Abstract
We performed a fragment screen on the dengue virus serotype 3 RNA-dependent RNA polymerase using x-ray crystallography. A screen of 1,400 fragments in pools of eight identified a single hit that bound in a novel pocket in the protein. This pocket is located in the polymerase palm subdomain and conserved across the four serotypes of dengue virus. The compound binds to the polymerase in solution as evidenced by surface plasmon resonance and isothermal titration calorimetry analyses. Related compounds where a phenyl is replaced by a thiophene show higher affinity binding, indicating the potential for rational design. Importantly, inhibition of enzyme activity correlated with the binding affinity, showing that the pocket is functionally important for polymerase activity. This fragment is an excellent starting point for optimization through rational structure-based design.
Keywords: crystallography, dengue virus (DENV), enzyme inhibitor, flavivirus, inhibitor, RNA polymerase, viral polymerase
Introduction
Dengue virus (DENV)3 is the most widespread mosquito-borne viral infection. Disease symptoms of DENV-infected patients range from a mild fever to severe plasma leakage and hemorrhagic shock (1). There are four serotypes of DENV (DENV-1 to -4) that concurrently circulate around the world in tropical and subtropical regions. The vast majority of clinical cases are not reported, and there are estimated to be approximately 390 million human cases of dengue worldwide per year (2). There is currently no licensed vaccine or antiviral to treat DENV infection, underlining the urgency for the development of safe therapeutics (3). DENV is a member of the Flavivirus genus that also includes other viruses that are pathogenic to humans such as West Nile virus (WNV), yellow fever virus, Japanese encephalitis virus (JEV), and tick-borne encephalitis virus.
One of the most attractive antiviral targets is the DENV RNA-dependent RNA polymerase (RdRp) because (i) viral polymerases are clinically proven therapeutic targets and (ii) the RdRp is the most conserved viral protein among the four serotypes of DENV so that the likelihood of a single compound with pan-serotype activity is higher than compounds targeting other viral proteins. The DENV RdRp activity resides in the C-terminal two-thirds of the viral nonstructural protein 5 (NS5) (reviewed in Ref. 4), whereas the N-terminal one-third of DENV NS5 encodes a methyltransferase (5).
Crystal structures of the RdRp domain and full-length NS5 have been determined (6, 7). The overall architecture of the polymerase resembles a right hand with fingers, palm, and thumb subdomains. DENV RdRp catalyzes de novo initiation, as well as elongation. Like other RdRps that perform de novo RNA synthesis, DENV RdRp has a fully encircled active site (8). These polymerases undergo a conformational change from a closed to an open conformation during the transition from de novo initiation to elongation. During this process, the initiation loop (also known as the priming loop) is thought to move out of the active site, in a manner similar to the hepatitis C virus polymerase (9).
A number of in vitro enzyme assays have been developed for DENV RdRp. These assays have been subjected to several high throughput screening campaigns (10–12). However, relatively few, if any, compounds have been demonstrated to bind to the RdRp protein. Two compound classes have been shown to target the RNA template entry tunnel of the DENV polymerase (11, 13). The interaction site for the first compound class was identified using UV-cross-linking and mapped to Met343 (11). For the second compound class, the binding site was defined by a co-crystal structure; the crystal structure showed that the compound forms a dimer and interacts with the large RNA tunnel of the protein, making lead optimization with this compound series challenging (13).
In addition to enzyme activity-based screening, fragment-based rational design can be an alternative approach to target the DENV RdRp. Here we report crystallography-based fragment screening to identify a novel pocket in the DENV polymerase that can be specifically targeted by small molecule inhibitors. The fragments bind to the new RdRp pocket with high ligand efficiency. Furthermore, analogs of the fragment inhibitors showed a correlation between binding affinity in solution, inhibition of enzyme activity, and binding by x-ray crystallography. The new RdRp pocket, the fragment inhibitors, the RdRp compound crystal structure, and the biophysical binding assays provide a solid foundation for rational drug discovery.
Experimental Procedures
Protein Expression in Escherichia coli and Purification
DENV-3 RdRp (residues 272–900 of NS5; GenBankTM accession number AY662691) was expressed and purified as described previously (13). DENV-4 NS5 was expressed and purified as described previously (10). DENV-3 and -4 RdRp constructs for surface plasmon resonance (SPR) analyses comprised N-terminal AviTag sequences (14) linked to residues 263–900 (DENV-3) or residues 264–900 (DENV-4) via PCR (primers are listed in supplemental Table S1) and cloned into the vector pET28a bearing a PreScission protease cleavage site (15) inserted downstream of the vector N-terminal His tag sequence. Expression plasmids for AviTag DENV3 or DENV4 RdRp proteins were co-transformed into BL21 cells with an expression plasmid for biotin protein ligase, BirA of E. coli, and proteins were purified as described previously (13). Biotinylation levels of AVI-tagged DENV RdRp proteins were determined with the biotin quantification kit (Pierce).
Compound Synthesis
The synthesis of PC-79-SH52 and FD-83-KI26 is summarized in supplemental Fig. S1. The Ardnt-Eistert reaction of 3-bromo-4-methoxybenzoic acid using trimethylsilyldiazomethane afforded the one carbon homologated ester. Subsequent palladium-catalyzed cross coupling with 2- and 3-thiophene boronic acids, followed by alkaline hydrolysis of the ester, provided the desired compounds.
