Abstract
Pioneer transcription factors initiate cell-fate changes by binding to silent target genes. They are among the first factors to bind key regulatory sites and facilitate chromatin opening. Here, we identify an additional role for pioneer factors. In early Caenorhabditis elegans foregut development, the pioneer factor PHA-4/FoxA binds promoters and recruits RNA polymerase II (Pol II), often in a poised configuration in which Pol II accumulates near transcription start sites. At a later developmental stage, PHA-4 promotes chromatin opening. We found many more genes with poised RNA polymerase than had been observed previously in unstaged embryos, revealing that early embryos accumulate poised Pol II and that poising is dynamic. Our results suggest that Pol II recruitment, in addition to chromatin opening, is an important feature of PHA-4 pioneer factor activity.
Embryonic development depends on precise patterns of gene expression that are orchestrated by key transcription factors such as pioneer transcription factors. Pioneer factors function at the earliest stage of transcriptional onset to facilitate chromatin opening at cis-regulatory sites, which enables additional factors to bind DNA (1). The founding pioneer factor is mammalian FoxA1, which associates with liver genes and promotes chromatin accessibility before transcriptional activation. In vitro, FoxA proteins bind nucleosomes and block chromatin compaction by H1 linker histones (1), and in vivo FoxA proteins open chromatin with the histone variant H2A.Z (2). It is unknown whether chromatin opening is the sole mechanism of transcriptional priming induced by pioneer transcription factors.
In Caenorhabditis elegans, pha-4 encodes a selector gene that specifies foregut fate (3). pha-4 is orthologous to FoxA proteins (4, 5) and interacts with H2A.Z (2), raising the question of whether pha-4 functions as a pioneer transcription factor in addition to its selector activities. We performed five tests that revealed that pha-4 had pioneer activity. First, PHA-4 associated with target genes M05B5.2, ceh-22, and myo-2 beginning at the 8E stage (“E” for endodermal cells), when PHA-4 was first detected (Fig. 1, A and B, and fig. S1A). We observed binding to promoters that are activated at early, mid-, or late embryogenesis and confirmed that the mid- (ceh-22) and late-stage (myo-2) genes were not expressed in our 8E sample (gastrulation stage). These data indicate that PHA-4 associates with endogenous foregut promoters hours before transcriptional onset, which is as expected for a pioneer factor. Second, we determined that PHA-4 bound nucleosomal DNA in vitro equivalently to its orthologs FoxA1 and FoxA2 (Fig. 1C). Third, chromatin sites bound by PHA-4 in vivo [measured with chromatin immunoprecipitation (ChIP)] (6) lacked stable nucleosomes [measured with formaldehyde-assisted isolation of regulatory elements (FAIRE)] (fig. S1C) (7), indicating PHA-4 association with open chromatin. Moreover, regions bound by PHA-4 were enriched for activating histone marks H3K4me2, H3K4me3, and acetylated H3K27 (fig. S1C) (8). Fourth, single-cell analysis with artificial chromosomes (Fig. 1D) revealed that chromatin was open in the foregut, where PHA-4 is expressed, but not in other cell types, which lack PHA-4, nor with a target promoter bearing a mutated PHA-4-binding site (Fig. 1E) (9). Fifth, we tracked PHA-4 association with chromatin during mitosis and observed that a portion of PHA-4 was retained on DNA in dividing foregut cells (fig. S2) (10). Together, the results reveal that PHA-4 fulfilled the criteria of a pioneer transcription factor (Fig. 1 and fig. S1B). It associated with binding sites early in development, bound DNA packaged in nucleosomes in vitro, and decompacted chromatin in vivo.
Fig. 1. PHA-4 is a pioneer factor.
