Abstract
Several classes of signaling proteins contain autoinhibitory domains that prevent unwarranted signaling and coordinate the induction of activity in response to external cues. CARD11, a scaffold protein critical for antigen receptor signaling to NF-κB, undergoes autoregulation by a poorly understood inhibitory domain (ID), which keeps CARD11 inactive in the absence of receptor triggering through inhibitory intramolecular interactions. This autoinhibitory strategy makes CARD11 highly susceptible to gain-of-function mutations that are frequently observed in diffuse large B cell lymphoma (DLBCL) and that disrupt ID-mediated autoinhibition, leading to constitutive NF-κB activity, which can promote lymphoma proliferation. Although DLBCL-associated CARD11 mutations in the caspase recruitment domain (CARD), LATCH domain, and coiled coil have been shown to disrupt intramolecular ID binding, surprisingly, no gain-of-function mutations in the ID itself have been reported and validated. In this study, we solve this paradox and report that the CARD11 ID contains an unusual array of four repressive elements that function cooperatively with redundancy to prevent spontaneous NF-κB activation. Our quantitative analysis suggests that potent oncogenic CARD11 mutations must perturb autoinhibition by at least three repressive elements. Our results explain the lack of ID mutations in DLBCL and reveal an unusual autoinhibitory domain structure and strategy for preventing unwarranted scaffold signaling to NF-κB.
Keywords: gene regulation, lymphoma, NF-κB (NF-kB), oncogene, scaffold protein, signal transduction, T-cell receptor (TCR), autoinhibitory domain, CARD11, diffuse large B cell lymphoma
Introduction
Many signaling pathways that control cell proliferation and differentiation have evolved inhibitory checkpoints that prevent uncontrolled signaling. Checkpoints can involve inhibitory proteins that provide negative feedback once signaling begins or that actively suppress signaling prior to receptor engagement. For several pathways, checkpoint function is achieved by an autoinhibitory domain, which governs the activity of the key pathway component in which it is found. Such autoinhibitory domains have been described for several classes of signaling proteins, including tyrosine (1, 2) and serine/threonine kinases (3–5), guanine nucleotide exchange factors (6), phospholipases (7, 8), and E3 ubiquitin ligases (9–11), among others (12, 13). Although an autoinhibitory domain can provide robust, concentration-independent regulation in cis, the presence of an autoinhibitory domain in a signaling protein can make it especially susceptible to mutations that disrupt autoinhibition and lead to constitutive signaling and potentially pathological cell behavior. A mechanistic understanding of autoinhibitory domain function is thus critical for understanding how the output of signaling pathways is determined and how the dysregulation of autoinhibition by mutation can promote oncogenic transformation.
CARD11 is a multidomain signaling scaffold whose activity is regulated by an autoinhibitory domain that is incompletely understood. CARD11 is required for antigen receptor signaling to the NF-κB transcription factor (14–22), a critical pathway for normal antigen-dependent lymphocyte activation in the adaptive immune response (23, 24). During signaling, CARD11 undergoes a transition from a closed inactive state to an open active scaffold that recruits several signaling cofactors into a complex to induce IKK kinase activity. This transition is controlled by an inhibitory domain (ID),2 which in the absence of receptor engagement participates in intramolecular interactions involving the caspase recruitment domain (CARD) and coiled-coil domains (25). Upon receptor triggering, the ID is phosphorylated on several serine residues (26–30), which somehow neutralizes the autoinhibitory activity of the ID and allows CARD11 to recruit several obligate pathway components, including Bcl10, MALT1, TRAF6, TAK1, caspase-8, and IKKγ, all of which require the CARD domain, coiled-coil domain, or both for CARD11 association (25).
CARD11 is frequently mutated in the activated B cell-like subtype of diffuse large B cell lymphoma (DLBCL), the most common form of non-Hodgkin lymphoma (31). The activated B cell-like DLBCL subtype is associated with the worst DLBCL prognosis and is characterized by constitutive, dysregulated activation of NF-κB, which is required for growth and proliferation of the lymphoma (32). DLBCL-associated CARD11 mutations are gain-of-function mutations that confer hyperactive signaling to NF-κB in the absence of antigen receptor triggering. Oncogenic CARD11 mutations were first reported in the coiled-coil domain (33), and two of the initially described mutations were found to cause hyperactivity by impairing autoinhibition by the ID (34). The mutations partially disrupted intramolecular binding by the ID and selectively enhanced the recruitment of Bcl10 to CARD11, but not that of other signaling cofactors, leading to Bcl10-dependent signaling to the IKK complex (34). A high-throughput signaling screen subsequently identified CARD11 gain-of-function mutations outside of the coiled coil, in the CARD and LATCH domains of CARD11 (35), which were also found to disrupt intramolecular interactions with the ID, selectively stimulate Bcl10 binding to CARD11, and spontaneously induce NF-κB activity (35). Gain-of-function mutations in the CARD and LATCH domains have also been reported in patient DLBCL biopsies, including several of the mutations identified in the high-throughput screen (36–40).
Although DLBCL-associated mutations in the CARD, LATCH, and coiled-coil domains appear to dysregulate CARD11 signaling by the common mechanism of interfering with ID-promoted autoinhibition, to our knowledge no mutations in the ID itself have been reported and validated in DLBCL. The lack of oncogenic CARD11 mutations in the ID is surprising because this domain should be the most obvious target for mutations that perturb autoinhibition. In this report, we resolve this apparent inconsistency and explain why the CARD11 ID is resistant to gain-of function mutations. We find that instead of functioning as a single autoinhibitory domain, the CARD11 ID is composed of an unusual array of repressive elements that function cooperatively with redundancy to keep CARD11 inactive prior to antigen receptor engagement.
Experimental Procedures
Cell Lines and Expression Constructs
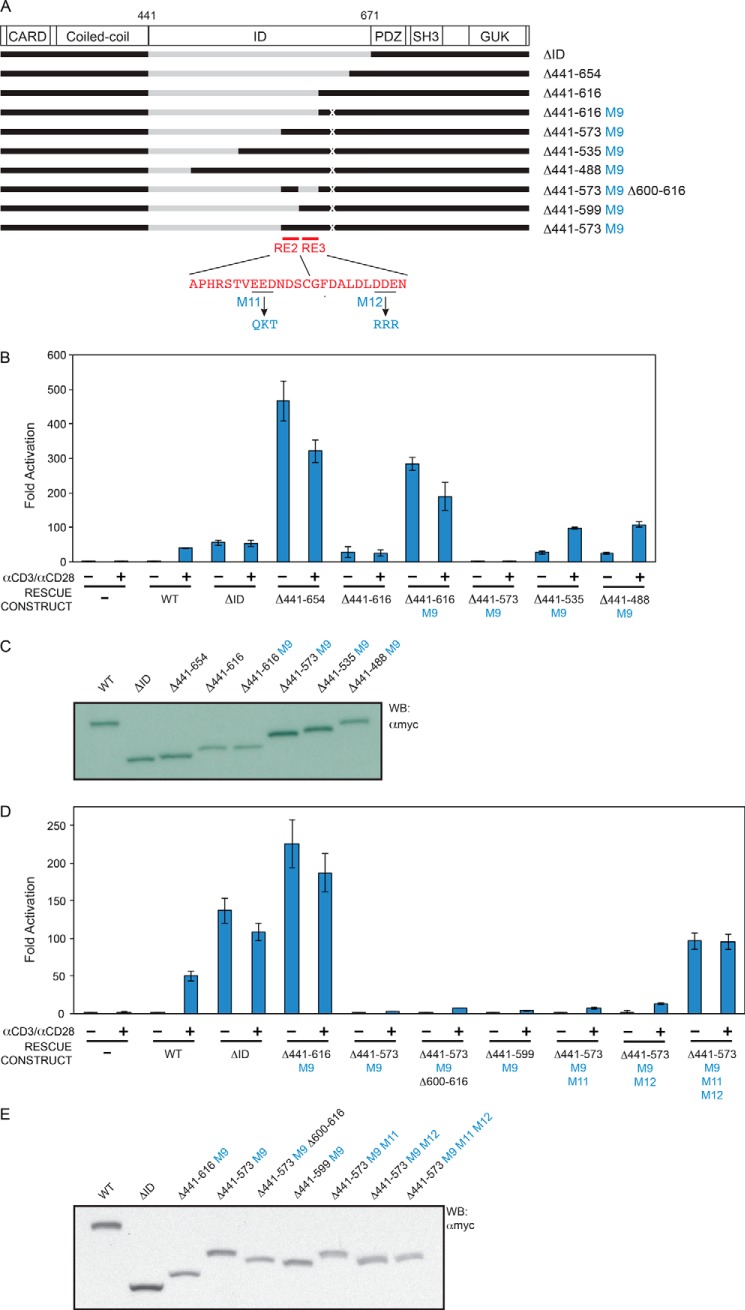
HEK293T and human Jurkat T cell lines were cultured as described previously (25). pc-CARD11, which expresses murine CARD11 with an N-terminal Myc tag, and CARD11ΔID have been described (25). Other CARD11 deletion constructs were cloned by standard molecular biology techniques and were sequence-verified. Constructs Δ441–573 M9, Δ441–535 M9, and Δ441–488 M9 in Fig. 5, B and C, and Δ441–599 M9 in Fig. 5, D and E, contain a substitution of residues 611–616 to DKTGRD to facilitate cloning. RE mutations were introduced into pc-CARD11 by PCR or QuikChange site-directed mutagenesis (Stratagene).
