Abstract
Many pathogenic microbes often release toxins that subvert the host's immune responses to render the environment suitable for their survival and proliferation. LeTx is one of the toxins causing immune paralysis by cleaving and inactivating the mitogen-activated protein kinase (MAPK) kinases (MEKs). Here, we show that inhibition of the histone deacetylase 8 (HDAC8) by either the HDAC8-specific inhibitor PCI-34051 or small interference (si)RNAs rendered LeTx-exposed murine macrophages responsive to LPS in pro-IL-1β production. HDAC8 selectively targeted acetylated histone H3 lysine 27 (H3K27Ac), which is known to associate with active enhancers. LeTx induced HDAC8 expression, in part through inhibiting p38 MAPK, which resulted in a decrease of H3K27Ac levels. Inhibition of HDAC8 increased H3K27Ac levels and enhanced NF-κB-mediated pro-IL-1β enhancer and messenger RNA production in LeTx-exposed macrophages. Collectively, this study demonstrates a novel role of HDAC8 in LeTx immunotoxicity and regulation of pro-IL-1β production likely through eRNAs. Targeting HDAC8 could be a strategy for enhancing immune responses in macrophages exposed to LeTx or other toxins that inhibit MAPKs.
Keywords: anthrax toxin, histone deacetylase (HDAC), interleukin 1 (IL-1), lipopolysaccharide (LPS), transcription enhancer
Introduction
The innate immune system provides protection from invading microbes by releasing antimicrobial and inflammatory mediators. However, infectious microbes subvert host immune responses by releasing various toxins. One of the most extreme cases is anthrax, which causes immune paralysis, toxic shock, and death of the host. The causative agent of anthrax is Bacillus anthracis, and LeTx is a main virulence factor that causes immune paralysis and mortality (1). LeTx is known to cause immune suppression by targeting the MAPK pathway that is involved in many aspects of immune responses (2–5). It includes two proteins, protective antigen (PA)2 and lethal factor (LF) (6–10). PA is a carrier protein that incorporates LF into the cytoplasm; LF is a metalloprotease that cleaves all MAPK kinases (MEK1–7), except MEK5 (11) and MEK7 (12). Inactivation of these MEKs results in almost complete inactivation of MAPKs, including the ERKs and p38 MAPK (p38), but partial or no effects on JNK/SAPKs in macrophages (13, 14) and other cell types (12, 15). Inhibition of ERKs and p38 by LeTx suppresses expression of various inflammatory cytokines in macrophages (16–18). Particularly, IL-1β, which is expressed as a pro-IL-1β form and proteolytically matured by caspase-1, plays an important role in mounting early immune responses to germinating B. anthracis (19), as well as multiple other bacterial pathogens (19–26). Severe immune suppression caused by LeTx is the main culprit for unrestricted proliferation of B. anthracis inside the host (3, 5, 27, 28).
Epigenetics is a cellular mechanism that inheritably regulates expression of genes without altering genomic DNA sequences in response to developmental and environmental cues. There are three distinct but inter-related mechanisms in epigenetics as follows: DNA methylation, chromatin structure modification, and non-coding RNAs. Among them, modifications of the N-terminal regions of histones by phosphorylation, methylation, and acetylation dynamically orchestrate chromatin structures and regulate gene expression. In macrophages, these epigenetic mechanisms are involved in activation (29), differentiation (30–32), and endotoxin tolerance (33–35). Various bacterial and viral microbes also epigenetically manipulate host immune responses to render environments suitable for their survival and proliferation. LeTx was shown to inhibit histone H3 serine 10 (H3S10) phosphorylation by targeting the ERK and p38 and to suppress recruitment of NF-κB to gene-specific promoters, including IL-6 and IL-8 (15). Acetylation on lysine residues is another key histone modification, mainly associated with transcriptional activation of genes. Levels of histone acetylation are regulated by two families of enzymes (36), the histone acetyltransferases and histone deacetylases (HDACs). We previously showed that LeTx up-regulates HDAC8 expression, which is involved in silencing the mitochondrial death genes, likely through targeting acetylated histone H3 lysine 27 (H3K27Ac) (37). HDAC8 was shown to target the core histones H2A/H2B, H3, and H4 in vitro (38–40); however, in vivo activity of HDAC8 toward histones and specific histone residues is unknown (41).
Enhancers are cis-regulatory elements that regulate gene expression at varying distances from their target genes (42). In mammalian cells, most active enhancers have the ability to recruit polymerase II and transcription factors and exhibit mono- or bi-directional transcription (43–47). These transcripts, referred to as eRNAs, are dynamically induced before or at the same time of the appearance of mRNAs (43, 48). In LPS-stimulated macrophages, the amount of eRNAs is also highly correlated with the amount of corresponding signal-dependent mRNA transcripts (47, 49). These enhancers are often demarcated by genomic regions that associate with mono-methylated histone H3 lysine 4 (H3K4me1) and histone acetyltransferases such as cAMP-response element-binding protein binding protein and p300; however, these markers do not distinguish active and poised enhancers. In fact, H3K27Ac-positive regions are a fraction of H3K4me1-positive regions and specific to active enhancers in a given cell type (50–52). Because H3K27Ac levels can be regulated by LeTx through HDAC8 (37), we examined whether LeTx regulates expression of inflammatory cytokines such as IL-1β by enhancers and whether HDAC8 inhibition protects macrophages from LeTx-induced immune suppression. Here, we show that induction of HDAC8 expression and activity by LeTx resulted in the decrease of H3K27Ac levels and subsequent suppression of pro-IL-1β eRNA and mRNA production. Also, inhibition of HDAC8 restored LPS responses in producing pro-IL-1β eRNA, mRNA, and protein in LeTx-exposed murine macrophages.
Experimental Procedures
Materials and Reagents
Anthrax lethal toxins (PA and LF) and Escherichia coli O111:B4 LPS were purchased from the List Biological Laboratories. Epigenetic chemical inhibitors used in this study are the following: apicidin (Santa Cruz Biotechnology); aza-2-deoxycytidine (azacitidine; Sigma); CAY10603 (Cayman Chemical); mocetinostat (MGCD0103; Selleck); MC1568 (APExBio Technology); PCI-34051 (Cayman Chemical); and panobinostat (LBH-589; Selleck). The ERKs, p38 MAPK inhibitors, U0126, SB203580, and NF-κB activation inhibitor, respectively, were purchased from Calbiochem. Anti-GFP and antibody raised against the N terminus of MEK1 (MEK1-NT) were obtained from Life Technologies, Inc., and StressGen Bioreagents, respectively. Antibodies for phospho-p38, phospho-ATF-2, phospho-IκB, and IκB were obtained from Cell Signaling. Monoclonal antibody for HDAC8 was purchased from Epigentek (catalog no. A-4008). Antibodies for H3K4Ac, H3K14Ac, H3K18Ac, H3K23Ac, H3K27Ac, H3K36Ac, H3K56Ac, H3K79Ac, and H3K27me3 were from Active Motif; pan-histone H3 was from BioVision; β-actin was from Rockland Inc.; and anti-NF-κB (p65) was from eBioscience. The pro-IL-1β antibody was a kind gift from Dr. Aurigemma (NCI-Frederick Cancer Research and Development Center, Frederick, MD). Predesigned siRNAs targeting HDAC8 (catalog no. SI1063902) and antisense oligonucleotides (ASO; LNATM GapmeRs, Exiqon) targeting pro-IL-1β eRNA (CAATCCTGGTTGATGA) were purchased from Qiagen and Exiqon, respectively.
Cell Culture and Transfection
RAW264.7 macrophages were cultured in Dulbecco's modified Eagle's medium as described previously (53). Primary bone marrow-derived macrophages (BMDMs) were prepared from 129/Svj mice or C57BL/6j mice as described previously (54). RAW264.7 cells stably transfected with pEGFP or pEGFP-HDAC8 were prepared and cultured as described previously (37). Transfections of RAW264.7 cells and BMDMs with siRNAs and ASOs were performed using the Lipofectamine RNAi Max kit (Invitrogen), according to the manufacturer's instructions.
