Abstract
Multiple neurodegenerative diseases are caused by the aggregation of the human α-Synuclein (α-Syn) protein. α-Syn possesses high structural plasticity and the capability of interacting with membranes. Both features are not only essential for its physiological function but also play a role in the aggregation process. Recently it has been proposed that α-Syn is able to form lipid-protein particles reminiscent of high-density lipoproteins. Here, we present a method to obtain a stable and homogeneous population of nanometer-sized particles composed of α-Syn and anionic phospholipids. These particles are called α-Syn lipoprotein (nano)particles to indicate their relationship to high-density lipoproteins formed by human apolipoproteins in vivo and of in vitro self-assembling phospholipid bilayer nanodiscs. Structural investigations of the α-Syn lipoprotein particles by circular dichroism (CD) and magic angle solid-state nuclear magnetic resonance (MAS SS-NMR) spectroscopy establish that α-Syn adopts a helical secondary structure within these particles. Based on cryo-electron microscopy (cryo-EM) and dynamic light scattering (DLS) α-Syn lipoprotein particles have a defined size with a diameter of ∼23 nm. Chemical cross-linking in combination with solution-state NMR and multiangle static light scattering (MALS) of α-Syn particles reveal a high-order protein-lipid entity composed of ∼8–10 α-Syn molecules. The close resemblance in size between cross-linked in vitro-derived α-Syn lipoprotein particles and a cross-linked species of endogenous α-Syn from SH-SY5Y human neuroblastoma cells indicates a potential functional relevance of α-Syn lipoprotein nanoparticles.
Keywords: alpha-synuclein (α-synuclein), apolipoprotein, high-density lipoprotein (HDL), lipid, membrane bilayer, Parkinson disease
Introduction
Synucleinopathies are neurodegenerative diseases; the three major ones being Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy; caused by the accumulation and aggregation of the α-Synuclein (α-Syn)2 protein (1, 2). Human α-Syn is an abundant brain protein possessing considerable structural plasticity. In aqueous solution, monomeric α-Syn is largely disordered (3–5) while in a membrane mimicking environment (5, 6) monomeric α-Syn undergoes a structural transition toward a helical state. During pathological aggregation α-Syn forms oligomeric species and ultimately mature amyloid fibrils, the latter consisting mainly of β-sheet structure (7–9). Binding of α-Syn to membranes is mediated by seven imperfect repeats in the N-terminal region of its primary sequence, which are reminiscent of those found in the amphipathic helices of apolipoproteins (3, 6, 10, 11). The interaction with biological membranes is at the basis of the multiple in vivo functions attributed to α-Syn so far: synaptic vesicle pool maintenance (12, 13), regulation of dopamine neurotransmission (14, 15), transport of lipids and fatty acids (16–20), membrane trafficking (21–23), synaptic plasticity (10, 24, 25), and assistance in SNARE complex formation (26–29). Finally, membranes play a role in α-Syn aggregation too, although their precise effect is still unclear as both inhibition and promotion have been reported (30–34).
α-Syn preferentially binds to membranes containing anionic phospholipids (6, 35) and to anionic detergents such as sodium dodecyl sulfate (SDS) (3). Upon binding, the N-terminal region of α-Syn undergoes a structural transition toward a helical state, while the C-terminal one remains substantially unstructured. Initial studies by solution-state nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR) spectroscopy performed in the presence of SDS or anionic small unilamellar vesicles (SUVs), respectively, suggested that the first ∼100 residues of α-Syn form a single extended helix (5, 36). Subsequent NMR data collected of α-Syn in complex with SDS proposed an alternative state, often termed horseshoe conformation, consisting of two distinct helices interrupted by a short break (11, 37–40). While some studies suggested that the horseshoe conformation results from the constraints imposed by the reduced size and higher curvature of detergent micelles (36, 41, 42), others reported evidence of helix breaking even with SUVs large enough to accommodate the extended helix (43, 44). Nowadays, it is accepted that multiple binding modes of α-Syn to lipids exist (45, 46) and that the horseshoe and extended helical states can coexist and even undergo inter-conversion (47–50). Not only does the interaction of α-Syn with lipid membranes induce changes in the protein conformation, but also cause a remodeling of the membranes due to membrane thinning (51–53) and membrane curvature changes (51, 54–57).
Interestingly, at high protein-to-lipid ratios, α-Syn was reported to be able to reshape giant lipid vesicles into lipoprotein nanoparticles (58) (7–10 nm size) having a morphology and shape similar to that of high-density lipoproteins and self-assembling phospholipid bilayer nanodiscs (59) formed by human apolipoproteins and membrane scaffolding proteins (MSPs). In these complexes, α-Syn appears to be multimeric and adopts a broken helical conformation. However, an exact determination of the molecular weight of the multimer and more generally of the properties of these particles is hampered by the heterogeneity of the sample preparations (58). In this work, stable and homogeneous populations of nanoparticles composed of phosphatidylserine lipids and α-Syn were obtained with the protocol typically used to prepare in vitro synthetic nanodiscs (60, 61). This method ensured a high extent of sample homogeneity, which in turn allowed a detailed biophysical characterization of the particles.
Experimental Procedures
Expression and Purification of α-Synuclein
Recombinant wild-type (WT) human α-Syn and the α-Syn variant α-Syn(C141) were overexpressed in the Escherichia coli strain BL21 StarTM (DE3) pLysS (Invitrogen) and purified as described previously (62). 2H, 15N-, and 13C,15N-labeled human α-Syn were produced using standard M9 minimal medium (63) based on D2O (Isotec) and H2O, respectively, supplemented with 3 g/liter 13C glucose (Isotec), and 1 g/liter 15NH4Cl (Isotec).
Preparation of α-Synuclein Lipoprotein Particles
Lipoprotein particles consisting of α-Syn and different types of lipids were produced using 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS), and sphingomyelin (brain, porcine) synthetic lipids (Avanti Polar Lipids). The lipids were solubilized in sodium cholate with a 2-fold molar excess of detergent and stored in a glass vial. α-Syn-derived lipoprotein particles were assembled following established protocols for the preparation of MSP nanodiscs (61). In details, α-Syn in 10 mm Na2HPO4 pH 7.0 buffer was mixed with the lipids dissolved in sodium cholate and the obtained sample was incubated overnight at 27 °C while shaking at 150 rpm. Different protein-to-lipid ratios were used as described under “Results” (see below). The formation of α-Syn nanodiscs was initiated with the addition of 80% w/v Biobeads SM2 (Bio-Rad) for 2 more hours at 27 °C while shaking at 150 rpm. The obtained preparations were injected into a Superdex 200 10/300GL size-exclusion chromatography (SEC) column (GE Healthcare) and eluted at a flow rate of 0.5 ml/min in (i) 20 mm Bis-Tris-HCl pH 7.0, 150 mm NaCl for transmission electron microscopy, (ii) 1× phosphate-buffered saline (PBS) pH 7.0 for multiangle static light scattering (MALS), and (iii) 20 mm HEPES pH 7.5, 150 mm NaCl for in vitro cross-linking experiments, respectively. The different buffers did not affect the position and the width of the α-Syn lipoprotein particle peak in the SEC elution profile (data not shown). The protein peak corresponding to the species of interest were merged and either buffer-exchanged for solution- and solid-state NMR in 20 mm Bis-Tris-HCl, pH 7.0, 20 mm NaCl using a PD10 desalting column (GE Healthcare), or used directly for subsequent analysis. When required, the concentration of the eluted sample was increased by using a 3-kDa molecular weight cut-off Centricon (Amicon) concentrator.
Circular Dichroism (CD)
CD spectra of monomeric α-Syn and α-Syn DOPS lipoprotein particles were collected using a Jasco J-815 CD spectrometer with a 1-mm quartz cell at 25 °C. Spectra were averaged from 10 replicates acquired at 0.2 nm step resolution from 195–260 nm with a spectral bandwidth of 1 nm, a scanning speed of 20 nm/min, and a data integration time of 2 s. The spectra of 9.0 μm monomeric α-Syn and 11.5 μm α-Syn DOPS lipoprotein particles were recorded in 9.9 mm and 8.6 mm Na2HPO4, pH 7, respectively, containing 0.1x PBS pH 7. Appropriate blanks under the same conditions were subtracted resulting in the final spectra.
Dynamic Light Scattering (DLS)
DLS measurements of α-Syn DOPS lipoprotein particles were performed directly after SEC on a Zetasizer Nano ZS (Malvern Instruments) Dynamic Light Scattering instrument equipped with a Peltier temperature controller set at room temperature. Disposable micro-cuvettes for size measurements at a scattering angle of 173° were used. Every sample was measured three times and the averaged intensity-size distribution is reported.