Crystallization, X-ray Structure Determination, and Fragment Screening
DENV-3 RdRp was crystallized as described previously (13). Crystals were soaked by adding 5 μl of the reservoir solution, supplemented with 10% DMSO/compound mixture directly to the crystallization drop and allowed to equilibrate for 4–6 h. For fragment screening, mixtures of eight compounds from a diverse collection of 1408 Novartis proprietary compounds with an average molecular mass of 210 Da were prepared from 50 mm solutions in DMSO, giving a final concentration of 6.25 mm per compound in the mixture. For soaking with single compounds, compounds were added from 50 mm solutions in DMSO. For cryoprotection, the crystals were transferred to the crystallization solution supplemented with 10% glycerol and 10% compound/DMSO.
Diffraction data were integrated using MOSFLM and scaled using SCALA, part of the CCP4 suite (16). The structures were refined using BUSTER, part of the global phasing suite, starting from the deposited DENV-3 RdRp structure (Protein Data Bank code 4HHJ) (13).
De Novo Initiation/Elongation Assay
The assay was performed as described previously, using full-length DENV-4 NS5 (12), except that compounds were tested in a 3-fold dilution from a top concentration of 1 mm. NTPs were included at 20 μm A/G/UTP and 5 μm ATTO-CTP. The RNA substrate (at 50 nm) was the DENV-4 5′-UTR-capsid (representing the first 166 nucleotides of viral genomic RNA) linked via a 19-nucleotide nonviral sequence followed by the 3′-UTR (nucleotides 10,264–10,653), as previously described (12). Error bars are standard deviations from three independent experiments.
Surface Plasmon Resonance
The experiments were performed using a Biacore T200 instrument. Approximately 5500 response units of biotinylated DENV-3 and -4 RdRp were captured in separate flow cells in 50 mm Tris, pH 7.5, 200 mm NaCl, 2 mm DTT, 0.05% Tween 20, and 3% DMSO at 4 °C. Flow cell 1 was left blank to serve as a reference. The compounds were tested in a 7-point 2-fold serial dilution, typically from 1,000 μm, and a zero concentration sample was subtracted from each run. Compounds were typically injected at a flow rate of 30 μl/min for 45 s with 60 s pf dissociation, starting from the 0 to the highest concentration. The data were analyzed using Biacore T200 evaluation software, version 2.0. For equilibrium analysis, the responses at steady state were plotted against analyte concentration and fitted to a simple 1:1 binding model. Ligand efficiency (LE) was calculated using the following equation: LE = 1.4(−logKd)/N, where N is the number of non-hydrogen atoms, and Kd is the equilibrium dissociation constant, determined using SPR. The error bars are standard deviations from three independent experiments.
Isothermal Titration Calorimetry
Experiments were performed using an iTC200 (GE Healthcare). DENV-4 RdRp (residues 274–900) in 20 mm HEPES, pH 7.0, 300 mm NaCl, and 2 mm TCEP was loaded into the cell, typically at 50 μm. The ligand was diluted from a 50 mm stock in DMSO into the same buffer (typically at 2.5 mm), and an equal concentration of DMSO was added to the RdRp sample. Experiments were run at 25 °C with the reference power set to 10. An initial injection of 0.2 μl was followed by 15 injections of 2.6 μl. A reference titration for the compound injected at the same concentration into buffer was subtracted. The injection peaks were integrated using Origin and fitted to a 1:1 binding isotherm. The error bars are standard deviations from three independent experiments.
Results
Identification of a Novel Ligand-binding Pocket in the DENV-3 Polymerase
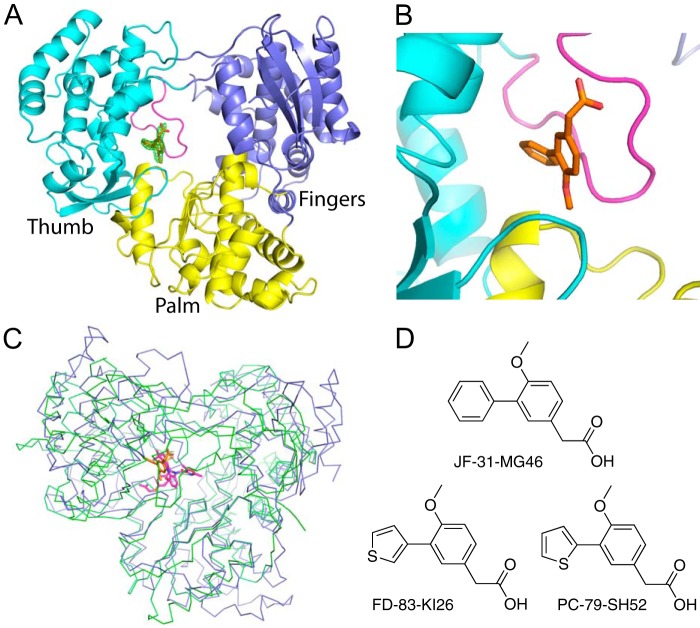
To identify inhibitors of the DENV RdRp, we performed a fragment screen of the DENV-3 RdRp using x-ray crystallography. A total of 1,408 compounds from a Novartis proprietary library, with an average molecular mass of 210 Da were screened in pools of 8, totaling 176 separate pools. RdRp crystals were soaked in drops containing compound mixtures for 6 h and then frozen in liquid nitrogen. Useful x-ray diffraction data were collected for 145 of176 pools (with resolution better than 2.5 Å). Pools were combined from 50 mm solutions in DMSO, and crystals were soaked with 10% compound/DMSO, giving a final concentration per compound of 625 μm. Only one data set, which was solved to 2.0 Å resolution from pool number 167, showed difference density that could be interpreted as a single contiguous compound. Models for the 8 possible fragments from the pool were built into the density, and only compound JF-31-MG46 fitted the data satisfactorily. To confirm that the single compound bound in the pocket, the DENV-3 RdRp was co-crystallized with the pure compound, and the structure was solved to 2.05 Å (Fig. 1 and Table 1). The co-crystal structure showed that the single compound bound at the same site and orientation with full occupancy. The difference density with the compound removed from the final round of refinement is shown in Fig. 1A. The compound bound in a novel site in the DENV-3 RdRp. Specifically, it bound between the thumb and palm subdomains of the RdRp and the priming loop. This site is similar to the palm I site in hepatitis C virus (17) (Fig. 1C).
