of the myogenic populations, paraffin sections were first stained with desmin antibody in combination with Cy5 secondary antibody and then hybridized with OTX2 probe labeled with SpectrumGreen and c-MYC probed labeled with SpectrumOrange. The subsequent hybridization procedure inevitably impaired the desmin immunofluorescent signals despite the treatment with the Clark’s fixative. Nonetheless, clear immunofluorescent signals of desmin were retained in MMB #1 and #2, where the myogenic populations are most pronounced. Figure 7B and Figure S6A showed the desmin-positive myoblasts with two copies of OTX2 (green) and 3–5 copies of c-MYC (white) in MMB #1. In MMB #2, three copies of OTX2 and 4–6 copies of c-MYC were found in desmin-positive myoblasts (Figure 7C & S6B). Thus, the desmin-positive myoblasts in these examined MMB samples share the same key cytogenetic signatures with the primitive neuroectodermal tumor cells and therefore should have arisen from the same origin. The loss of OTX2 expression may occur in certain tumor cell populations during tumor progression, which, following the model of OTX2 knockdown in various medulloblastoma cell lines, can lead to extensive myogenic differentiation along with the induction of neuronal markers of those tumor cells.
Figure 7. Myogenic cells shares the same cytogenetic signatures with the primitive medulloblastoma tumor cells in MMBs.
(A) Hybridization of a normal human leukocyte in metaphase with the OTX2 probe showed two copies of OTX2 locus (left picture). Paraffin-embedded samples of MMB #1–3 were hybridized with OTX2-probe labeled in green and c-MYC probed labeled in orange and rendered in red color. DAPI staining revealed the nuclei.
(B and C) MMB #1 and #2 samples were first stained with mouse desmin antibody and anti-mouse Cy5 (red) secondary antibody. Subsequently, the sections were hybridized with OTX2 and c-MYC probes and the nuclei were visualized by DAPI (blue). OTX2 signals were rendered in green and c-MYC in white. Yellow arrows indicate the myogenic cells with desmin staining. Enlarged images of the two nuclei indicated by the yellow arrows in B are displayed on the right side of B.
Discussion
OTX2 is involved in defining vertebrate brain structure during embryogenesis (6). On molecular level, OTX2 controls neuron subtype identity as a repressing factor in coordination with other transcriptional regulators (9, 11, 12, 48). In this study, we identified that the OTX2 HD domain mediates transcriptional repression of OTX2 in medulloblastoma cells independently of its DNA-binding ability. It is possible that this repression is mediated by a binding partner with OTX2 HD. HD includes aa 35–95 of OTX2, distinct from the en1-like motif (aa 151–158) identified as the docking site of Tle4 repressor (49), and could serve as the binding site of other transcriptional regulators, such as the TALE-homeodomain transcriptional co-activator Meis2 (50). Given the HDAC-independent nature of the OTX2 repression of myogenic pathway in medulloblastoma cells, in contrast to the REST/HDAC-dependent suppression of neuronal markers mediated also by OTX2, an unknown repressor independent of HDAC could be recruited by OTX2 HD and act as a key element in controlling the multiple differentiation pathways in medulloblastoma cells. Further understanding of this molecular mechanism could be of therapeutic significance for medulloblastoma.
Recently, it was reported that the Tet-on Dox-inducible knockdown of OTX2 activated cell cycle regulators and neuronal genes (21). As our microarray experiment was performed with the transient transfection of OTX2 siRNA and the knockdown experiments can be affected by different factors such as the various knockdown efficacies and culture conditions, we assessed the potential impact of Dox in myogenic differentiation by using the mouse C2C12 myoblasts in light of this new report (Supplemental Figure S7). C2C12 cells can maintain the fibroblast-like phenotype with the growth media and undergo myogenic differentiation into multinuclear elongated myotubes in the differentiation media. While the impact of 0.1 µg/ml Dox, a concentration used in this study, is not obvious in myogenic conversion, the higher concentrations above 1–2 µg/ml Dox clearly impaired the myogenic conversion in C2C12 cells. Thus, the Tet-on system could carry potential risk of reducing myogenic differentiation especially at higher Dox concentration.