Negative-Stain Electron Microscopy (EM)
α-Syn lipoprotein particles were diluted to ∼10 μm in 20 mm Bis-Tris-HCl, pH 7.0, 150 mm NaCl. An aliquot of 5 μl was deposited for 1 min on a glow-discharged carbon-coated copper grid for EM. The grids were blotted and washed twice with drops of water before staining with 2% w/v uranyl acetate for 15 s. EM images were acquired using a FEI Morgagni 268 transmission electron microscope operated at 100 kV.
Cryo-Electron Microscopy (cryo-EM)
α-Syn lipoprotein particles were diluted to a concentration of ∼13.8 μm in 20 mm Bis-Tris-HCl pH 7.0, 150 mm NaCl. Aliquots of 3 μl were applied to glow-discharged holey-carbon film coated copper grids (Quantifoil GmbH, Jena, Germany), blotted, and vitrified in a FEI Vitrobot Mark IV freezing device. Grids were transferred to a Philips CM200FEG cryo-EM instrument, operated at 200 kV, and images were recorded on a TVIPS F416 CMOS camera, using an electron dose of 20 electrons/Å2 and a nominal magnification of 50 kx.
Image Processing
For images of negatively stained α-Syn lipoprotein particles, 7650 particles were manually selected using Boxer from the EMAN2 suit (64), aligned, and classified using EMAN2.
For cryo-EM images of α-Syn lipoprotein particles, 9300 particles were manually selected from 108 images using Boxer. Particle images were aligned and classified into 100 2D classes using EMAN2.
Solution-State NMR Spectroscopy
All solution-state NMR spectra were recorded at 30 °C on a Bruker 700 MHz Avance III spectrometer equipped with a triple resonance cryoprobe. Two-dimensional [15N,1H]-TROSY (65) spectra (time domain data size 256*2048 complex points, t1max(15N) = 44.8 ms and t2max(1H) = 204.9 ms) were collected from samples consisting of different unlabeled lipids and 2H, 15N-labeled α-Syn prepared with the procedure described above. The lipid-free state of α-Syn was measured using a 1.5-mm sample of 13C, 15N-labeled α-Syn. three-dimensional constant time (ct)-TROSY-HNCA, three-dimensional ct-TROSY-HNCACB (66), and three-dimensional ct-TROSY-HN(CO)CA (67) experiments were acquired for sequential assignment (68) of 1H, 13C, and 15N backbone resonances. The spectra were processed with the program PROSA (69) and analyzed with the program XEASY (70). 15N-edited diffusion measurements with increasing diffusion coding gradient strengths (4 ms total duration) and a 105-ms diffusion delay were recorded for a total of 2048 transients per gradient strength.
Solid-State NMR Spectroscopy
A sample of uniformly 13C, 15N-labeled WT human α-Syn DOPS lipoprotein particles was filled into a ZrO2 3.2-mm rotor (Bruker) using a home-made filling tool (71) in a SW40 TI rotor and an optima L90-K ultracentrifuge (Beckman). Solid-state NMR experiments were performed at 15 °C on a Bruker 850 MHz Avance II spectrometer using a Bruker triple-resonance 3.2 mm Efree probe. A 20-ms DARR spectrum was recorded (time domain data size 1024*3072 complex points, t1max(13C) = 10.2 ms and t2max(13C) = 15.4 ms) and processed using topspin 2.1 (Bruker).
In Vitro Cross-Linking
Purified α-Syn DOPS lipoprotein particles in 20 mm HEPES pH 7.5, 150 mm NaCl were cross-linked using disuccinimidyl glutarate (DSG) purchased from Thermo Scientific. Immediately before usage, DSG was dissolved at 25 mm in DMSO. At a final concentration of 83 μm α-Syn DOPS lipoprotein particles and 0.249–4.2 mm DSG cross-linker, the samples were incubated for 30 min at 37 °C while shaking at 600 rpm. The cross-linking reaction was quenched by adding 1 m Tris-HCl, pH 7.5 to a final concentration of 50 mm and mixing the samples for additional 15 min at 25 °C at 600 rpm.
In Vivo Cross-Linking
SH-SY5Y human neuroblastoma cells expressing endogenous α-Syn were purchased from ATCC and cultured in DMEM medium supplemented with 10% fetal calf serum and 1% penicillin-streptomycin-glutamine. Five 10-cm dishes were grown at 80% confluence, then harvested in PBS supplemented with protease inhibitor mixture (Roche) (72) and washed twice with the same buffer. Samples containing ∼8.106 cells in 200 μl PBS buffer supplemented with protease inhibitor mixture were cross-linked with 2 mm DSG (final concentration) using the in vitro cross-linking protocol (see above). After quenching the reaction, the cells were lysed by 15-s sonication using a microtip at 39% power followed by centrifugation at 25,000 × g. The supernatant was used for the subsequent immunoblotting (see below).
Gel Electrophoresis and Immunoblotting
The samples obtained from the cross-linking experiments were mixed with Laemmli buffer at ratio 1:1 and subsequently analyzed by SDS-PAGE. For the analysis of in vitro cross-linking reactions, 4–12% gradient NuPAGE Bis-Tris gels (Invitrogen) were used and stained with Coomassie Brilliant Blue. Immunoblotting was carried out for detection of in vitro and in vivo cross-linked samples using a 15% polyacrylamide gel. Nitrocellulose membranes were incubated in PBS containing 0.4% paraformaldehyde for 30 min at room temperature (72), rinsed twice with PBS, and blocked in PBS-T buffer (PBS containing 0.05% Tween-20 (Sigma)) supplemented with 5% fat-free milk for 1 h at room temperature. After blocking, membranes were incubated with the primary anti-α-Synuclein antibody LB509 (at 1:5000 dilution, from Abcam) for 1 h at room temperature followed by three washes for 10 min with PBS-T. Subsequently, the washed membranes were incubated in blocking buffer containing the goat anti-mouse antibody conjugated to horseradish peroxidase (at 1:3000 dilution from Bio-Rad) for 1 h at room temperature. The membranes were then washed three times for 10 min in PBS-T and developed with enhanced chemiluminescence (ECL Prime, GE Healthcare) according to the manufacturer's instructions.
Multiangle Static Light Scattering (MALS)
The oligomeric state of α-Syn in DOPS lipoprotein particles was determined by multiangle static light scattering coupled with SEC and refractive index measurements. An additional C-terminal cysteine at position 141, denoted α-Syn(C141), was introduced in human α-Syn and labeled with 4-dimethylaminophenylazophenyl-4′maleimide (DABMI) (Invitrogen) according to manufacturer's instructions. The average molar weights of the protein-lipid complex, the protein, and the lipid fraction in the α-Syn DOPS lipoprotein particles were then determined at the absorbance of DABMI (454 nm) using a protein extinction coefficient of 1666 ml/(g cm), protein dn/dc = 0.185 ml/g, and DOPS dn/dc = 0.141 ml/g, respectively.
Results
Preparation, Morphology, and Size of α-Synuclein-derived Lipoprotein Particles
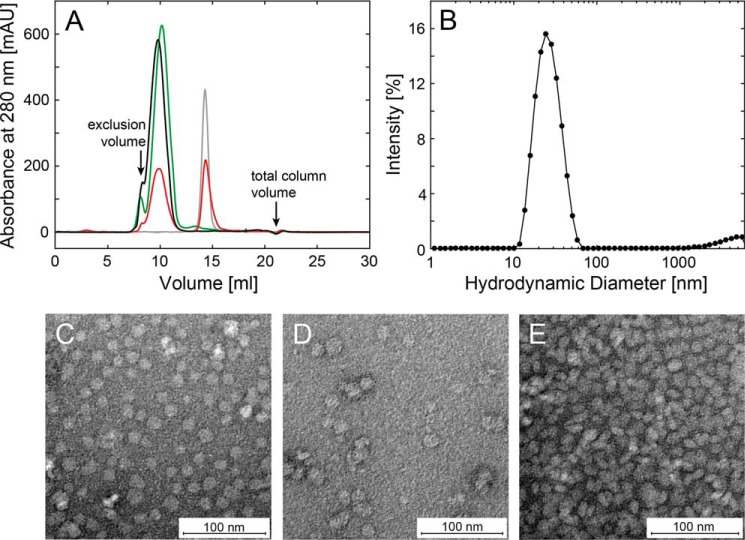
Nanometer-sized particles consisting of lipids and α-Syn, hereafter called α-Syn lipoprotein particles, were produced following protocols previously used to obtain in vitro nanodiscs from lipids and MSPs (60, 61). Briefly, the protein was first mixed with the desired lipids dissolved in sodium cholate. After incubation, the detergent was removed, and the phospholipid bilayer was encapsulated by α-Syn molecules. To separate lipid-bound α-Syn from residual monomeric protein, the sample was subjected to size exclusion chromatography (SEC). Several ratios of α-Syn to the lipids of interest were tested. The formation of α-Syn lipoprotein particles and the extent of incorporation of α-Syn into these complexes were then monitored by SEC (Fig. 1A). The elution profiles in the presence of DOPS or POPS phospholipids showed that at a molar protein-to-lipid ratio of 1 to 40 full incorporation of α-Syn and lipids into α-Syn lipoprotein particles were obtained (Fig. 1A, please note that α-Syn monomer elutes at 14.7 ml). Additionally, sphingomyelin was used to assess whether the binding of α-Syn to a natural, zwitterionic lipid can also result in the formation of α-Syn lipoprotein particles. Sphingomyelin is a component of synaptic vesicles and is the most abundant sphingolipid in human cell membranes. Interestingly, when α-Syn was incubated with sphingomyelin at the same protein-to-lipid ratio as used for the negatively charged lipids, a residual amount of monomeric α-Syn (peak at 14.7 ml) was detected in the SEC indicating that the formation of α-Syn lipoprotein particles is more comprehensive with negatively charged lipids (Fig. 1A).
