Abstract
T cell activation following antigen binding to the T cell receptor (TCR) involves the mobilization of intracellular Ca2+ to activate the key transcription factors nuclear factor of activated T lymphocytes (NFAT) and NF-κB. The mechanism of NFAT activation by Ca2+ has been determined. However, the role of Ca2+ in controlling NF-κB signaling is poorly understood, and the source of Ca2+ required for NF-κB activation is unknown. We demonstrate that TCR- but not TNF-induced NF-κB signaling upstream of IκB kinase activation absolutely requires the influx of extracellular Ca2+ via STIM1-dependent Ca2+ release-activated Ca2+/Orai channels. We further show that Ca2+ influx controls phosphorylation of the NF-κB protein p65 on Ser-536 and that this posttranslational modification controls its nuclear localization and transcriptional activation. Notably, our data reveal that this role for Ca2+ is entirely separate from its upstream control of IκBα degradation, thereby identifying a novel Ca2+-dependent distal step in TCR-induced NF-κB activation. Finally, we demonstrate that this control of distal signaling occurs via Ca2+-dependent PKCα-mediated phosphorylation of p65. Thus, we establish the source of Ca2+ required for TCR-induced NF-κB activation and define a new distal Ca2+-dependent checkpoint in TCR-induced NF-κB signaling that has broad implications for the control of immune cell development and T cell functional specificity.
Keywords: calcium, calcium release-activated calcium channel protein 1 (ORAI1), NF-κB transcription factor, posttranslational modification (PTM), PKC, stromal interaction molecule 1 (STIM1), T-cell receptor (TCR)
Introduction
Activation of T cells following antigen binding to the T cell antigen receptor (TCR)3 induces diverse lineage- and fate-specific proinflammatory and immune-modulatory responses. Central to these responses is the induction of quantitatively distinct intracellular Ca2+ signals and their selective activation of the key transcription factors NFAT and NF-κB (1–6). The mechanism by which Ca2+ controls NFAT activation in lymphocytes is well established (7). In contrast, although Ca2+ has been implicated in TCR-induced NF-κB signaling (8–10), how Ca2+ regulates NF-κB activity is largely unexplored and represents a significant gap in our understanding of transcriptional control of T cell development, activation, and functional specificity.
In resting T cells, classical NF-κB consists of heterodimers of p50/p65 or p50/c-Rel that are retained in the cytosol by members of the inhibitory family of IκB proteins (11, 12). Following TCR engagement, IκB kinase (IKK)-mediated phosphorylation triggers the ubiquitination and proteasomal degradation of IκBα, releasing p50/p65 and p50/c-Rel, which localize to the nucleus to initiate transcription of crucial immune-regulatory, proinflammatory, and proproliferative genes (13–30). Although TCR-mediated Ca2+ mobilization has been implicated in proximal steps of NF-κB activation (8–10), the precise mechanisms and source of Ca2+ that regulate nuclear localization and transcriptional activation of NF-κB are poorly defined. It is well established that TCR signaling induces inositol 1,4,5-trisphosphate-mediated depletion of Ca2+ from the endoplasmic reticulum (ER). A resulting Ca2+ dissociation from the ER membrane protein stromal interaction molecule 1 (STIM1) triggers its oligomerization and relocalization to ER membrane domains juxtaposed to the plasma membrane (31–33), where STIM1 physically gates Orai (also known as Ca2+ release-activated Ca2+) channels, allowing extracellular Ca2+ to enter the cell (34, 35). However, it is not known whether Ca2+ control of TCR-induced NF-κB signaling requires STIM1- and Orai1-mediated Ca2+ influx or whether the initial release of Ca2+ from the ER is sufficient for classical NF-κB activation.
In this study, we sought to determine both the source and mechanism of Ca2+ control of antigen receptor-induced NF-κB activation in T cells. We show that influx of extracellular Ca2+ via STIM1 and Orai is critical for TCR- but not TNF-induced IκBα degradation and NF-κB activation. Importantly, we also demonstrate that Ca2+-dependent, PKCα-mediated phosphorylation of p65 critically regulates its nuclear localization and transcriptional activation following TCR engagement. Thus, our findings define important new proximal and distal Ca2+-dependent checkpoints in TCR-induced NF-κB signaling that have broad implications for the control of immune cell development and functional specificity.
Materials and Methods
Cells and Cell Culture
Primary human T cells were obtained from the University of Pennsylvania Immunology Core facility. Jurkat T cells were from the ATCC, and Jurkat T cells stably expressing E106A Orai1 were a gift from Dr. Jonathan Soboloff (Temple University, Philadelphia, PA). All cells were cultured in RPMI 1640 medium (BioWhittaker, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, Thermo Scientific, Logan, UT), 2 mm l-glutamine, penicillin (50 units/ml), and streptomycin (50 units/ml).
Antibodies and Reagents
Antibodies recognizing p65-Ser(P)-536 (3033S) and IκBα (4814S) were purchased from Cell Signaling Technology (Danvers, MA). Anti-p65 (372R/G) was purchased from Santa Cruz Biotechnology (Dallas, TX). Anti-α-tubulin (T1568) was from Sigma-Aldrich (St. Louis, MO). Anti-STIM1 (610954) was from BD Transduction Laboratories (Franklin Lakes, NJ). Anti-human TCR (c305 clone) was a gift from Dr. Gary Koretzky (Cornell University, New York, NY), and anti-CD28 was from Invitrogen. Protein A horseradish peroxidase-conjugated antibody (18-160) was from Millipore (Danvers, MA). Alexa Fluor 488 goat anti-rabbit (A11008) and Alexa Fluor 546 goat anti-rabbit (A11010) used for immunofluorescence were obtained from Invitrogen. Recombinant human TNF-α was purchased from R&D Systems (Minneapolis, MN), PMA, and ionomycin were from Sigma-Aldrich, and the luciferase reporter assay system was obtained from Promega (Fitchburg, WI).
Plasmids and Transfections
A cDNA construct expressing full-length p65 N-terminally tagged with EGFP was obtained from Addgene (Cambridge, MA). Mutant p65 constructs were generated using a site-directed mutagenesis kit (Stratagene, La Jolla, CA) to convert serine 536 to alanine or aspartic acid. Short hairpin STIM1 suppression and rescue constructs were generated in the laboratory of Dr. Dan Billadeau (Mayo Clinic, Rochester, MN), as were EGFP-shPKCα and EGFP-pCMS2 (control vector). For transfection, Jurkat T cells were suspended at 20 million cells/ml in RPMI 1640 medium, and 10 million cells were electroporated with 10 μg of DNA (for overexpression or mutant expression) or 40 μg of DNA (for suppression assays) at 315 V for 10 ms using a BTX ECM 830 electroporator (Harvard Apparatus, Holliston, MA). STIM1 and PKCα suppression assays were performed 48 h post-transfection, and EGFP-p65 and p65 mutant expression assays were performed 16–24 h post-transfection.
Immunoblotting
Cells were harvested and lysed using Nonidet P-40 lysis buffer consisting of 50 mm Tris-HCl (pH 7.5), 20 mm EDTA, 1% Nonidet P-40, and complete inhibitors (1 mm sodium orthovanadate, 1 mm PMSF, 10 μg/ml leupeptin, and 5 μg/ml aprotinin). Protein concentrations in cell lysates were determined using Bio-Rad reagent and quantified in a Cary 50 Bio UV-visible spectrophotometer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis (4–15%, Bio-Rad) and then transferred onto PVDF membranes (Millipore, Billerica, MA). Membranes were probed with the respective primary anti-human antibodies and then incubated with protein A HRP secondary antibodies. Blots were developed with enhanced chemiluminescence using Pierce ECL Western blotting substrate. All immunoblots presented are from a single experiment representative of at least three independent experiments.
Luciferase Reporter Analysis
Luciferase-based transcriptional analysis was performed on Jurkat T cells transfected with 2 μg of total DNA (PBXII κB firefly luciferase and pRL TK Renilla luciferase in a 20:1 ratio) per transfection (5 × 106 cells in 500 μl of medium) using a square-wave BTX electroporator at 315 V for 10 ms. Twenty-four hours after transfection, cells were treated with PMA (200 nm), PMA (200 nm) and ionomycin (1 μg/ml), anti TCR (0.5 μg/ml), and anti-CD28 (1:50) or TNF (10 ng/ml) for 4 h. Cells were then lysed in passive lysis buffer (Promega), and luciferase activity was measured using a Luminoscan 96-well automated luminometer (Thermo LabSystems, Franklin, MA). Firefly/Renilla luciferase ratios were calculated using Ascent software (Thermo LabSystems), and the mean ratio from at least three independent experiments (3–4 replicates/experiment) for each condition was compared.