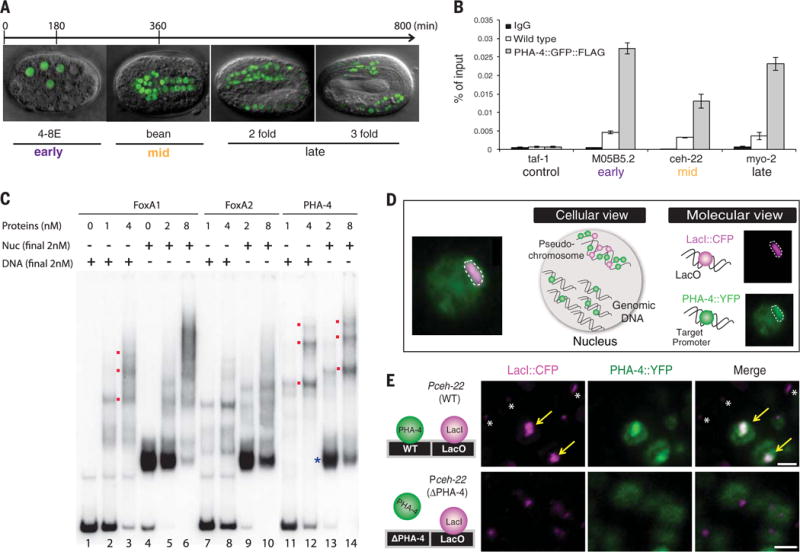
(A) PHA-4∷green fluorescent protein (GFP) (green) during stages of embryogenesis. Early embryos are enriched for 8E stage, and mid-embryos are enriched for bean stage (11). (B) PHA-4∷GFP∷FLAG (11) binding endogenous targets M05B5.2 (expressed early), ceh-22 (mid), and myo-2 (late), detected with ChIP–quantitative PCR. Wild-type embryos lack FLAG, a negative control. taf-1 is not a PHA-4 target. n = 3 replicates, mean ± SEM. (C) PHA-4 binds nucleosomes. Shown is recombinant PHA-4 compared with FoxA proteins incubated with the albumin enhancer containing a FoxA1 binding site as free DNA (DNA) or nucleosomal (Nuc) DNA. Bound PHA-4 generated slow migrating bands (red). (D) Artificial chromosomes with PHA-4∷yellow fluorescent protein (YFP) bound target promoters in single cells. (E) PHA-4∷YFP (green) bound artificial chromosomes (purple, LacI) bearing the ceh-22 promoter (arrows). PHA-4∷YFP binding was abolished when ceh-22 carried a mutated PHA-4 binding site (ΔPHA-4; bottom). Asterisks mark artificial chromosomes in nonforegut cells. Scale bar, 2 μm.
To examine the role of PHA-4 in transcription, we mapped Pol II occupancy by means of genome-wide ChIP-sequencing (ChIP-seq). Previous studies with C. elegans Pol II had focused on relatively late time points, after transcription was established for many genes (6, 11). Our interest was earlier stages, before transcriptional onset. We analyzed early embryos after PHA-4 bound to target genes but before their transcription (~8E stage) and compared those embryos to mid-stage, transcriptionally active embryos (bean stage) (staging is provided in fig. S3). To localize Pol II, we mapped its position relative to the transcription start site (TSS) (11) and calculated three scores: promoter occupancy for Pol II spanning the TSS (Fig. 2A), Pol II within gene bodies (Fig. 2B), and the poising index as the ratio of the promoter to the gene body values (Fig. 2C). The poising index reflects the relative quantity of Pol II close to the site of transcriptional initiation (12). Poising has been detected in diverse organisms including, to a degree, C. elegans (12, 13).
Fig. 2. Early embryos accumulate poised Pol II.