Western Blotting
Preparation of total cell lysates and immunoblotting were performed as reported previously (37).
Chromatin Immunoprecipitation (ChIP) Analysis
ChIP analysis was conducted as described previously (37), using H3K27Ac antibody (Active Motif), anti-NF-κB (p65; eBioscience), or rabbit IgG antibody (Sigma) as a control. Purified DNAs were subjected to quantitative real time PCR analysis using Power SYBR Green PCR Master Mix (Applied Biosystems) and primers targeting up to ∼10 kb upstream of the IL-1β transcription starting site. Primers used for ChIP analysis are as follows: primer 1, forward (F), CTTCTTTACTTATCCTCTTGCTCAG, and reverse (R), TGAGAGGGAAAGAACAGACCC; primer 2, F, AAACCCTTGAGCTGATGCCT, and R, TCATTGCCTCCTCCCAGACA; primer 3, F, CAGGGTGGGCTCAAGCATTA, and R, GGATCGGCCTACTGACCTTG. Data are presented as percentage of the precipitated target sequence as compared with input DNA.
Quantitative Real Time PCR
eRNA or mRNA expression was quantified by quantitative real time PCR (qPCR) as reported previously (37). Briefly, total cellular RNAs were isolated using TRIzol (Life Technologies, Inc.) according to the manufacturer's instructions and reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (New England Biotechnology) with random hexamers (for eRNA) or oligo(dT) primers (for mRNA). qPCR analyses were performed with a Rotor-Gene RG3000 quantitative multiplex PCR instrument (Montreal Biotech) using Power SYBR Green PCR Master Mix (Applied Biosystems). In eRNA analysis, qPCR was also carried out on RNA, which was not reverse-transcribed, to control for the lack of genomic DNA contamination. The data were normalized by expression of the GAPDH housekeeping gene. Primers used for qPCR are as follows: IL-1β, F, GTGGACCTTCCAGGATGAGG, and R. GCTTGGGATCCACACTCTCC; tumor necrosis factor-α, F, ATGAGAAGTTCCCAAATGGCC, and R, TCCACTTGGTGGTTTGCTACG; and GAPDH, F, GCATTGTGGAAGGGCTCATG, and R, TTGCTGTTGAAGTCGCAGGAG. Primers for eRNA and NF-κB ChIP assays are as follows: e1, F, ATGGAGCCCATCCCAGAG, and R, AGTTACCAGCAGGGCCACTC; e2, F, AATCACGAACAGACGACCATC, and R, GCCTCCCTATCTCCCTACCTT; e3, F, ACAGTCTCGCCACAGAAAGAA, and R, CCATCAAAAGGACAACTGCAT; e4, F, CTATGGCCTATGGCTTCTGC, and R, TTTTGCCACATGGCTGATAA; e5, F, CTAGTCCCAGGGAGTTCTGC, and R, AGGGTTAGGCGCTATGGTCT; e6, F, AGTGCATGTTCCAACGTCAA, and R, GACCATCAAGAACAGCAGCA; e7, F, ACTTGGGGAGGAAAGGATGT, and R, ATGAGGAGCAAGCCCAGTGAG; e8, F, GCTAAGCAATGACTGTCCTCA, and R, ATGAGAGGGAAAGAACAGACCC; e9, F, AGGAGGTTTGTCTGGGAGGA, and R, ATGTTGTGCAACTTGCCTGC; e10, F, GGGAGCTCTTCTTGCTTGGA, and R, TACTGCCTGCATCCATCTGC; e11, F, CCTGACCCACACAAGGAAGT, and R, ATGTGCGGAACAAAGGTAGG.
HDAC8 Immunoprecipitation and HDAC Assay
Histone deacetylase enzyme activity of endogenous or ectopically expressed HDAC8 was measured using purified histones as the substrate. Substrate histones were prepared from wild-type RAW264.7 cells as described in the Abcam protocol. For HDAC8 immunoprecipitation, cells were suspended and incubated on ice for 10 min in the RIPA lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Igepal CA-630 (Nonidet P-40), 0.5% sodium deoxycholate, 0.1%) containing cOmplete Protease Inhibitor Mixture (Roche Applied Science) and PMSF (1 mm) and incubated for 10 min on ice. Cell lysates were centrifuged for 10 min at 3500 × g at 4 °C, and the supernatants were transferred to a new tube (tube A). Insoluble pellets were resuspended in the RIPA buffer containing 1.5 mm MgCl2 and incubated on ice for another 10 min with vortexing every minute. Resuspended mixtures were centrifuged at 15,500 × g for 10 min, and supernatants were collected (tube B). Tube A and B were combined and used for immunoprecipitation, as described previously (38, 55). Briefly, total lysates were mixed with anti-HDAC8 or anti-EGFP antibody and rotated for 4 h at 4 °C. Protein G-SepharoseTM fast flow beads (Sigma) were added and incubated for 1 h. The immunocomplex was washed three times with the RIPA lysis buffer and then washed with 100 μl of HDAC assay buffer (50 mm Tris-HCl, pH 8.0, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2). Sepharose beads-HDAC8 complexes were mixed with purified histones in HDAC assay buffer and incubated in a shaking incubator at 37 °C for the indicated times. After incubation, reaction mixtures were centrifuged for 1 min at 10,700 × g at 4 °C, and supernatants were collected and prepared for Western blot by adding 4× SDS sample buffer (125 mm Tris-HCl, pH 6.8, 4% SDS, 50% glycerol, 0.08% bromphenol blue, and 5% β–mercaptoethanol) to measure histone deacetylation levels.
Statistical Analysis
Data were analyzed using GraphPad Prism 4.0 (GraphPad Software). The results are presented as the mean ± S.D. of three independent repeats. Data were analyzed as indicated in the figure legends. Statistical significance was defined as *, p < 0.05.