Quantitative Real-time PCR
To quantify IκBα expression, cDNA was synthesized from RNA isolated (RNeasy Plus mini kit, Qiagen) from PMA- or PMA and ionomycin-stimulated cells with a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). cDNA was amplified with IκBα (forward, 5′-CCGAGAC TTTCGAGGA AATACC-3′; reverse, 5′-ACGTGTGGCCATTGTAGTT-3′) and β-actin-specific primers (forward, 5′-TCAGCAAGCAGGAGTATGACGAG-3′; reverse, 5′-ATTGTGAACTTTGGGGGATGC-3′) on a 7500 Fast real-time PCR system (Applied Biosciences, Warrington, UK) using Power SYBR Green PCR Master Mix (Applied Biosciences). Ct values were obtained in triplicate for each target and analyzed with instrument software v1.3.1 (Applied Biosystems).
Microarray Analysis
RNA was isolated using an RNeasy Plus kit (Qiagen). Biotin-labeled cRNA was generated using the Illumina TotalPrep RNA amplification kit, and a Bioanalyzer (Agilent Technologies, Wilmington, DE) was used to assess total RNA and cRNA quality. Illumina HumanHT-12 version 4 expression bead chips were hybridized with cRNA from two biological replicates per condition and scanned on an Illumina BeadStation 500GX. Scanned images were converted to raw expression values using GenomeStudio v1.8 software (Illumina). Data analysis was performed using the statistical computing environment R (v3.2.3), the Bioconductor suite of packages for R, and RStudio (v0.98). Raw data were background-subtracted, variance-stabilized, and normalized by robust spline normalization using the Lumi package (36). Differentially expressed genes were identified by linear modeling and Bayesian statistics using the Limma package (37, 38). Probe sets that were differentially regulated (≥1.5-fold change between all treatments, false detection rate ≦ 5% after controlling for multiple testing using the Benjamini-Hochberg method (39, 40)) were used for heatmap generation in R. Clusters of co-regulated genes were identified by Pearson correlation using the hclust function of the stats package in R. Differentially expressed NF-κB-dependent genes were identified using a list of validated and putative NF-κB target genes curated by the laboratory of Dr. Thomas Gilmore at Boston University. All microarray data have been deposited in the GEO database for public access (GSE76804).
Chromatin Immunoprecipitation
Jurkat T cells (10 × 106) transfected with either EGFP-shPKCα or control EGFP-pCMS2 vector (48 h) were stimulated with PMA (200 nm) and ionomycin (1 μm) for 30 min at 37 °C. Chromatin was prepared using a Covaris truChIP chromatin shearing kit (Covaris Inc., Woburn, MA). Briefly, cells were fixed in 1% methanol-free formaldehyde for 5 min at room temperature, and then fixation was quenched with 0.125 m glycine at room temperature for 5 min. Cells were washed twice with cold PBS and then lysed at 4 °C with rocking for 10 min. Nuclei were then washed and transferred to an AFA milliTUBE for ultrasonication. Samples were sheared using a Covaris S220 focused ultrasonicator for 1500 s in a 6 °C bath at a duty cycle of 5%, an intensity of 4, a peak incident power of 140 Watt, and at 200 cycles/burst. p65 was precipitated from sheared chromatin (200–1000 bp) with anti-p65 (5 μg, Santa Cruz Biotechnology, rabbit 372X) or normal rabbit IgG (5 μg, Cell Signaling Technology, 2729) for 12–16 h at 4 °C. Immunoprecipitated chromatin was then incubated with protein G Dynabeads (Life Technologies) for 2 h at 4 °C, and chromatin was eluted (50 mm Tris (pH 8.0) and 10 mm EDTA) at 65 °C on a thermomixer (1200 rpm) for 30 min. Cross-linking was reversed by incubating recovered chromatin at 65 °C for 12 h, followed by incubation with RNase A for 2 h at 37 °C and proteinase K for 30 min at 55 °C. DNA was then purified using a ChIP DNA clean and concentrator kit (Zymo Research, Irvine, CA), and quantitative PCR was performed to quantify p65 binding to IκBα, CXCL8, and TNF promotors using IκBα-specific (forward, 5′-TTGGGATCTCAGCAGCCGAC-3′; reverse, 5′-GCCACTAGGGTCACGGACAG-3′), CXCL8-specific (forward, 5′-CAGGTTTGCCCTGAG GGG ATG-3′; reverse, 5′-GGAGTGCTCCG GTGG CTTTT-3′), and TNF-specific (forward, 5′-CCCGCGATGGAG AAGAAACC-3′; reverse, 5′-GTCCTTGCTGAGGGAGCGTC-3′) primers.
Quantitation of p65 Nuclear Translocation
Jurkat T cells transfected with EGFP-shPKCα or EGFP-pCMS2 (48 h) or untransfected cells suspended in medium containing 2 mm Ca2+ or Ca2+ free equivalent solution were adhered to Cell-Tak-treated coverslips for 10–15 min. Cells were then stimulated at 37 °C as indicated. For PKCα suppression experiments, cells were stimulated in the presence of 2 mm Ca2+. At the indicated times, cells were fixed in formaldehyde (3.7%) for 30 min, permeabilized with 0.2% Triton-X-100 for 15 min, and blocked overnight in 2% BSA at 4 °C. Fixed and blocked cells were incubated with rabbit anti-p65 primary antibody (Santa Cruz Biotechnology, catalog no. 372, 1 μg/ml) for 1 h at 37 °C or overnight at 4 °C degrees, washed three times for 5 min each in 1% BSA in PBS, and incubated with Alexa 488 or 546 goat anti-rabbit secondary antibody (4 μg/ml) for 1 h at 37 °C. Nuclei were then labeled with Hoechst 33342 (Life Technologies, catalog no. H3570, 4 μg/ml), washed three times for 5 min each in 1% BSA in PBS, and mounted in Fluoromount (Fisher). Images of p65 localization were obtained with a Yokagawa spinning disk confocal system (Tokyo, Japan) mounted on a Leica DMI4000 microscope (Leica Microsystems, Wetzlar, Germany), and imaging parameters were optimized independently for each channel to maintain fluorescence within the linear range while maximizing intensity resolution. Images of p65 and Hoechst were overlaid, and cytoplasmic/nuclear p65 localization was determined using the Multiwavelength Cell Scoring application (Molecular Devices, Downingtown, PA). Average nuclear and cytoplasmic p65 fluorescence intensities were quantified within cytoplasmic and nuclear compartments, and intensity ratios were determined for each cell.
Real-time Localization of WT and Mutant p65
Jurkat T cells expressing WT and p65 Ser-536 mutants (16–24 h) were adhered to Cell-Tak-coated (BD Biosciences) coverslips and maintained in culture medium (RPMI 1640 medium, 10% FBS, 1% Glutamax) in a temperature- and CO2-controlled chamber for 1 h during imaging. GFP-WT and GFP-p65 mutants were visualized every 10 s after stimulation with PMA (200 nm) with ionomycin (1 μm) and/or PMA (200 nm) with and without the delayed addition of ionomycin (1 μm).
Calcium Imaging
Jurkat T cells (3 million cells/ml) were loaded with 3 μm Fura-2 acetoxymethyl ester (Molecular Probes, Eugene, OR) in external solution containing 145 mm NaCl, 4.5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm glucose, 10 mm HEPES, 2 mm glutamine, and 2% fetal bovine serum (Hyclone, Thermo Scientific) for 10 min at 25 °C. Cells were adhered to coverslips coated with Cell-Tak (BD Biosciences), mounted on the stage of a Leica DMI6000 microscope configured with a Photometrics Evolve 512 camera (Tucson, AZ) using an Olympus ×40 oil objective (Shinjuku, Tokyo, Japan), and images were acquired with MetaFluor software (Molecular Devices). During imaging, cells were perfused with Ca2+-free bath solution before activation with PMA (200 nm) and ionomycin (1 μm), thapsigargin (1 μm), anti-TCR (0.5 μg/ml) and CD28 (1:50) antibodies, or TNF (10 ng/ml) to evaluate stimulus-dependent Ca2+ release from the ER. The cells were then perfused with bath solution containing 2 mm Ca2+ to assess Ca2+ entry via activated Orai channels. In some experiments, cells were pretreated for 15 min with the Orai-1 inhibitor Synta66 (50 μm, Aobious, Gloucester, MA) prior to stimulation. Ca2+ mobilization was analyzed by plotting the emission ratio of 340/380-nm excitation for each cell. Each plot is the averaged ratio from at least 30 cells.
Statistical Analysis
Significance for all statistical tests was determined at p < 0.05 and is shown as *, p < 0.05; **, p < 0.01; and ***, p < 0.001 in all figures. Average firefly/Renilla luciferase ratios were calculated from three to four independent experiments and analyzed using two-tailed Welch's t test. Western blot protein intensities were quantified using ImageJ (http://imagej.nih.gov/ij/), and average protein intensity values were compared using two-tailed Welch's t test. p65 nuclear-to-cytoplasmic fluorescence intensity ratios were assessed for normality using probability plots and Kolmogorov-Smirnov test for normality. Normal distributions were compared using two-tailed Student's t test, and non-normal data were compared using Wilcoxon rank-sum test. Quantitative PCR relative quantification (RQ) values and percent input values were compared using two-tailed Welch's t test.