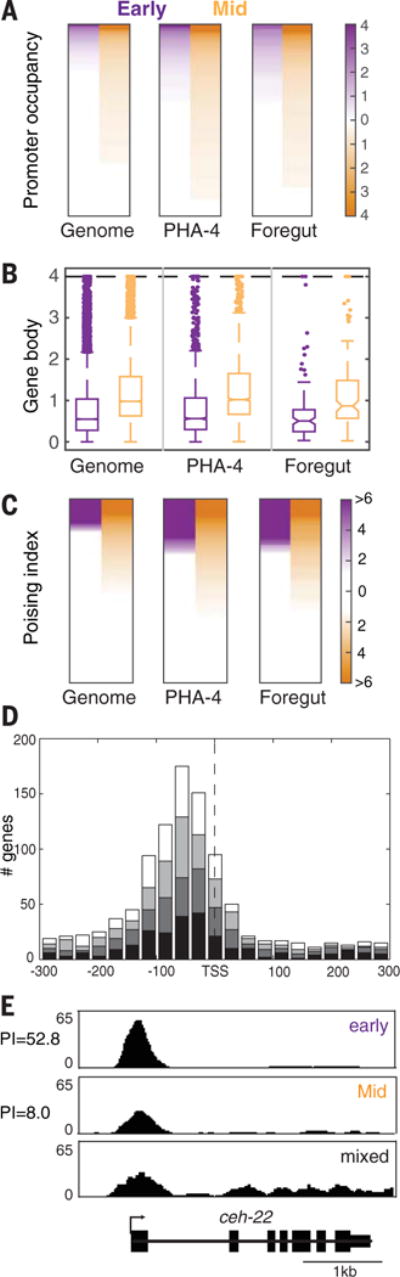
(A) Pol II occupancy at promoters (enrichment over input). (B) Normalized gene activity scores for Pol II occupancy within gene bodies. (C) Poising index. For (A) to (C), numbers are provided in table S3; “Early” and “Mid” denote stages. (D) Pol II is enriched upstream of the TSS. Genes with Pol II peaks near defined TSSs (11) were divided into four quartiles according to gene body activity scores (from low, black, to high, white) and graphed for Pol II across the gene. (E) Pol II at the ceh-22 locus in early, mid-, and late or mixed stage embryos. Mixed population sample is from (6).
We began by surveying the whole genome. In early embryos, most genes showed little Pol II at either promoters or gene bodies (Fig. 2, A and B), suggesting that most of the genome was inactive. However, ~20% of genes had Pol II near the TSS and little Pol II within gene bodies, leading to a high poising index (≥2.5) (Fig. 2C). As development progressed, the Pol II signal for both promoters and gene bodies increased, resulting in a broad range of poising values (Fig. 2C and fig. S4C). This result suggested that the mid-stage poising scores reflected a surge in Pol II activity at the level of initiation and elongation, and that poising in C. elegans is temporally regulated, similar to other animals (12). Most genes had docked Pol II, in which Pol II bound just upstream of the TSS (14) (Fig. 2D) (11). Pol II “pausing” was also observed 3 to the TSS, like other species (12, 14), but we observed fewer cases of pausing as compared with docking. We suggest that poising in C. elegans is more prevalent than had been previously recognized. Earlier studies observed some poising in starved larvae and in samples bearing mixtures of stages (6, 13, 14). In our samples, poising was associated with both early and mid-stages, with index values typically higher in early embryos because occupancy of Pol II within gene bodies was low. Our analysis gives a picture of Pol II loading and transcriptional onset during embryogenesis.
We next examined Pol II at foregut-associated genes. We observed an enrichment of poised Pol II: 27% of foregut genes were poised early compared with 17% for the whole genome (Fig. 2C). At the bean stage, 36% of PHA-4–bound promoters had a poising index >2.5, compared with 29% for the whole genome. We confirmed the ChIP-seq result by means of ChIP–quantitative polymerase chain reaction (PCR) for four foregut genes exhibiting different Pol II poising scores (11). For example, Pol II was poised at the ceh-22 promoter in early embryos before transcriptional onset, but it subsequently decreased at the TSS and increased in the gene body (Fig. 2E and fig. S4B). Quantitative reverse transcriptase–PCR (RT-PCR) analysis demonstrated that ceh-22 mRNA was activated in mid-embryos, as expected (Fig. 3, B and C) (11, 15). These data suggest that poised Pol II often reflects preparation for transcriptional activation (12). Consistent with this idea, genes with poised Pol II were associated with Gene Ontology (GO) terms “embryonic development” and “embryonic morphogenesis” (table S1). However, poising likely has additional roles because we also detected embryonic poised Pol II at a subset of genes not expressed in embryos (such as mex-3) or associated with GO terms such as “post-embryonic development.”
Fig. 3. PHA-4 is required for Pol II occupancy at foregut genes.