FIGURE 5.
Identification of RE2 and RE3. A, schematic of the constructs assayed. B and D, Jurkat T cells in which CARD11 was stably knocked down were transfected with CSK-LacZ and Igκ2-IFN-LUC in the presence of expression vectors for the indicated Myc-tagged CARD11 variants and stimulated with anti-CD3/anti-CD28 cross-linking for 5 h as indicated. B, two-tailed unpaired Student's t test with unequal variance resulted in p values <0.0015 for the values obtained under unstimulated conditions with the following constructs as compared with that observed with Δ441–616 M9: Δ441–573 M9, Δ441–535 M9, and Δ441–488 M9. D, same test resulted in p values <0.007 for the values obtained under unstimulated conditions with Δ441–573 M9Δ600–616 and Δ441–599 M9 as compared with that observed with Δ441–616 M9. The same test resulted in the following p values in the comparison with Δ441–573 M9: Δ441–573 M9, M11: p = 0.13, Δ441–573 M9, M12: p = 0.25, Δ441–573 M9, M11, M12: p = 0.004. C and E, HEK293T cells were transfected with the same amounts of each expression vector used in B and D, and lysates were probed by Western blot (WB) using anti-Myc primary antibody to indicate the relative expression level of each variant. β-Galactosidase activity, driven by CSK-LacZ, was used to calculate equivalent amounts of lysate for Western analysis.
HEK293T Protein Expression Assay
Cells were plated at 5 × 105 cells in 2 ml of media into each well of a 6-well plate 22.5–26 h prior to transfection. Cells were transfected with 200 ng of pCSK-LacZ, 1500 ng of Igκ2-IFN-LUC, and 65–610 ng of expression construct for each Myc-tagged CARD11 variant. In each experiment, samples were supplemented with empty pcDNA3 vector to keep the total amount of DNA per transfection constant at 3 μg. The media were changed 22–24 h post-transfection and harvested 39–42 h post-transfection. Cells were incubated with 500 μl of lysis buffer (Promega) on ice for at least 10 min and scraped from the plate, and cell debris was cleared by centrifugation at 17,970 × g for 5 min at 4 °C. Three microliters of lysate were used to measure β-galactosidase activity in a chemiluminescent β-gal reporter gene assay (Roche Applied Science) according to the manufacturer's instructions. Chemiluminescence was measured by a luminometer (Berthold Technologies TriStar LB 941 Multimode Microplate Reader) integrating for 10 s after a 2-s delay. β-Galactosidase activity was used to normalize lysates for transfection efficiency and recovery. Normalized lysates were resolved on an 8% SDS-polyacrylamide gel, transferred to polyvinylidene difluoride membrane, and analyzed by Western blot with anti-Myc (sc-40; Santa Cruz Biotechnology) antibodies to determine relative expression and standardize the nanogram amounts of expression vector required to achieve comparable amounts of all CARD11 variants in each experiment.
Determination of CARD11 Activity in Jurkat T Cells
The production of stable CARD11 knockdown Jurkat T cells (KD-CARD11) and control cells expressing the shNT non-target hairpin (NT) were performed as described previously (35). NT and KD-CARD11 Jurkat cells were transiently transfected with 200 ng of pCSK-LacZ, 1500 ng of Igκ2-IFN-LUC, and 65–610 ng of each pc-CARD11 variant, stimulated with anti-CD3/anti-CD28 as indicated, and harvested as described previously (25, 41). Fold activation was calculated for each sample by dividing the background-corrected luciferase activity, normalized to background-corrected β-gal activity, by that observed in the sample containing only empty expression vector in the absence of stimulation. All results shown are the average (±S.D.) of triplicate samples from a single experiment, representative of two or three that were performed.
Jurkat T Cell Protein Expression Assay
Reporter assays of KD-CARD11 Jurkat T cells were scaled up by a factor of 9. For each sample, 2.7% of cells were harvested in 36 μl of Promega Lysis Buffer for determination of relative transfection efficiency by β-gal reporter gene assay (Roche Applied Science), whereas 97.3% of cells were harvested in 1314 μl of IP Lysis Buffer (25). The amounts normalized to transfection efficiency were precleared with a 10-μl bed volume of protein G-Sepharose for 30 min at 4 °C with rotation. 2 μg of anti-Myc antibody (sc-40; Santa Cruz Biotechnology) was added to cleared lysates and incubated overnight at 4 °C, with rotation. 10 μl of beads that had been blocked with insulin were added, incubated for 2 h at 4 °C with rotation, washed four times for 5 min with IP Lysis Buffer, and then boiled in the presence of SDS loading buffer. Eluates were resolved on SDS-PAGE for Western blotting with anti-CARD11 primary antibody (ProSci 3189).
Results
Identification of Repressive Elements (REs) and Activating Elements (AEs) in the CARD11 ID
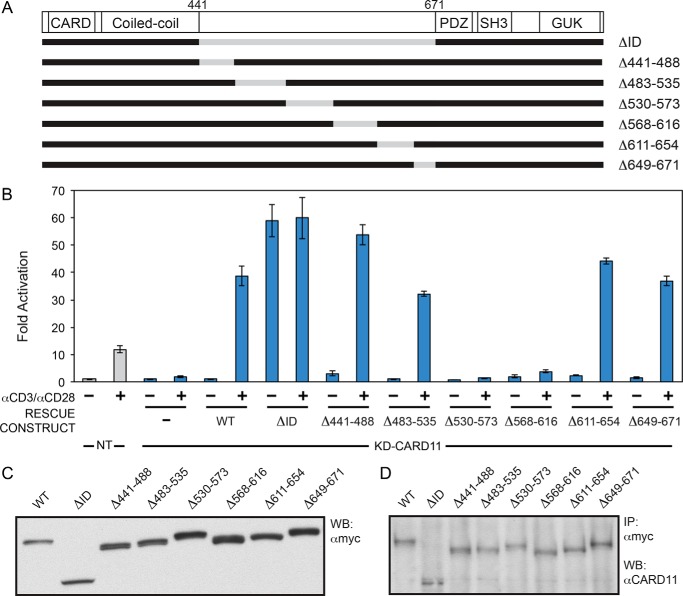
The CARD11 ID, which resides between the coiled-coil and PDZ domains (Fig. 1), is predicted to have little or no secondary structure (42–45) and instead consist of an intrinsically unstructured 231-residue region (46). Several studies have demonstrated that the deletion of the ID results in constitutive robust CARD11 signaling in the basal unstimulated state, comparable with that achieved by the wild-type protein upon antigen receptor engagement (25, 27, 47). This constitutive activity upon ID deletion results from the removal of autoinhibition and indicates that the ID does not contribute to the folding or function of CARD11 domains outside of the ID.