FIGURE 1.
Crystal structure of the DENV-3 polymerase bound to JF-31-MG46. A, the structure of the DENV-3 polymerase is shown as a cartoon, with the palm, fingers, and thumb subdomains colored yellow, blue, and cyan, respectively, and the priming loop colored magenta. The compound, JF-31-MG46, is shown as orange sticks. The difference density, where the compound is removed from the final round of refinement is shown as a green mesh contoured at 3 σ. B, alternative view, colored as in A showing the compound binds between the priming loop and the thumb and palm subdomains. C, alignment of the DENV-3 polymerase shown as blue ribbons with the hepatitis C virus polymerase (Protein Data Bank code 3HKY) bound to a palm site I inhibitor, shown as green ribbons. The compounds bound to the DENV and hepatitis C virus RdRps are shown as sticks colored orange and magenta, respectively. D, the chemical structures of the compounds used in these experiments.
TABLE 1.
Data collection and refinement statistics
Statistics for the highest resolution shell are shown in parentheses.
| Compound | JF-31-MG46 | PC-79-SH52 | FD-83-KI26 |
|---|---|---|---|
| Wavelength (Å) | 1.0 | 1.0 | 1.0 |
| Resolution range (Å) | 40.4–2.05 (2.123–2.05) | 29.28–2.0 (2.071–2.0) | 40–2.0 (2.071–2.0) |
| Space group | C 2 2 21 | C 2 2 21 | C 2 2 21 |
| Unit cell | 161.58, 177.09, 57.88, 90, 90, 90 | 161.3, 176.22, 57.84, 90, 90, 90 | 161.7, 178.33, 57.93, 90, 90, 90 |
| Total reflections | 103,529 (10,024) | 106,090 (9,406) | 108,118 (9,647) |
| Unique reflections | 52,275 (5,106) | 55,362 (5,271) | 56,680 (5,465) |
| Multiplicity | 2.0 (2.0) | 1.9 (1.8) | 1.9 (1.8) |
| Completeness (%) | 99.42 (98.82) | 98.62 (94.94) | 99.35 (96.85) |
| Mean I/σ(I) | 9.89 (1.81) | 10.70 (1.79) | 10.89 (1.96) |
| Wilson B-factor | 29.14 | 29.00 | 27.36 |
| Rmerge | 0.05843 (0.4776) | 0.04979 (0.4321) | 0.0513 (0.4516) |
| Rmeas | 0.08263 | 0.07041 | 0.07254 |
| CC1/2 | 0.997 (0.701) | 0.997 (0.714) | 0.997 (0.671) |
| CC* | 0.999 (0.908) | 0.999 (0.913) | 0.999 (0.896) |
| Rwork | 0.1928 (0.3066) | 0.1881 (0.2996) | 0.1883 (0.2891) |
| Rfree | 0.2307 (0.3594) | 0.2247 (0.3441) | 0.2285 (0.3249) |
| Number of non-hydrogen atoms | 5,175 | 5,236 | 5,182 |
| Macromolecules | 4,780 | 4,780 | 4,780 |
| Ligands | 20 | 19 | 19 |
| Water | 375 | 437 | 383 |
| Protein residues | 585 | 585 | 585 |
| RMS bonds | 0.013 | 0.014 | 0.014 |
| RMS angles | 1.67 | 1.71 | 1.69 |
| Ramachandran favored (%) | 97 | 96 | 97 |
| Ramachandran outliers (%) | 0.17 | 0.52 | 0.17 |
| Clashscore | 1.79 | 1.37 | 1.48 |
| Average B-factor | 41.80 | 41.10 | 39.70 |
| Macromolecules | 41.50 | 40.60 | 39.40 |
| Ligands | 40.80 | 31.90 | 30.90 |
| Solvent | 45.70 | 47.00 | 43.50 |
| Protein Data Bank code | 5F3T | 5F3Z | 5F41 |
The DENV RdRp Palm Site Is Conserved across the Four Serotypes
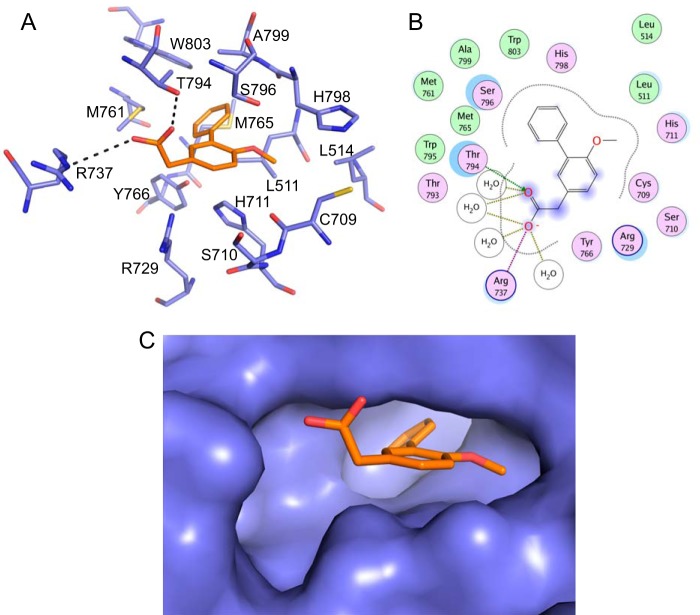
An analysis of the ligand interaction map (Fig. 2) showed that the compound directly contacts a number of residues from the RdRp. The carboxylic acid interacts with the side chain OH of Thr794 and forms a salt bridge with the side chain of Arg737. It is also in close proximity to Arg729 and makes water-mediated interactions with Arg737, the carbonyl of Thr793, and the amide NH of Trp795. The methoxy group of the compound sits in a small lipophilic pocket created by Cys709, Leu514, His798, and Leu511. The core phenyl sits between Ser796 on one side and Tyr766 and His711 on the