Results
HDAC8 Inhibition Restores the Production of Pro-IL-1β in Response to LPS in LeTx-exposed Murine Macrophages
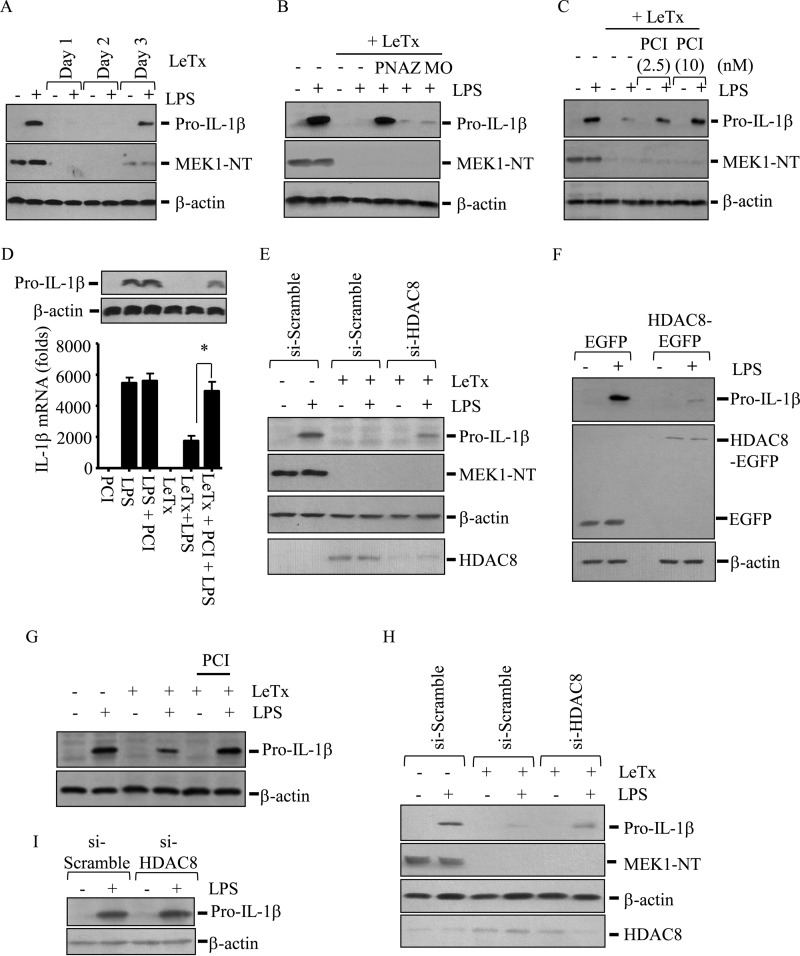
LeTx suppresses expression of various inflammatory cytokines by inactivating MEKs (3–5). Consistent with these reports, a sub-lethal dose of LeTx (100 ng/ml PA and LF, each for 4 h) completely cleaved MEK1 and inhibited expression of pro-IL-1β induced by LPS (100 ng/ml) for more than 2 days in RAW264.7 macrophages (Fig. 1A). Our previous studies have shown that these LeTx-exposed macrophages are epigenetically reprogrammed through DNA methylation and histone acetylation (37, 53). When we examined whether epigenetic reprogramming is also involved in the inhibition of pro-Il-1β production, the pan-specific HDAC inhibitor panobinostat also restored expression of pro-IL-1β induced by LPS in LeTx-exposed RAW264.7 macrophages (Fig. 1B). However, the DNA methyltransferase 1 inhibitor azacitidine or the HDAC1–3- and −11-specific inhibitor mocetinostat had no such effects. Further examination on isoform-specific HDAC inhibitors showed that the HDAC8-specific inhibitor PCI-34051 (PCI) restored the expression of pro-IL-1β protein and mRNA (Fig. 1C) in LeTx-exposed macrophages, whereas other isoform-specific HDAC inhibitors, such as apicidin (HDAC3-specific), CAY10603 (HDAC6-specific), and MC1568 (HDAC7-specific), failed to show such effects (data not shown). Interestingly, PCI had no effect on the overall pro-IL-1β mRNA and protein production in response to LPS in non-LeTx-exposed cells (Fig. 1D). To rule out any off-target effects of PCI, we used small interference RNAs targeting HDAC8 (si-HDAC8) and found that si-HDAC8 also restored pro-IL-1β production in LeTx-exposed cells (Fig. 1E). To further confirm the role of HDAC8, EGFP-conjugated HDAC8 (HDAC8-EGFP) was stably overexpressed, and the expression of pro-IL-1β in response to LPS was examined. Clearly, LPS failed to produce pro-IL-1β in HDAC8-EGFP-overexpressing cells (Fig. 1F) but not EGFP vector-overexpressing cells. To further examine whether HDAC8 inhibition renders similar effects on primary macrophages, bone marrow-derived macrophages from two different strains were treated with PCI or si-HDAC8. PCI-treated primary BMDMs from C57BL/6j mice (Fig. 1G) and si-HDAC8-treated BMDMs from 129SvJ mice (Fig. 1H) were also able to express pro-IL-1β by LPS even after LeTx treatments. Consistent with Fig. 1D, knockdown of HDAC8 did not have any effects on overall production of pro-IL-1β in 129SvJ BMDMs (Fig. 1I). Collectively, these results suggest that HDAC8 inhibition prevents the inhibitory effects of LeTx on the expression of pro-IL-1β in murine immortalized and primary macrophages.
FIGURE 1.
HDAC8 inhibition prevents the inhibitory effects of LeTx on the expression of pro-IL-1β induced by LPS. A–F, RAW264.7 cells were treated with a sub-lethal dose of LeTx (100 ng/ml LF and 100 ng/ml PA) for 4 h and further cultured in fresh media for the time indicated. A, LeTx-exposed (day 1–3) or non-exposed cells were stimulated with LPS (100 ng/ml) for 4 h. B, cells cultured for 16–20 h after LeTx treatments were incubated with or without pan-specific HDAC inhibitor PN (1 nm), DNA methyltransferase 1 (DNMT1), inhibitor azacitidine (AZ; 200 nm), and HDAC1–3- and -11-specific inhibitor mocetinostat (MO; 150 nm) for an additional 20 h. These cells were then treated with LPS (100 ng/ml) for 4 h. C, cells were incubated with or without the HDAC8-specific inhibitor PCI for an additional 20 h and treated with LPS (100 ng/ml) for 4 h. D, cells were treated with LeTx and/or PCI (20 nm) and treated with LPS (100 ng/ml) for 3 h. Pro-IL-1β protein (upper panel) and mRNA (lower panel) levels were measured by qPCR. Data are expressed as means ± S.D. (n = 3; *, p < 0.05, Student's t test). E, RAW264.7 cells were transfected with scramble (si-Scramble) or HDAC8-targeting (si-HDAC8) siRNAs. After 10 h of transfection, cells were treated with LeTx for 20 h and stimulated with LPS for 4 h. F, cells stably transfected with pEGFP or pEGFP-HDAC8 were treated with LPS for 4 h. HDAC8-EGFP expression was measured by immunoblotting using anti-EGFP (middle panel). G–I, primary BMDMs from B57/BL6j (G) and 129SvJ (H and I) mice were treated with PCI, scramble (si-scramble) or HDAC8 targeting (si-HDAC8) siRNAs for 10 h. Cells were then treated with LeTx (100 ng/ml LF and 100 ng/ml PA) for 3 h and further cultured for 20 h in fresh culture media. (A–C and E–I) Expression of pro-IL-1β protein and MEK1 degradation in total cell lysates were analyzed by Western blot. Immunoblotting for β-actin was used as the loading control. Data are representative images of three independent experiments.
HDAC8 Deacetylates H3K27 in Vitro
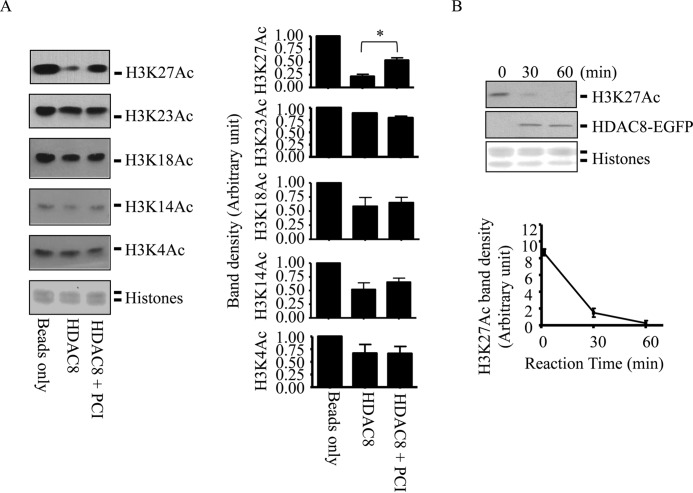
HDAC8 was shown to target histone H3 in vitro (56); however, their specific target sites of H3 are unknown. To examine whether HDAC8 deacetylates specific residues of H3, HDAC8 was immunoprecipitated from RAW264.7 cells, and the immunocomplex was incubated with histones prepared from nuclear extracts of RAW264.7 cells in the presence or absence of PCI for 60 min. As shown in Fig. 2A, the immunoprecipitated HDAC8 significantly deacetylated H3K27Ac, which was inhibited by PCI, but not other residues, including H3K23, H3K18, H3K14, and H3K4. To further confirm, HDAC8-EGFP was ectopically expressed in RAW264.7 macrophages and precipitated using anti-EGFP antibody. The HDAC8-EGFP immunocomplex also deacetylated H3K27Ac within 30 min of the reaction (Fig. 2B).
FIGURE 2.
HDAC8 deacetylates H3K27 in vitro. A, immunoprecipitated endogenous HDAC8 was prepared from wild-type RAW264.7 cells by immunoprecipitation using anti-HDAC8. Beads only or HDAC8 immunocomplex was incubated with histones purified from wild-type RAW264.7 cells in HDAC assay buffer in the presence or absence of PCI (20 nm) at 37 °C for 60 min. H3K27Ac levels were analyzed by Western blots (left panel). B, similarly, HDAC8-EGFP complexes were prepared from RAW264.7 cells stably transfected with pEGFP-HDAC8 using EGFP antibody. HDAC8-EGFP immune complexes were incubated with histones for the time indicated, and levels of H3K27Ac were analyzed by Western blot. Immunoblot for HDAC8-EGFP was used to visualize the immunoprecipitated HDAC8-EGFP. Histones used for substrates were visualized by Ponceau S staining. Images shown are representative results of three independent experiments. Band intensities compared with loading controls were analyzed by the ImageJ program (National Institutes of Health) and are expressed as means ± S.D. (n = 3); Student's t test; *, p < 0.05.