FIGURE 1.
CARD11 ID contains more than one redundant repressive element. A, schematic of the constructs assayed. B, Jurkat T cells in which CARD11 was stably knocked down (KD-CARD11) or control cells expressing a control shRNA (NT) were transfected with CSK-LacZ and Igκ2-IFN-LUC in the presence of expression vectors for the indicated Myc-tagged CARD11 variants and stimulated with anti-CD3/anti-CD28 cross-linking for 5 h as indicated. A two-tailed unpaired Student's t test with unequal variance resulted in p values >0.05 for the values obtained under unstimulated conditions with the following constructs as compared with that observed with wild-type CARD11: Δ441–488, Δ483–535, Δ530–573, Δ568–616, and Δ649–671. C, HEK293T cells were transfected with the same amounts of each expression vector used in B, and lysates were probed by Western blot (WB) using anti-Myc primary antibody to indicate the relative expression level of each variant. β-Galactosidase activity, driven by CSK-LacZ, was used to calculate equivalent amounts of lysate for Western analysis. D, KD-CARD11 Jurkat T cell transfections in B were scaled up 9-fold, and each Myc-tagged CARD11 variant was immunoprecipitated (IP) by anti-Myc antibody and assayed by Western blot using anti-CARD11 antibody as described under “Experimental Procedures.” β-Galactosidase activity, driven by CSK-LacZ, was used to calculate equivalent amounts of lysate for Western analysis.
To identify regions of the CARD11 ID that are prone to gain-of-function mutations, we generated six deletion constructs that span the ID (Fig. 1A) and assayed them for derepressed constitutive signaling to NF-κB, which would be manifested by enhanced signaling, as compared to wild-type CARD11, in the absence of antigen receptor triggering. These constructs were assayed in a previously established RNAi-rescue assay (25, 34, 35) in which Jurkat T cells with stable CARD11 knockdown are transfected with murine CARD11 variants and assayed for anti-CD3/anti-CD28-mediated activation of Igκ2-IFN-LUC, an NF-κB-responsive luciferase reporter. We have previously shown using a panel of CARD11 variants with a range of activities that the induction of Igκ2-IFN-LUC reporter in Jurkat T cells quantitatively correlates with the induction of nuclear NF-κB DNA binding activity (35). The RNAi-rescue assay was employed to specifically avoid the overexpression of CARD11 variants and any associated artifacts. Because of the low levels of CARD11 variant expression under the transfection conditions of this assay, HEK293T cells were used to assess the relative protein expression of CARD11 variants using the same amounts of expression vectors as those used to transfect Jurkat T cells, an approach that has been previously validated (25, 34, 35).
As reported previously, CARD11 knockdown resulted in defective NF-κB induction by TCR cross-linking, which was rescued with expression of a wild-type hairpin-resistant CARD11 cDNA (Fig. 1B). As expected, rescue with CARD11 ΔID, which lacks the entire ID, led to constitutive signaling in the absence of TCR triggering (Fig. 1B). Thus, as reported previously, the RNAi-rescue assay preserves two essential features of CARD11 behavior in the antigen receptor signaling pathway, a lack of signaling in the absence of TCR triggering due to the autoinhibitory role of the ID, and robust inducible signaling once the TCR is triggered with anti-CD3/anti-CD28 cross-linking.
Surprisingly, none of the six ID deletions we assayed resulted in constitutive NF-κB activation in the absence of anti-CD3/anti-CD28 treatment, even though the panel of deletions covers the entire ID (Fig. 1B). These results suggested that the ID must harbor two or more redundant REs, such that when one is deleted another is sufficient for autoinhibition of CARD11 signaling activity prior to antigen receptor triggering.
To further validate the use of HEK293T cells to assess the relative level of CARD11 variant expression elicited by expression constructs in Jurkat T cells, we scaled up the transient transfection in Fig. 1B in Jurkat T cells by 9-fold and assayed CARD11 variant expression level by Western blotting anti-Myc immunoprecipitates with anti-CARD11 antibody. As shown in Fig. 1, C and D, the same relative quantities of expression vectors yielded the same relative protein variant expression levels in Jurkat T cells (Fig. 1D) as in HEK293T cells (Fig. 1C), thus validating the use of HEK293T cells to assess the amount of CARD11 protein expression produced from a given amount of expression vector.
The deletion of regions 530–573 and 568–616 within the ID resulted in a lack of TCR-induced CARD11-dependent signaling to NF-κB (Fig. 1, A and B). This was not surprising because these deletions remove serines 564, 567, and 577, which are known to be required for antigen receptor-induced derepression of CARD11 signaling activity via their signal-dependent phosphorylation (26, 27, 29, 30).
To map the redundant REs in the CARD11 ID, we added back portions of the ID to the ΔID and used the RNAi-rescue assay to assess basal repression of CARD11 activity. Fig. 2 presents a schematic summary of the elements we discovered using this approach. In addition to mapping REs in this manner, we also used mutagenesis to destroy inhibitory RE activity while preserving the relative spacing of N- and C-terminal domains in the respective mutant and parental CARD11 constructs. For mutagenesis, we sought to change the chemical character of each targeted amino acid side chain without any desire to preserve putative local RE structure because our intention was to destroy potentially redundant RE activity in the context of ID regions predicted to be intrinsically unstructured.
FIGURE 2.
Schematic of the elements identified in the ID in this study and their associated mutations. Top, CARD11 domain structure is depicted with the relative location of repressive and activating elements in the ID. The LATCH domain lies between the CARD and the coiled coil, L3 between PDZ and Src homology 3, and L4 between Src homology 3 and GUK. Bottom, amino acid sequence of the ID is depicted. Repressive and activating elements are indicated in red and green, respectively. Mutations used in the study are indicated in blue.
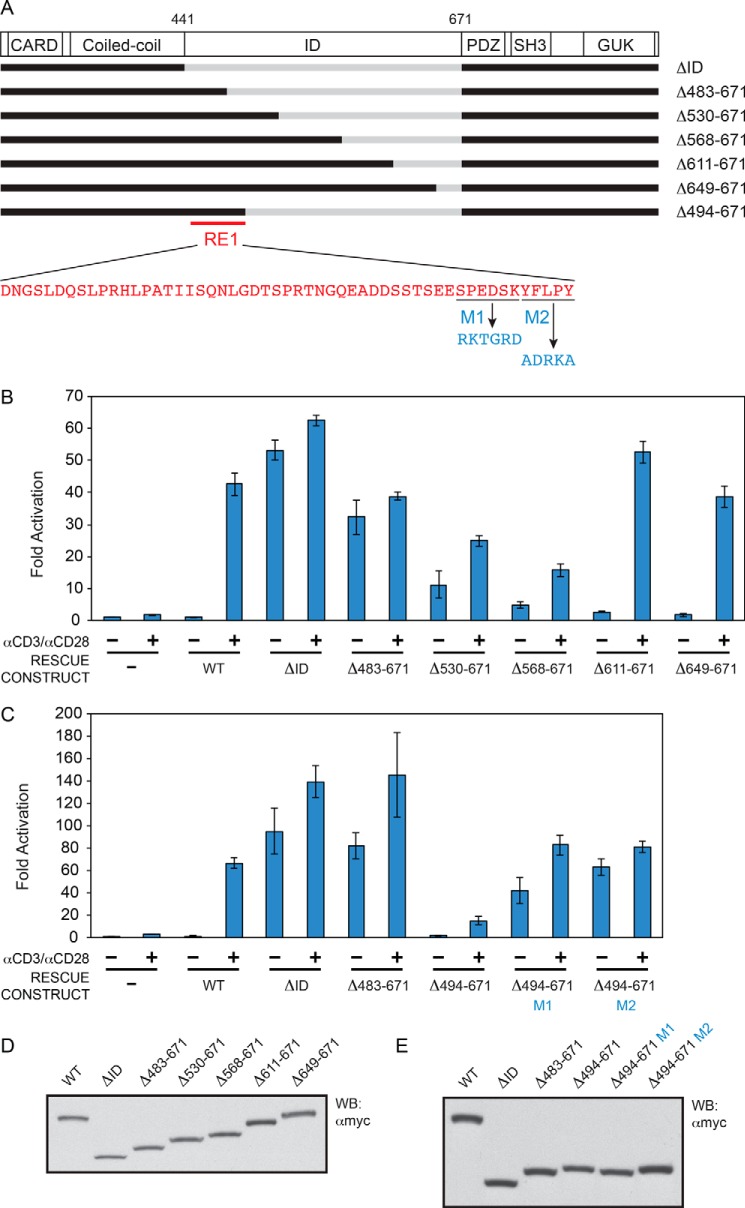
The successive addition of regions extending from the N-terminal end of the ID suggested the presence of a repressive element between residues 441 and 529 (Fig. 3, A, B, and D), based upon the ability of this region to reduce the level of activity of the ΔID under basal conditions in the absence of TCR cross-linking. The repressive element in this region was mapped more finely to the Asp-441 to Tyr-493 region and named RE1. The addition of RE1 to the ΔID was sufficient to confer autoinhibition in the basal state, as indicated by the comparison of activities between the ΔID and the Δ494–671 construct in the absence of anti-CD3/anti-CD28 treatment (Fig. 3, A, C, and E). We also defined two mutations within RE1 that disrupted its repressive activity, M1 and M2, each of which mutate residues at the C-terminal end of RE1 (Fig. 3, A, C, and E).