other. The pendent phenyl sits at the back of the pocket that is formed by several hydrophobic residues, including Trp803, Met761, Met765, Ala799, and Leu511. Additionally, Ser710 sits at the mouth of the pocket. Importantly, all these residues are conserved across the DENV-1 to DENV-4 serotypes; most residues are also conserved in the other flaviviruses WNV, yellow fever virus, and JEV, with the exception of Met765 (Leu in JEV and WNV), Leu511 (Val in JEV), and His711 (Asn in JEV and WNV). This suggests that the residues lining the pocket are functionally important for flavivirus RdRp activity.
FIGURE 2.
Interactions between JF-31-MG46 and the DENV-3 RdRp. A, the residues lining the DENV palm pocket are shown as blue sticks, with the compound JF-31-MG46 shown as orange sticks. The individual residues are labeled according to their numbering in the DENV-3 polymerase. B, two-dimensional ligand interaction map generated using a molecular operating environment. Polar residues are colored light purple, and basic residues also have a blue ring. Hydrophobic residues are green. The degree of solvent exposure is shown by the blue halos. H-bond interactions to the amino acid side chain are shown as dashed green arrows pointing toward the H-bond acceptor. The ionic interaction is shown as a dashed purple line, and water-mediated contacts are shown as gold dashed lines. C, Surface representation of the DENV-3 palm pocket with the compound shown as orange sticks.
JF-31-MG46 Binds to DENV-3 and DENV-4 RdRp by SPR
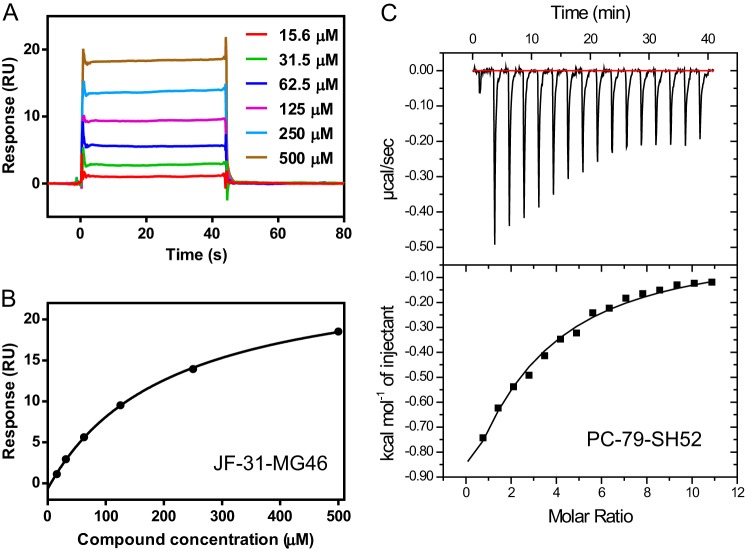
Next, we determined experimentally whether JF-31-MG46 bound to both DENV-3 and DENV-4 RdRps in solution. We developed a SPR binding assay. The extended DENV-3 and DENV-4 RdRp domains, which contain additional residues at the N terminus of the protein, were expressed with an N-terminal AVI-tag for site-specific biotinylation during expression in E. coli. The additional residues at the N terminus of the RdRp domain improve the stability and enzymatic activity of the domain (15). The additional residues also improved the stability of the protein on the chip surface and reduced nonspecific binding (data not shown). The purified biotinylated proteins were captured directly onto the surface of a biosensor chip precoated with streptavidin until the baseline had increased by ∼5,500 response units. The compounds were then injected across the surface in a 2-fold serial dilution, starting from the lowest concentration. An example titration is shown in Fig. 3A. The binding and dissociation rates were too fast to measure, as expected for a fragment, so the steady-state binding level was measured to determine the affinity (Fig. 3B and Table 2). JF-31-MG46 bound in a saturable manner to both the DENV-3 and DENV-4 RdRps, although there was a slight difference in the binding affinity to the two different serotypes, with Kd values of 209 and 613 μm, respectively. As a negative control, the compound did not bind to the methyltransferase domain of DENV-4 (data not shown). These data suggest that the binding in solution is related to the binding mode seen in the DENV-3 crystals. Repeat injections of JF-31-MG46 showed that the DENV-3 and DENV-4 RdRp proteins were stable on the chip surface at 4 °C for >24 h, with minimal decrease in the binding signal over this time, indicating that this assay can be adapted to HTS screening for compounds or fragments that bind directly to the RdRp (data not shown).