LeTx Decreases H3K27Ac Levels at Least in Part by Inducing HDAC8 Expression
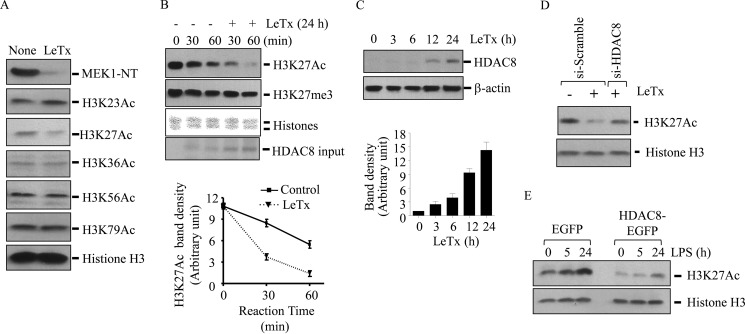
Previously, we showed that LeTx-exposed cells harbor lower levels of H3K27Ac (37). We confirmed that LeTx only caused an apparent decrease in H3K27Ac levels but not in H3K23, H3K36, H3K56, and H3K79 (Fig. 3A). We then examined whether LeTx-treated macrophages had higher HDAC8 deacetylation activity toward H3K27Ac levels. To this end, endogenous HDAC8 was immunoprecipitated from LeTx-exposed RAW264.7 cells, and their activities toward H3K27Ac were examined in vitro. As shown in Fig. 3B, immunoprecipitated HDAC8 from LeTx-exposed cells had higher deacetylating activities toward H3K27Ac when compared with those from non-treated (control) cells. In contrast, immunoprecipitated HDAC8 had no effects on H3K27 methylation levels in either non-exposed or LeTx-exposed cells. Next, we examined how LeTx induced HDAC8 activity. First, LeTx induced a gradual increase of HDAC8 protein levels over 24 h (Fig. 3C), which was consistent with higher HDAC8 inputs in LeTx-exposed samples (Fig. 3B, bottom lane). To further confirm the role of HDAC8 in H3K27 deacetylation, H3K27Ac levels were examined after either ectopically overexpressing or knocking down HDAC8. As expected, si-HDAC8 prevented LeTx-induced H3K27 deacetylation (Fig. 3D), and HDAC8-EGFP overexpression caused deacetylation of H3K27Ac (Fig. 3E). These results suggest that LeTx decreased H3K27Ac levels at least in part by inducing expression of HDAC8.
FIGURE 3.
LeTx decreased H3K27Ac levels in an HDAC8-dependent manner. A, RAW264.7 cells were treated with a sub-lethal dose of LeTx (100 ng/ml LF and 100 ng/ml PA) for 4 h and further cultured for an additional 20 h in fresh media. Cell lysates were prepared for immunoblotting against various residue-specific histone H3 acetylation antibodies. Anti-MEK1-NT was used for indication of LeTx incorporation into the cytosol, and histone H3 was used as loading controls. B, HDAC8 immunocomplexes were prepared from non- or LeTx (100 ng/ml LF and 100 ng/ml PA)-exposed RAW264.7 macrophages, and their activities toward H3K27Ac were measured at 30 and 60 min after incubating HDAC8 immunocomplexes and substrates. Immunoblotting against H3K27me3 was used as negative controls. Histones used for substrates were visualized by Ponceau S staining, and Western blots for HDAC8 were used to visualize input of immunoprecipitated HDAC8. Images shown are representative results of three independent experiments. C, RAW264.7 cells were exposed with LeTx (100 ng/ml LF and 100 ng/ml PA) for 2 h and further cultured with fresh media for the time indicated. HDAC8 protein was analyzed by Western blot. Immunoblots are representative data of three independent experiments. B and C, H3K27Ac band intensities were analyzed by the ImageJ program (National Institutes of Health) (lower panels). Data are expressed as means ± S.D. (n = 3). D, RAW264.7 cells were transfected with scramble (si-Scramble) or HDAC8 siRNA (si-HDAC8) for 10 h. Cells were then treated with a sub-lethal dose of LeTx (100 ng/ml LF and 100 ng/ml PA) for 20–24 h, and H3K27Ac levels in cell lysates were analyzed by Western blots. Results are representative data of three independent experiments. E, cell lysates were prepared from RAW264.7 cells stably transfected with pEGFP or pEGFP-HDAC8 and primed by LPS for the time indicated, and immunoreactivities of H3K27Ac were analyzed by Western blot using H3K27Ac-specific antibody.
HDAC8 Inhibition Reverses the Inhibitory Effect of LeTx on Pro-IL-1β eRNA Production Induced by LPS
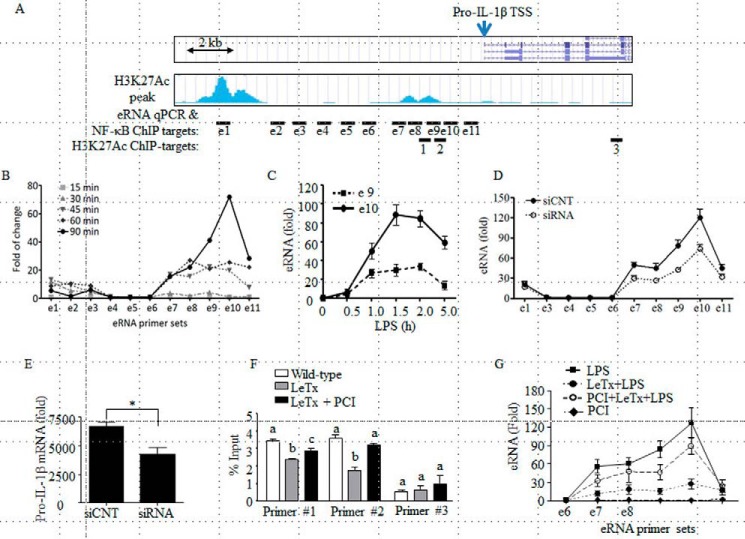
H3K27Ac is a marker for active enhancers (49). Active enhancers produce eRNAs, which are dynamically regulated by signaling events and highly correlated with corresponding signal-dependent transcription changes in promoters of nearby genes (49, 57). Therefore, we examined whether production of pro-IL-1β eRNAs induced by LPS was regulated by LeTx. Based on the ENCODE/UCSD bone marrow-derived macrophage H3K27Ac chromatin immunoprecipitation sequencing (ChIP-seq) database, two peaks of DNA sequencing frequency were found in area, ∼10 kb (peak 1) and ∼2 kb (peak 2) upstream of the pro-IL-1β transcription start site (TSS) (Fig. 4A). These peaks also coincided with the genomic regions highly associated with H3K4me1 but not with H3Kme3, indicating active enhancers (50–52). Therefore, we designed 11 qPCR primer sets encompassing the genomic area to analyze the production of pro-IL-1β-associated eRNAs. Consistent with the database, LPS rapidly induced the transcription of eRNAs related to the two H3K27Ac peaks, apparently higher levels from peak 2 than peak 1 (Fig. 4B). Kinetics in the induction of these eRNAs were distinct; the production of peak 1 eRNA reached the maximum level in 45 min and diminished to a basal level in 90 min, whereas the production of peak 2 eRNA reached the maximum level in 90 min, which gradually decreased in 5 h of LPS treatments (Fig. 4, B and C). Knockdown of peak 2 eRNA by using ASO (LNATM GapmeRs) (Fig. 4D) lowered the production of pro-IL-1β mRNA and protein induced by LPS to a similar degree of eRNA suppression by siRNA (Fig. 4E). However, the same ASO treatments had no effects on the production of tumor necrosis factor-α mRNA (Fig. 4E, right panel), ruling out nonspecific effects of ASO. These results suggest that the production of peak 2 eRNA was required for the optimal production of pro-IL-1β mRNA. Furthermore, H3K27Ac ChIP-qPCR experiments showed that LeTx significantly decreased association of H3K27Ac with the peak 2 region, which was reversed by PCI (Fig. 4F). Consistent with these results, production of peak 2 eRNA by LPS was inhibited by LeTx, which was then prevented by PCI treatment (Fig. 4G). These results suggest that HDAC8 was involved in the inhibition of pro-IL-1β enhancer activity in LeTx-exposed macrophages.