FIGURE 3.
Identification of RE1. A, schematic of the constructs assayed. B and C, Jurkat T cells in which CARD11 was stably knocked down were transfected with CSK-LacZ and Igκ2-IFN-LUC in the presence of expression vectors for the indicated Myc-tagged CARD11 variants and stimulated with anti-CD3/anti-CD28 cross-linking for 5 h as indicated. B, two-tailed unpaired Student's t test with unequal variance resulted in p values <0.008 for the values obtained under unstimulated conditions with the following constructs as compared with that observed with CARD11 ΔΙD: Δ483–671, Δ530–671, Δ568–671, Δ611–671, and Δ649–671. C, same test resulted in a p value <0.02 for the values obtained under unstimulated conditions with Δ494–671 as compared with that observed with CARD11 ΔΙD, and p values <0.006 for the comparison between Δ494–671 and either Δ494–671 M1 or Δ494–671 M2. D and E, HEK293T cells were transfected with the same amounts of each expression vector used in B and C, and lysates were probed by Western blot using anti-Myc primary antibody to indicate the relative expression level of each variant. β-Galactosidase activity, driven by CSK-LacZ, was used to calculate equivalent amounts of lysate for Western analysis.
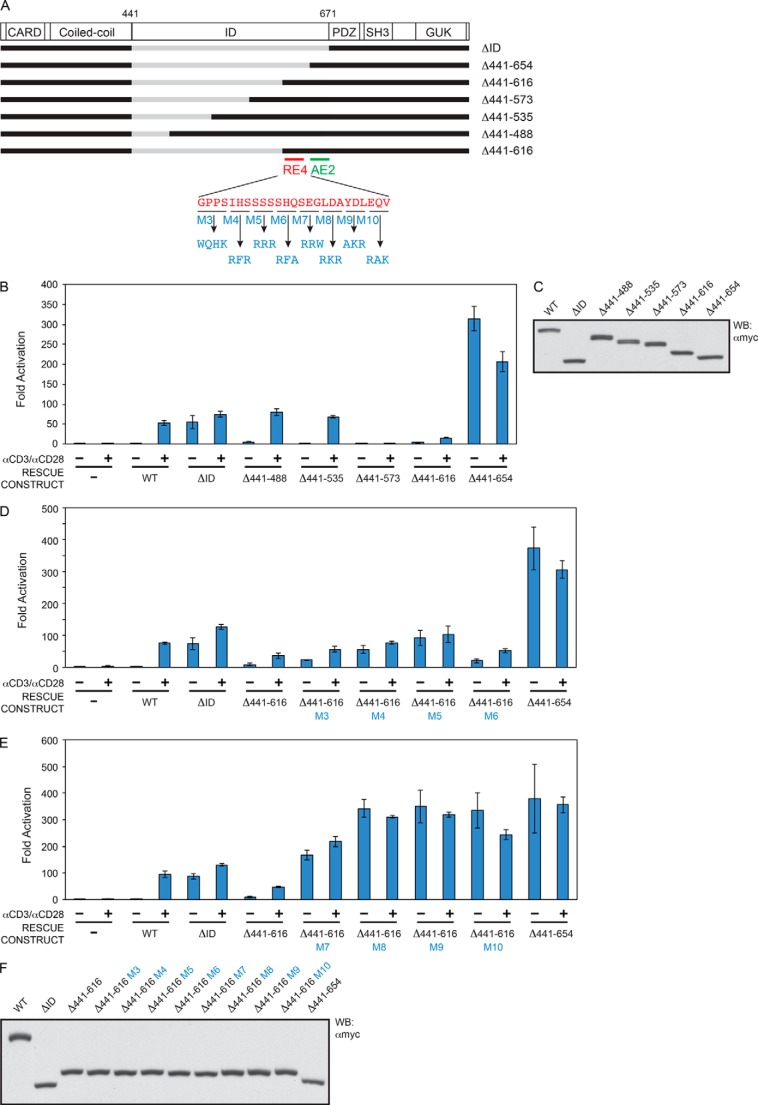
The successive addition of regions extending from the C-terminal end of the ID surprisingly identified a region between residues 655 and 671 that enhanced the specific activity of the ΔID by 5.7-fold; we refer to this region as activating element 2 (AE2) (Fig. 4, A–C). Further addition of the 617–654 region resulted in complete repression of CARD11 signaling in the basal state, as indicated by the dramatic difference in activities between the Δ441–654 and Δ441–616 constructs in the absence of TCR cross-linking (Fig. 4, A–C). We further mapped the RE in this region to span Gly-617 to Val-641 and named it RE4 (Fig. 2, 4, B and C). We used the construct containing RE4 and AE2 elements (Δ441–616) to identify several RE4 mutations that could deleteriously affect RE4 repressive activity (Fig. 4, A and D–F), including the M9 mutation toward the C-terminal end of RE4.
FIGURE 4.
Identification of RE4 and AE2. A, schematic of the constructs assayed. B, D, and E, Jurkat T cells in which CARD11 was stably knocked down were transfected with CSK-LacZ and Igκ2-IFN-LUC in the presence of expression vectors for the indicated Myc-tagged CARD11 variants and stimulated with anti-CD3/anti-CD28 cross-linking for 5 h as indicated. B, two-tailed unpaired Student's t test with unequal variance resulted in p values <0.004 for the values obtained under unstimulated conditions with the following constructs as compared with that observed with CARD11 Δ441–654: ΔID, Δ441–488, Δ441–535, Δ441–573, and Δ441–616. D, same test resulted in p values <0.05 for the values obtained under unstimulated conditions with the following constructs as compared with that observed with Δ441–616: Δ441–616 M3, Δ441–616 M4, and Δ441–616 M5. E, same test resulted in p values <0.015 for the values obtained under unstimulated conditions with the following constructs as compared with that observed with Δ441–616: Δ441–616 M7, Δ441–616 M8, Δ441–616 M9, and Δ441–616 M10. C and F, HEK293T cells were transfected with the same amounts of each expression vector used in B, D, and E, and lysates were probed by Western blot (WB) using anti-Myc primary antibody to indicate the relative expression level of each variant. β-Galactosidase activity, driven by CSK-LacZ, was used to calculate equivalent amounts of lysate for Western analysis.