FIGURE 3.
Biophysical characterization of compound binding to the DENV RdRp. A, individual sensorgrams for the DENV-3 polymerase binding to increasing concentrations of JF-31-MG46, measured by surface plasmon resonance. B, the steady-state binding level in A is plotted versus the compound concentration, and the data are fitted to a single-site binding equation to calculate the equilibrium dissociation constant. C, isothermal titration calorimetry plot for PC-79-SH52 binding to the DENV-4 RdRp. The area under each of the peaks (an individual injection) was fitted to a single-site binding equation to determine the equilibrium association constant and binding enthalpy.
TABLE 2.
SPR binding affinities
| DENV-3 RdRp SPR |
DENV-4 RdRp SPR |
|||||
|---|---|---|---|---|---|---|
| Kd | Rmax | Ligand efficiency | Kd | Rmax | Ligand efficiency | |
| μm | % | μm | % | |||
| JF-31-MG46 | 210 ± 21 | 140 ± 25 | 0.29 | 610 ± 49 | 140 ± 40 | 0.25 |
| PC-79-SH52 | 29 ± 4 | 120 ± 18 | 0.38 | 230 ± 28 | 200 ± 54 | 0.30 |
| FD-83-KI26 | 67 ± 13 | 130 ± 43 | 0.34 | 240 ± 12 | 180 ± 39 | 0.30 |
Related Compounds Bind to DENV-3 and DENV-4 RdRp
To show that related compounds can also bind to the DENV palm site, we synthesized compounds that contained a thiophene replacement for the phenyl (Fig. 1D). Thiophene is commonly used as a bioisostere replacement for phenyl groups. These compounds showed a higher affinity for both the DENV-3 and DENV-4 RdRp proteins using SPR (Table 2), and the compounds bound with excellent ligand efficiency. The increased affinity was particularly apparent for DENV-3, where the 2-thiophene (PC-79-SH52) showed >7 times increase in affinity (from a Kd of 209 μm for JF-31-MG46 to a Kd of 27.8 μm for PC-79-SH52) measured by SPR. The increase for the 3-thiophene (FD-83-KI26) was ∼3-fold (from a Kd of 209 μm for JF-31-MG46 to a Kd of 67.1 μm for FD-83-KI26). However, all these compounds bound with lower affinity to the DENV-4 polymerase than the DENV-3 polymerase. This was most striking for the 2-thiophene compound PC-79-SH52 (Kd of 27.8 and 226 μm, to DENV-3 and DENV-4 RdRp, respectively), where the difference between the two serotypes was approximately eight times. Nevertheless, the best fragment (PC-79-SH52) still bound with a ligand efficiency of 0.3 to the DENV-4 polymerase, which is considered a good level for hit optimization.
The Binding Affinity Was Confirmed Using Isothermal Titration Calorimetry
To confirm the binding affinities seen by SPR for the DENV-4 polymerase, we used isothermal titration calorimetry (Fig. 3C and Table 3). No binding was detected to JF-31-MG46, presumably because of the lack of sufficient sensitivity at the maximum compound solubility. However, saturable binding was detected to both the thiophene-containing compounds. The measured affinities were the same as that seen by SPR, within experimental error. This confirms that the compounds bind to the RdRp in solution and indicates that the affinities measured by SPR are reliable.
TABLE 3.
ITC binding affinities and enzyme inhibition
| DENV-4 RdRp ITC |
DENV-4 NS5 de novo initiation |
||||
|---|---|---|---|---|---|
| Kd | ΔH | S | IC50 | Hill coefficient | |
| μm | cal·mol−1 | cal·mol−1·degree−1 | μm | ||
| JF-31-MG46 | NBa | 730 ± 8 | 0.94 ± 0.17 | ||
| PC-79-SH52 | 270 ± 31 | −5,960 ± 490 | −3.9 ± 1.9 | 140 ± 7 | 0.94 ± 0.10 |
| FD-83-KI26 | 320 ± 58 | −7,700 ± 1,700 | −9.9 ± 2.7 | 210 ± 8 | 0.96 ± 0.12 |
a NB, no binding detected.
Blocking the Palm Site in the DENV-3 RdRp Inhibits Polymerase Activity
We have identified a novel compound-binding site in the DENV polymerase. However, it is important to show that this pocket is functionally important and that binding to this site inhibits the enzymatic activity of the DENV RdRp. Because the compounds bind between the priming loop and the thumb and palm subdomains and the priming loop is expected to move away during elongation, we tested the inhibitory activity of these compounds in a de novo initiation assay (Table 3). The DENV-4 polymerase was used because this enzyme was considerably more active than the DENV-3 polymerase at de novo initiation and because we had binding data using two different methods to the DENV-4 polymerase, so we wanted to understand whether the binding affinity agreed with the level of inhibition. Also, the fragments bound to the DENV-4 polymerase with lower affinity, indicating that it would be more difficult to inhibit this serotype. In addition, we tested inhibition of DENV-4 polymerase because we wanted to identify compounds that inhibited all four serotypes of DENV polymerase, not just DENV-3. Compounds were preincubated with the enzyme before adding the RNA and NTP substrates in the same manner as our high throughput screening campaign (12). The concentration required for 50% inhibition in enzyme activity (IC50) for JF-31-MG46, PC-79-SH52, and FD-83-KI26 was 734, 144, and 205 μm, respectively, which is consistent with the affinities measured by SPR and ITC (Table 3). This demonstrates that binding of these fragments to the DENV RdRp palm site inhibits the RdRp activity.