FIGURE 4.
HDAC8 inhibition reverses the inhibitory effects of LeTx on pro-IL-1β eRNA production following LPS stimulation. A, snapshot image of the pro-IL-1β genomic region and H3K27Ac ChIP-seq read frequency in BMDMs in the ENCODE database is presented. Lines with “e”-prefixed numeric number (e1–11) indicate primer targets for eRNA-qPCR and NF-κB ChIP-qPCR, and lines with number (1–3) indicate primer targets for H3K27Ac ChIP-qPCR. B, RAW264.7 cells were stimulated with LPS (100 ng/ml) for the time indicated, and eRNA productions were analyzed by qPCR using the 11 primer sets. C, similarly, production of eRNAs in response to LPS (100 ng/ml) at various time points was analyzed by qPCR using eRNA qPCR primer set e9 and e10. Data are expressed as means ± S.D. (n = 3). D and E, RAW264.7 macrophage cells were transfected with control random or pro-IL-1β eRNA antisense oligonucleotides (LNATM GapmeRs) for 24 h. Cells were then stimulated with LPS (100 ng/ml) for 1.5 h (eRNA) or 3.0 h (mRNA and protein), and production of eRNA (D) or mRNA/protein (E) was analyzed by qPCR and Western blots. Data are expressed as means ± S.D. (n = 3); *, p < 0.05, Student's t test; images shown are representative results of three independent experiments. F and G, RAW264.7 cells were pre-treated with LeTx (100 ng/ml LF and 100 ng/ml PA) for 4 h and further cultured with fresh media for 16–20 h. Surviving cells were then incubated with or without PCI (20 nm) for an additional 20 h. F, ChIP-qPCR analysis was performed in these cells using anti-H3K27Ac and H3K27Ac ChIP primers (H3K27Ac ChIP-target primers 1–3) targeting the H3K27Ac associated enhancer-like region located ∼2.0 kb upstream of the pro-IL-1β TSS and pro-IL-1β intragenic region. Data are expressed as means ± S.D. (n = 3). The same annotation above the bars indicates statistically insignificant within the primer sets (one-way analysis of variance, and Tukey's post hoc test). G, production of eRNAs in response to LPS (100 ng/ml) at 1.5 h were analyzed by qPCR using eRNA qPCR primer sets from e6 to e11. Data are expressed as means ± S.D. (n = 3).
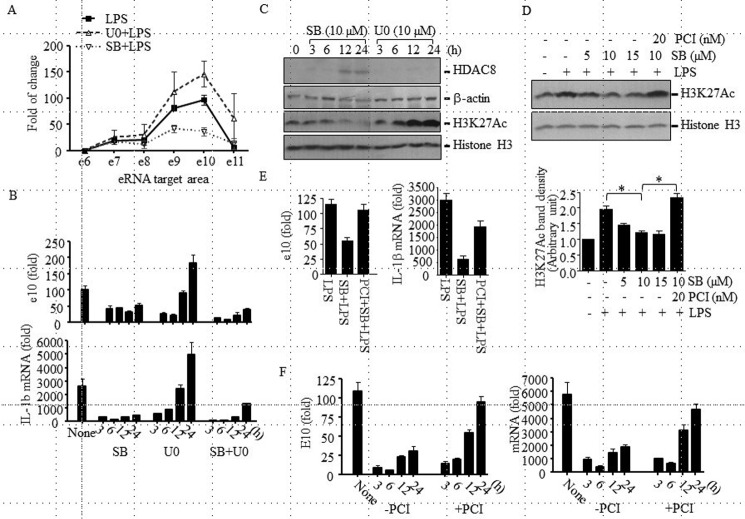
HDAC8 Is Involved in the p38-mediated Inhibition of Pro-IL-1β eRNA and mRNA Production at the Late Stage of LeTx Intoxication
To further examine which MAPK inhibition is involved in the LeTx-induced suppression of pro-IL-1β enhancer activity, RAW264.7 macrophages were pre-treated with the MEK1/2- and p38-specific inhibitors, U0126 (U0) and SB203580 (SB), respectively, for 24 h, and production of peak 2 eRNA by LPS was analyzed. Between the two inhibitors, only SB substantially suppressed eRNA production but not U0 (Fig. 5A). In fact, U0 transiently (up to 6 h) inhibited the eRNA and mRNA production and even tended to further enhance the production after 24 h of treatment (Fig. 5, A and B). However, concomitant treatments of SB and U0 suppressed the eRNA and mRNA production (Fig. 5B). Loss of the inhibitory effect of U0 was not due to an incomplete inhibition, because U0 was added each time before LPS treatments, and single treatment of U0 was able to inhibit ERK phosphorylation for 24 h (data not shown). Consistent with these results, only SB was able to induce HDAC8 production and subsequent hypo-acetylation of H3K27 in a PCI-sensitive manner after 12–24 h of treatments (Fig. 5, C and D). U0 had no effects on HDAC8 expression (Fig. 5C) and rather induced hyper-acetylation of H3K27 (Fig. 5C, lane 3). As expected, inhibition of pro-IL-1β eRNA and mRNA production by SB was prevented by PCI (Fig. 5E). Because induction of HDAC8 expression and H3K27Ac deacetylation was a late event, occurring ∼12 h after p38 inhibition, we examined the kinetics of PCI effects on pro-IL-1β eRNA and mRNA productions in LeTx-exposed cells. In line with the results shown in Fig. 5, B and C, PCI gradually prevented the inhibitory effects of LeTx only after 12 h of LeTx treatments (Fig. 5F). These results suggest that HDAC8 inhibition restored the LPS response only at the late stage of LeTx intoxication.
FIGURE 5.
HDAC8 involvement in the LeTx-induced inhibition of pro-IL-1β eRNA and mRNA expression in a p38 inhibition-dependent manner. A, RAW264.7 cells were pretreated with SB203580 (SB; 10 μm) or U0126 (U0; 10 μm) for 18–20 h and then treated with LPS (100 ng/ml) in the absence or presence of inhibitors for 1.5 h. Pro-IL-1β eRNA production was analyzed by qPCR. Data are expressed as means ± S.D. (n = 3). B, RAW264.7 cells were pretreated with SB (10 μm) or U0 (10 μm) or SB and U0 for the time indicated and stimulated with LPS (100 ng/ml) for 1.5 h. Pro-IL-1 β eRNA and mRNA productions were analyzed by qPCR. C, RAW264.7 cells were treated with SB (10 μm) or U0 (10 μm), and HDAC8 protein and H3K27Ac levels were analyzed by Western blot. Western blots for β-actin and histone 3 (H3) were used as loading controls. D, RAW264.7 cells were cultured in the presence or absence of SB (5–15 μm) ± PCI (20 nm) for 20 h and then treated with LPS (100 ng/ml) for 3 h. H3K27Ac levels were analyzed by Western blot. Western blots for histone H3 were used as loading controls. Bands intensities compared with loading controls were analyzed by the ImageJ program (National Institutes of Health) and are expressed as means ± S.D. (n = 3); Student's t test; *, p < 0.05. E, similarly, cells were pretreated with SB (10 μm) ± PCI (20 nm) for 20 h and then treated with LPS (100 ng/ml) for 90 min. Pro-IL-1β eRNA and mRNA productions were analyzed by qPCR. F, RAW264.7 cells were treated with LeTx (100 ng/ml LF and 100 ng/ml PA) for 2 h and further cultured in the fresh media with or without PCI (20 nm). LeTx-exposed or non-exposed cells were treated with LPS (100 ng/ml) for 1.5 h at the time indicated after LeTx treatment. Pro-IL-1β eRNA and mRNA levels were measured by qPCR. Data are expressed as means ± S.D. (n = 3).