Next, we used the construct containing AE2 and M9-mutated RE4 (Δ441–616 M9) to look for other redundant REs that may lie N-terminal to RE4 (Fig. 5A). We found that residues 574–610 could confer autoinhibition when added to the Δ441–616 M9 construct (Fig. 5, B and C, cf. Δ441–616 M9 to Δ441–573 M9). We further mapped two REs within this region, RE2, which spans Ala-586 to Cys-599 (Fig. 2, 5, A, D, and E), and RE3, which spans Gly-600 to Asn-610 (Figs. 2 and 5, A, D, and E). Either RE2 or RE3 was sufficient to repress activity of the Δ441–616 M9 construct, as indicated by the low activity of the Δ441–573 M9 Δ600–616 construct and the Δ441–599 M9 construct in the absence of anti-CD3/anti-CD28 treatment (Fig. 5, D and E). The simultaneous mutation of RE2 and RE3 was required to reverse the inhibitory effect of the 574–610 region, consistent with the presence of two redundant REs in this small portion of the ID (Fig. 5, A, D, and E, cf. Δ441–573 M9, Δ441–573 M9 M11, Δ441–573 M9 M12, and Δ441–573 M9 M11 M12).
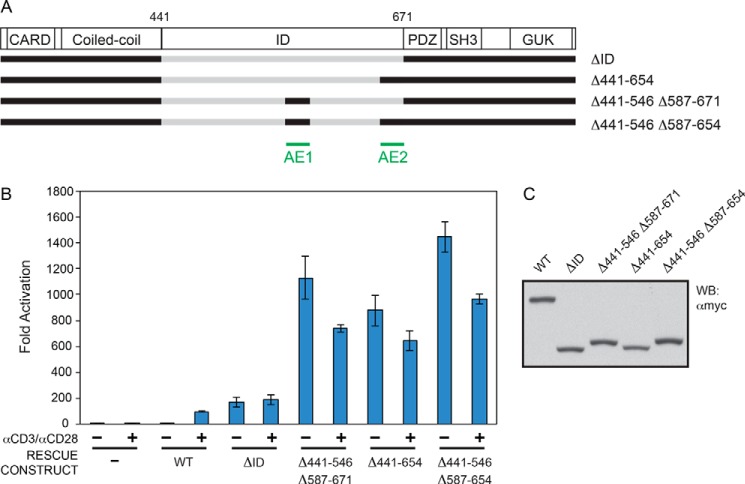
This analysis revealed the presence of repressive elements RE1, RE2, RE3, and RE4 within the ID (Fig. 2). We did not obtain evidence of any further redundant REs in the ID. However, the enhanced activity of the Δ441–535 M9 construct as compared with the Δ441–573 M9 construct (Fig. 5, A–C) suggested the possibility of an activating element residing between residue 536 and RE2. We mapped this enhancing activity between Leu-547 and Ala-586, and we named it AE1 (Figs. 2 and 6). The addition of AE1 to the ΔID construct resulted in 6.6-fold increased activity (Fig. 6, A–C). While AE1 and AE2 could each enhance the activity of the ΔID, their combined presence did not have an additive effect in this assay (Fig. 6, A–C), suggesting that they enhance activity by redundant mechanisms.
FIGURE 6.
Relative activities conferred by AE1 and AE2. A, schematic of the constructs assayed. B, Jurkat T cells in which CARD11 was stably knocked down were transfected with CSK-LacZ and Igκ2-IFN-LUC in the presence of expression vectors for the indicated Myc-tagged CARD11 variants and stimulated with anti-CD3/anti-CD28 cross-linking for 5 h as indicated. A two-tailed unpaired Student's t test with unequal variance resulted in p values <0.0085 for the values obtained under unstimulated conditions with the following constructs as compared with that observed with CARD11 ΔID: Δ441–546 Δ587–671, Δ441–654, Δ441–546, and Δ587–654. C, HEK293T cells were transfected with the same amounts of each expression vector used in B, and lysates were probed by Western blot (WB) using anti-Myc primary antibody to indicate the relative expression level of each variant. β-Galactosidase activity, driven by CSK-LacZ, was used to calculate equivalent amounts of lysate for Western analysis.
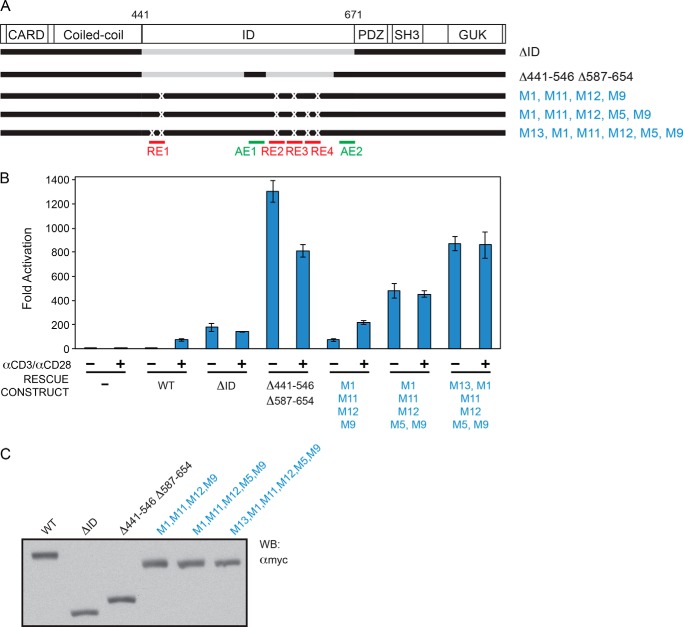
We next wanted to verify the repressive roles of RE1, RE2, RE3, and RE4 in the context of full-length CARD11. We introduced mutations M1, M11, M12, and M9 into full-length CARD11 to simultaneously perturb the repressive functions of all four REs. In the RNAi-rescue assay, we found that the combined presence of these mutations did not result in as much of an enhancement of activity as we expected; this construct was not even as active as the ΔID, despite the fact that it contained both AE1 and AE2 (Fig. 7, A–C). We suspected that the M1, M11, M12, and M9 substitutions may not completely disrupt all RE repressive activity and that the residual RE activity of four mutated REs in cis might afford significant autoinhibition. We therefore introduced further mutations into RE1 and RE4 because the M1 mutation only partially disrupted RE1 function in the experiments in Fig. 3 and because RE4 appeared to be a bipartite element based on the mutational analysis in Fig. 4. Addition of the M5 mutation in RE4 significantly enhanced the activity of the full-length mutated CARD11 construct in the absence of TCR cross-linking, and this was further enhanced by the addition of the M13 mutation in RE1 (Figs. 2 and 7, A–C). The activity of this most active construct (M1, M5, M9, M11, M12, and M13) approached that observed with the construct in which AE1 and AE2 were added to the ΔID (Δ441–546 Δ587–654) (Fig. 7, A–C). Thus, the combined mutation of RE1 (M1 and M13), RE2 (M11), RE3 (M12), and RE4 (M5 and M9) in the context of full-length CARD11 resulted in significant derepression of CARD11 in the basal state and verified an autoinhibitory role of these REs in the full-length CARD11 context.
FIGURE 7.
RE1, RE2, RE3, and RE4 function in the context of full-length CARD11. A, schematic of the constructs assayed. B, Jurkat T cells in which CARD11 was stably knocked down were transfected with CSK-LacZ and Igκ2-IFN-LUC in the presence of expression vectors for the indicated Myc-tagged CARD11 variants and stimulated with anti-CD3/anti-CD28 cross-linking for 5 h as indicated. A two-tailed unpaired Student's t test with unequal variance resulted in p values <0.015 for the values obtained under unstimulated conditions with the following constructs as compared with that observed with wild-type CARD11: Δ441–546 Δ587–654; M1, M11, M12, M9; M1, M11, M12, M5, M9; and M1, M13, M11, M12, M5, M9. C, HEK293T cells were transfected with the same amounts of each expression vector used in B, and lysates were probed by Western blot (WB) using anti-Myc primary antibody to indicate the relative expression level of each variant. β-Galactosidase activity, driven by CSK-LacZ, was used to calculate equivalent amounts of lysate for Western analysis.