FIGURE 1.
α-Syn-containing lipoprotein particles are well defined in size. A, size-exclusion gel chromatography (Superdex 200 10/300GL) of α-Syn DOPS (black), α-Syn POPS (green), and α-Syn sphingomyelin (red) lipoprotein particles. Monomeric (gray) α-Syn elutes at ∼14.3 ml. B, dynamic light scattering of α-Syn-containing DOPS lipoprotein particles. The size-exclusion chromatography fraction at the maximum of the absorbance of 280 nm (9.5–10 ml) consists of particles with an average hydrodynamic radius of 25.8 ± 0.8 nm. C–E, negatively stained electron micrographs containing the maxima of the size-exclusion chromatography fractions at the absorbance of 280 nm of purified (C) α-Syn DOPS, (D) α-Syn POPS, and (E) α-Syn sphingomyelin lipoprotein particles. The radius varies from ∼19–28 nm, scale bar 100 nm.
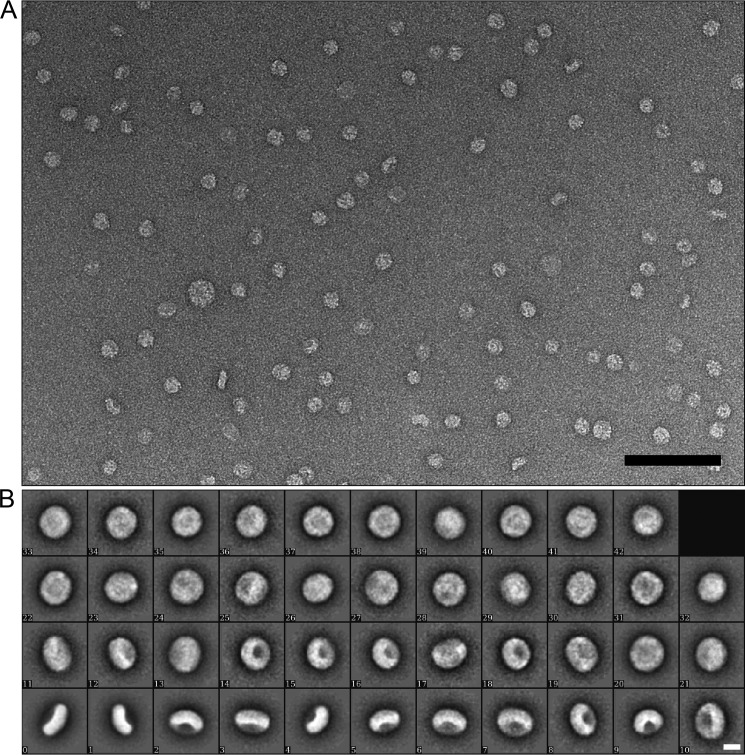
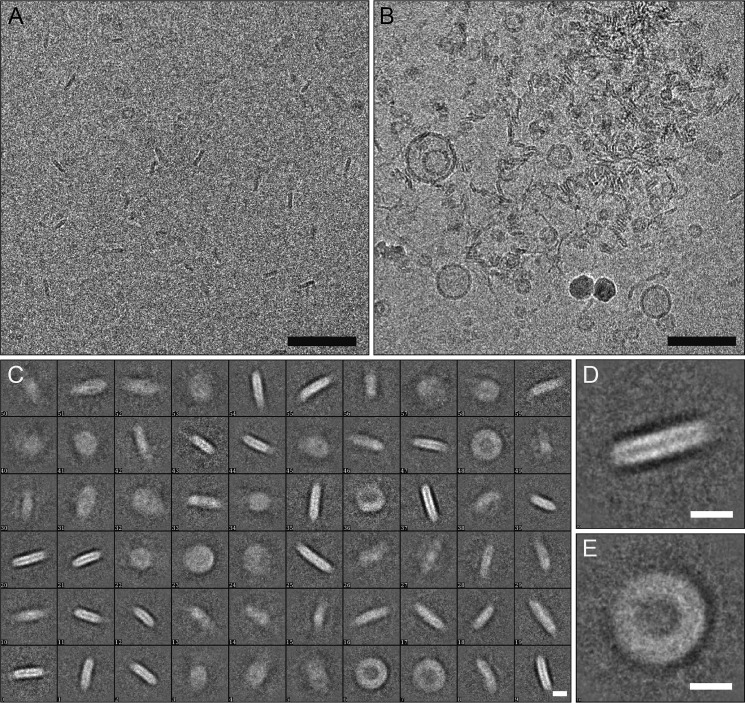
Subsequently, the morphology of the α-Syn lipoprotein particles prepared with either of the lipids (i.e. DOPS, POPS, and sphingomyelin) eluting at ∼9.8 ml in the SEC were assessed by negative-stain EM (Fig. 1, C–E, Fig. 2). From EM images of negatively stained particles, 7650 particles were manually selected, aligned, and classified. A population of circular particles having a diameter of ∼19–28 nm was observed. In line with this finding are the results from DLS measurements on α-Syn DOPS lipoprotein particles, where the most abundant particles present in the sample appeared to have a Z-average size of 25.8 ± 0.8 nm (from cumulate analysis of three repeated measurements of the same sample) and a poly-dispersion index of 0.20 ± 0.02 (Fig. 1B). Next, the α-Syn DOPS lipoprotein particles were examined in detail by cryo-EM (Fig. 3). A total of 9300 α-Syn DOPS particles were selected from electron micrographs and subjected to classification and class-averaging procedures. The characteristic class averages (Fig. 3C) showed roundish/ellipsoidal particles with a low-density inner region attributed to the DOPS bilayer of ∼10 nm in diameter, which is surrounded by a ∼6–7 nm wide α-Syn belt of higher density (Fig. 3, D and E). Similar to the architecture of lipoprotein particles formed by the apolipoprotein A1 (73, 74) and α-Syn induced nanoparticles (58), the observed higher density features at the periphery of the discs are compatible with the interpretation that α-Syn molecules are wrapped around the DOPS lipids in a ring-like manner (Fig. 3E).
FIGURE 2.
Electron microscopy of α-Syn DOPS lipoprotein particles. A, electron micrograph of negatively stained α-Syn lipoprotein particles. Both top and side views of the nanoparticles are visible (scale bar 100 nm). B, two-dimensional classification of α-Syn lipoprotein particles (scale bar 10 nm).
FIGURE 3.
Cryo-electron microscopy images of α-Syn DOPS lipoprotein particles. A, on the cryo-electron micrograph mostly side views of α-Syn DOPS lipoprotein particles are well visible. B, α-Syn DOPS lipoprotein particles have a tendency to aggregate. C, two-dimensional classification of α-Syn DOPS lipoprotein particles. Three main populations of nanoparticles with diameters of 19–21 nm, 23–24 nm, 27–28 nm, respectively, were observed. D, side and E, top view of two-dimensional nanoparticle classes with a diameter of ∼23 nm. The scale bars indicate 100 nm in (A, B) and 10 nm in (C–E).
The collected data indicate that our protocol allows obtaining nanometer-sized particles composed of α-Syn and anionic phospholipids, and that such samples have a uniform size as determined by the relatively small width of both the SEC elution profile peak and the DLS size distribution poly-dispersion index (Fig. 1, A and B). The samples eluted from the SEC appear to be quite stable, since increasing their concentration and re-injecting them in the same SEC column did not change the elution profile (data not shown). Storage at 4 °C over a couple of weeks did not alter the morphology as evidenced by EM (data not shown).
Helical Secondary Structure of α-Synuclein in the Lipoprotein Particles
Circular dichroism (CD) was used to compare the secondary structure of monomeric α-Syn versus that of the lipid-bound state prepared as described above (Fig. 4A). In line with previous studies by many authors, the measured CD spectrum of monomeric α-Syn had the features of highly disordered proteins, with a maximum negative ellipticity per residue ([Θ]MRW) at ∼199 nm (3–6). In contrast, the CD spectrum of α-Syn DOPS lipoprotein particles, which eluted in the SEC at ∼9.8 ml, had the characteristics of α-helical proteins, with two negative [Θ]MRW peaks at ∼210 and 221 nm, respectively, and one positive [Θ]MRW peak at ∼195 nm. This spectrum resembles those obtained upon binding of α-Syn to anionic lipid vesicles and anionic detergents (3, 5, 6).