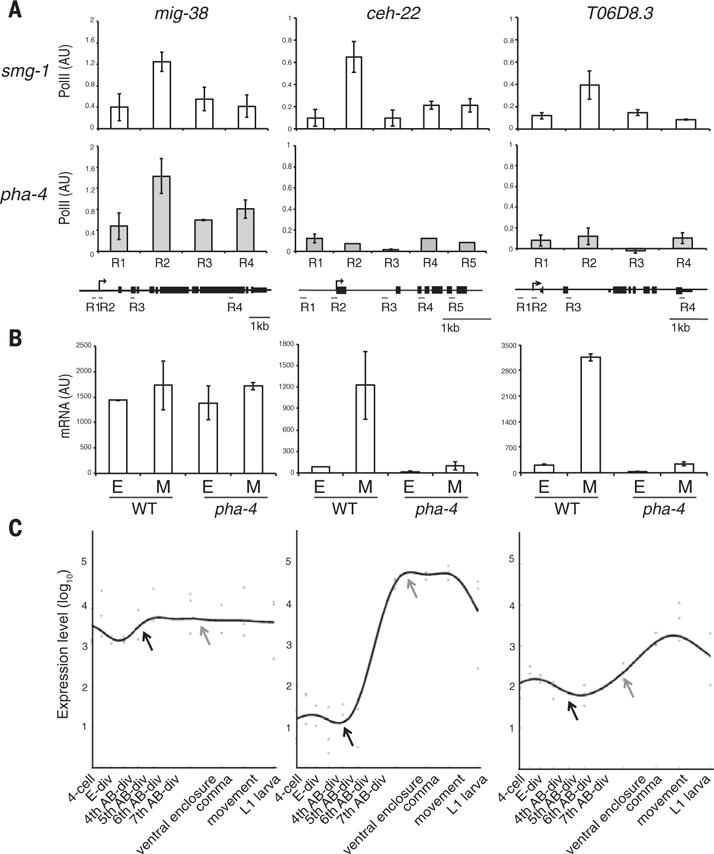
(A) Pol II occupancy at mig-38 (poised, ubiquitous) versus ceh-22 and T06D8.3 (poised, foregut) in smg-1 control versus pha-4(ts) embryos, normalized to eft-3 (set at 1) and srw-99 (set at 0). Error bars indicate n = 3 replicates, mean ± SEM. (B) mRNA abundance (quantitative RT-PCR) for mig-38, ceh-22, and T06D8.3 for wild-type or pha-4(ts) embryos early (E) or mid (M). n = 3 replicates, mean ± SEM. (C) Gene expression profiles from (20). Early stage (dark gray arrowhead) is at the fifth and sixth AB-div. Mid-stage (light gray arrowhead) is equivalent to ventral enclosure.
Pol II poising at developmentally expressed genes has been observed in Drosophila embryos (16), but poising in C. elegans had been associated predominantly with starvation (6, 13). A likely explanation for the difference between the prior studies and ours is the embryonic stage (11). For example, we found that mixtures of embryos with a range of ages lacked poised Pol II for ceh-22, underscoring the importance of staging (fig. S4A).
To test whether pha-4 affects Pol II occupancy, we performed Pol II ChIP with a pha-4(ts) temperature-sensitive strain that combines pha-4(zu225nonsense) with smg-1(cc546ts) (fig. S6A) (17). smg-1(cc546) alone served as a control. Growth of pha-4(ts) was complicated because pha-4 is an essential gene, and therefore we relied on ChIP–quantitative PCR, which requires less material.
At restrictive temperature, pha-4 mutants failed to accumulate poised Pol II at three tested loci (ceh-22, T06D8.3, and K10D3.4) or elongating Pol II at one (M05B5.2) (Fig. 3A and fig. S6B) (11). mig-38, which has PHA-4 bound but is expressed broadly, was not affected by pha-4(ts) (Fig. 3A). Recent studies found that Pol II poising was not tissue-specific for Drosophila muscle (16), but our analysis indicates that PHA-4 helps Pol II associate with its target foregut genes in worms.