Results
Extracellular Ca2+ Is Required for TCR-induced NF-κB Signaling
Ca2+ regulates proximal TCR signaling upstream of IKK activation (8–10). However, the precise function of Ca2+ and the source of Ca2+ required for NF-κB activation are unknown. To address these questions, we first asked whether the initial release of Ca2+ from ER stores was sufficient or whether sustained influx of extracellular Ca2+ is required for NF-κB activation in T cells. To distinguish between these pools of Ca2+, we activated T cells in the presence or absence of extracellular Ca2+ with either anti-CD3 and anti-CD28 (3/28) to co-engage the TCR and CD28 or with the diacylglycerol analog PMA together with ionomycin (P/I), which activate PKCθ and release ER-stored Ca2+, respectively, and we compared these responses to those induced by the proinflammatory cytokine TNF. In Ca2+-free medium, 3/28 and P/I, but not TNF, induced a transient rise in cytoplasmic Ca2+ concentration because of release from the ER. Reintroduction of extracellular Ca2+ led to a sustained secondary increase in cytoplasmic Ca2+ levels via entry through activated Orai1/Ca2+ release-activated Ca2+ channels in 3/28- and P/I-stimulated cells (35) (Fig. 1A). Thus, stimulating cells in the absence of extracellular Ca2+ allows us to specifically determine whether release from ER stores alone is sufficient for NF-κB signal activation.
FIGURE 1.
Extracellular Ca2+ regulates TCR- but not TNF-induced classical NF-κB activation in human T lymphocytes. A, Jurkat T cells loaded with Fura-2/AM were initially bathed in Ca2+-free medium and stimulated with PMA (200 nm) and ionomycin (1 μm) (left panel), anti-CD3/CD28 (center panel), or TNF (10 ng/ml, right panel) to assess release from intracellular (ER) stores. Subsequent perfusion with Ca2+-containing (2 mm) medium was performed to assess the extent of Orai activation. B, Jurkat T cells bathed in Ca2+-containing (2 mm) (top panels) or Ca2+ free (0 mm Ca2+, bottom panels) medium were activated by cross-linking CD3 and CD28 with P/I or TNF (10 ng/ml) for the times indicated, and IκBα levels were determined by immunoblotting. C, NF-κB transcriptional activity was measured under identical conditions as the immunoblot analyses in B in Jurkat T cells expressing an NF-κB firefly luciferase reporter. Firefly luciferase activity is expressed relative to a Renilla luciferase control before (0 h) and after (4 h) stimulation with anti-CD3/28 (left), P/I (center), or TNF (right) in the presence (+) or absence (−) of extracellular Ca2+. Mean firefly/Renilla luciferase ratios ± S.E. from three independent experiments (4 replicates/experiment) are displayed, and statistical significance was evaluated using Welch's t test. ***, p < 0.001; N.S., not significant. D, primary human CD4+ T cells were stimulated in Ca2+-replete (0.4 mm) or Ca2+-free (0 mm) medium, and then IκBα levels were determined by immunoblotting. Blots were probed with anti-α-tubulin as a loading control.
As shown in Fig. 1B, top panels, all three stimuli induced the expected degradation and resynthesis of IκBα in Jurkat T cells, consistent with activation of the IKK complex and the classical NF-κB pathway. In contrast, neither 3/28 nor P/I induced IκBα degradation in Ca2+-free medium, whereas IκBα degradation and resynthesis in response to TNF remained intact (Fig. 1B, bottom panels). Consistent with the effects on IκBα degradation, 3/28-stimulated NF-κB transcriptional activity was completely inhibited and P/I-induced transcriptional activation was significantly reduced in Ca2+-free medium (Fig. 1C). In contrast, TNF-induced NF-κB reporter activity was unaltered in the presence or absence of extracellular Ca2+ (Fig. 1C). A similar regulation of IκBα expression was observed in primary human CD4+ T cells (Fig. 1D). Collectively, these findings reveal that transient release of Ca2+ from ER stores is not sufficient and that extracellular Ca2+ is required for TCR-induced NF-κB activation.
TCR-induced NF-κB Activation Requires STIM1 and Orai1
The extracellular Ca2+ requirement for TCR-induced NF-κB activation implies a crucial role for STIM1-operated Orai1 channel-mediated Ca2+ influx. To explore this, we expressed a STIM1 shRNA construct or a bicistronic variant for concomitant re-expression of shRNA resistant STIM1 to normal levels in Jurkat T cells (Fig. 2A). STIM1 suppression inhibited 3/28- and P/I-induced extracellular Ca2+ influx, and this was rescued by re-expression of STIM1 (Fig. 2B). Consistent with the lack of TCR-induced NF-κB signaling in Ca2+-free medium (Fig. 1, B and C), STIM1 suppression prevented 3/28- and P/I-induced IκBα degradation in T cells (Fig. 2C, left versus center panel). In contrast, IκBα degradation and re-expression were normal in STIM1-rescued cells, confirming that the inhibition was due to loss of STIM1 (Fig. 2D, right panel). Furthermore, both 3/28- and P/I-induced NF-κB transcriptional activity was reduced in STIM1-suppressed cells, and this was again rescued by concomitant STIM1 re-expression (Fig. 2C). Notably, TNF-induced IκBα degradation and NF-κB transcriptional activity were unaffected by suppressing STIM1 (Fig. 2, C and D).
FIGURE 2.
STIM1-dependent Ca2+ entry is required for TCR- but not TNF-induced NF-κB activation. A, immunoblot analysis of STIM1 and α-tubulin levels in Jurkat T cells transfected with either vector alone, shSTIM1, or shSTIM1-STIM1 suppression and rescue vectors. **, p < 0.01; n = 4 experiments. B, the role of STIM1 in P/I- and 3/28-mediated Ca2+ entry was similarly assessed in control (vector-transfected) Jurkat T cells, STIM1-suppressed cells, and STIM1 suppression with STIM1 rescue as described in A. Each trace represents the average response of at least 30 cells and is representative of at least three separate experiments. C, Jurkat T cells were transfected with control vector (left panel), shSTIM1 (center panel), or shSTIM1-STIM1 (right panel) and then incubated for the times indicated with 3/28, P/I, or TNF. Lysates were immunoblotted with either anti-IκBα or α-tubulin loading control. D, Jurkat T cells were transfected with the NF-κB luciferase reporter construct together with either vector alone, shSTIM1, or shSTIM1-STIM1 and then activated with 3/28, P/I, or TNF for 4 h. Mean firefly:Renilla luciferase ratios ± S.E. pooled from at least three independent experiments (3 replicates/experiment) are shown and were compared using Welch's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; N.S., not significant.
To confirm the role of Orai in Ca2+-dependent NF-κB activation, we examined the consequence of inhibiting Orai-mediated Ca2+ influx with the Orai inhibitor Synta66 (Fig. 3A) and by expressing a mutant Orai1 (glutamic acid at position 106 mutated to alanine, E106A) that exerts a dominant negative effect on the Ca2+ permeability of endogenous Orai channels (Fig. 3C). In the presence of Synta66, the sarcoplasmic/endoplasmic reticulum Ca2+ ATPase inhibitor thapsigargin triggered Ca2+ release from the ER, evident as a small transient increase in cytoplasmic Ca2+ (in Ca2+-free medium), but no subsequent sustained increase in cytoplasmic Ca2+ (Fig. 3A, compare the top and bottom panels) following perfusion with Ca2+-containing medium. Consistent with the effects of STIM1 suppression (Fig. 2), Synta66 (Fig. 3B) inhibited 3/28- but not TNF-induced IκBα degradation. A similar block in stimulus induced Ca2+ entry, and IκBα degradation was observed in permeation-defective Orai1-E106A cells (Fig. 3, C and D) Taken together, these results reveal an obligate role for STIM1-operated Orai1-mediated Ca2+ entry in TCR- but not TNF-induced IκBα degradation and NF-κB activation.
FIGURE 3.
Orai1-mediated Ca2+ entry is required for TCR- but not TNF-induced NF-κB activation. A, Jurkat T cells were incubated with (bottom panel) and without (top panel) Synta66 (50 μm) and then activated with thapsigargin (1 μm) under Ca2+-free (0 mm) and Ca2+-replete (2 mm) conditions. B, Jurkat T cells were either untreated (−) or incubated for 15 min (+) with Synta66 and then stimulated with anti-CD3/28 or TNF for the times indicated. Time-dependent changes in IκBα were determined by immunoblotting, and anti-α-tubulin was used as a loading control. C, Ca2+ traces in Jurkat T cells stably overexpressing the dominant negative E106A Orai1 that were stimulated with P/I, 3/28, or TNF as shown. D, Jurkat T cells stably overexpressing either wild-type Orai (Orai-cyan fluorescent protein, Orai-CFP) or the dominant negative Orai1-E106A were stimulated with 3/28, P/I, or TNF for the times indicated, and immunoblot analysis was performed to quantify IκBα and α-tubulin.