FIGURE 4.
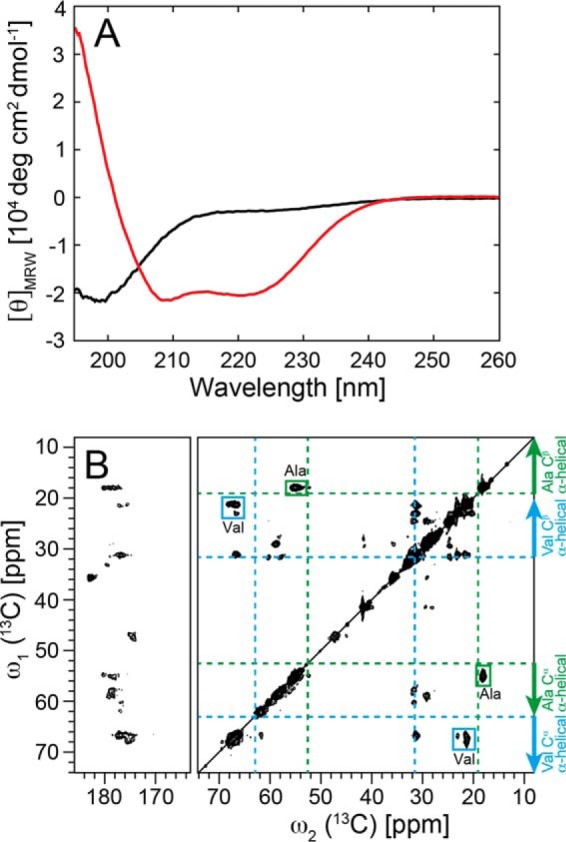
Helical secondary structure of α-Syn in lipoprotein particles. A, conformational change of α-Syn determined by CD from a predominantly unfolded state (black) free in solution to an α-helical state in α-Syn DOPS lipoprotein particles (red). For the α-Syn DOPS lipoprotein particles, the size exclusion chromatography fraction 9.5–10 ml (Fig. 1A) was used. B, two-dimensional 13C DARR solid-state NMR spectra (at a magnetic field of 850 MHz 1H frequency) of uniformly 13C, 15N-labeled α-Syn DOPS lipoprotein particles. The assigned cross peaks from Ala and Val residues are indicated by green and cyan boxes, respectively. Indicated with dotted green and cyan lines are the random coil 13Cα, 13Cβ chemical shifts for the amino acid residues Ala and Val, respectively. Arrows show the chemical shift area typically observed for α-helical secondary structure.
In addition to CD, the α-helical secondary structure of α-Syn in α-Syn DOPS lipoprotein particles was verified by magic angle solid-state NMR (MAS SS-NMR). Observed 13Cα-13Cβ cross-peaks for alanine and valine amino acid residues in the 13C-13C DARR correlation spectrum of 13C, 15N-labeled α-Syn DOPS lipoprotein particles fell within the regions typical detected for α-helices (75) (Fig. 4B). Thus, CD together with chemical shift MAS SS-NMR analyses indicated that α-Syn DOPS lipoprotein particles contain α-helical secondary structure elements.
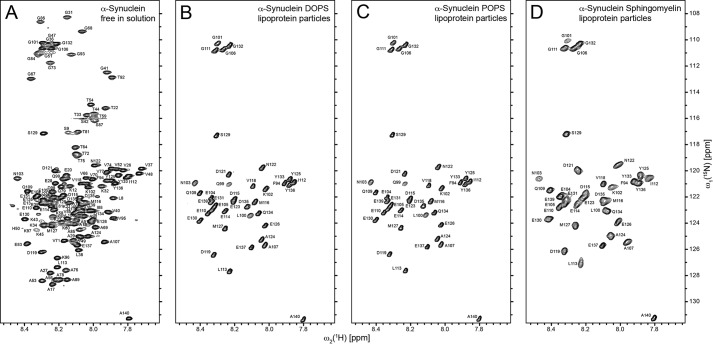
Next, we collected two-dimensional [15N,1H]-TROSY solution-state NMR spectra of 2H, 15N-labeled α-Syn DOPS lipoprotein particles and free 13C, 15N-labeled α-Syn (Fig. 5). As expected for a largely disordered protein, the two-dimensional [15N,1H]-TROSY spectrum of free α-Syn has a resonance dispersion of ∼1 ppm in the proton dimension. Although often overlapped, all expected peaks were found and assigned (Fig. 5A) in agreement with others (5, 37). Compared with that of the free protein, the spectra of α-Syn DOPS, α-Syn POPS, and α-Syn sphingomyelin lipoprotein particles showed instead cross peaks of only the ∼40 C-terminal residues (Fig. 5, B–D). Increasing the temperature by 20 °C or lowering the pH to 4.5 did not increase the number of cross peaks visible in the two-dimensional [15N,1H]-TROSY spectra of α-Syn lipoprotein particles (data not shown). These studies indicate that the ∼40 C-terminal residues are flexible and do not take part in the core structure of the α-Syn lipoprotein particle. Similar observations have been documented also for α-Syn studies in presence of SDS micelles or SUVs containing anionic phospholipids (5, 37).
FIGURE 5.
The C-terminal ∼40 residues of α-Syn are flexible in α-Syn lipoprotein particles as evidenced by two-dimensional [15N,1H]-TROSY spectra. The spectra of (A) 13C, 15N-labeled α-Syn free in solution and 2H, 15N-labeled (B) α-Syn DOPS, (C) α-Syn POPS, and (D) α-Syn sphingomyelin lipoprotein particles in a buffer containing 20 mm Bis-Tris-HCl, pH 7, 20 mm NaCl were measured at a magnetic field of 700 MHz 1H frequency at 30 °C. The cross peaks are labeled with the one-letter amino acid residue code. The slight peak doubling observed is attributed to 2H isotope effects.
In summary, taken the results from CD, MAS SS-NMR, and solution-state NMR together, residues 1–100 of WT human α-Syn adopt an α-helical structure while the ∼40 C-terminal residues remain unstructured in α-Syn lipoprotein particles.
The Protein-Lipid Composition of α-Synuclein Lipoprotein Particles
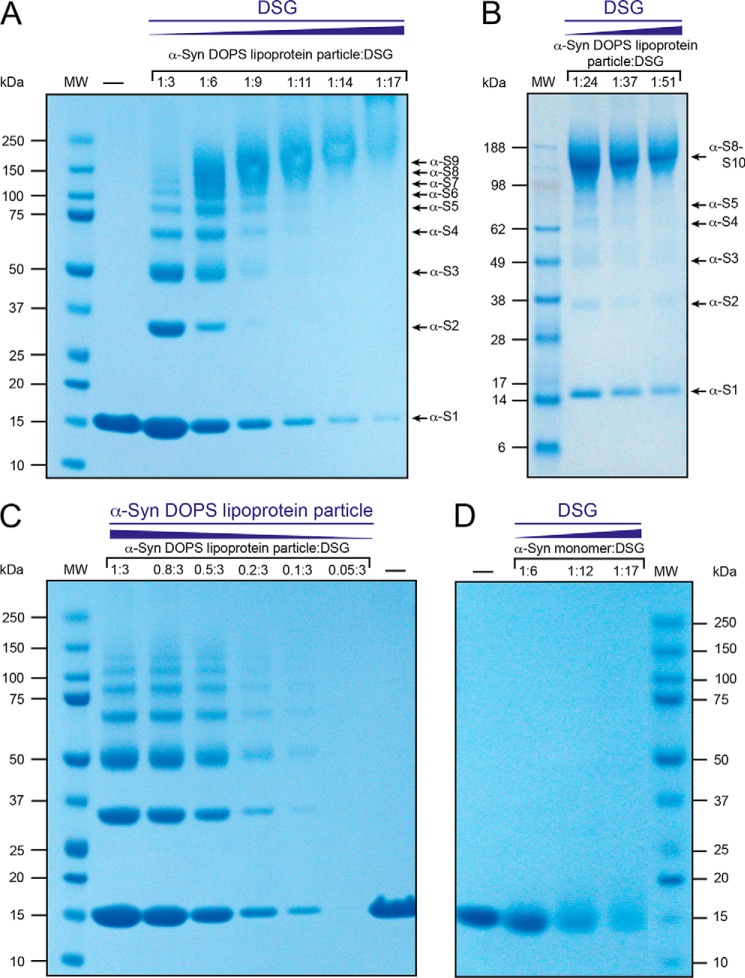
To address the composition of the membrane-bound state of α-Syn in DOPS lipoprotein particles, and in particular whether α-Syn stays monomeric in the complex or adopts a multimeric form, we performed chemical cross-linking experiments with the disuccinimidyl glutarate (DSG, spacer length 7.7 Å) linker. By doing so, we would like to note that interpretations of chemical cross-linking experiments of protein complexes have to be taken with care because they may be inherently inefficient and depend on the size of the cross-linking molecule, which can also nonspecifically link proteins to each other in a diffusion-controlled reaction at high cross-linker concentrations (28, 76, 77) or the cross-linker may covalently attach phospholipids to α-Syn (28). We treated our samples with the DSG cross-linker, because its high efficiency was reported in recent in vivo cross-linking experiments (72) of α-Syn. We observed well-resolved monomeric, dimeric, trimeric up to octa-decameric bands by SDS-PAGE (Fig. 6, A and B). At higher DSG concentration, a predominant single ∼150 kDa species was detected (Fig. 6B). Intramolecular cross-linking was investigated by varying the α-Syn DOPS lipoprotein particle concentration keeping thereby the used DSG amount constant (Fig. 6C). We observed weaker bands on SDS-PAGE gels with decreasing concentration of α-Syn DOPS lipoprotein particles (Fig. 6C) indicating that α-Syn molecules were not cross-linked between different lipoprotein particles. In the absence of DSG, α-Syn DOPS lipoprotein particles ran as a monomer on a SDS-PAGE. Importantly, free α-Syn remained monomeric in presence of different DSG cross-linker concentrations (Fig. 6D). The amino acid sequence of α-Syn contains fifteen Lys residues that are targeted by the DSG cross-linker. An increasing concentration of DSG results in a larger number of DSG-modified Lys residues and therefore in a larger molecular weight distribution of unfolded monomeric DSG-bound α-Syn that is manifested by broader and weaker bands on a SDS-PAGE (Fig. 6D). In contrast to the studies by Dettmer et al. (72), we find no evidence that unfolded monomeric α-Syn favors in vitro interactions with other α-Syn molecules forming a stable tetramer (78). These data indicate that α-Syn DOPS lipoprotein particles are composed of 8–10 α-Syn molecules.