Crystal Structures of Thiophene Fragments Bound to the Palm Pocket
In an attempt to understand the reason for the increased affinity of the thiophene compounds to the DENV-3 polymerase, we determined the co-crystal structures of these compounds in the RdRp (Fig. 4). The compounds both bound into the same palm site on the polymerase. In both cases, the thiophene bound in a specific orientation that could be unambiguously assigned based on the greater electron density of the sulfur atom. The difference density is shown at 10 σ in Fig. 4B, after the compound was removed from the final round of refinement. The structures show that there is almost no movement of the amino acid side chains that line the pocket. The sulfur atom of the 2-thiophene (PC-79-SH52), which produced the greatest increase in potency, points to a small pocket lined by the side chains of Ala799, Ser796, and Leu511. The hydroxyl of Ser796 picks up an additional noncovalent interaction with the sulfur atom, which may be responsible for much of the increased affinity (18). The sulfur atom of the 3-thiophene points toward a hydrophobic patch at the back of the pocket, lined by Trp803, Met761, and Met765. The increased hydrophobicity of the sulfur in the 3-thiophene likely explains its increased binding affinity. The sulfur atom contacts the sulfur of Met761. The above results of ligand-RdRp crystal structures and biophysical binding represent the first example of a discrete compound-binding site on the DENV polymerase where there is agreement between IC50 for enzymatic inhibition, binding affinity in solution, and binding by x-ray crystallography.
FIGURE 4.
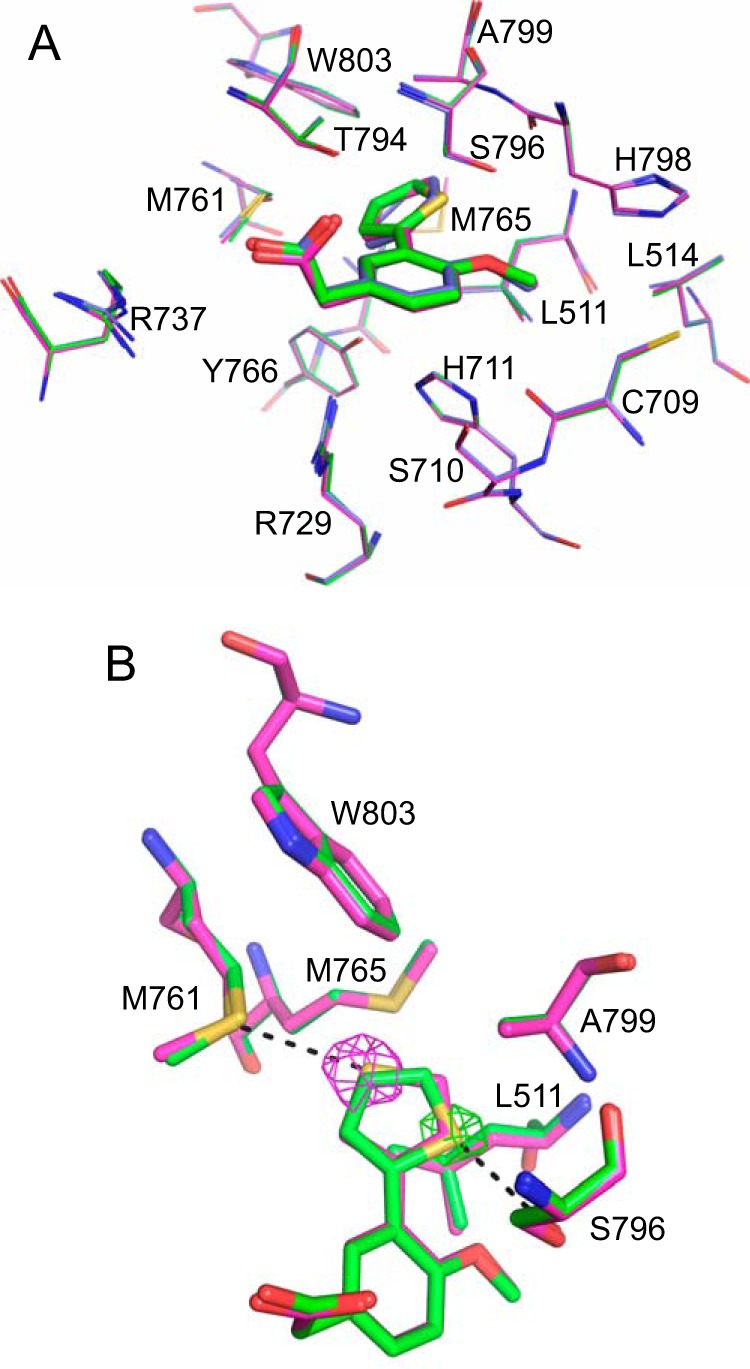
Crystal structures of the DENV-3 RdRp bound to PC-79-SH52 and FD-83-KI26. A, comparison of the bound conformations of JF-31-MG46 (blue), PC-79-SH52 (green), and FD-83-KI26 (magenta). The individual residues lining the palm pocket are labeled and shown as lines. The compounds are shown as sticks in the same color as the residues lining the pocket. B, the specific orientation of PC-79-SH52 (green) and FD-83-KI26 (magenta), with interacting residues all shown as sticks. The difference density where the compound is removed from the final round of refinement is shown at 10 σ and shown as a mesh in the same color as the compound that it corresponds to.