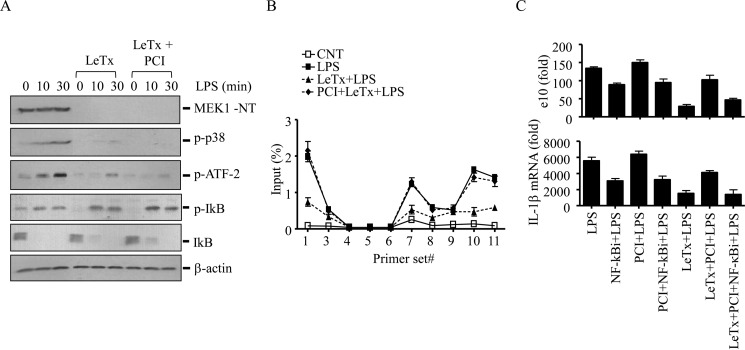
HDAC8 Inhibition Increases LPS-induced Recruitment of NF-κB to the Pro-IL-1β Enhancer Regions in LeTx-treated Macrophages
Because inhibition of p38 was involved in the prolonged inhibition of pro-IL-1β production, we examined whether HDAC8 inhibition reversed p38 signaling events prevented by LeTx. LeTx abolished phosphorylation of p38 and its substrate transcription factor ATF-2, and PCI had no effects on these inhibitory effects (Fig. 6A). Unlike MAPKs, LeTx had no effects on the activation of NF-κB induced by LPS based on phosphorylation and degradation of the inhibitor κB (IκB; Fig. 6A). Therefore, we examined the recruitment of NF-κB to the enhancer and promoter regions of pro-IL-1β by NF-κB ChIP-qPCR analysis. As shown in Fig. 6B, LPS increased recruitment of NF-κB to the H3K27Ac-associated enhancer regions (peaks 1 and 2) and the promoter region adjacent to the pro-IL-1β TSS (primer sets 10 and 11). LeTx inhibited NF-κB recruitment to these regions, which was prevented by PCI pre-treatments (Fig. 6B). Furthermore, inhibition of NF-κB by the chemical inhibitor (NF-κBi) partially suppressed the production of pro-IL-1β eRNA and mRNA (Fig. 6C). However, unlike in LeTx-exposed cells, PCI failed to restore the production in NF-κBi-treated cells. Also, NF-κBi was able to prevent the effect of PCI in reversing the inhibitory effects of LeTx. Collectively, these results suggest that NF-κB was involved in the activation of pro-IL-1β enhancer through a signaling event residing downstream of HDAC8/H3K27Ac, and that the inhibition of HDAC8 likely reinstated LPS responses through enhancing recruitment of NF-κB to the enhancer regions in LeTx-exposed macrophages (Fig. 7).
FIGURE 6.
HDAC8 inhibition increases LPS-induced NF-κB association into the pro-IL-1β enhancer region in LeTx-exposed cells. RAW264.7 cells were treated with a sub-lethal dose of LeTx (100 ng/ml LF and 100 ng/ml PA) for 4 h and further cultured in fresh media for 16 h. These cells were then incubated with or without the HDAC8-specific inhibitor PCI (20 nm) for an additional 20 h and treated with LPS (100 ng/ml) for the time indicated. A, levels of phospho-p38, phospho-ATF-2, phospho-IκB, total IκB, and MEK1 were analyzed by Western blots. Immunoblotting for β-actin was used as loading controls. B, cells were stimulated with LPS (100 ng/ml) for 45 min, and the levels of NF-κB association with the enhancer and promoter areas of pro-IL-1β were analyzed by ChIP-qPCR. C, cells were pretreated with NF-κB inhibitor (0.5 μm) 2 h before LPS treatment and exposed to LPS (100 ng/ml) for 1.5 or 3 h for eRNA or mRNA analysis. Pro-IL-1β eRNA and mRNA levels were measured by qPCR. Data are expressed as means ± S.D. (n = 3).
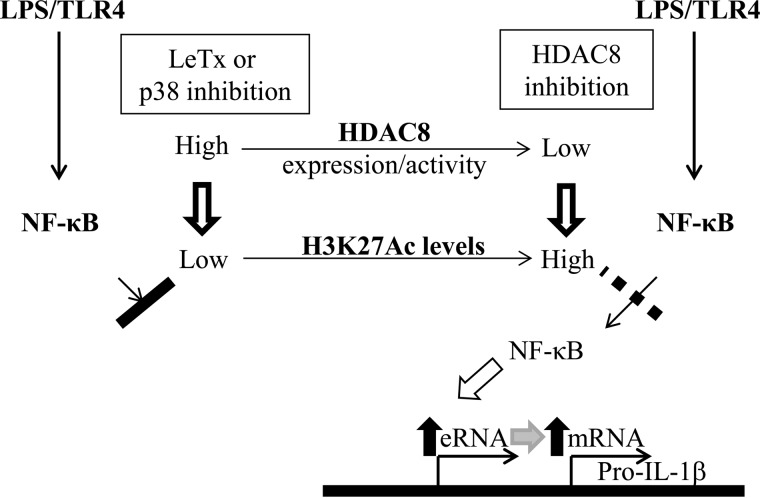
FIGURE 7.
Schematic presentation of the proposed role of HDAC8 and H3K27Ac in regulating LPS-induced pro-IL-1β eRNA production in LeTx-exposed macrophages. LeTx or p38 inhibition causes hypo-acetylation of H3K27 and limits accessibility of NF-κB to the pro-IL-1β enhancer region, which leads to decrease of pro-IL-1β eRNA and subsequent mRNA production. Inhibition of HDAC8 induces hyper-acetylation of H3K27, which allows NF-κB access to the enhancer region in response to LPS, and reverses the inhibitory effects of LeTx or p38 inhibition.
Discussion
Activation of the microbial sensing pattern recognition receptors, such as TLR4 by LPS, activates latent and signal-dependent transcription factors that induce expression of various inflammatory genes by activating promoters (58, 59). A large number of enhancers are also activated by LPS, producing eRNAs before or at the same time as the promoter-driven mRNA production (42, 43, 49, 60). These eRNAs have shown to be involved in the formation of enhancer-promoter chromatin loops and enhancement of mRNA transcription in cis (61, 62). Here, we showed that LPS induced two peaks of eRNA production within ∼15 kb upstream to the pro-IL-1β TSS in RAW264.7 macrophages (Fig. 4B). In line with a previous study in human monocyte THP-1 cells (60), antisense LNATM GapmeRs were able to knock down the eRNA, which resulted in the inhibition of pro-IL-1β mRNA production induced by LPS (Fig. 4, D and E). These results suggest that the eRNA is indeed involved in the production of pro-IL-1β mRNA by LPS in murine macrophages. To date, little is known about the signaling pathways involved in regulating enhancer activities. A recent study showed that transcription factors activated by divergent signaling pathways, including ERKs, p38 and NF-κB are involved in eRNA production in LPS-stimulated THP-1 monocytes (63). We also showed that all ERKs, p38 and NF-κB pathways, were required for optimal production of pro-IL-1β eRNA and mRNA production but in different kinetics (Figs. 5B and 6C).