FIGURE 6.
α-Syn DOPS lipoprotein particles comprise 8–10 copies of α-Syn as evidenced by cross-linking studies. SDS-PAGE, 4–12% NuPAGE Bis-Tris gel (Invitrogen), of cross-linked α-Syn DOPS lipoprotein particles. A, lane 1, molecular weight marker (MW, Precision Plus ProteinTM Dual Color Standard, BIORAD). Lane 2, (−) Non-cross-linked α-Syn DOPS lipoprotein particles (control). Lanes 3–8, α-Syn DOPS lipoprotein particles (final concentration 83 μm) were exposed to increasing concentrations (molar ratios) of the amine-reactive cross-linker DSG. Arrowheads indicate presumed α-Syn monomer (α-S1) and oligomers (such as α-S2 for dimeric α-Syn). B, lane 1, molecular weight marker (MW, SeeBlue plus2 prestained Standard, Invitrogen). Lanes 2–4, cross-linked α-Syn DOPS lipoprotein particles (final concentration 83 μm) with increasing concentrations of DSG as indicated. Presumed α-Syn monomer and oligomers are indicated by arrowheads. C, lane 1, MW, Precision Plus ProteinTM Dual Color Standard, BIORAD). Lanes 2–7, decreasing concentrations of α-Syn DOPS lipoprotein particles were exposed to a constant concentration of DSG (final concentration 249 μm). Lane 8, (−) non cross-linked α-Syn DOPS lipoprotein particles for control. D, approximately 9 μg monomeric α-Syn was loaded per well. Lane 1, (−) non cross-linked monomeric α-Syn control. Lanes 2–4, cross-linked monomeric α-Syn (final concentration 83 μm) with increasing concentrations of DSG. Lane 5, MW (Precision Plus ProteinTM Dual Color Standard, Biorad).
Next, one-dimensional (1D) 15N-filtered solution-state NMR diffusion experiments were used to assess the molecular weight of the α-Syn lipoprotein particles. The measured translational diffusion coefficient of 2.3.10−7 cm2 s−1 is similar for DOPS, POPS, and sphingomyelin α-Syn lipoprotein particles. From the diffusion coefficient a rough estimate for the apparent molecular weight of the α-Syn lipoprotein particles of ∼400 kDa is obtained.
An exact mass and composition of a protein-lipid entity can however only be determined by size exclusion coupled multiangle static light scattering (MALS) combined with refraction index measurements (79). In the case of α-Syn lipoprotein particles a fluorophore-labeled α-Syn(C141) variant, which was labeled at the additionally introduced C-terminal residue Cys-141 with the dye DABMI, was used for the protein detection in the MALS studies because the large molecular weight of the complex may cause potential scattering issues. By doing so we observed a molecular weight of ∼982 kDa for the total α-Syn DOPS lipoprotein particle complex, ∼865 kDa for the DOPS lipids, and ∼116 kDa for the protein component (Fig. 7). These measures indicate that α-Syn DOPS lipoprotein particles are composed of eight α-Syn and ∼1070 DOPS molecules with a protein-to-lipid mass ratio of ∼1:8 (Fig. 7). By comparison, a maximum of ∼620 lipid molecules are observed in nanodiscs assembled with DPPC and two apolipoprotein A-1 derived MSP2N2 constructs (61). The α-Syn-to-DOPS ratio is in line with a subsequent comparison of the integrated α-Syn 1HN signals (7.8 - 8.5 ppm) with the signal from the polar head methyl group of DOPS at 3.2 ppm extracted from a one-dimensional 1H NMR spectrum of α-Syn DOPS lipoprotein particles showing a protein-to-lipid ratio in the range of ∼1:10 (data not shown).
FIGURE 7.
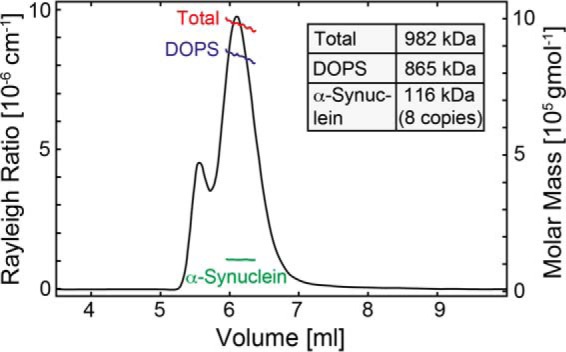
Analysis of the α-Syn and DOPS composition in α-Syn DOPS lipoprotein particles. Molecular weight analysis of the α-Syn DOPS lipoprotein complex performed by multiangle static light scattering coupled with size-exclusion gel chromatography and refractive index measurements. The black line corresponds to the static light scattering signal at 454 nm of DABMI-labeled α-Syn(C141) in the presence of DOPS lipids; red, blue, and green lines show average molar masses of the complex, the lipid component, and the protein component in the lipoprotein particle, respectively. Following these investigations, the protein mass is ∼116 kDa indicating that α-Syn is of octameric nature in DOPS lipoprotein particles.
α-Synuclein Adopts an Oligomeric State in Vivo Similar to the Lipoprotein Particles
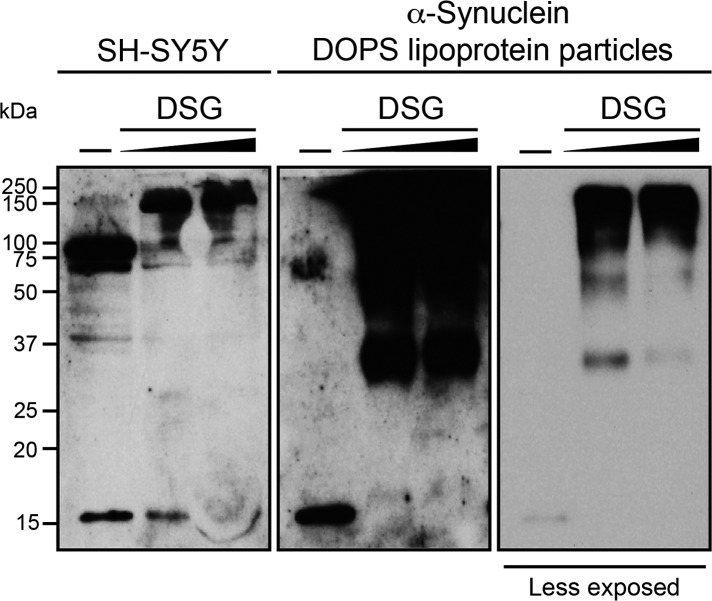
To investigate whether α-Syn lipoprotein particles may exist in a natural cellular environment, we used SH-SY5Y human neuroblastoma cells that express endogenous human α-Syn. Native assemblies of α-Syn were trapped through the cell-permeable DSG cross-linker (72). In addition to monomeric α-Syn, predominant SDS-stable high molecular weight α-Syn species of ∼60 and 90 kDa were detected in the supernatant of unpurified non-cross-linked SH-SY5Y cells (Fig. 8; left panel, first lane). The presence of SDS-stable high molecular weight α-Syn species (72) have been described before suggesting that they may be building blocks for larger α-Syn oligomers, which cause toxicity by aggregation to the cell membrane (80, 81). Upon addition of DSG cross-linker we observed a trapped high molecular weight ∼150 kDa α-Syn species in neuroblastoma cells accompanied with a decrease of the monomeric and 60–90 kDa α-Syn species (Fig. 8; left panel, second and third lane). The discrete pronounced ∼150-kDa band indicates specific interactions between multiple α-Syn monomers since a completely diffusion-controlled (nonspecific) cross linking would have resulted in diffuse continuous bands from α-Syn cross-linked to various cytosolic proteins. The observation that the size of the in vivo cross-linked α-Syn species is similar to the one of in vitro cross-linked α-Syn DOPS lipoprotein particles (Fig. 8; right panel, second and third lane) indicates that the 150-kDa species corresponds to α-Syn lipoprotein particles. In agreement with this notion, it has been demonstrated that a 150-kDa homo-multimeric α-Syn species can be present in vivo (28) suggesting that α-Syn lipoprotein particles may indeed exist in vivo. However, it cannot be excluded that the ∼150-kDa band in Fig. 8 is a lipid-free α-Syn multimer as the reported toxic α-Syn oligomers that accumulate during the process of amyloid formation (81).