Discussion
We have identified a novel compound-binding pocket in the DENV-3 RdRp using fragment-based screening by x-ray crystallography. The hit compound and related compounds bind to the polymerase in solution, as well as by x-ray crystallography, and inhibit the polymerase activity. This is the first example of a compound series that is known to bind to a discrete pocket in the DENV polymerase. The fact that the compounds inhibit the polymerase activity of the enzyme suggests that this pocket is functionally important. X-ray crystal soaking experiments showed that a GTP nucleotide bound to the DENV-3 polymerase at a similar position (6). Density was only seen for the triphosphate, but the phosphates bound at the mouth of the pocket, suggesting that this pocket may have a role in binding one of the initiating nucleotides during initiation. An ATP or GTP nucleotide was shown to bind to a similar position in the JEV RdRp crystal structure (19). However, again only a single initiating nucleotide is bound; it is not clear whether this is the correct position for initiation or whether it is a crystallization artifact. We propose that this palm pocket is conserved to stabilize the binding of one of the initiating nucleotides.
A detailed characterization of the mode of inhibition of the compounds presented here is not possible given their potency. However, the more potent compounds (PC-79-SH52 and FD-83-KI26) do show complete inhibition at the highest concentrations tested (data not shown), indicating that they completely block RNA synthesis rather than slowing the rate of incorporation.
The bound fragments are small molecules (molecular masses of 242–248 Da) and bind with a good ligand efficiency of 0.29–0.38 to DENV-3 RdRp and 0.25–0.30 to DENV-4 RdRp. This suggests that it is possible to further optimize the affinity and potency of the compound by growing the compound to make additional contacts inside and outside the pocket. In particular the pocket has a narrow hydrophobic opening at the back of the pocket bordered by Trp803, Met765, Leu511, and Ala799 that is occupied by a single water molecule (Fig. 5). Growing the fragment in this direction would be expected to significantly increase the binding affinity. Also, the compounds only occupy part of the front of the pocket. There is another opening between Met761 and Arg737 to reach Trp762, where there is the potential to gain additional binding affinity. This region of the pocket is occupied by five water molecules.
FIGURE 5.
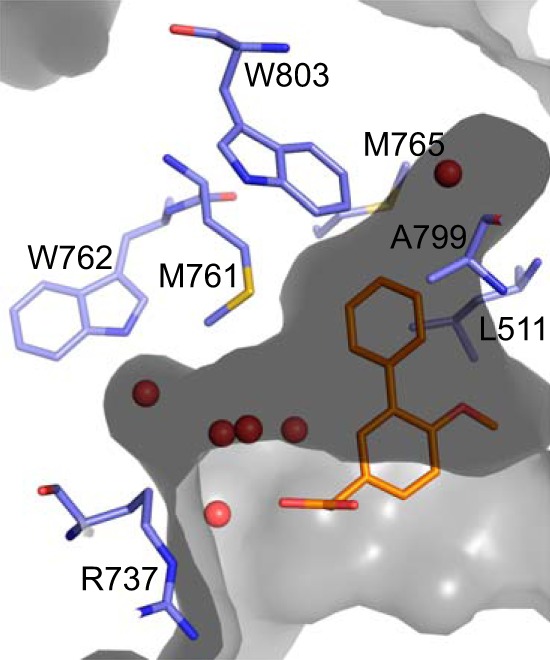
Shape of the DENV RdRp palm pocket. The surface of the palm pocket is shown in gray with the water molecules discussed in the text shown as red spheres. Key residues to gain potency are shown as blue sticks and labeled. The compound, JF-31-MG46, is shown as orange sticks.
The reason for the difference in affinity between DENV-3 and DENV-4 RdRp is not clear considering that all of the residues lining this pocket are conserved. It is likely that distant residue differences modestly affect the conformation of the pocket. Despite extensive efforts from our group and others, there is still no crystal structure available for the DENV-4 polymerase. Attempts to co-crystallize the DENV-4 polymerase with these fragments have so far been unsuccessful. It will be necessary to determine the crystal structure of the DENV-4 polymerase to understand the difference in affinity between the two serotype RdRps. Several of the residues are also conserved in the WNV and JEV polymerases; the crystal structures of these RdRps show similar pockets between the priming loop and palm and thumb subdomains (7, 19, 20). It may be possible that similar fragments are capable of binding these pockets to inhibit these enzymes. The fragments and ligand-binding sites presented here represent a real opportunity for structure-based hit to lead and lead optimization.
Author Contributions
C. G. N. performed the crystallographic and biophysics experiments and wrote the manuscript. T. E. B., R. A., and S. K. W. conceived and planned the SPR data presented in Fig. 3 and Table 2. C. C. S. performed the biochemical experiments presented in Table 3. F. Y. and S. N. analyzed the x-ray data and designed the compounds PC-79-SH52 and FD-83-KI26. C. G. N., S. P. L., F. Y., S. N., P.-Y. S., and P. W. S. conceived the project. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank the beamline scientists at PXII and PXIII of the Swiss Light Source for assistance with data collection. We thank Sanny Ing for assistance with parallel synthesis of compounds.
The authors were employees of Novartis when the experiments were performed.

This article contains supplemental Table S1 and Fig. S1.