In general, HDACs suppress gene transcription through deacetylating N-terminal tails of core histones and interacting with co-repressors, resulting in the formation of heterochromatin and transcriptional deactivation, respectively (64). HDAC8 is a member of the class I HDACs that are mainly localized in the nucleus and deacetylate core histones (40). However, unlike others, HDAC8 is localized both in the cytoplasm and nucleus and was shown to deacetylate the core histones H2A/H2B, H3, and H4 in vitro (38–40) but not yet in vivo (41, 56, 65, 66). Failure in detecting histones as in vivo substrates of HDAC8 could be due to highly complex post-translational modifications at the N terminus of histones and limitations in sensitivity and specificity of the experimental approaches. Our approaches using antibodies that can detect specific acetylation sites of histone H3 found a strong reverse-correlation between the level of HDAC8 expression and H3K27Ac (Fig. 3, D and E). Immunoprecipitated endogenous HDAC8 or ectopically expressed EGFP-HDAC8 also selectively deacetylated H3K27Ac in a PCI-sensitive manner (Fig. 2). In addition, H3K27Ac ChIP-qPCR analysis suggested that HDAC8 inhibition enhanced association of H3K27Ac with the enhancer region of pro-IL-1β but not with the intronic region (Fig. 4F). This selective activity of HDAC8 on H3K27Ac probably explains the failure in detecting changes in global histone H3 acetylation in PCI-treated cells (67) and preference of HDAC8 for specific gene regions in H3 deacetylation (68). Collectively, these results suggest that HDAC8 is involved in the regulation of pro-IL-1β enhancer activity through deacetylation of H3K27Ac.
Although this study established a good correlation between HDAC8 and the level of H3K27Ac, it is still possible that HDAC8 regulates pro-IL-1β eRNA/mRNA independent of H3K27Ac. HDAC8 was shown to be involved in adenoviral E1A-12 protein-mediated gene suppression (69) and transcriptional repression by the inversion-16 fusion gene products in acute myeloid leukemia cells (70). However, contribution of these genes to LPS immune responses is unknown. In addition, several proteins, including protein phosphatase (PP) 1, heat shock proteins, cofilin, α-actin, the structural maintenance of chromosome-3 (SMC3), ARID1A, MLL2, and the estrogen-related receptor-α, have been shown to interact with HDAC8 (65, 71–75). Involvement of these proteins in immune responses has yet to be explored. Interestingly, association of HDAC8 with PP1 inhibits cAMP-response element-binding protein activity (76), which could influence IL-1β production. However, other HDACs also have the same effect (73); thus, the HDAC8-specific effect on LeTx-exposed macrophages (Fig. 1B) suggests a distinct mechanism. Of interest, HDAC8 was shown to target the cohesin subunit SMC3 and lysine-specific demethylase MLL2. The cohesin complex is known to promote registration of promoter-enhancer DNA elements and to recruit or stabilize transcription factors (42). MLL2 is involved in demethylating mono-methylated H3K4 (77), which is a marker for enhancers (77–80). Thus, it is possible that HDAC8 regulates eRNA and mRNA expression through these molecules. In addition, HDAC8 was also shown to target histone H4 (41). Because we have not fully examined deacetylation activity of HDAC8 toward histone H4, we could not rule out the contributions of histone H4 deacetylation to the effect of HDAC8 inhibition. Further extensive studies are required to address these issues.
Here, we showed that HDAC8 was involved in the suppression of IL-1β eRNA/mRNA production only at the late stage (after 12 h exposure) of LeTx intoxication (Fig. 5F). Cleavage of MEKs by LeTx causes complete inactivation of both ERKs and p38 for over 48 h (Figs. 1A and 6A). Inactivation of p38 caused persistent inhibition of pro-IL-1β expression, whereas inactivation of ERKs only led to transient (<12 h) inhibition and even further enhanced the production 24 h after treatments (Fig. 5B). The cause of the transient and/or enhancing effects of ERK inhibition is unknown at this moment. Because ERK inhibition induced a robust increase of H3K27Ac levels 12 h after the treatment (Fig. 5C), the levels of H3K27Ac could be a compensatory mechanism induced by the lack of ERKs activation. Because the levels of HDAC8 expression did not change by ERK inhibition (Fig. 5C), the increase of H3K27Ac levels could be due to enhanced H3K27 acetyltransferase activity. Previously, inhibition of ERKs was shown to prevent the H3K27 deacetylase p300 (81) from degradation and increased histone H3 acetylation (82).
Interestingly, dual inhibition of ERKs and p38 persistently suppressed the production of pro-IL-1β eRNA and mRNA (Fig. 5B), which was consistent with LeTx treatment (Figs. 1D, 4G, and 5F). Because p38 inhibition alone was able to mimic the dual inhibition or LeTx effects (Fig. 5B), we believe that p38 inhibition was the main contributor to the prolonged inhibitory effect of LeTx. Because the inhibitory effects were prevented by PCI and si-HDAC8 (Fig. 5, E and F), HDAC8 likely played a key role in the effect. Further examination on the mechanism of HDAC8 in the regulation of eRNA/mRNA production showed that HDAC8 inhibition by PCI facilitated the recruitment of NF-κB to the enhancer and promoter regions of pro-IL-1β (Fig. 6B). However, PCI was not able to reverse the effects of the NF-κB inhibition (Fig. 6C). Based on these results, we believe that HDAC8 induced by LeTx or p38 inhibition leads to hypo-acetylation of H3K27 and limits accessibility of NF-κB to the pro-IL-1β enhancer and promoter regions in macrophages; however, hyper-acetylation of H3K27 induced by HDAC8 inhibition reverses the effects of LeTx and p38 inhibition (Fig. 7).
To date, the role of HDAC8 in immune regulation has been controversial. Previously, the inhibition of HDAC8 improves immune responses by increasing interferon regulatory factor 3 (IRF3) and NF-κB activities (83); however, the HDAC8-selective inhibitor ITF3056 suppresses TNF-α and IL-1β production (84). Contradicting results on the role of HDAC8 in immune response are likely due to specificities of inhibitors, particularly at high doses of inhibitors, and/or multiple targets of HDAC8 (both in the cytoplasm and nucleus). Our study clearly suggested that HDAC8 is involved in the suppression of macrophage immune responses when p38 is inhibited; however, further detailed studies are required for the role of HDAC8 in immune responses.
LeTx is a key B. anthracis virulence factor inhibiting immune responses (1). During inhalation anthrax infections, LeTx is produced by germinating spores at the early phase of infection (85), and systemic production of LeTx and bacteremia exhibit a multiphasic kinetic profile as follows: low levels at 24 h, increased at 48 h, declined at 72 h, and then increased at 96–120 h of infection when the host's condition deteriorates (86). Therefore, the delayed immune suppression possibly mediated by HDAC8 may play a key role in the later stage immune evasion and subsequent fulminant infection. In addition, other bacterial toxins such as listeriolysine O and OspF, released by Listeria monocytogenes and Shigella flexneri, respectively, also inhibit the MAPK pathway and host immune responses (15, 87–89). Although the involvement of HDAC8 in immune suppression during these infections remains to be explored, we speculate that HDAC8 inhibition could be beneficial for these infectious diseases. In summary, this study for the first time demonstrated that inhibition of HDAC8 restored the expression of pro-IL-1β in LeTx-exposed macrophages, likely by activating H3K27Ac-mediated enhancer activity.
Author Contributions
S. O. K. conceived and coordinated the study and wrote the paper. S. D. H. designed, performed, and analyzed the experiments shown in Figs. 1–6. C. R. designed, performed, and analyzed the experiments shown in Fig. 4F. S. M. provided technical assistance and contributed to the preparation of the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Dr. Aurigemma (NCI-Frederick Cancer Research and Development Center, Frederick, MD) for providing pro-IL-1β antibody.
This work was supported by Natural Sciences and Engineering Research of Canada Grant RGPIN 312482-2013 (to S. O. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- PA
- protective antigen
- LF
- lethal factor
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- EGFP
- enhanced GFP
- BMDM
- bone marrow-derived macrophage
- F
- forward
- R
- reverse
- qPCR
- quantitative real time PCR
- TSS
- transcription start site
- SB
- SB203580
- U0
- U0126
- ASO
- antisense oligonucleotide
- PCI
- PCI-34051.