REs Cooperatively Repress CARD11-mediated NF-κB Activation
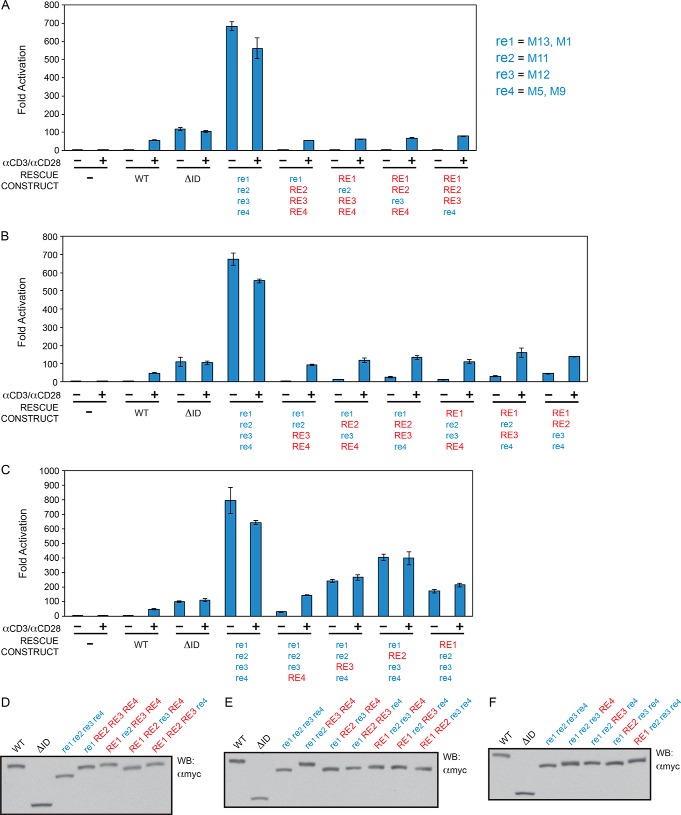
To quantitatively demonstrate the functional redundancy of the four REs in repressing spontaneous CARD11 signaling to NF-κB, we compared the activities of CARD11 constructs containing mutations in all combinations of REs. RE1 was mutated with simultaneous M1 and M13 mutations, RE2 with the M11 mutation, RE3 with the M12 mutation, and RE4 with simultaneous M5 and M9 mutations. In the RNAi-rescue assay, each construct was directly compared with wild-type CARD11, CARD11ΔID, and the construct containing simultaneous mutations in all four REs (Fig. 8 and Table 1). For ease of representation, the presence of a mutated RE in a construct is indicated by lowercase letters (e.g. re1), whereas the presence of a wild-type RE is indicated by capital letters (e.g. RE1). The re1 re2 re3 re4 quadruple RE mutant displayed an activity that was 640-fold higher than wild-type CARD11 and 6.7-fold higher than the ΔID (Fig. 8, A and D) in the absence of TCR engagement.
FIGURE 8.
RE1, RE2, RE3, and RE4 cooperatively repress CARD11 signaling to NF-κB. A–C, Jurkat T cells in which CARD11 was stably knocked down (KD-CARD11) were transfected with CSK-LacZ and Igκ2-IFN-LUC in the presence of expression vectors for the indicated Myc-tagged CARD11 variants and stimulated with anti-CD3/anti-CD28 cross-linking for 5 h as indicated. A two-tailed unpaired Student's t test with unequal variance resulted in the following p values in comparison with wild-type CARD11 under unstimulated conditions: re1 re2 re3 re4, p = 0.0004; re1 RE2 RE3 RE4, p = 0.397; RE1 re2 RE3 RE4, p = 0.054; RE1 RE2 re3 RE4, p = 0.0016; RE1 RE2 RE3 re4, p = 0.0054; re1 re2 RE3 RE4, p = 0.0397; re1 RE2 re3 RE4, p = 0.0008; re1 RE2 RE3 re4, p = 0.011; RE1 re2 re3 RE4, p = 0.016; RE1 re2 RE3 re4, p = 0.005; RE1 RE2 re3 re4, p = 0.0012; re1 re2 re3 RE4, p = 0.005; re1 re2 RE3 re4, p = 0.0007; re1 RE2 re3 re4, p = 0.001; RE1 re2 re3 re4, p = 0.0016. D–F, HEK293T cells were transfected with the same amounts of each expression vector used in A–C, and lysates were probed by Western blot using anti-Myc primary antibody to indicate the relative expression level of each variant. β-Galactosidase activity, driven by CSK-LacZ, was used to calculate equivalent amounts of lysate for Western analysis. For ease of representation, the presence of a mutated RE in a construct is indicated by blue lowercase letters (e.g. re1), whereas the presence of a wild-type RE is indicated by red capital letters (e.g. RE1).
TABLE 1.
Relative Activities of RE mutants

a The relative activity was determined by normalizing the mean fold activation determined in Figure 8 to that observed with wild-type CARD11 at comparable levels of expression in the absence of anti-CD3/anti-CD28 treatment.
As shown in Fig. 8, A and D, and Table 1, the mutation of any single RE had little effect on the basal activity of CARD11 in the absence of anti-CD3/anti-CD28 treatment, consistent with the redundant action of the array of REs and the resistance of the ID to potentially gain-of-function point mutations. The mutation of RE4 had the largest effect, but it only increased activity 3.5-fold over that of wild type.
Variants containing the mutation of two REs displayed higher activities in the basal state, ranging from 4.0-fold (re1 re2 RE3 RE4) to 40-fold (RE1 RE2 re3 re4) over wild-type CARD11 (Fig. 8, B and E, and Table 1), but they were all significantly repressed (16- to 160-fold) as compared with the re1 re2 re3 re4 quadruple RE mutant. Double RE mutants containing RE4 mutations were more active than those with wild-type RE4.
The triple RE mutants ranged from 28-fold (re1 re2 re3 RE4) to 390-fold (re1 RE2 re3 re4) more active than wild-type CARD11 (Fig. 8, C and F, and Table 1), and when compared with the re1 re2 re3 re4 quadruple RE mutant, they suggested the relative potential of each wild-type RE to individually repress in the full-length CARD11 context in the absence of TCR triggering. In this view, RE4 alone provided 23-fold repression; RE1 alone provided 3.8-fold repression; RE3 alone provided 2.7-fold repression, and RE2 alone provided 1.6-fold repression.
The results indicated that in all combinations the REs cooperate with each other to potentiate their repressive effects on CARD11 signaling to NF-κB prior to antigen receptor engagement. For example, for each individual RE, the addition of any of the other three REs increases the quantitative extent of repression. Similarly, for each pair of REs, the addition of either of the other two REs increases repression. For the triplet RE combinations, the addition of the fourth RE had relatively mild effects, consistent with the redundant properties of the array. The cooperativity we observed in the assay of these variants supported our assumption that the mutations we introduced to kill the activity of an individual RE do not have inadvertent negative effects on the folding or function of the other REs. In addition, the signaling output of the single RE mutants following TCR triggering (Fig. 8A), which was comparable to that elicited by wild-type CARD11, confirmed that the RE mutations we introduced do not affect the folding or function of other CARD11 domains outside of the ID.
Discussion
We have defined four REs in the CARD11 ID that function cooperatively to autoinhibit CARD11 signaling to NF-κB in the basal state. Their cooperative action results in a redundancy of repressive function that makes the ID resistant to gain-of-function CARD11 mutations and explains why potent mutations in the ID have not been identified in DLBCL. DLBCL-associated mutations instead occur in the CARD, LATCH, or coiled coil, where single amino acid mutations have the potential to activate the protein 80–160-fold in lymphocytes (34, 35). Our quantitative analysis suggests that to achieve this level of hyperactivity, three or more REs would have to be disabled, which would require a combination of mutations in the ID that would only occur with extremely low probability. DLBCLs instead appear to select for single amino acid mutations in the CARD, LATCH, and coiled coil that presumably disrupt or bypass repression by three or more REs. In the accompanying paper (48), we discuss how oncogenic CARD11 point mutations can disrupt autoinhibition mediated by multiple REs.