FIGURE 8.
In vivo cross-linked endogenous α-Syn in SH-SY5Y human neuroblastoma cells shows a major high molecular weight species of similar size as cross-linked α-Syn DOPS lipoprotein particles. Left panel, SH-SY5Y cells were exposed to increasing concentrations of the amine-reactive cross-linker DSG: (−) non cross-linked as control (lane 1), 1 and 2 mm DSG cross-linked SH-SY5Y cells (lanes 2 and 3). Middle and right panel, (−) non cross-linked (lane 1), 1 and 2 mm DSG cross-linked α-Syn DOPS lipoprotein particles (lanes 2 and 3).
Discussion
There is a significant sequence similarity between α-Syn and apolipoproteins (58). In particular, the seven imperfect repeats in the N-terminal region of α-Syn are reminiscent of those found in the amphipathic helices of apolipoproteins (3, 6, 10, 11). Moreover, in presence of giant vesicles composed of phosphatidylglycerol at high protein-to-lipid ratios α-Syn can form lipoprotein nanoparticles (58) with a diameter of 7–10 nm having a morphology and shape similar to that of the high-density lipoproteins (HDL) and self-assembling phospholipid bilayer nanodiscs (59) formed by human apolipoproteins and membrane scaffolding proteins (MSPs), respectively. Here, we established the preparation of stable and homogeneous samples of nanoparticles composed of phosphatidylserine lipids and α-Syn using a protocol typically applied for the preparation of synthetic nanodiscs (60, 61). This method ensures a high extent of sample homogeneity allowing for a detailed characterization of the α-Syn lipoprotein particles. Multiangle as well as dynamic light scattering (MALS and DLS), electron microscopy (EM), cross-linking, and NMR have been used to determine the size, composition and morphology of these particles. They are ellipsoidal discoid-like entities with a size of ∼19–28 nm, comprising 8–10 α-Syn molecules and a protein-to-lipid ratio of 1:8 to 1:10. This data is in support with theoretical calculations following a procedure established for MSP nanodiscs (82). Using the average surface area per one DOPS lipid molecule of 65.3 Å2 (83), and a lipid cylinder of 20.8 nm, ∼9 α-Syn molecules are requested for wrapping around the lipid cylinder with helical secondary structure. The size and composition of our α-Syn lipoprotein particles differ from recently reported values of α-Syn lipoprotein nanoparticles (58). Having a similar ellipsoidal shape, the particles are approximately three times larger (∼19–28 nm compared to ∼7–10 nm) with a doubled protein-to-lipid ratio (∼1:10 compared to ∼1:20). We explain these variations by a significant different mode of formation using either the self-assembling nanodisc approach that starts with a detergent lipid mixture (61) or the extraction of lipids by α-Syn from phosphatidylglycerol vesicles into nanoparticles (58).
Circular dichroism (CD) and magic angle solid-state NMR (MAS SS-NMR) measurements confirmed the helical fold of α-Syn inside the nanoparticles while solution-state NMR determined that the ∼40 C-terminal residues remain flexible. Finally, comparing in vitro with in vivo cross-linking results it has been suggested that α-Syn lipoprotein particles may occur in vivo.
Based on the role of high-density lipoproteins (HDL) in extra cellular cholesterol storage (84) and the high concentration of α-Syn in synapse (85), it is straight forward to propose that the α-Syn lipoprotein particle may be a storage container of negatively charged lipids, that may serve as a lipid molecule source during expansion and remodeling of the synaptic membrane surface. While this hypothesis is sound, we failed so far to demonstrate that α-Syn lipoprotein particles are able to fuse with a plasma membrane mimetic through the action of the SNARE complex assembly together with synaptobrevin (data not shown) (28).
In conclusion, our presented data describe a method to prepare defined nanometer-sized lipid particles of α-Syn and anionic phospholipids. In vivo cross-linked multimeric α-Syn appeared to be similar in molecular weight as in vitro cross-linked α-Syn lipoprotein particles suggesting that they may be of functional relevance. In our view, α-Syn lipoprotein particles may serve as a lipid molecule source during expansion and remodeling of the synaptic membrane surface, a hypothesis that we will further pursue.
Author Contributions
C. E. produced α-Synuclein protein samples, prepared α-Synuclein lipoprotein particles, performed and analyzed CD, DLS, negative-stain EM, solution-state NMR, and cross-linking experiments. S. C. produced α-Synuclein protein samples, analyzed CD and DLS data. J. K. and H. S. collected and analyzed EM data. I. M. performed MALS and analyzed the data. J. G. performed immunoblotting of cross-linked endogenous α-Synuclein expressed in SH-SY5Y human neuroblastoma cells. X. L. and J. R. designed and performed lipid fusion experiments. J. V. and B. H. M. collected solid-state NMR spectra of α-Synuclein lipoprotein particles. N. N. provided technical assistance in negative-stain EM. C. E., S. C., J. K., I. M., J. G., J. V., S. C., B. H. M., P. P., J. R., H. S., and R. R. contributed to scientific discussions and prepared the manuscript.
This work was supported by a Swiss National Foundation Sinergia grant. The authors declare that they have no conflicts of interest with the contents of this article.
- α-Syn
- α-Synuclein
- SUV
- small unilamellar vesicle
- MSP
- membrane scaffolding protein
- CD
- circular dichroism
- SEC
- size exclusion chromatography
- MAS SS-NMR
- magic angle solid-state NMR
- MALS
- multiangle static light scattering
- DLS
- dynamic light scattering
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phospho-l-serine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- DSG
- disuccinimidyl glutarate
- DABMI
- 4-dimethylaminophenylazophenyl-4′maleimide.