Ca2+ Controls the Transcriptional Activity of TCR-induced NF-κB
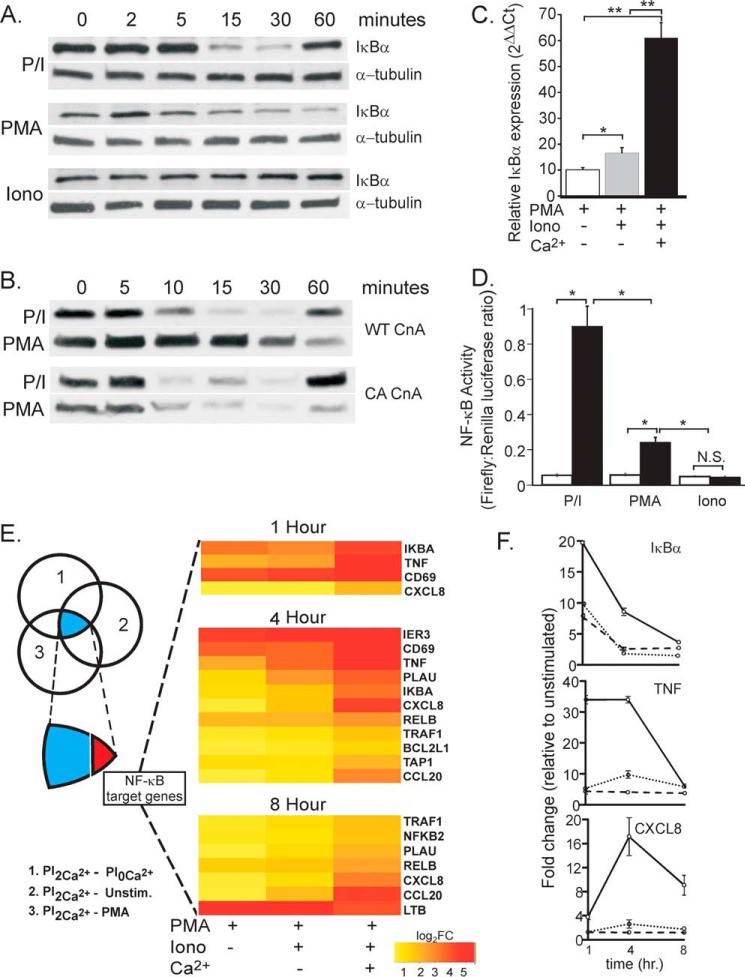
P/I treatment mimics TCR signaling upstream of IKK activation because PMA activates the strictly diacylglycerol-dependent and Ca2+-independent “novel” PKC isoform PKCθ (41), and ionomycin-mediated Ca2+ release from the ER activates STIM1-dependent Orai activation (Figs. 1A and 2B). Thus, both 3/28 and P/I stimulation of T cells induce rapid PKCθ-mediated IKK-dependent degradation of IκBα, followed by resynthesis of IκBα via NF-κB-driven transcription (Figs. 1, 2, 3 and 4A). In seeking to determine the precise contribution of Ca2+ release from the ER to NF-κB activation, we found that PMA alone induces substantial IκBα degradation (Fig. 4A, center panel), suggesting that strong pharmacological activation of PKCθ can circumvent the requirement for Ca2+ upstream of IKK activation. However, treatment with ionomycin alone had no effect on IκBα levels (Fig. 4A, bottom panel), indicating that Ca2+ mobilization in the absence of PKCθ activation is not sufficient to activate the IKK complex.
FIGURE 4.
PKC activation is sufficient for IκBα degradation, but Ca2+ is required for NF-κB transcriptional activation. A, Jurkat T cells were stimulated as indicated with P/I, PMA alone, or ionomycin (Iono) alone, and IκBα and α-tubulin levels were measured by immunoblotting. B, Jurkat T cells stably expressing either WT CnA or constitutively active (CA) CnA were stimulated as indicated with P/I or PMA alone, and IκBα and α-tubulin levels were detected by immunoblotting. C, Jurkat T cells were stimulated with PMA or P/I in the absence and presence of extracellular Ca2+ for 60 min, and IκBα mRNA was quantified by quantitative RT-PCR. IκBα Ct values were subtracted from β-actin and compared between stimulated and unstimulated samples using Welch's t test. *, p < 0.05; **, p < 0.01. D, NF-κB transcriptional activity in Jurkat T cells expressing the NF-κB luciferase reporter stimulated 4 h with P/I, PMA alone, or ionomycin alone (mean firefly:Renilla luciferase ratios ± S.E. of three independent experiments (4 replicates/experiment) were compared using Welch's t test. *, p < 0.05; N.S., not significant. E, microarray analysis of gene expression at 1, 4, and 8 h from PMA, P/I in 2 mm Ca2+ (PI2Ca2+)-treated, P/I in 0 mm Ca2+ (PI0Ca2+)-treated, and unstimulated (Unstim, 1 h) cells. Genes differentially expressed (log2 -fold change (FC), >0.59; false detection rate, <0.05) between PI2Ca2+ and PI2Ca2+ (1), PI2Ca2+ and unstimulated cells (2), and PI2Ca2+ and PMA (3) were identified (blue), and, for each time point, the log2 -fold change in expression of validated NF-κB target genes (red) is listed relative to levels in unstimulated cells. F, time course of IκBα (top panel), TNF (center panel), and CXCL8 (bottom panel) mRNA expression (-fold change) from the same microarray following stimulation with PMA alone (dotted lines), P/I in 0 mm Ca2+ (dashed lines), and P/I in 2 mm Ca2+ (solid lines).
Strikingly, although PMA in the absence of ionomycin induced IκBα degradation, this was not followed by IκBα resynthesis (Fig. 4A, compare IκBα levels at 60 min). Moreover, the kinetics of PMA-induced IκBα degradation were delayed compared with the response to P/I. We reasoned that the delayed IκBα degradation following stimulation with PMA alone likely reflects a cooperative role previously identified for the Ca2+ regulated phosphatase calcineurin A (CnA) in CBM complex formation, IKK activation, and IκBα degradation (10, 42). Indeed, overexpression of a Ca2+-independent, constitutively active CnA rescued the delay in PMA-induced IκBα degradation so that the rate and extent of degradation were indistinguishable from those in P/I-stimulated cells (Fig. 4B). Importantly, although constitutively active CnA rescued the modulatory role of Ca2+ in proximal steps of NF-κB activation (i.e. IKK activation), it did not rescue IκBα re-expression in PMA-stimulated cells, indicating that a separate Ca2+-dependent mechanism regulates the distal steps of NF-κB activation.
Supporting this conclusion and consistent with the lack of resynthesis of IκBα in the absence of Ca2+ (Fig. 4A), analysis of IκBα mRNA levels revealed a limited induction of IκBα transcription when PKC was activated with PMA or in response to PMA and ionomycin under Ca2+ free conditions. In contrast, in the presence of extracellular Ca2+, IκBα mRNA expression was significantly induced by PMA and ionomycin (Fig. 4C).
Because IκBα re-expression is driven by activated NF-κB as a negative feedback loop to limit NF-κB signaling (43), we questioned whether Ca2+-dependent IκBα protein re-expression reflects a global requirement for Ca2+ in NF-κB transcriptional activation. As shown in Fig. 4D, ionomycin alone failed to activate NF-κB reporter activity. PMA alone triggered a small but significant increase in activity compared with baseline; however, PMA and ionomycin together significantly enhanced (∼4-fold) NF-κB-driven transcriptional activation. Together, these data indicate that Ca2+ controls the distal transcriptional activation of NF-κB following P/I stimulation of T cells.
To extend these findings, we performed an unbiased transcriptional analysis to fully assess the extent of Ca2+ control over NF-κB-regulated gene expression. Microarray analysis was performed on T cells stimulated in the absence or presence of extracellular Ca2+ with either PMA alone or PMA and ionomycin. Transcriptional analyses identified 20, 96, and 112 differentially expressed genes (false detection rate, <0.05, log2 -fold change, >0.59) at 1, 4, and 8 h, respectively, between unstimulated, PMA-treated, and P/I-treated T cells (Fig. 4E, blue). This list of differentially expressed genes was further refined to include only validated and putative NF-κB target genes on the basis of known targets (44–47). In total, we found that 10–20% of all Ca2+-regulated genes were NF-κB targets (Fig. 4E, red). Strikingly, 11 of 12 of these NF-κB target genes were dramatically increased by Ca2+ entry (2- to 20-fold increase, false detection rate, <0.05) relative to PMA stimulation (no Ca2+ mobilization) or stimulation with P/I in 0 mm Ca2+ (ER release but no Ca2+ entry). This analysis also revealed that, among this cohort, three classical NF-κB regulated genes (IκBα, CXCL8, and TNF) were among the top differentially expressed Ca2+-dependent genes (Fig. 4, E and F). Together, these data reveal an entirely novel function for Ca2+ in regulating TCR-induced, NF-κB-dependent gene activation.