The origin of the muscle elements in MMB has been debated for many years (3, 25). The very few genetic features of some MMBs described so far include the alterations in 17q and low level c-MYC amplification (3). In this study, with all four MMBs expressing OTX2, MMB #1–3 were investigated on a cytogenetic level and MMB #1&2 were further examined for myogenic marker desmin. We demonstrated that in both MMBs, the same cytogenetic signatures of OTX2 and c-Myc copy number profile exist in desmin-positive myogenic cells and primitive neuroectodermal cells. OTX2 knockdown in D425 and other medulloblastoma cells led to myogenic and neuronal differentiation, closely resembling the myogenic and neuronal elements mixed in MMBs (24, 25). Immunofluorescent staining of MMB #1–3 revealed the mutually exclusive nature of OTX2 expression in undifferentiated cells and myogenic phenotypes, which was modeled in vitro in Figure 4 by the loss of OTX2 expression following myogenic conversion of D425 cells co-cultured with C2C12 myoblasts. Thus, the differentiated myogenic cells appear to share common origin with the primitive neuroectodermal cells and the loss of OTX2 expression in certain primitive populations within MMB could have triggered the myogenic differentiation and the formation of striated muscle elements. Micro-environmental factors, genomic or epigenetic instability of the tumor could all possibly contribute to the loss of OTX2 expression in some tumor cell populations. When co-cultured with the myoblast or injected in the mouse muscle, neural stem cells can differentiate to myotubes (45). In our co-culturing assay of D425 cells with C2C12 myoblasts, D425 cells could differentiate to myotubes bearing multinuclear features and muscle markers, accompanied by the loss of OTX2 expression. Although the potential factors triggering this kind of myogenic differentiation remain undetermined, these data indicated the general myogenic potential of some medulloblastoma cells and a reprogramming in gene expression that silenced OTX2.
During the normal brain development, OTX2 regulates the fate and represses the differentiation of OTX2-expressing neural progenitors through a network of transcription factors and external effectors (10–12). OTX2 is expressed in a widespread pattern in forebrain, midbrain and hindbrain at least till P13 in the rat brain, and becomes silenced or largely reduced after P30 with the exception of the pineal gland and VTA neuron (7–9). In the rat cerebellum, OTX2 expression was observed from E16 through P18 and remained weakly detectable until P30, with expression in EGL and germinal layer from P2 to P18 (8). This window of OTX2 expression corresponds with the period of brain development till the completion of neuronal differentiation. During that time frame, OTX2 guides neural progenitors/precursors through their differentiation into specific neuron subtypes, such as the glutamatergic neurons in the thalamus and meso-diencephalic dopaminergic (mdDA) neurons, in a highly coordinated manner together with a complex network of transcriptional regulators (12, 48). In the normal brain development, this may prevent the display of the myogenic potential existing in the neural stem cells/progenitors until the neuronal terminal differentiation, whereas the loss of the aberrant expression of OTX2 in medulloblastoma cells might give rise to the myogenic differentiation in MMB.
The finding that OTX2 represses transcription via its HD and blocks differentiation in the OTX2-positive medulloblastoma provides an opportunity for therapies designed to interfere with OTX2 and trigger differentiation and growth arrest in the tumor. The inducible OTX2 knockdown in the intracranial D425 xenograft tumor resulted in a marked improvement of animal survival, indicating a promising therapeutic potential. Following OTX2 knockdown, genes responsible for growth arrest such as p21 and p57, as well as apoptotic gene caspase 9, were induced. Flow cytometry revealed a minor population (10.53%) of dead cells upon OTX2 knockdown in D425 cells. Thus, it is likely that therapeutic benefits observed with OTX2 knockdown in vitro and in the mouse xenograft model were the results of a combination of anti-proliferation and pro-apoptotic effects. Our previous study evaluated the possible use of retinoic acid (RA), a natural regulator of neural development and a repressor of OTX2 promoter, in treating OTX2-positive medulloblastoma xenograft tumor in mice (19). We determined that although RA was capable of repressing the OTX2 expression, inducing neuronal differentiation and suppressing the growth of medulloblastoma cells, the opposing bFGF signaling in the brain could render retinoic acid ineffective. In this line of investigation, an approach more targeted on the HD-mediated transcriptional repression of OTX2, possibly a small molecule drug, could offer a better chance of therapeutic success.