References
- 1. Ulusoy A., and Di Monte D. A. (2013) α-Synuclein elevation in human neurodegenerative diseases: Experimental, pathogenetic, and therapeutic implications. Mol. Neurobiol. 47, 484–494 [DOI] [PubMed] [Google Scholar]
- 2. Spillantini M. G., and Goedert M. (2000) The α-Synucleinopathies: Parkinson's Disease, Dementia with Lewy Bodies, and Multiple System Atrophy. Ann. N.Y. Acad. Sci. 920, 16–27 [DOI] [PubMed] [Google Scholar]
- 3. Weinreb P. H., Zhen W., Poon A. W., Conway K. A., and Lansbury P. T. (1996) NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry 35, 13709–13715 [DOI] [PubMed] [Google Scholar]
- 4. Uversky V. N. (2003) A protein-chameleon: conformational plasticity of α-synuclein, a disordered protein involved in neurodegenerative disorders. J. Biomol. Struct. Dyn. 21, 211–234 [DOI] [PubMed] [Google Scholar]
- 5. Eliezer D., Kutluay E., Bussell R. Jr., and Browne G. (2001) Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 307, 1061–1073 [DOI] [PubMed] [Google Scholar]
- 6. Davidson W. S., Jonas A., Clayton D. F., and George J. M. (1998) Stabilization of α-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 273, 9443–9449 [DOI] [PubMed] [Google Scholar]
- 7. Heise H., Hoyer W., Becker S., Andronesi O. C., Riedel D., and Baldus M. (2005) Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 102, 15871–15876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vilar M., Chou H.-T., Lührs T., Maji S. K., Riek-Loher D., Verel R., Manning G., Stahlberg H., and Riek R. (2008) The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. 105, 8637–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gath J., Bousset L., Habenstein B., Melki R., Böckmann A., and Meier B. H. (2014) Unlike Twins: An NMR Comparison of Two α-Synuclein Polymorphs Featuring Different Toxicity. PloS One 9, e90659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. George J. M., Jin H., Woods W. S., and Clayton D. F. (1995) Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron 15, 361–372 [DOI] [PubMed] [Google Scholar]
- 11. Bussell R. Jr., and Eliezer D. (2003) A structural and functional role for 11-mer repeats in α-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 329, 763–778 [DOI] [PubMed] [Google Scholar]
- 12. Murphy D. D., Rueter S. M., Trojanowski J. Q., and Lee V. M.-Y. (2000) Synucleins are developmentally expressed, and α-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J. Neurosci. 20, 3214–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cabin D. E., Shimazu K., Murphy D., Cole N. B., Gottschalk W., McIlwain K. L., Orrison B., Chen A., Ellis C. E., Paylor R., Lu B., and Nussbaum R. L. (2002) Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking α-synuclein. J. Neurosci. 22, 8797–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abeliovich A., Schmitz Y., Fariñas I., Choi-Lundberg D., Ho W.-H., Castillo P. E., Shinsky N., Verdugo J. M. G., Armanini M., Ryan A., Hynes M., Phillips H., Sulzer D., and Rosenthal A. (2000) Mice Lacking α-Synuclein Display Functional Deficits in the Nigrostriatal Dopamine System. Neuron 25, 239–252 [DOI] [PubMed] [Google Scholar]
- 15. Nemani V. M., Lu W., Berge V., Nakamura K., Onoa B., Lee M. K., Chaudhry F. A., Nicoll R. A., and Edwards R. H. (2010) Increased expression of α-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron 65, 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Halliday G. M., Ophof A., Broe M., Jensen P. H., Kettle E., Fedorow H., Cartwright M. I., Griffiths F. M., Shepherd C. E., and Double K. L. (2005) α-Synuclein redistributes to neuromelanin lipid in the substantia nigra early in Parkinson's disease. Brain 128, 2654–2664 [DOI] [PubMed] [Google Scholar]
- 17. Golovko M. Y., Barceló-Coblijn G., Castagnet P. I., Austin S., Combs C. K., and Murphy E. J. (2009) The role of α-synuclein in brain lipid metabolism: a downstream impact on brain inflammatory response. Mol. Cell Biochem. 326, 55–66 [DOI] [PubMed] [Google Scholar]
- 18. Sharon R., Goldberg M. S., Bar-Josef I., Betensky R. A., Shen J., and Selkoe D. J. (2001) α-Synuclein occurs in lipid-rich high molecular weight complexes, binds fatty acids, and shows homology to the fatty acid-binding proteins. Proc. Natl. Acad. Sci. 98, 9110–9115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharon R., Bar-Joseph I., Mirick G. E., Serhan C. N., and Selkoe D. J. (2003) Altered fatty acid composition of dopaminergic neurons expressing α-synuclein and human brains with α-synucleinopathies. J. Biol. Chem. 278, 49874–49881 [DOI] [PubMed] [Google Scholar]
- 20. Barceló-Coblijn G., Golovko M. Y., Weinhofer I., Berger J., and Murphy E. J. (2007) Brain neutral lipids mass is increased in α-synuclein gene-ablated mice. J. Neurochem. 101, 132–141 [DOI] [PubMed] [Google Scholar]
- 21. Cooper A. A., Gitler A. D., Cashikar A., Haynes C. M., Hill K. J., Bhullar B., Liu K., Xu K., Strathearn K. E., Liu F., Cao S, Caldwell K. A., Caldwell G. A., Marsischky G., Kolodner R. D., Labaer J., Rochet J. C., Bonini N. M., and Lindquist S. (2006) α-Synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science 313, 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gitler A. D., Bevis B. J., Shorter J., Strathearn K. E., Hamamichi S., Su L. J., Caldwell K. A., Caldwell G. A., Rochet J.-C., McCaffery J. M., Barlowe C., and Lindquist S. (2008) The Parkinson's disease protein α-synuclein disrupts cellular Rab homeostasis. Proc. Natl. Acad. Sci. 105, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee H. J., Kang S. J., Lee K., and Im H. (2011) Human α-synuclein modulates vesicle trafficking through its interaction with prenylated Rab acceptor protein 1. Biochem. Biophys. Res. Commun. 412, 526–531 [DOI] [PubMed] [Google Scholar]
- 24. Clayton D. F., and George J. M. (1998) The synucleins: a family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci. 21, 249–254 [DOI] [PubMed] [Google Scholar]
- 25. Hartman V. N., Miller M. A., Clayton D. F., Liu W.-C., Kroodsma D. E., and Brenowitz E. A. (2001) Testosterone regulates α-synuclein mRNA in the avian song system. Neuroreport 12, 943–946 [DOI] [PubMed] [Google Scholar]
- 26. Chandra S., Gallardo G., Fernández-Chacón R., Schlüter O. M., and Südhof T. C. (2005) α-Synuclein cooperates with CSPα in preventing neurodegeneration. Cell 123, 383–396 [DOI] [PubMed] [Google Scholar]
- 27. Burré J., Sharma M., Tsetsenis T., Buchman V., Etherton M. R., and Südhof T. C. (2010) α-Synuclein promotes SNARE-Complex assembly in vivo and in vitro. Science 329, 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burré J., Sharma M., and Südhof T. C. (2014) α-Synuclein assembles into higher-order multimers upon membrane binding to promote SNARE complex formation. Proc. Natl. Acad. Sci. U.S.A. 111, E4274–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lai Y., Kim S., Varkey J., Lou X., Song J.-K., Diao J., Langen R., and Shin Y.-K. (2014) Nonaggregated α-synuclein influences SNARE-dependent vesicle docking via membrane binding. Biochemistry 53, 3889–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu M., and Fink A. L. (2003) Lipid binding inhibits α-synuclein fibril formation. J. Biol. Chem. 278, 16873–16877 [DOI] [PubMed] [Google Scholar]
- 31. Necula M., Chirita C. N., and Kuret J. (2003) Rapid anionic micelle-mediated α-synuclein fibrillization in vitro. J. Biol. Chem. 278, 46674–46680 [DOI] [PubMed] [Google Scholar]
- 32. Martinez Z., Zhu M., Han S., and Fink A. L. (2007) GM1 specifically interacts with α-synuclein and inhibits fibrillation. Biochemistry 46, 1868–1877 [DOI] [PubMed] [Google Scholar]
- 33. Haque F., Pandey A. P., Cambrea L. R., Rochet J.-C., and Hovis J. S. (2010) Adsorption of α-synuclein on lipid bilayers: Modulating the structure and stability of protein assemblies. J. Phys. Chem. B. 114, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reynolds N. P., Soragni A., Rabe M., Verdes D., Liverani E., Handschin S., Riek R., and Seeger S. (2011) Mechanism of membrane interaction and disruption by α-synuclein. J. Am. Chem. Soc. 133, 19366–19375 [DOI] [PubMed] [Google Scholar]
- 35. Jo E., McLaurin J., Yip C. M., George-Hyslop P. S., and Fraser P. E. (2000) α-Synuclein membrane interactions and lipid specificity. J. Biol. Chem. 275, 34328–34334 [DOI] [PubMed] [Google Scholar]
- 36. Jao C. C., Der-Sarkissian A., Chen J., and Langen R. (2004) Structure of membrane-bound α-synuclein studied by site-directed spin labeling. Proc. Natl. Acad. Sci. U.S.A. 101, 8331–8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chandra S., Chen X., Rizo J., Jahn R., and Südhof T. C. (2003) A broken α-helix in folded α-synuclein. J. Biol. Chem. 278, 15313–15318 [DOI] [PubMed] [Google Scholar]
- 38. Ulmer T. S., Bax A., Cole N. B., and Nussbaum R. L. (2005) Structure and dynamics of micelle-bound human α-synuclein. J. Biol. Chem. 280, 9595–9603 [DOI] [PubMed] [Google Scholar]
- 39. Bisaglia M., Tessari I., Pinato L., Bellanda M., Giraudo S., Fasano M., Bergantino E., Bubacco L., and Mammi S. (2005) A topological model of the interaction between α-synuclein and sodium dodecyl sulfate micelles. Biochemistry 44, 329–339 [DOI] [PubMed] [Google Scholar]