Ca2+ Is Required for TCR-induced p65 Nuclear Localization
The failure of PMA alone to induce IκBα resynthesis following its degradation (Fig. 4A) implies that Ca2+ controls NF-κB activity distal to IKK activation. To address the mechanism of this regulation, we first asked whether Ca2+ entry was required for NF-κB nuclear localization following IκBα degradation. As expected, P/I triggered rapid nuclear translocation of p65, which peaked 30 min after stimulation (Fig. 5, A and B). PMA alone had an effect at 30 and 60 min of stimulation, but the extent was significantly less than that induced by P/I at all time points (Fig. 5B). In contrast, ionomycin alone did not induce p65 nuclear localization at any time point. To confirm this apparent role for extracellular Ca2+ in p65 nuclear localization, we examined the effects of PMA and ionomycin in the presence and absence of extracellular Ca2+. Again, ionomycin alone failed to trigger p65 nuclear localization in either the absence or presence of Ca2+ (Fig. 5, C and D). However, p65 was strongly driven to the nucleus by the combination of P/I in the presence of extracellular Ca2+, whereas, in Ca2+-free medium, ionomycin failed to synergize with PMA, and the extent of p65 nuclear localization was identical to that triggered by PMA alone (Fig. 5D). Taken together, these findings reveal an essential role for Orai-mediated entry of extracellular Ca2+ in nuclear translocation of p65 following its release from IκBα in response to TCR signaling.
FIGURE 5.
Ca2+ controls TCR-induced p65 nuclear localization. A, Jurkat cells were stimulated with PMA, ionomycin (Iono), or both for 15, 30, or 60 min, and then nuclear localization of p65 was determined by confocal imaging. B, the data are presented as a mean ± S.E. ratio of nuclear:cytoplasmic p65 from three independent experiments and were compared using Student's t test. *, p < 0.05; **, p < 0.01. C, Jurkat T cells were stimulated as in A for 30 min in the absence or presence of extracellular Ca2+, and p65 localization was determined by confocal imaging. D, the ratio of nuclear:cytoplasmic p65 was determined from three independent experiments, are presented as the mean ± S.E. ratio of nuclear:cytoplasmic p65, and were compared using Student's t test. *, p < 0.05; **, p < 0.01.
Ca2+ Regulates TCR-induced Phosphorylation of p65 at Ser-536
At least 12 p65 serine or threonine residues have been identified whose phosphorylation regulates its nuclear localization and/or transcriptional activation (13–15, 22, 23, 26–28, 30, 48–52). We therefore asked whether Ca2+ regulates p65 nuclear translocation by controlling the phosphorylation of any of these residues. Among these, we focused on signal-induced phosphorylation of p65 Ser-536 (25, 53) because this has been implicated in TNF-driven NF-κB activation (15). We found that neither PMA nor ionomycin alone induces Ser-536 phosphorylation (Fig. 6A). However, activation with both together (P/I) induced a robust transient increase in phosphorylation at this residue that peaked at 15 min (Fig. 6A). Furthermore, the synergistic effect of PMA and ionomycin on Ser-536 phosphorylation requires extracellular Ca2+ because no phospho-p65 was detected in cells stimulated in Ca2+-free medium (Fig. 6B). Notably, TNF also induced transient Ser-536 phosphorylation, although this occurred more rapidly than the response to P/I (peak at 5 min), suggesting distinct regulatory mechanisms. Moreover, TNF stimulated robust Ser-536 phosphorylation in the absence of extracellular Ca2+ (Fig. 6C), again consistent with an obligate role for Ca2+ entry in TCR but not TNF-induced NF-κB activation in T cells.
FIGURE 6.
Ca2+ controls the phosphorylation of p65 at Ser536. A, Jurkat T cells were stimulated with P/I, PMA alone, or ionomycin (Iono) alone for the times shown, and then lysates were immunoblotted to determine the amounts of total p65 and Ser-536 phosphorylation. B, cells were treated with P/I in the absence or presence of 2 mm Ca2+, and then p65 or Ser(P)-536 amounts were determined by immunoblotting. C, Jurkat cells were incubated with TNF for the times shown in either Ca2+-containing or Ca2+-free extracellular bath solution, and then lysates were prepared and immunoblotted using the antibodies indicated. D, WT p65-GFP, and p65-GFP with serine 536-to-alanine (S536A) and serine 536-to-aspartate (S536D) point mutations were expressed in Jurkat T cells to determine the role of Ser-536 phosphorylation in p65 nuclear localization. WT and mutant p65-GFP localization was visualized over a time course of 60 min in live cells by spinning disk confocal microscopy. In each instance, cells were first stimulated for 30 min with PMA alone to trigger IκBα degradation and then were treated in the continued presence of PMA with ionomycin after 30 min to assess the role of Ca2+ in p65 nuclear localization. E, Jurkat cells were treated for the times indicated with either P/I (left panel) or PMA alone, followed by addition of ionomycin after 30 min (right panel), and then the amounts of IκBα were determined by immunoblotting.
Phosphorylation of p65 at Ser-536 has been shown to alter the kinetics of p65 nuclear translocation (24, 25). To determine whether Ca2+-dependent Ser-536 phosphorylation regulates p65 nuclear localization, we expressed wild-type p65, a non-phosphorylatable serine-to-alanine (S536A) mutant, and a serine-to-aspartate (S536D) phosphomimic mutant in T cells. We then visualized nuclear translocation in real time by confocal imaging. Cells were first stimulated for 30 min with PMA alone to induce IκBα degradation. Then ionomycin was added, and cells were observed for an additional 30 min. Consistent with the role for Ca2+ in p65 nuclear localization, in cells expressing WT p65, PMA alone did not promote p65 nuclear migration, whereas robust nuclear translocation was observed within 10 min of Ca2+ mobilization by exposure to ionomycin (Fig. 6D, top panel, 40-min time point). In contrast, after PMA treatment, ionomycin did not induce nuclear localization of S536A, whereas S536D exhibited Ca2+-independent nuclear localization during the initial 30 min of PMA stimulation (Fig. 6D, center and bottom panels) without any requirement for Ca2+ mobilization with ionomycin. Together, these results establish that Ca2+ entry is required for TCR-induced phosphorylation of p65 at Ser-536, and our mutational analysis indicates that this phosphorylation licenses the nuclear localization of p65 following its release from IκBα.
PKCα Regulates Ca2+-dependent p65-Ser-536 Phosphorylation
Several kinases, including IKKα, IKKβ, IKKϵ, TBK1, and PKA, have been implicated in the phosphorylation of p65 at specific serine residues that regulate its transcriptional activation (26, 54), but none of these kinases are known to be Ca2+ dependent. Because PKCα is a Ca2+-dependent Ser/Thr kinase (55, 56) and is the predominant conventional PKC isoform in T cells, we used shRNA knockdown (Fig. 7) to determine whether PKCα plays a role in TCR-induced NF-κB signaling and, specifically, whether it mediates Ca2+-dependent phosphorylation of p65 at Ser-536. PKCα suppression did not affect IκBα degradation induced by P/I, indicating no apparent role for PKCα upstream of IKK activation (Fig. 7B). However, similar to incubating cells with PMA alone (Figs. 4A), PKCα suppression prevented the resynthesis of IκBα normally observed 60 min after stimulation with P/I (Fig. 7B, compare lanes 5 and 10). In contrast, PKCα suppression did not affect TNF-induced IκBα degradation or resynthesis. Consistent with a role in Ser-536 phosphorylation, PKCα suppression significantly reduced P/I-induced Ser-536 phosphorylation but had no significant effect on phosphorylation induced by TNF (Fig. 7, C and D). Thus, these data confirm that Ca2+-dependent activation of PKCα regulates TCR-induced phosphorylation of p65 at Ser-536.
FIGURE 7.
PKCα mediates Ca2+-dependent but not TNF-induced p65 nuclear localization and promotor binding. A, PKCα levels were suppressed >90% 48 h after transfection of Jurkat T cells with a PKCα suppression construct as measured by immunoblot analysis. The densitometry plot represents the mean ± S.E. from five independent experiments. B, Jurkat cells transfected with either vector alone (Control) or shPKCα were stimulated with either P/I or TNF for the times shown. Lysates were prepared, and IκBα or α-tubulin was measured by immunoblot analysis. A representative example of four separate experiments is shown. C, cells were treated as described in B, and then p65 and Ser(P)-536 levels were quantified by immunoblot analysis. D, densitometric analysis of Ser-536 phosphorylation relative to the total p65 amount. Each value represents the mean ± S.E. of normalized values from four independent experiments. **, p < 0.01; Welch's t test. E, Jurkat cells were stimulated for the indicated time with PMA and ionomycin, and p65 nuclear and cytoplasmic localization in control and PKCα-suppressed cells was analyzed by confocal microscopy. The distribution of nuclear:cytoplasmic ratios is plotted for the indicated time points (the example is representative of three separate experiments). F, box plot of the p65 nuclear to cytoplasmic ratio. Wilcoxon rank-sum test revealed significant inhibition of p65 nuclear localization at each time point after stimulation. G, chromatin immunoprecipitation analysis of p65 promotor binding in cells stimulated for 60 min with PMA and ionomycin. IκBα, TNF, and CXCL8 abundance relative to chromatin input was compared between WT and PKCα-suppressed cells. Promoter binding (mean ± S.D.) relative to chromatin input is shown for IgG (control) and p65 immunoprecipitates (representative of three independent experiments. **, p < 0.01, Welch's t test.