Pioneer transcription factors promote chromatin opening at target genes to modulate gene expression. We therefore wondered whether chromatin opening by PHA-4 affected Pol II loading. We adapted FAIRE (7, 11) to track regions of the C. elegans genome with absent or unstable nucleosomes. In older, wild-type embryos, we found a strong correlation between PHA-4 binding and open chromatin, characterized by a high FAIRE signal, at both promoters (H2A.Z+, H3K4me3+, and H3K27ac+) and enhancers (H3K4me1/2+) (fig. S7). Conversely, Pol II occupancy was only weakly correlated with open chromatin (fig. S7A), which is similar to Drosophila (18). To determine the contribution of pha-4 to chromatin opening, we surveyed three foregut genes by means of FAIRE–quantitative PCR at early and mid-stages after pha-4 inactivation (Fig. 4A). At the 8E stage, pha-4(ts) embryos had a FAIRE signal equivalent to wild-type embryos for ceh-22 and K10D3.4, suggesting that PHA-4 did not contribute to chromatin opening at these early stages. T06D8.3, however, showed a ~10% decrease in FAIRE, suggesting nucleosomes depended on pha-4 for at least some opening. In mid-stage embryos, reduction of pha-4 lead to a decrease in open FAIRE regions spanning the PHA-4 binding site and the Pol II binding site for T06D8.3 and K10D3.4. The effect was less dramatic for ceh-22, with a small reduction at the PHA-4 binding region. Nevertheless, PHA-4 still promoted ceh-22 opening. These data suggest that PHA-4 promotes chromatin opening at mid-stages, at least for the surveyed genes, but has less of an effect early.
Fig. 4. PHA-4 promotes chromatin openness during mid-stage embryogenesis.
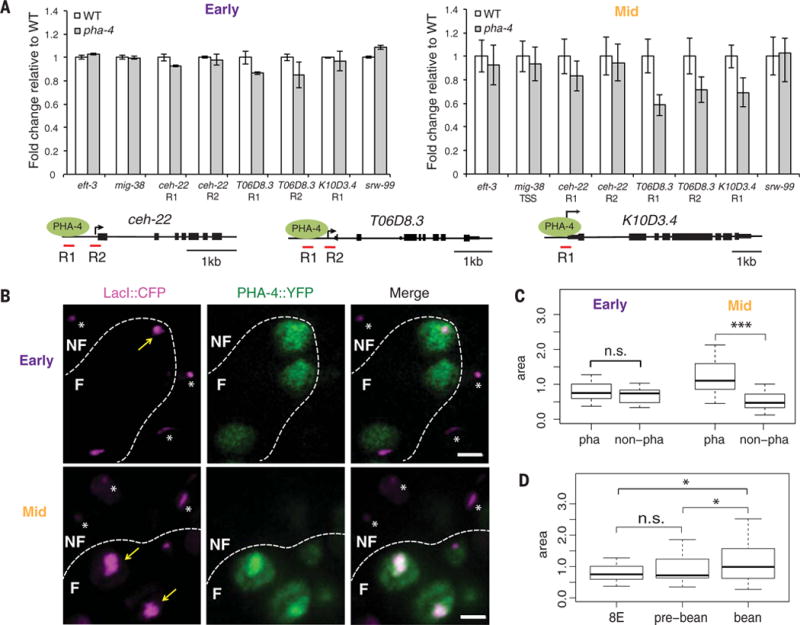
(A) Chromatin opening tracked by FAIRE–quantitative PCR. Early (left), FAIRE signals for three poised foregut genes were similar between wild-type (white) and pha-4(ts) (gray). Mid- (right), FAIRE signals were reduced at poised foregut genes in pha-4(ts) embryos. The gene structures show positions for PHA-4 binding (R1) and TSS (R2) sites. n = 3 replicates, mean ± SEM. (B) Artificial chromosomes (CFP∷LacI, purple) bearing the ceh-22 promoter bound by PHA-4∷YFP (green) in early and mid-stage embryos. Dotted lines distinguish foregut (F) from nonforegut (NF). Scale bar, 2 μm. (C) Areas of artificial chromosomes carrying the ceh-22 promoter in foregut (pha) versus nonforegut (non-pha). (D) Areas of artificial chromosomes carrying the ceh-22 promoter in the foregut at different stages. *P = 0.01 to 0.05; ***P < 0.001.