- 40. Bussell R. Jr., Ramlall T. F., and Eliezer D. (2005) Helix periodicity, topology, and dynamics of membrane-associated α-Synuclein. Protein Sci. 14, 862–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jao C. C., Hegde B. G., Chen J., Haworth I. S., and Langen R. (2008) Structure of membrane-bound α-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. 105, 19666–19671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trexler A. J., and Rhoades E. (2009) α-Synuclein binds large unilamellar vesicles as an extended helix†. Biochemistry 48, 2304–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Drescher M., Veldhuis G., van Rooijen B. D., Milikisyants S., Subramaniam V., and Huber M. (2008) Antiparallel arrangement of the helices of vesicle-bound α-synuclein. J. Am. Chem. Soc. 130, 7796–7797 [DOI] [PubMed] [Google Scholar]
- 44. Bortolus M., Tombolato F., Tessari I., Bisaglia M., Mammi S., Bubacco L., Ferrarini A., and Maniero A. L. (2008) Broken helix in vesicle and micelle-bound α-synuclein: Insights from site-directed spin labeling-EPR experiments and MD simulations. J. Am. Chem. Soc. 130, 6690–6691 [DOI] [PubMed] [Google Scholar]
- 45. Bodner C. R., Dobson C. M., and Bax A. (2009) Multiple tight phospholipid-binding modes of α-synuclein revealed by solution NMR spectroscopy. J. Mol. Biol. 390, 775–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Högen T., Levin J., Schmidt F., Caruana M., Vassallo N., Kretzschmar H., Bötzel K., Kamp F., and Giese A. (2012) Two different binding modes of α-synuclein to lipid vesicles depending on its aggregation state. Biophys. J. 102, 1646–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Georgieva E. R., Ramlall T. F., Borbat P. P., Freed J. H., and Eliezer D. (2008) Membrane-bound α-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J. Am. Chem. Soc. 130, 12856–12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferreon A. C. M., Gambin Y., Lemke E. A., and Deniz A. A. (2009) Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. 106, 5645–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robotta M., Braun P., van Rooijen B., Subramaniam V., Huber M., and Drescher M. (2011) Direct evidence of coexisting horseshoe and extended helix conformations of membrane-bound α-Synuclein. ChemPhysChem. 12, 267–269 [DOI] [PubMed] [Google Scholar]
- 50. Lokappa S. B., and Ulmer T. S. (2011) α-Synuclein populates both elongated and broken helix states on small unilamellar vesicles. J. Biol. Chem. 286, 21450–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Braun A. R., Sevcsik E., Chin P., Rhoades E., Tristram-Nagle S., and Sachs J. N. (2012) α-Synuclein induces both positive mean curvature and negative Gaussian curvature in membranes. J. Am. Chem. Soc. 134, 2613–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pfefferkorn C. M., Jiang Z., and Lee J. C. (2012) Biophysics of α-synuclein membrane interactions. Biochim. Biophys. Acta BBA-Biomembr. 1818, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ouberai M. M., Wang J., Swann M. J., Galvagnion C., Guilliams T., Dobson C. M., and Welland M. E. (2013) α-Synuclein senses lipid packing defects and induces lateral expansion of lipids leading to membrane remodeling. J. Biol. Chem. 288, 20883–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Varkey J., Isas J. M., Mizuno N., Jensen M. B., Bhatia V. K., Jao C. C., Petrlova J., Voss J. C., Stamou D. G., Steven A. C., and Langen R. (2010) Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. J. Biol. Chem. 285, 32486–32493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pandey A. P., Haque F., Rochet J.-C., and Hovis J. S. (2011) α-Synuclein-induced tubule formation in lipid bilayers. J. Phys. Chem. B. 115, 5886–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mizuno N., Varkey J., Kegulian N. C., Hegde B. G., Cheng N., Langen R., and Steven A. C. (2012) Remodeling of lipid vesicles into cylindrical micelles by α-synuclein in an extended α-helical conformation. J. Biol. Chem. 287, 29301–29311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Westphal C. H., and Chandra S. S. (2013) Monomeric synucleins generate membrane curvature. J. Biol. Chem. 288, 1829–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Varkey J., Mizuno N., Hegde B. G., Cheng N., Steven A. C., and Langen R. (2013) α-Synuclein oligomers with broken helical conformation form lipoprotein nanoparticles. J. Biol. Chem. 288, 17620–17630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bayburt T. H., Grinkova Y. V., and Sligar S. G. (2002) Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2, 853–856 [Google Scholar]
- 60. Nath A., Atkins W. M., and Sligar S. G. (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 61. Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., and Sligar S. G. (2009) Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Campioni S., Carret G., Jordens S., Nicoud L., Mezzenga R., and Riek R. (2014) The Presence of an Air–Water Interface Affects Formation and Elongation of α-Synuclein Fibrils. J. Am. Chem. Soc. 136, 2866–2875 [DOI] [PubMed] [Google Scholar]
- 63. Sambrook J. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press [Google Scholar]
- 64. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., and Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 65. Pervushin K., Riek R., Wider G., and Wüthrich K. (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Salzmann M., Wider G., Pervushin K., Senn H., and Wüthrich K. (1999) TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J. Am. Chem. Soc. 121, 844–848 [Google Scholar]
- 67. Bayrhuber M., and Riek R. (2011) Very simple combination of TROSY, CRINEPT and multiple quantum coherence for signal enhancement in an HN(CO)CA experiment for large proteins. J. Magn. Reson. 209, 310–314 [DOI] [PubMed] [Google Scholar]
- 68. Cavanagh J., Fairbrother W. J. III, A. G. P., Skelton N. J., and Rance M. (2010) Protein NMR Spectroscopy: Principles and Practice, Academic Press [Google Scholar]
- 69. Güntert P., Dötsch V., Wider G., and Wüthrich K. (1992) Processing of multi-dimensional NMR data with the new software PROSA. J. Biomol. NMR. 2, 619–629 [Google Scholar]
- 70. Bartels C., Xia T. H., Billeter M., Güntert P., and Wüthrich K. (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 6, 1–10 [DOI] [PubMed] [Google Scholar]
- 71. Böckmann A., Gardiennet C., Verel R., Hunkeler A., Loquet A., Pintacuda G., Emsley L., Meier B. H., and Lesage A. (2009) Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR. 45, 319–327 [DOI] [PubMed] [Google Scholar]
- 72. Dettmer U., Newman A. J., Luth E. S., Bartels T., and Selkoe D. (2013) In vivo cross-linking reveals principally oligomeric forms of α-Synuclein and β-Synuclein in neurons and non-neural cells. J. Biol. Chem. 288, 6371–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., and Segrest J. P. (2010) Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285, 4652–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mishra V. K., Palgunachari M. N., Datta G., Phillips M. C., Lund-Katz S., Adeyeye S. O., Segrest J. P., and Anantharamaiah G. M. (1998) Studies of synthetic peptides of human apolipoprotein A-I containing tandem amphipathic α-helixes. Biochemistry 37, 10313–10324 [DOI] [PubMed] [Google Scholar]
- 75. Wishart D. S., and Sykes B. D. (1994) The 13C Chemical-Shift Index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 76. Melcher K. (2004) New chemical crosslinking methods for the identification of transient protein-protein interactions with multiprotein complexes. Curr. Protein Pept. Sci. 5, 287–296 [DOI] [PubMed] [Google Scholar]
- 77. Migneault I., Dartiguenave C., Bertrand M. J., and Waldron K. C. (2004) Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTechniques 37, 790–796, 798–802 [DOI] [PubMed] [Google Scholar]
- 78. Bartels T., Choi J. G., and Selkoe D. J. (2011) α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477, 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Maslennikov I., Krupa M., Dickson C., Esquivies L., Blain K., Kefala G., Choe S., and Kwiatkowski W. (2009) Characterization of protein detergent complexes by NMR, light scattering, and analytical ultracentrifugation. J. Struct. Funct. Genomics 10, 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Winner B., Jappelli R., Maji S. K., Desplats P. A., Boyer L., Aigner S., Hetzer C., Loher T., Vilar M., Campioni S., Tzitzilonis C., Soragni A., Jessberger S., Mira H., Consiglio A., Pham E., Masliah E., Gage F. H., and Riek R. (2011) In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. U.S.A. 108, 4194–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen S. W., Drakulic S., Deas E., Ouberai M., Aprile F. A., Arranz R., Ness S., Roodveldt C., Guilliams T., De-Genst E. J., Klenerman D., Wood N. W., Knowles T. P. J., Alfonso C., Rivas G., Abramov A. Y., Valpuesta J. M., Dobson C. M., and Cremades N. (2015) Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc. Natl. Acad. Sci. U.S.A. 112, E1994–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Denisov I. G., Grinkova Y. V., Lazarides A. A., and Sligar S. G. (2004) Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 83. Petrache H. I., Tristram-Nagle S., Gawrisch K., Harries D., Parsegian V. A., and Nagle J. F. (2004) Structure and fluctuations of charged phosphatidylserine bilayers in the absence of salt. Biophys. J. 86, 1574–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Navab M., Reddy S. T., Van Lenten B. J., and Fogelman A. M. (2011) HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat. Rev. Cardiol. 8, 222–232 [DOI] [PubMed] [Google Scholar]
- 85. Lashuel H. A., Overk C. R., Oueslati A., and Masliah E. (2013) The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 14, 38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]