Why has CARD11 evolved this array of cooperative REs? Our quantitative analysis indicates that there is much more potential signaling activity in CARD11 than previously appreciated. The quadruple RE mutant re1 re2 re3 re4 is 640-fold more active than wild-type CARD11, more active than any previously described CARD11 variant. The latent activity in CARD11 is due in part to the presence of the activating elements AE1 and AE2, which have not been previously identified and which increase CARD11 specific activity by an undetermined mechanism. CARD11 has likely evolved its array of REs to ensure that high levels of CARD11 signaling do not occur in normal cells unless and until the antigen receptor is engaged. Clearly, hyperactive CARD11 signaling in the 80–160-fold range above wild-type can promote the aberrant lymphocyte proliferation and survival that is seen in DLBCL (33, 35). In addition, in the absence of lymphocyte transformation, constitutive hyperactive CARD11 signaling in the 80–100-fold over wild-type range (e.g. G123D and C49Y (35)) can perturb lymphocyte homeostasis and promote BENTA disease in humans, a disorder that results from germline gain-of-function CARD11 mutation, involves B cell expansion and T cell anergy, and presents with features of immunodeficiency (49–52). We also speculate that the array of REs in the CARD11 ID may be important in determining the kinetics of signal induction after antigen receptor engagement, or for the kinetics by which CARD11 returns to the basal inactive state after antigen receptors are triggered. Consistent with the latter notion, the quadruple RE mutant re1 re2 re3 re4 results in much higher levels of NF-κB activity than what is observed after 5 h of TCR cross-linking (Fig. 8).
In conclusion, our data reveal a remarkable and unique strategy by which the CARD11 signaling scaffold achieves autoinhibition to prevent unwarranted signaling, an array of four small REs that range in length from 11 to 53 residues and that function cooperatively with redundancy. Clearly, autoinhibition by the ID is significantly more elaborate than previously modeled (25–27). The REs have evolved not only to autoinhibit in the basal state but also to be neutralized enough during antigen receptor signaling to promote normal signaling to the IKK complex and NF-κB. Their neutralization occurs by an undefined mechanism that requires the phosphorylation of serines 564, 567, 577, and 657 in the ID (26, 27, 29, 30), and further work will be required to determine precisely how this occurs. Redundant REs may also regulate other proteins that signal to NF-κB or other targets that regulate cellular proliferation and survival. We note that their redundancy may allow them to escape the strict evolutionary conservation that would facilitate their recognition by sequence homology.
Author Contributions
R. P. J. and J. M. T. designed, performed, and analyzed the experiments. J. L. P. conceived of the study and designed and analyzed the experiments. All authors approved the final version of the manuscript.
Acknowledgments
We thank John Bettridge, Corinne Hamblet, deMauri Mackie, Sarah Pedersen, and Alyssa Ward for critical reading of the manuscript and Mollie Meffert and Stephen Desiderio for helpful discussions and advice.
This work was supported by National Institutes of Health Grant RO1CA177600 and funds from The Johns Hopkins University Institute for Cell Engineering. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as a Paper of the Week.
- ID
- inhibitory domain
- DLBCL
- diffuse large B cell lymphoma
- RE
- repressive element
- TCR
- T cell receptor
- NT
- non-target
- AE
- activating element
- CARD
- caspase recruitment domain
- IKK
- IκB kinase.
References
- 1. Xu W., Harrison S. C., and Eck M. J. (1997) Three-dimensional structure of the tyrosine kinase c-Src. Nature 385, 595–602 [DOI] [PubMed] [Google Scholar]
- 2. Sicheri F., Moarefi I., and Kuriyan J. (1997) Crystal structure of the Src family tyrosine kinase Hck. Nature 385, 602–609 [DOI] [PubMed] [Google Scholar]
- 3. Goldberg J., Nairn A. C., and Kuriyan J. (1996) Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I. Cell 84, 875–887 [DOI] [PubMed] [Google Scholar]
- 4. Chen L., Jiao Z. H., Zheng L. S., Zhang Y. Y., Xie S. T., Wang Z. X., and Wu J. W. (2009) Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature 459, 1146–1149 [DOI] [PubMed] [Google Scholar]
- 5. Xu J., Shen C., Wang T., and Quan J. (2013) Structural basis for the inhibition of Polo-like kinase 1. Nat. Struct. Mol. Biol. 20, 1047–1053 [DOI] [PubMed] [Google Scholar]
- 6. Aghazadeh B., Lowry W. E., Huang X. Y., and Rosen M. K. (2000) Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102, 625–633 [DOI] [PubMed] [Google Scholar]
- 7. DeBell K., Graham L., Reischl I., Serrano C., Bonvini E., and Rellahan B. (2007) Intramolecular regulation of phospholipase C-γ1 by its C-terminal Src homology 2 domain. Mol. Cell. Biol. 27, 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bunney T. D., Esposito D., Mas-Droux C., Lamber E., Baxendale R. W., Martins M., Cole A., Svergun D., Driscoll P. C., and Katan M. (2012) Structural and functional integration of the PLCγ interaction domains critical for regulatory mechanisms and signaling deregulation. Structure 20, 2062–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kobashigawa Y., Tomitaka A., Kumeta H., Noda N. N., Yamaguchi M., and Inagaki F. (2011) Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc. Natl. Acad. Sci. U.S.A. 108, 20579–20584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duda D. M., Olszewski J. L., Schuermann J. P., Kurinov I., Miller D. J., Nourse A., Alpi A. F., and Schulman B. A. (2013) Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21, 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mari S., Ruetalo N., Maspero E., Stoffregen M. C., Pasqualato S., Polo S., and Wiesner S. (2014) Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure 22, 1639–1649 [DOI] [PubMed] [Google Scholar]
- 12. Pufall M. A., and Graves B. J. (2002) Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 18, 421–462 [DOI] [PubMed] [Google Scholar]
- 13. Peterson J. R., and Golemis E. A. (2004) Autoinhibited proteins as promising drug targets. J. Cell. Biochem. 93, 68–73 [DOI] [PubMed] [Google Scholar]
- 14. Gaide O., Favier B., Legler D. F., Bonnet D., Brissoni B., Valitutti S., Bron C., Tschopp J., and Thome M. (2002) CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-κB activation. Nat. Immunol. 3, 836–843 [DOI] [PubMed] [Google Scholar]
- 15. Wang D., You Y., Case S. M., McAllister-Lucas L. M., Wang L., DiStefano P. S., Nuñez G., Bertin J., and Lin X. (2002) A requirement for CARMA1 in TCR-induced NF-κB activation. Nat. Immunol. 3, 830–835 [DOI] [PubMed] [Google Scholar]
- 16. Pomerantz J. L., Denny E. M., and Baltimore D. (2002) CARD11 mediates factor-specific activation of NF-κB by the T cell receptor complex. EMBO J. 21, 5184–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hara H., Wada T., Bakal C., Kozieradzki I., Suzuki S., Suzuki N., Nghiem M., Griffiths E. K., Krawczyk C., Bauer B., D'Acquisto F., Ghosh S., Yeh W. C., Baier G., Rottapel R., and Penninger J. M. (2003) The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity 18, 763–775 [DOI] [PubMed] [Google Scholar]
- 18. Jun J. E., Wilson L. E., Vinuesa C. G., Lesage S., Blery M., Miosge L. A., Cook M. C., Kucharska E. M., Hara H., Penninger J. M., Domashenz H., Hong N. A., Glynne R. J., Nelms K. A., and Goodnow C. C. (2003) Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity 18, 751–762 [DOI] [PubMed] [Google Scholar]
- 19. Egawa T., Albrecht B., Favier B., Sunshine M. J., Mirchandani K., O'Brien W., Thome M., and Littman D. R. (2003) Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr. Biol. 13, 1252–1258 [DOI] [PubMed] [Google Scholar]
- 20. Newton K., and Dixit V. M. (2003) Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr. Biol. 13, 1247–1251 [DOI] [PubMed] [Google Scholar]