Given the role we have identified for p65 Ser-536 phosphorylation in p65 nuclear localization (Fig. 6D) and the role for PKCα in p65 Ser-536 phosphorylation, we investigated whether PKCα controls p65 nuclear localization. Consistent with the role of PKCα-dependent p65 phosphorylation, PKCα suppression significantly decreased the extent of p65 nuclear localization 15, 30, 60, and 90 min post-stimulation with P/I in 2 mm Ca2+ (Fig. 7, E and F). Importantly, p65 localization, expressed as the nuclear to cytoplasmic ratio, in unstimulated T cells was identical regardless of the level of PKCα expression (median nuclear to cytoplasmic ratio−PKC, 0.54; median nuclear to cytoplasmic ratio+PKCα, 0.53).
We next asked to what extent this phosphorylation of p65 impacts p65 binding to promotors of Ca2+-dependent genes identified in our transcriptional analysis. We performed ChIP analyses to assess p65 binding to three genes found by transcriptional analysis to exhibit the strongest Ca2+ dependent induction (IκBα, CXCL8, and TNF) and to assess the role of PKCα-dependent p65 phosphorylation in promotor binding (Fig. 7G). Quantification of p65 binding to IκBα, CXCL8, and TNF promoters in Jurkat T cells demonstrated that PKCα suppression significantly reduced p65 binding to IκBα, TNF, and CXCL8 promoters (Fig. 7G). These results are also consistent with the observed reduction in IκBα protein re-expression (Fig. 7B), confirming that reduced promoter binding directly impacted subsequent transcription and protein expression. Together, this comprehensive analysis establishes the critical importance of Ca2+ dependent PKCα activation in p65 nuclear localization, promotor binding, and transcriptional activation of a cohort of key NF-κB target genes.
Discussion
The need to precisely determine how Ca2+ regulates distinct transcriptional responses in T cells is underscored by that fact that almost 60% of TCR-induced genes are subject to Ca2+-dependent control (57). The notion that Ca2+ regulates NF-κB activation in lymphocytes is rooted in decades-old work demonstrating that NFAT and NF-κB activity is tuned to distinct calcium dynamics (3, 5, 6). Specifically, NFAT activation requires steady-state Ca2+ elevations (2, 3, 5, 6), although the amplitude of steady-state Ca2+ signals may further dictate which NFAT isoform is activated (58, 59). In contrast, selective activation of NF-κB has been linked to low-frequency spikes in cytoplasmic Ca2+ (3, 5). However, little is known about the nature of these Ca2+ signals, and the source of Ca2+ required to activate NF-κB has not previously been explored. Thus, in comparison with the established role and mechanism of Ca2+-dependent NFAT activation, the mechanisms by which Ca2+ regulates TCR-induced NF-κB activation remain undefined.
We first asked whether the relatively infrequent Ca2+ spikes that selectively activate NF-κB in lymphocytes (3, 5, 6) could be generated from ER release without a need for extracellular influx (60). Unexpectedly, we found that ER release is insufficient and that Ca2+ influx via Orai is required to activate NF-κB. We then focused on the mechanism by which Orai-mediated Ca2+ entry regulates NF-κB activation.
Engagement of the TCR triggers canonical NF-κB activation by PKCθ-driven formation of the CBM complex (containing CARMA1, Bcl10, and MALT1) (8, 9, 61, 62). During formation of the CBM complex, Ca2+ has been implicated in CARMA1 and Bcl10 phosphorylation via Calmodulin kinase II (63–65) and Bcl10 dephosphorylation by Calcineurin A (10, 42). Our data confirm this general modulatory role for Ca2+ in steps proximal to IKK activation and IκBα degradation. However, we also found that pharmacological activation of PKCθ using either PMA stimulation alone (Fig. 4A) or PMA plus ionomycin in Ca2+-free medium (Fig. 1B) triggers substantial IκBα degradation in the absence of Ca2+ mobilization. Thus, rather than exhibiting an absolute requirement for Ca2+, our data suggest that Ca2+ cooperates with PKCθ to accelerate the rate and, possibly, extent of IκBα degradation. Hence, although PKCθ activation is sufficient for CBM complex formation and IKK activation, Ca2+ serves a modulatory role via CnA upstream of IKK activation. An additional finding here is the obligate role for Ca2+ in NF-κB activation distal to IκBα degradation, where it controls p65 phosphorylation, nuclear localization, target gene promotor binding, and transcriptional activation.
This regulatory role for Ca2+ in IKK-distal signaling is entirely novel and establishes Ca2+ as a critical regulator at multiple checkpoints of NF-κB activity. Notably, although TNF signaling involves IKK activation and induction of p65 phosphorylation, our data establish that this occurs independently of any requirement for Ca2+. Most importantly, we show that TCR-induced p65 phosphorylation on Ser-536 definitively involves Ca2+-dependent activation of PKCα and that the kinetics of this phosphorylation are distinct from the regulation of p65 phosphorylation in response to TNF. A number of separate kinases have been described that control TNF-driven and TNF-independent p65 phosphorylation (13, 15, 20, 22–28, 30, 49, 50, 66–69). However, none of these require Ca2+, and our experiments show that PKCα plays no role in TNF signaling. Thus, we established pathway-specific nodes of control for TCR versus TNF-induced NF-κB signaling in which Ca2+ regulation of PKCα represents a novel but crucial regulatory step in TCR-induced transcriptional activation of NF-κB in T cells. Furthermore, we delineated two separate Ca2+-dependent checkpoints, one proximal and one distal to IKK activation, that modulate TCR-induced NF-κB signaling.
Our results have far-reaching implications concerning the mechanisms controlling T cell development and cell fate specification. In this regard, recent work has highlighted fundamental roles for TCR-induced Ca2+ entry in the development of immunity (34). For example, individuals with functional defects in STIM or Ca2+ release-activated Ca2+/Orai are profoundly immune-deficient, and mice conditionally lacking STIM in T lymphocytes develop autoreactive disorders in part because of defective thymic natural regulatory T cell induction by high-affinity self-agonists (70). A similar and selective defect in natural regulatory T cell development occurs in mice selectively lacking either c-Rel (71–76) or upstream mediators of NF-κB activation, including BCL10, PKCθ, CARMA1, CnAβ, and IKKβ (77–81). Although our study focuses on p65-dependent transcriptional activation, the dual sensitivity of proximal and distal Ca2+ signals we identified and the role of c-Rel in natural regulatory T cell development raises the intriguing possibility that c-Rel transcriptional activation is also Ca2+-dependent. If this were the case, then one would speculate that p65 and c-Rel regulation, like NFAT isoforms, might be tuned to quantitatively or qualitatively distinct Ca2+ dynamics. Thus, although we have shown that p65 nuclear localization and transcriptional activation are regulated by Ca2+-dependent PKCα-mediated phosphorylation of p65 Ser-536, distinct Ca2+-activated kinases could control c-Rel activity. Hence, critical goals of future studies will be to quantify the Ca2+ threshold of IKK activation and p65 phosphorylation, identify the range of Ca2+-dependent Ser/Thr kinases activated following TCR engagement, and elucidate the role of Ca2+ in c-Rel activation.
Author Contributions
B. D. F., M. J. M., C. M. G., C. T. B., L. A. M., and U. H. designed the experiments. B. F., M. M., and C. B. wrote the manuscript. X. L., C. B., K. M., L. M., C. G., and K. A. M. performed the immunoblot assays. X. L. and C. B. performed the calcium measurements. X. L. performed the luciferase assays. G. R. and C. B. performed the p65 localization experiments. C. B., J. J. M., X. L., L. M., C. G., and D. P. B. performed the transcriptional assays. C. B. and G. R. performed the statistical analyses. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Patrick Doonan, Arindam Basu, Igor Brodsky, Naomi Philip, and Alexandra Delaney for assistance and helpful discussions.
The work was supported by National Institutes of Health Grants RO1AI060921 (to B. F.) and RO1HL096642 (to M. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TCR
- T cell receptor
- IKK
- IκB kinase
- ER
- endoplasmic reticulum
- PMA
- phorbol 12-myristate 13-acetate
- EGFP
- enhanced GFP
- P/I
- phorbol 12-myristate 13-acetate/ionomycin
- 3/28
- CD3/CD28
- CnA
- calcineurin A
- CBM
- CARMA1-Bcl10-MALT1.