Supplementary Material
Acknowledgement
We thank Raluca Yonescu for excellent cytogenetic work and Jennifer Edwards for her contribution in SAGE analysis. We thank Dr. Christopher Cheadle of the Johns Hopkins Illumina core laboratory for Illumina microarray analysis. We appreciate the technical advice and help by Betty Tyler, I-Mei Siu, Gary Gallia and Colette M. apRhys.
Grant Support
This project was supported by NIH Grant R01 NS052507, the Virginia and D.K. Ludwig Fund for Cancer Research, and the Children’s Cancer Foundation. GJR is the recipient of the Irving J. Sherman M.D. Research Professorship.
Footnotes
Conflict of interest: The authors declare that they have no affiliations that would constitute a financial conflict of interest relating to the subject matter of this study.
References
- 1.Klesse LJ, Bowers DC. Childhood medulloblastoma: current status of biology and treatment. CNS Drugs. 24:285–301. doi: 10.2165/11530140-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2.Eberhart CG. Molecular diagnostics in embryonal brain tumors. Brain Pathol. 2011;21:96–104. doi: 10.1111/j.1750-3639.2010.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helton KJ, Fouladi M, Boop FA, Perry A, Dalton J, Kun L, et al. Medullomyoblastoma: a radiographic and clinicopathologic analysis of six cases and review of the literature. Cancer. 2004;101:1445–1454. doi: 10.1002/cncr.20450. [DOI] [PubMed] [Google Scholar]
- 4.Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamson DC, Shi Q, Wortham M, Northcott PA, Di C, Duncan CG, et al. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 2010;70:181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simeone A. Otx1 and Otx2 in the development and evolution of the mammalian brain. Embo J. 1998;17:6790–6798. doi: 10.1093/emboj/17.23.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14:5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rath MF, Munoz E, Ganguly S, Morin F, Shi Q, Klein DC, et al. Expression of the Otx2 homeobox gene in the developing mammalian brain: embryonic and adult expression in the pineal gland. J Neurochem. 2006;97:556–566. doi: 10.1111/j.1471-4159.2006.03773.x. [DOI] [PubMed] [Google Scholar]
- 9.Di Salvio M, Di Giovannantonio LG, Omodei D, Acampora D, Simeone A. Otx2 expression is restricted to dopaminergic neurons of the ventral tegmental area in the adult brain. Int J Dev Biol. 2010;54:939–945. doi: 10.1387/ijdb.092974ms. [DOI] [PubMed] [Google Scholar]
- 10.Simeone A, Di Salvio M, Di Giovannantonio LG, Acampora D, Omodei D, Tomasetti C. The role of otx2 in adult mesencephalic-diencephalic dopaminergic neurons. Mol Neurobiol. 2011;43:107–113. doi: 10.1007/s12035-010-8148-y. [DOI] [PubMed] [Google Scholar]
- 11.Puelles E, Annino A, Tuorto F, Usiello A, Acampora D, Czerny T, et al. Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development. 2004;131:2037–2048. doi: 10.1242/dev.01107. [DOI] [PubMed] [Google Scholar]
- 12.Puelles E, Acampora D, Gogoi R, Tuorto F, Papalia A, Guillemot F, et al. Otx2 controls identity and fate of glutamatergic progenitors of the thalamus by repressing GABAergic differentiation. J Neurosci. 2006;26:5955–5964. doi: 10.1523/JNEUROSCI.1097-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boon K, Eberhart CG, Riggins GJ. Genomic amplification of orthodenticle homologue 2 in medulloblastomas. Cancer Res. 2005;65:703–707. [PubMed] [Google Scholar]
- 14.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The Genetic Landscape of the Childhood Cancer Medulloblastoma. Science. 2011 doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, et al. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919–924. [PubMed] [Google Scholar]