We extended these results in three ways. First, we determined that pha-4(ts) had no impact on three nonforegut genes (eft-3, mig-38, and srw-99) (Fig. 4A). Second, we used artificial chromosomes bearing PHA-4 target promoters and fluorescently tagged PHA-4 to examine chromatin opening in single cells (9). We observed decompaction of artificial chromosomes in the foregut of mid-stage embryos but not early embryos (Fig. 4, B to D) (9). Artificial chromosomes in nonforegut cells failed to decompact at either stage (Fig. 4, B and C). Third, a comparison of wild-type 4E embryos (with little to no PHA-4) and 8E embryos (with detectable PHA-4) revealed a decrease in FAIRE values at the 8E for both foregut and nonforegut genes (fig. S7). Thus, PHA-4 binding did not induce detectable decompaction at the 8E stage compared with the 4E. The data suggest that PHA-4 induces decompaction predominantly at mid-stages, after Pol II binding.
This study reveals widespread poised Pol II during C. elegans development and shows that the pioneer transcription factor PHA-4 contributes to Pol II recruitment at poised and transcribed genes within the foregut. PHA-4 activity is critical during early embryonic stages when we observe Pol II recruitment, suggesting these early events are essential for proper organogenesis (17). C. elegans embryos develop in 13 hours, with rapid changes in gene expression. Pol II poising may accommodate these dynamics by promoting rapid and/or synchronous transcriptional onset (12, 19). Recruitment of poised Pol II is followed by decompaction of chromatin. One appealing hypothesis is that deposition of Pol II at TSS regions may participate in chromatin opening, along with PHA-4 (18). This scenario predicts that Pol II binds to regions that would otherwise contain stable nucleosomes, a prediction that is borne out by our FAIRE analysis, and that Pol II interferes with the construction of nucleosomes. It will be of interest to see whether other pioneer factors or FoxA proteins also poise Pol II.
Acknowledgments
We thank J. Whetstine for ChIP advice, A. Schier and S. von Stetina for comments, and G. Marnellos and M. Clamp for informatics. Some strains were obtained from the Caenorhabditis Genetics Center (CGC), funded by NIHP40OD010440. S.E.M. received support from the MacArthur Foundation and grant NIHR37GM056264, E.L. received support from grant NSFMCB-1413134, and K.S.Z. received support from grant NIHR37GM36477. Sequencing data are accessible from the National Center for Biotechnology Information Gene Expression Omnibus: GSM1666978 R1.CE.8E.ChIP GSM1666979, R1.CE.8E.Input GSM1666980, R2.CE.8E.ChIP GSM1666981, R2.CE.8E.Input GSM1666982, R1.CE.BE.ChIP GSM1666983, and R1.CE.BE.Input.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/348/6241/1372/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S7
Tables S1 to S3
References
REFERENCES AND NOTES
- 1.Iwafuchi-Doi M, Zaret KS. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Updike DLDL, Mango SESE. PLOS Genet. 2006;2:e161. doi: 10.1371/journal.pgen.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mango SE. Annu Rev Cell Dev Biol. 2009;25:597–628. doi: 10.1146/annurev.cellbio.24.110707.175411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horner MA, et al. Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalb JM, et al. Development. 1998;125:2171–2180. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- 6.Zhong M, et al. PLOS Genet. 2010;6:e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho JWK, et al. Nature. 2014;512:449–452. [Google Scholar]
- 9.Fakhouri THI, Stevenson J, Chisholm AD, Mango SE. PLOS Genet. 2010;6:e1001060. doi: 10.1371/journal.pgen.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caravaca JM, et al. Genes Dev. 2013;27:251–260. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supplementary materials on Science Online.
- 12.Adelman K, Lis JT. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baugh LR, Demodena J, Sternberg PW. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 14.Maxwell CSCS, et al. Cell Reports. 2014;6:455–466. doi: 10.1016/j.celrep.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuchenthal CA, Chen W, Okkema PG. Genesis. 2001;31:156–166. doi: 10.1002/gene.10018. [DOI] [PubMed] [Google Scholar]
- 16.Gaertner B, et al. Cell Reports. 2012;2:1670–1683. doi: 10.1016/j.celrep.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiefer JC, Smith PA, Mango SE. Dev Biol. 2007;303:611–624. doi: 10.1016/j.ydbio.2006.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilchrist DA, et al. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boettiger AN, Levine M. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin M, Hashimshony T, Wagner F, Yanai I. Dev Cell. 2012;22:1101–1108. doi: 10.1016/j.devcel.2012.04.004. [DOI] [PubMed] [Google Scholar]