References
- 1. Lewis R. S. (2003) Calcium oscillations in T-cells: mechanisms and consequences for gene expression. Biochem. Soc. Trans. 31, 925–929 [DOI] [PubMed] [Google Scholar]
- 2. Healy J. I., Dolmetsch R. E., Lewis R. S., and Goodnow C. C. (1998) Quantitative and qualitative control of antigen receptor signalling in tolerant B lymphocytes. Novartis Found. Symp. 215, 137–144, discussion 144–145, 186–190 [DOI] [PubMed] [Google Scholar]
- 3. Dolmetsch R. E., Lewis R. S., Goodnow C. C., and Healy J. I. (1997) Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386, 855–858 [DOI] [PubMed] [Google Scholar]
- 4. Healy J. I., Dolmetsch R. E., Timmerman L. A., Cyster J. G., Thomas M. L., Crabtree G. R., Lewis R. S., and Goodnow C. C. (1997) Different nuclear signals are activated by the B cell receptor during positive versus negative signaling. Immunity 6, 419–428 [DOI] [PubMed] [Google Scholar]
- 5. Dolmetsch R. E., Xu K., and Lewis R. S. (1998) Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392, 933–936 [DOI] [PubMed] [Google Scholar]
- 6. Li W., Llopis J., Whitney M., Zlokarnik G., and Tsien R. Y. (1998) Cell-permeant caged InsP3 ester shows that Ca2+ spike frequency can optimize gene expression. Nature 392, 936–941 [DOI] [PubMed] [Google Scholar]
- 7. Li H., Rao A., and Hogan P. G. (2011) Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 21, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trushin S. A., Pennington K. N., Algeciras-Schimnich A., and Paya C. V. (1999) Protein kinase C and calcineurin synergize to activate IκB kinase and NF-κB in T lymphocytes. J. Biol. Chem. 274, 22923–22931 [DOI] [PubMed] [Google Scholar]
- 9. Steffan N. M., Bren G. D., Frantz B., Tocci M. J., O'Neill E. A., and Paya C. V. (1995) Regulation of IκBα phosphorylation by PKC- and Ca2+-dependent signal transduction pathways. J. Immunol. 155, 4685–4691 [PubMed] [Google Scholar]
- 10. Palkowitsch L., Marienfeld U., Brunner C., Eitelhuber A., Krappmann D., and Marienfeld R. B. (2011) The Ca2+-dependent phosphatase calcineurin controls the formation of the Carma1-Bcl10-Malt1 complex during T cell receptor-induced NF-κB activation. J. Biol. Chem. 286, 7522–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baker R. G., Hayden M. S., and Ghosh S. (2011) NF-κB, inflammation, and metabolic disease. Cell Metab. 13, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vallabhapurapu S., and Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 13. Hochrainer K., Racchumi G., and Anrather J. (2013) Site-specific phosphorylation of the p65 protein subunit mediates selective gene expression by differential NF-κB and RNA polymerase II promoter recruitment. J. Biol. Chem. 288, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clavijo P. E., and Frauwirth K. A. (2012) Anergic CD8+ T lymphocytes have impaired NF-κB activation with defects in p65 phosphorylation and acetylation. J. Immunol. 188, 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buss H., Handschick K., Jurrmann N., Pekkonen P., Beuerlein K., Müller H., Wait R., Saklatvala J., Ojala P. M., Schmitz M. L., Naumann M., and Kracht M. (2012) Cyclin-dependent kinase 6 phosphorylates NF-κB p65 at serine 536 and contributes to the regulation of inflammatory gene expression. PLoS ONE 7, e51847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xing D., Gong K., Feng W., Nozell S. E., Chen Y. F., Chatham J. C., and Oparil S. (2011) O-GlcNAc modification of NF-κB p65 inhibits TNFα-induced inflammatory mediator expression in rat aortic smooth muscle cells. PLoS ONE 6, e24021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Itatsu K., Sasaki M., Harada K., Yamaguchi J., Ikeda H., Sato Y., Ohta T., Sato H., Nagino M., Nimura Y., and Nakanuma Y. (2009) Phosphorylation of extracellular signal-regulated kinase 1/2, p38 mitogen-activated protein kinase and nuclear translocation of NF-κB are involved in upregulation of matrix metalloproteinase-9 by tumour necrosis factor-α. Liver Int. 29, 291–298 [DOI] [PubMed] [Google Scholar]
- 18. Gutierrez H., O'Keeffe G. W., Gavaldà N., Gallagher D., and Davies A. M. (2008) NF-κB signaling either stimulates or inhibits neurite growth depending on the phosphorylation status of p65/RelA. J. Neurosci. 28, 8246–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wittwer T., and Schmitz M. L. (2008) NIK and Cot cooperate to trigger NF-κB p65 phosphorylation. Biochem. Biophys. Res. Commun. 371, 294–297 [DOI] [PubMed] [Google Scholar]
- 20. Sánchez-Valdepeñas C., Punzón C., San-Antonio B., Martin A. G., and Fresno M. (2007) Differential regulation of p65 and c-Rel NF-κB transactivating activity by Cot, protein kinase C ζ and NIK protein kinases in CD3/CD28 activated T cells. Cell. Signal. 19, 528–537 [DOI] [PubMed] [Google Scholar]
- 21. Harris J., Olière S., Sharma S., Sun Q., Lin R., Hiscott J., and Grandvaux N. (2006) Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK ϵ. J. Immunol. 177, 2527–2535 [DOI] [PubMed] [Google Scholar]
- 22. Mattioli I., Geng H., Sebald A., Hodel M., Bucher C., Kracht M., and Schmitz M. L. (2006) Inducible phosphorylation of NF-κB p65 at serine 468 by T cell costimulation is mediated by IKK ϵ. J. Biol. Chem. 281, 6175–6183 [DOI] [PubMed] [Google Scholar]
- 23. Doyle S. L., Jefferies C. A., and O'Neill L. A. (2005) Bruton's tyrosine kinase is involved in p65-mediated transactivation and phosphorylation of p65 on serine 536 during NF-κB activation by lipopolysaccharide. J. Biol. Chem. 280, 23496–23501 [DOI] [PubMed] [Google Scholar]
- 24. Sasaki C. Y., Barberi T. J., Ghosh P., and Longo D. L. (2005) Phosphorylation of RelA/p65 on serine 536 defines an IκBα-independent NF-κB pathway. J. Biol. Chem. 280, 34538–34547 [DOI] [PubMed] [Google Scholar]
- 25. Mattioli I., Sebald A., Bucher C., Charles R. P., Nakano H., Doi T., Kracht M., and Schmitz M. L. (2004) Transient and selective NF-κB p65 serine 536 phosphorylation induced by T cell costimulation is mediated by IκB kinase β and controls the kinetics of p65 nuclear import. J. Immunol. 172, 6336–6344 [DOI] [PubMed] [Google Scholar]
- 26. Campbell K. J., and Perkins N. D. (2004) Post-translational modification of RelA(p65) NF-κB. Biochem. Soc. Trans. 32, 1087–1089 [DOI] [PubMed] [Google Scholar]
- 27. Buss H., Dörrie A., Schmitz M. L., Frank R., Livingstone M., Resch K., and Kracht M. (2004) Phosphorylation of serine 468 by GSK-3β negatively regulates basal p65 NF-κB activity. J. Biol. Chem. 279, 49571–49574 [DOI] [PubMed] [Google Scholar]
- 28. Vermeulen L., De Wilde G., Van Damme P., Vanden Berghe W., and Haegeman G. (2003) Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 22, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhong H., May M. J., Jimi E., and Ghosh S. (2002) The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9, 625–636 [DOI] [PubMed] [Google Scholar]
- 30. Jang M. K., Goo Y. H., Sohn Y. C., Kim Y. S., Lee S. K., Kang H., Cheong J., and Lee J. W. (2001) Ca2+/calmodulin-dependent protein kinase IV stimulates NF-κB transactivation via phosphorylation of the p65 subunit. J. Biol. Chem. 276, 20005–20010 [DOI] [PubMed] [Google Scholar]
- 31. Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E. Jr., and Meyer T. (2005) STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., and Stauderman K. A. (2005) STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang S. L., Yu Y., Roos J., Kozak J. A., Deerinck T. J., Ellisman M. H., Stauderman K. A., and Cahalan M. D. (2005) STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437, 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feske S., Skolnik E. Y., and Prakriya M. (2012) Ion channels and transporters in lymphocyte function and immunity. Nat. Rev. Immunol. 12, 532–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hogan P. G., Lewis R. S., and Rao A. (2010) Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu. Rev. Immunol. 28, 491–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du P., Kibbe W. A., and Lin S. M. (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 [DOI] [PubMed] [Google Scholar]
- 37. Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article 3 [DOI] [PubMed] [Google Scholar]
- 38. Gentleman R., Carey V., Huber W., Irizarry R., and Dutoit S. (2005) Limma: Linear Models for Microarray Data, Springer, New York [Google Scholar]
- 39. Reiner A., Yekutieli D., and Benjamini Y. (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 40. Benjamini Y., and Hochberg Y. (1995) Controlling false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 [Google Scholar]