- 17.de Haas T, Oussoren E, Grajkowska W, Perek-Polnik M, Popovic M, Zadravec-Zaletel L, et al. OTX1 and OTX2 expression correlates with the clinicopathologic classification of medulloblastomas. J Neuropathol Exp Neurol. 2006;65:176–186. doi: 10.1097/01.jnen.0000199576.70923.8a. [DOI] [PubMed] [Google Scholar]
- 18.Wortham M, Jin G, Sun JL, Bigner DD, He Y, Yan H. Aberrant Otx2 expression enhances migration and induces ectopic proliferation of hindbrain neuronal progenitor cells. PLoS One. 2012;7:e36211. doi: 10.1371/journal.pone.0036211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai R, Siu IM, Tyler BM, Staedtke V, Gallia GL, Riggins GJ. Evaluation of retinoic acid therapy for OTX2-positive medulloblastomas. Neuro Oncol. 2010;12:655–663. doi: 10.1093/neuonc/nop062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunt J, de Haas TG, Hasselt NE, Zwijnenburg DA, Koster J, Versteeg R, et al. Regulation of cell cycle genes and induction of senescence by overexpression of OTX2 in medulloblastoma cell lines. Mol Cancer Res. 2010;8:1344–1357. doi: 10.1158/1541-7786.MCR-09-0546. [DOI] [PubMed] [Google Scholar]
- 21.Bunt J, Hasselt NE, Zwijnenburg DA, Hamdi M, Koster J, Versteeg R, et al. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer. 2011 doi: 10.1002/ijc.26474. [DOI] [PubMed] [Google Scholar]
- 22.Leonard JR, Cai DX, Rivet DJ, Kaufman BA, Park TS, Levy BK, et al. Large cell/anaplastic medulloblastomas and medullomyoblastomas: clinicopathological and genetic features. J Neurosurg. 2001;95:82–88. doi: 10.3171/jns.2001.95.1.0082. [DOI] [PubMed] [Google Scholar]
- 23.Sachdeva MU, Vankalakunti M, Rangan A, Radotra BD, Chhabra R, Vasishta RK. The role of immunohistochemistry in medullomyoblastoma--a case series highlighting divergent differentiation. Diagn Pathol. 2008;3:18. doi: 10.1186/1746-1596-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergmann M, Pietsch T, Herms J, Janus J, Spaar HJ, Terwey B. Medullomyoblastoma: a histological, immunohistochemical, ultrastructural and molecular genetic study. Acta Neuropathol. 1998;95:205–212. doi: 10.1007/s004010050788. [DOI] [PubMed] [Google Scholar]
- 25.Kido M, Ueno M, Onodera M, Matsumoto K, Imai T, Haba R, et al. Medulloblastoma with myogenic differentiation showing double immunopositivity for synaptophysin and myoglobin. Pathol Int. 2009;59:255–260. doi: 10.1111/j.1440-1827.2009.02360.x. [DOI] [PubMed] [Google Scholar]
- 26.Friedman HS, Burger PC, Bigner SH, Trojanowski JQ, Wikstrand CJ, Halperin EC, et al. Establishment and characterization of the human medulloblastoma cell line and transplantable xenograft D283 Med. J Neuropathol Exp Neurol. 1985;44:592–605. doi: 10.1097/00005072-198511000-00005. [DOI] [PubMed] [Google Scholar]
- 27.Pietsch T, Scharmann T, Fonatsch C, Schmidt D, Ockler R, Freihoff D, et al. Characterization of five new cell lines derived from human primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1994;54:3278–3287. [PubMed] [Google Scholar]
- 28.Chatelain G, Fossat N, Brun G, Lamonerie T. Molecular dissection reveals decreased activity and not dominant negative effect in human OTX2 mutants. J Mol Med. 2006;84:604–615. doi: 10.1007/s00109-006-0048-2. [DOI] [PubMed] [Google Scholar]