- 21. Stepensky P., Keller B., Buchta M., Kienzler A. K., Elpeleg O., Somech R., Cohen S., Shachar I., Miosge L. A., Schlesier M., Fuchs I., Enders A., Eibel H., Grimbacher B., and Warnatz K. (2013) Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J. Allergy Clin. Immunol. 131, 477–485 [DOI] [PubMed] [Google Scholar]
- 22. Greil J., Rausch T., Giese T., Bandapalli O. R., Daniel V., Bekeredjian-Ding I., Stütz A. M., Drees C., Roth S., Ruland J., Korbel J. O., and Kulozik A. E. (2013) Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. J. Allergy Clin. Immunol. 131, 1376–1383 [DOI] [PubMed] [Google Scholar]
- 23. Schulze-Luehrmann J., and Ghosh S. (2006) Antigen-receptor signaling to nuclear factor κB. Immunity 25, 701–715 [DOI] [PubMed] [Google Scholar]
- 24. Jiang C., and Lin X. (2012) Regulation of NF-κB by the CARD proteins. Immunol. Rev. 246, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCully R. R., and Pomerantz J. L. (2008) The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-kappaB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association. Mol. Cell. Biol. 28, 5668–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsumoto R., Wang D., Blonska M., Li H., Kobayashi M., Pappu B., Chen Y., Wang D., and Lin X. (2005) Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-κB activation. Immunity 23, 575–585 [DOI] [PubMed] [Google Scholar]
- 27. Sommer K., Guo B., Pomerantz J. L., Bandaranayake A. D., Moreno-García M. E., Ovechkina Y. L., and Rawlings D. J. (2005) Phosphorylation of the CARMA1 linker controls NF-κB activation. Immunity 23, 561–574 [DOI] [PubMed] [Google Scholar]
- 28. Wegener E., Oeckinghaus A., Papadopoulou N., Lavitas L., Schmidt-Supprian M., Ferch U., Mak T. W., Ruland J., Heissmeyer V., and Krappmann D. (2006) Essential role for IκB kinase β in remodeling Carma1-Bcl10-Malt1 complexes upon T cell activation. Mol. Cell 23, 13–23 [DOI] [PubMed] [Google Scholar]
- 29. Shinohara H., Maeda S., Watarai H., and Kurosaki T. (2007) IκB kinase β-induced phosphorylation of CARMA1 contributes to CARMA1 Bcl10 MALT1 complex formation in B cells. J. Exp. Med. 204, 3285–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thome M., Charton J. E., Pelzer C., and Hailfinger S. (2010) Antigen receptor signaling to NF-κB via CARMA1, BCL10, and MALT1. Cold Spring Harb. Perspect. Biol. 2, a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim K. H., Yang Y., and Staudt L. M. (2012) Pathogenetic importance and therapeutic implications of NF-κB in lymphoid malignancies. Immunol. Rev. 246, 359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davis R. E., Brown K. D., Siebenlist U., and Staudt L. M. (2001) Constitutive nuclear factor κB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 194, 1861–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lenz G., Davis R. E., Ngo V. N., Lam L., George T. C., Wright G. W., Dave S. S., Zhao H., Xu W., Rosenwald A., Ott G., Muller-Hermelink H. K., Gascoyne R. D., Connors J. M., Rimsza L. M., et al. (2008) Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319, 1676–1679 [DOI] [PubMed] [Google Scholar]
- 34. Lamason R. L., McCully R. R., Lew S. M., and Pomerantz J. L. (2010) Oncogenic CARD11 mutations induce hyperactive signaling by disrupting autoinhibition by the PKC-responsive inhibitory domain. Biochemistry 49, 8240–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chan W., Schaffer T. B., and Pomerantz J. L. (2013) A quantitative signaling screen identifies CARD11 mutations in the CARD and LATCH domains that induce Bcl10 ubiquitination and human lymphoma cell survival. Mol. Cell. Biol. 33, 429–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Compagno M., Lim W. K., Grunn A., Nandula S. V., Brahmachary M., Shen Q., Bertoni F., Ponzoni M., Scandurra M., Califano A., Bhagat G., Chadburn A., Dalla-Favera R., and Pasqualucci L. (2009) Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature 459, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lohr J. G., Stojanov P., Lawrence M. S., Auclair D., Chapuy B., Sougnez C., Cruz-Gordillo P., Knoechel B., Asmann Y. W., Slager S. L., Novak A. J., Dogan A., Ansell S. M., Link B. K., Zou L., et al. (2012) Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 3879–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Montesinos-Rongen M., Schmitz R., Brunn A., Gesk S., Richter J., Hong K., Wiestler O. D., Siebert R., Küppers R., and Deckert M. (2010) Mutations of CARD11 but not TNFAIP3 may activate the NF-κB pathway in primary CNS lymphoma. Acta Neuropathol. 120, 529–535 [DOI] [PubMed] [Google Scholar]
- 39. Bu R., Bavi P., Abubaker J., Jehan Z., Al-Haqawi W., Ajarim D., Al-Dayel F., Uddin S., and Al-Kuraya K. S. (2012) Role of NF-κB regulators-TNFAIP3 and CARD11 in Middle Eastern diffuse large B cell lymphoma. Leuk. Lymphoma 53, 1971–1977 [DOI] [PubMed] [Google Scholar]
- 40. Dong G., Chanudet E., Zeng N., Appert A., Chen Y. W., Au W. Y., Hamoudi R. A., Watkins A. J., Ye H., Liu H., Gao Z., Chuang S. S., Srivastava G., and Du M. Q. (2011) A20, ABIN-1/2, and CARD11 mutations and their prognostic value in gastrointestinal diffuse large B-cell lymphoma. Clin. Cancer Res. 17, 1440–1451 [DOI] [PubMed] [Google Scholar]
- 41. Lamason R. L., Kupfer A., and Pomerantz J. L. (2010) The dynamic distribution of CARD11 at the immunological synapse is regulated by the inhibitor kinesin GAKIN. Mol. Cell 40, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 43. Cuff J. A., and Barton G. J. (1999) Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins 34, 508–519 [DOI] [PubMed] [Google Scholar]
- 44. Ouali M., and King R. D. (2000) Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 9, 1162–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rost B. (2001) Review: protein secondary structure prediction continues to rise. J. Struct. Biol. 134, 204–218 [DOI] [PubMed] [Google Scholar]
- 46. Dosztányi Z., Csizmók V., Tompa P., and Simon I. (2005) The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 347, 827–839 [DOI] [PubMed] [Google Scholar]
- 47. Blonska M., Joo D., Zweidler-McKay P. A., Zhao Q., and Lin X. (2012) CARMA1 controls Th2 cell-specific cytokine expression through regulating JunB and GATA3 transcription factors. J. Immunol. 188, 3160–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jattani R. P., Tritapoe J. M., and Pomerantz J. L. (2016) Intramolecular interactions and regulation of cofactor binding by the four repressive elements in the Caspase recruitment domain-containing protein 11 (CARD11) inhibitory domain. J. Biol. Chem. 291, 8338–8348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Snow A. L., Xiao W., Stinson J. R., Lu W., Chaigne-Delalande B., Zheng L., Pittaluga S., Matthews H. F., Schmitz R., Jhavar S., Kuchen S., Kardava L., Wang W., Lamborn I. T., Jing H., et al. (2012) Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J. Exp. Med. 209, 2247–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Turvey S. E., Durandy A., Fischer A., Fung S. Y., Geha R. S., Gewies A., Giese T., Greil J., Keller B., McKinnon M. L., Neven B., Rozmus J., Ruland J., Snow A. L., Stepensky P., and Warnatz K. (2014) The CARD11-BCL10-MALT1 (CBM) signalosome complex: Stepping into the limelight of human primary immunodeficiency. J. Allergy Clin. Immunol. 134, 276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brohl A. S., Stinson J. R., Su H. C., Badgett T., Jennings C. D., Sukumar G., Sindiri S., Wang W., Kardava L., Moir S., Dalgard C. L., Moscow J. A., Khan J., and Snow A. L. (2015) Germline CARD11 mutation in a patient with severe congenital B cell lymphocytosis. J. Clin. Immunol. 35, 32–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Buchbinder D., Stinson J. R., Nugent D. J., Heurtier L., Suarez F., Sukumar G., Dalgard C. L., Masson C., Parisot M., Zhang Y., Matthews H. F., Su H. C., Durandy A., Fischer A., Kracker S., and Snow A. L. (2015) Mild B-cell lymphocytosis in patients with a CARD11 C49Y mutation. J. Allergy Clin. Immunol. 136, 819–821 [DOI] [PMC free article] [PubMed] [Google Scholar]