- 41. Barouch-Bentov R., and Altman A. (2006) Protein kinase Cθ: new perspectives on its functions in T cell biology. Adv. Exp. Med. Biol. 584, 1–13 [DOI] [PubMed] [Google Scholar]
- 42. Frischbutter S., Gabriel C., Bendfeldt H., Radbruch A., and Baumgrass R. (2011) Dephosphorylation of Bcl-10 by calcineurin is essential for canonical NF-κB activation in Th cells. Eur. J. Immunol. 41, 2349–2357 [DOI] [PubMed] [Google Scholar]
- 43. Sun S. C., Ganchi P. A., Ballard D. W., and Greene W. C. (1993) NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science 259, 1912–1915 [DOI] [PubMed] [Google Scholar]
- 44. Wang B., Wei H., Prabhu L., Zhao W., Martin M., Hartley A. V., and Lu T. (2015) Role of novel serine 316 phosphorylation of the p65 subunit of NF-κB in differential gene regulation. J. Biol. Chem. 290, 20336–20347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pahl H. L. (1999) Activators and target genes of Rel/ NF-κB transcription factors. Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 46. Oh H., and Ghosh S. (2013) NF-κB: roles and regulation in different CD4+ T-cell subsets. Immunol. Rev. 252, 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gilmore F. (2015) NF-κB transcription factors, Boston University Department of Biology [Google Scholar]
- 48. Takeuchi H., Hirano T., Whitmore S. E., Morisaki I., Amano A., and Lamont R. J. (2013) The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-κB RelA/p65. PLoS Pathog. 9, e1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Geng H., Wittwer T., Dittrich-Breiholz O., Kracht M., and Schmitz M. L. (2009) Phosphorylation of NF-κB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 10, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmitz M. L., Mattioli I., Buss H., and Kracht M. (2004) NF-κB: a multifaceted transcription factor regulated at several levels. ChemBioChem 5, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 51. Fujita T., Nolan G. P., Ghosh S., and Baltimore D. (1992) Independent modes of transcriptional activation by the p50 and p65 subunits of NF-κB. Genes Dev. 6, 775–787 [DOI] [PubMed] [Google Scholar]
- 52. Schmitz M. L., and Baeuerle P. A. (1991) The p65 subunit is responsible for the strong transcription activating potential of NF-κB. EMBO J. 10, 3805–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oeckinghaus A., Hayden M. S., and Ghosh S. (2011) Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 [DOI] [PubMed] [Google Scholar]
- 54. Campbell K. J., and Perkins N. D. (2006) Regulation of NF-κB function. Biochem. Soc. Symp. 73, 165–180 [DOI] [PubMed] [Google Scholar]
- 55. Steinberg S. F. (2008) Structural basis of protein kinase C isoform function. Physiol. Rev. 88, 1341–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Altman A., Mally M. I., and Isakov N. (1992) Phorbol ester synergizes with Ca2+ ionophore in activation of protein kinase C PKCα and PKCβ isoenzymes in human T cells and in induction of related cellular functions. Immunology 76, 465–471 [PMC free article] [PubMed] [Google Scholar]
- 57. Feske S., Giltnane J., Dolmetsch R., Staudt L. M., and Rao A. (2001) Gene regulation mediated by calcium signals in T lymphocytes. Nat. Immunol. 2, 316–324 [DOI] [PubMed] [Google Scholar]
- 58. Podtschaske M., Benary U., Zwinger S., Höfer T., Radbruch A., and Baumgrass R. (2007) Digital NFATc2 activation per cell transforms graded T cell receptor activation into an all-or-none IL-2 expression. PLoS ONE 2, e935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tomida T., Hirose K., Takizawa A., Shibasaki F., and Iino M. (2003) NFAT functions as a working memory of Ca2+ signals in decoding Ca2+ oscillation. EMBO J. 22, 3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dolmetsch R. E., and Lewis R. S. (1994) Signaling between intracellular Ca2+ stores and depletion-activated Ca2+ channels generates [Ca2+]i oscillations in T lymphocytes. J. Gen. Physiol. 103, 365–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Coudronniere N., Villalba M., Englund N., and Altman A. (2000) NF-κB activation induced by T cell receptor/CD28 costimulation is mediated by protein kinase Cθ. Proc. Natl. Acad. Sci. U.S.A. 97, 3394–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ghosh S., and Baltimore D. (1990) Activation in vitro of NF-κB by phosphorylation of its inhibitor IκB. Nature 344, 678–682 [DOI] [PubMed] [Google Scholar]
- 63. Oruganti S. R., Edin S., Grundström C., and Grundström T. (2011) CaMKII targets Bcl10 in T-cell receptor induced activation of NF-κB. Mol. Immunol. 48, 1448–1460 [DOI] [PubMed] [Google Scholar]
- 64. Ishiguro K., Ando T., Goto H., and Xavier R. (2007) Bcl10 is phosphorylated on Ser138 by Ca2+/calmodulin-dependent protein kinase II. Mol. Immunol. 44, 2095–2100 [DOI] [PubMed] [Google Scholar]
- 65. Ishiguro K., Green T., Rapley J., Wachtel H., Giallourakis C., Landry A., Cao Z., Lu N., Takafumi A., Goto H., Daly M. J., and Xavier R. J. (2006) Ca2+/calmodulin-dependent protein kinase II is a modulator of CARMA1-mediated NF-κB activation. Mol. Cell. Biol. 26, 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Y., Mo X., Piper M. G., Wang H., Parinandi N. L., Guttridge D., and Marsh C. B. (2011) M-CSF induces monocyte survival by activating NF-κB p65 phosphorylation at Ser276 via protein kinase C. PLoS ONE 6, e28081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hu J., Haseebuddin M., Young M., and Colburn N. H. (2005) Suppression of p65 phosphorylation coincides with inhibition of IκBα polyubiquitination and degradation. Mol. Carcinog. 44, 274–284 [DOI] [PubMed] [Google Scholar]
- 68. Hu J., Nakano H., Sakurai H., and Colburn N. H. (2004) Insufficient p65 phosphorylation at S536 specifically contributes to the lack of NF-κB activation and transformation in resistant JB6 cells. Carcinogenesis 25, 1991–2003 [DOI] [PubMed] [Google Scholar]
- 69. Zhong H., Voll R. E., and Ghosh S. (1998) Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1, 661–671 [DOI] [PubMed] [Google Scholar]
- 70. Oh-Hora M., Yamashita M., Hogan P. G., Sharma S., Lamperti E., Chung W., Prakriya M., Feske S., and Rao A. (2008) Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9, 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schmidt A. M., Zou T., Joshi R. P., Leichner T. M., Pimentel M. A., Sommers C. L., and Kambayashi T. (2013) Diacylglycerol kinase ζ limits the generation of natural regulatory T cells. Sci. Signal. 6, ra101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lee H. M., Bautista J. L., and Hsieh C. S. (2011) Thymic and peripheral differentiation of regulatory T cells. Adv. Immunol. 112, 25–71 [DOI] [PubMed] [Google Scholar]
- 73. Lio C. W., and Hsieh C. S. (2011) Becoming self-aware: the thymic education of regulatory T cells. Curr. Opin. Immunol. 23, 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hori S. (2010) c-Rel: a pioneer in directing regulatory T-cell lineage commitment? Eur. J. Immunol. 40, 664–667 [DOI] [PubMed] [Google Scholar]
- 75. Deenick E. K., Elford A. R., Pellegrini M., Hall H., Mak T. W., and Ohashi P. S. (2010) c-Rel but not NF-κB1 is important for T regulatory cell development. Eur. J. Immunol. 40, 677–681 [DOI] [PubMed] [Google Scholar]
- 76. Long M., Park S. G., Strickland I., Hayden M. S., and Ghosh S. (2009) NF-κB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity 31, 921–931 [DOI] [PubMed] [Google Scholar]
- 77. Medoff B. D., Sandall B. P., Landry A., Nagahama K., Mizoguchi A., Luster A. D., and Xavier R. J. (2009) Differential requirement for CARMA1 in agonist-selected T-cell development. Eur. J. Immunol. 39, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Molinero L. L., Yang J., Gajewski T., Abraham C., Farrar M. A., and Alegre M. L. (2009) CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J. Immunol. 182, 6736–6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jana S., Jailwala P., Haribhai D., Waukau J., Glisic S., Grossman W., Mishra M., Wen R., Wang D., Williams C. B., and Ghosh S. (2009) The role of NF-κB and Smad3 in TGFβ-mediated Foxp3 expression. Eur. J. Immunol. 39, 2571–2583 [DOI] [PubMed] [Google Scholar]
- 80. Gupta S., Manicassamy S., Vasu C., Kumar A., Shang W., and Sun Z. (2008) Differential requirement of PKCθ in the development and function of natural regulatory T cells. Mol. Immunol. 46, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Coutinho A., Caramalho I., Seixas E., and Demengeot J. (2005) Thymic commitment of regulatory T cells is a pathway of TCR-dependent selection that isolates repertoires undergoing positive or negative selection. Curr. Top. Microbiol. Immunol. 293, 43–71 [DOI] [PubMed] [Google Scholar]