- 29.Goldhamer DJ, Brunk BP, Faerman A, King A, Shani M, Emerson CP., Jr Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121:637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Suzuki T, Mori N, Greengard P. Identification of a functional silencer element involved in neuron-specific expression of the synapsin I gene. Proc Natl Acad Sci U S A. 1993;90:1460–1464. doi: 10.1073/pnas.90.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai RY, Dieter P, Peschel C, Morris SW, Duyster J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-gamma to mediate its mitogenicity. Mol Cell Biol. 1998;18:6951–6961. doi: 10.1128/mcb.18.12.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wehle D, Yonescu R, Long PP, Gala N, Epstein J, Griffin CA. Fluorescence in situ hybridization of 12p in germ cell tumors using a bacterial artificial chromosome clone 12p probe on paraffin-embedded tissue: clinical test validation. Cancer Genet Cytogenet. 2008;183:99–104. doi: 10.1016/j.cancergencyto.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Nolen LD, Amor D, Haywood A, St Heaps L, Willcock C, Mihelec M, et al. Deletion at 14q22-23 indicates a contiguous gene syndrome comprising anophthalmia, pituitary hypoplasia, and ear anomalies. Am J Med Genet A. 2006;140:1711–1718. doi: 10.1002/ajmg.a.31335. [DOI] [PubMed] [Google Scholar]
- 34.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 35.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 37.Asakura A, Lyons GE, Tapscott SJ. The regulation of MyoD gene expression: conserved elements mediate expression in embryonic axial muscle. Dev Biol. 1995;171:386–398. doi: 10.1006/dbio.1995.1290. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto M, Watt CD, Schmidt RJ, Kuscuoglu U, Miesfeld RL, Goldhamer DJ. Cloning and characterization of a novel MyoD enhancer-binding factor. Mech Dev. 2007;124:715–728. doi: 10.1016/j.mod.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 40.Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 41.Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol Cell Biol. 2001;21:2404–2412. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 43.Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci U S A. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer SL, Reddick WE, Gajjar A. Understanding the Cognitive Impact on Children Who Are Treated for Medulloblastoma. J Pediatr Psychol. 2007 doi: 10.1093/jpepsy/jsl056. [DOI] [PubMed] [Google Scholar]
- 45.Galli R, Borello U, Gritti A, Minasi MG, Bjornson C, Coletta M, et al. Skeletal myogenic potential of human and mouse neural stem cells. Nat Neurosci. 2000;3:986–991. doi: 10.1038/79924. [DOI] [PubMed] [Google Scholar]
- 46.Fife RS, Sledge GW, Jr, Roth BJ, Proctor C. Effects of doxycycline on human prostate cancer cells in vitro. Cancer Lett. 1998;127:37–41. doi: 10.1016/s0304-3835(98)00003-2. [DOI] [PubMed] [Google Scholar]
- 47.Onoda T, Ono T, Dhar DK, Yamanoi A, Nagasue N. Tetracycline analogues (doxycycline and COL-3) induce caspase-dependent and -independent apoptosis in human colon cancer cells. Int J Cancer. 2006;118:1309–1315. doi: 10.1002/ijc.21447. [DOI] [PubMed] [Google Scholar]
- 48.Omodei D, Acampora D, Mancuso P, Prakash N, Di Giovannantonio LG, Wurst W, et al. Anterior-posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development. 2008;135:3459–3470. doi: 10.1242/dev.027003. [DOI] [PubMed] [Google Scholar]
- 49.Heimbucher T, Murko C, Bajoghli B, Aghaallaei N, Huber A, Stebegg R, et al. Gbx2 and Otx2 interact with the WD40 domain of Groucho/Tle corepressors. Mol Cell Biol. 2007;27:340–351. doi: 10.1128/MCB.00811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agoston Z, Schulte D. Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development. 2009;136:3311–3322. doi: 10.1242/dev.037770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.