The atomic coordinates and structure factors (codes 5F3T, 5F3Z, and 5F41) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- DENV
- dengue virus
- WNV
- West Nile virus
- JEV
- Japanese encephalitis virus
- RdRp
- RNA-dependent RNA polymerase
- SPR
- surface plasmon resonance
- ITC
- isothermal titration calorimetry.
References
- 1. Whitehorn J., and Simmons C. P. (2011) The pathogenesis of dengue. Vaccine 29, 7221–7228 [DOI] [PubMed] [Google Scholar]
- 2. Bhatt S., Gething P. W., Brady O. J., Messina J. P., Farlow A. W., Moyes C. L., Drake J. M., Brownstein J. S., Hoen A. G., Sankoh O., Myers M. F., George D. B., Jaenisch T., Wint G. R., Simmons C. P., Scott T. W., Farrar J. J., and Hay S. I. (2013) The global distribution and burden of dengue. Nature 496, 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simmons C. P., Wolbers M., Nguyen M. N., Whitehorn J., Shi P. Y., Young P., Petric R., Nguyen V. V., Farrar J., and Wills B. (2012) Therapeutics for dengue: recommendations for design and conduct of early-phase clinical trials. PLoS Negl. Trop. Dis. 6, e1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lim S. P., Noble C. G., and Shi P. Y. (2015) The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 119, 57–67 [DOI] [PubMed] [Google Scholar]
- 5. Dong H., Fink K., Züst R., Lim S. P., Qin C. F., and Shi P. Y. (2014) Flavivirus RNA methylation. J. Gen. Virol. 95, 763–778 [DOI] [PubMed] [Google Scholar]
- 6. Yap T. L., Xu T., Chen Y. L., Malet H., Egloff M. P., Canard B., Vasudevan S. G., and Lescar J. (2007) Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 81, 4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao Y., Soh T. S., Zheng J., Chan K. W., Phoo W. W., Lee C. C., Tay M. Y., Swaminathan K., Cornvik T. C., Lim S. P., Shi P. Y., Lescar J., Vasudevan S. G., and Luo D. (2015) A crystal structure of the dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog. 11, e1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caillet-Saguy C., Lim S. P., Shi P. Y., Lescar J., and Bressanelli S. (2014) Polymerases of hepatitis C viruses and flaviviruses: Structural and mechanistic insights and drug development. Antivir. Res. 105, 8–16 [DOI] [PubMed] [Google Scholar]
- 9. Appleby T. C., Perry J. K., Murakami E., Barauskas O., Feng J., Cho A., Fox D. 3rd, Wetmore D. R., McGrath M. E., Ray A. S., Sofia M. J., Swaminathan S., and Edwards T. E. (2015) Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347, 771–775 [DOI] [PubMed] [Google Scholar]
- 10. Niyomrattanakit P., Abas S. N., Lim C. C., Beer D., Shi P. Y., and Chen Y. L. (2011) A fluorescence-based alkaline phosphatase-coupled polymerase assay for identification of inhibitors of dengue virus RNA-dependent RNA polymerase. J. Biomol. Screen. 16, 201–210 [DOI] [PubMed] [Google Scholar]
- 11. Niyomrattanakit P., Chen Y. L., Dong H., Yin Z., Qing M., Glickman J. F., Lin K., Mueller D., Voshol H., Lim J. Y., Nilar S., Keller T. H., and Shi P. Y. (2010) Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J. Virol. 84, 5678–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith T. M., Lim S. P., Yue K., Busby S. A., Arora R., Seh C. C., Wright S. K., Nutiu R., Niyomrattanakit P., Wan K. F., Beer D., Shi P. Y., and Benson T. E. (2015) Identifying initiation and elongation inhibitors of dengue virus RNA polymerase in a high-throughput lead-finding campaign. J. Biomol. Screen. 20, 153–163 [DOI] [PubMed] [Google Scholar]
- 13. Noble C. G., Lim S. P., Chen Y. L., Liew C. W., Yap L., Lescar J., and Shi P. Y. (2013) Conformational flexibility of the dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 87, 5291–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beckett D., Kovaleva E., and Schatz P. J. (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim S. P., Koh J. H., Seh C. C., Liew C. W., Davidson A. D., Chua L. S., Chandrasekaran R., Cornvik T. C., Shi P. Y., and Lescar J. (2013) A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J. Biol. Chem. 288, 31105–31114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nyanguile O., Devogelaere B., Vijgen L., Van den Broeck W., Pauwels F., Cummings M. D., De Bondt H. L., Vos A. M., Berke J. M., Lenz O., Vandercruyssen G., Vermeiren K., Mostmans W., Dehertogh P., Delouvroy F., Vendeville S., VanDyck K., Dockx K., Cleiren E., Raboisson P., Simmen K. A., and Fanning G. C. (2010) 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus. J. Virol. 84, 2923–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beno B. R., Yeung K. S., Bartberger M. D., Pennington L. D., and Meanwell N. A. (2015) A survey of the role of noncovalent sulfur interactions in drug design. J. Med. Chem. 58, 4383–4438 [DOI] [PubMed] [Google Scholar]
- 19. Surana P., Satchidanandam V., and Nair D. T. (2014) RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state. Nucleic Acids Res. 42, 2758–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malet H., Egloff M. P., Selisko B., Butcher R. E., Wright P. J., Roberts M., Gruez A., Sulzenbacher G., Vonrhein C., Bricogne G., Mackenzie J. M., Khromykh A. A., Davidson A. D., and Canard B. (2007) Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 282, 10678–10689 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.