References
- 1. Hicks C. W., Cui X., Sweeney D. A., Li Y., Barochia A., and Eichacker P. Q. (2011) The potential contributions of lethal and edema toxins to the pathogenesis of anthrax associated shock. Toxins 3, 1185–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agrawal A., Lingappa J., Leppla S. H., Agrawal S., Jabbar A., Quinn C., and Pulendran B. (2003) Impairment of dendritic cells and adaptive immunity by anthrax lethal toxin. Nature 424, 329–334 [DOI] [PubMed] [Google Scholar]
- 3. Turk B. E. (2007) Manipulation of host signalling pathways by anthrax toxins. Biochem. J. 402, 405–417 [DOI] [PubMed] [Google Scholar]
- 4. Ha S. D., Ng D., Pelech S. L., and Kim S. O. (2007) Critical role of the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase-3 signaling pathway in recovery from anthrax lethal toxin-induced cell cycle arrest and MEK cleavage in macrophages. J. Biol. Chem. 282, 36230–36239 [DOI] [PubMed] [Google Scholar]
- 5. Baldari C. T., Tonello F., Paccani S. R., and Montecucco C. (2006) Anthrax toxins: a paradigm of bacterial immune suppression. Trends Immunol. 27, 434–440 [DOI] [PubMed] [Google Scholar]
- 6. Milne J. C., Blanke S. R., Hanna P. C., and Collier R. J. (1995) Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol. Microbiol. 15, 661–666 [DOI] [PubMed] [Google Scholar]
- 7. Milne J. C., and Collier R. J. (1993) pH-dependent permeabilization of the plasma membrane of mammalian cells by anthrax protective antigen. Mol. Microbiol. 10, 647–653 [DOI] [PubMed] [Google Scholar]
- 8. Klimpel K. R., Arora N., and Leppla S. H. (1994) Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol. Microbiol. 13, 1093–1100 [DOI] [PubMed] [Google Scholar]
- 9. Tonello F., Ascenzi P., and Montecucco C. (2003) The metalloproteolytic activity of the anthrax lethal factor is substrate-inhibited. J. Biol. Chem. 278, 40075–40078 [DOI] [PubMed] [Google Scholar]
- 10. Ascenzi P., Visca P., Ippolito G., Spallarossa A., Bolognesi M., and Montecucco C. (2002) Anthrax toxin: a tripartite lethal combination. FEBS Lett. 531, 384–388 [DOI] [PubMed] [Google Scholar]
- 11. Vitale G., Bernardi L., Napolitani G., Mock M., and Montecucco C. (2000) Susceptibility of mitogen-activated protein kinase kinase family members to proteolysis by anthrax lethal factor. Biochem. J. 352, 739–745 [PMC free article] [PubMed] [Google Scholar]
- 12. Xu L., Fang H., and Frucht D. M. (2008) Anthrax lethal toxin increases superoxide production in murine neutrophils via differential effects on MAPK signaling pathways. J. Immunol. 180, 4139–4147 [DOI] [PubMed] [Google Scholar]
- 13. Kassam A., Der S. D., and Mogridge J. (2005) Differentiation of human monocytic cell lines confers susceptibility to Bacillus anthracis lethal toxin. Cell. Microbiol. 7, 281–292 [DOI] [PubMed] [Google Scholar]
- 14. Ha S. D., Ng D., Lamothe J., Valvano M. A., Han J., and Kim S. O. (2007) Mitochondrial proteins Bnip3 and Bnip3L are involved in anthrax lethal toxin-induced macrophage cell death. J. Biol. Chem. 282, 26275–26283 [DOI] [PubMed] [Google Scholar]
- 15. Raymond B., Batsche E., Boutillon F., Wu Y. Z., Leduc D., Balloy V., Raoust E., Muchardt C., Goossens P. L., and Touqui L. (2009) Anthrax lethal toxin impairs IL-8 expression in epithelial cells through inhibition of histone H3 modification. PLoS Pathog. 5, e1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ribot W. J., Panchal R. G., Brittingham K. C., Ruthel G., Kenny T. A., Lane D., Curry B., Hoover T. A., Friedlander A. M., and Bavari S. (2006) Anthrax lethal toxin impairs innate immune functions of alveolar macrophages and facilitates Bacillus anthracis survival. Infect. Immun. 74, 5029–5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Erwin J. L., DaSilva L. M., Bavari S., Little S. F., Friedlander A. M., and Chanh T. C. (2001) Macrophage-derived cell lines do not express proinflammatory cytokines after exposure to Bacillus anthracis lethal toxin. Infect. Immun. 69, 1175–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pellizzari R., Guidi-Rontani C., Vitale G., Mock M., and Montecucco C. (1999) Anthrax lethal factor cleaves MKK3 in macrophages and inhibits the LPS/IFNγ-induced release of NO and TNFα. FEBS Lett. 462, 199–204 [DOI] [PubMed] [Google Scholar]
- 19. Terra J. K., Cote C. K., France B., Jenkins A. L., Bozue J. A., Welkos S. L., LeVine S. M., and Bradley K. A. (2010) Cutting edge: resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. J. Immunol. 184, 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lara-Tejero M., Sutterwala F. S., Ogura Y., Grant E. P., Bertin J., Coyle A. J., Flavell R. A., and Galán J. E. (2006) Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J. Exp. Med. 203, 1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedra J. H., Sutterwala F. S., Sukumaran B., Ogura Y., Qian F., Montgomery R. R., Flavell R. A., and Fikrig E. (2007) ASC/PYCARD and caspase-1 regulate the IL-18/IFN-γ axis during Anaplasma phagocytophilum infection. J. Immunol. 179, 4783–4791 [DOI] [PubMed] [Google Scholar]
- 22. Raupach B., Peuschel S. K., Monack D. M., and Zychlinsky A. (2006) Caspase-1-mediated activation of interleukin-1β (IL-1β) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect. Immun. 74, 4922–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mariathasan S., Weiss D. S., Dixit V. M., and Monack D. M. (2005) Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J. Exp. Med. 202, 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsuji N. M., Tsutsui H., Seki E., Kuida K., Okamura H., Nakanishi K., and Flavell R. A. (2004) Roles of caspase-1 in Listeria infection in mice. Int. Immunol. 16, 335–343 [DOI] [PubMed] [Google Scholar]
- 25. Sansonetti P. J., Phalipon A., Arondel J., Thirumalai K., Banerjee S., Akira S., Takeda K., and Zychlinsky A. (2000) Caspase-1 activation of IL-1β and IL-18 are essential for Shigella flexneri-induced inflammation. Immunity 12, 581–590 [DOI] [PubMed] [Google Scholar]
- 26. Fortier A., Diez E., and Gros P. (2005) Naip5/Birc1e and susceptibility to Legionella pneumophila. Trends Microbiol. 13, 328–335 [DOI] [PubMed] [Google Scholar]
- 27. Fukao T. (2004) Immune system paralysis by anthrax lethal toxin: the roles of innate and adaptive immunity. Lancet Infect. Dis. 4, 166–170 [DOI] [PubMed] [Google Scholar]
- 28. Coggeshall K. M., Lupu F., Ballard J., Metcalf J. P., James J. A., Farris D., and Kurosawa S. (2013) The sepsis model: an emerging hypothesis for the lethality of inhalation anthrax. J. Cell Mol. Med. 17, 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Santa F., Totaro M. G., Prosperini E., Notarbartolo S., Testa G., and Natoli G. (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 30. Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., Honma K., Matsuyama T., Yui K., Tsujimura T., Standley D. M., et al. (2010) The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 [DOI] [PubMed] [Google Scholar]
- 31. Ishii M., Wen H., Corsa C. A., Liu T., Coelho A. L., Allen R. M., Carson W. F. 4th., Cavassani K. A., Li X., Lukacs N. W., Hogaboam C. M., Dou Y., and Kunkel S. L. (2009) Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yasui T., Hirose J., Tsutsumi S., Nakamura K., Aburatani H., and Tanaka S. (2011) Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J. Bone Miner. Res. 26, 2665–2671 [DOI] [PubMed] [Google Scholar]
- 33. Foster S. L., Hargreaves D. C., and Medzhitov R. (2007) Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 [DOI] [PubMed] [Google Scholar]
- 34. Chen X., El Gazzar M., Yoza B. K., and McCall C. E. (2009) The NF-κB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J. Biol. Chem. 284, 27857–27865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu T. F., Yoza B. K., El Gazzar M., Vachharajani V. T., and McCall C. E. (2011) NAD+-dependent SIRT1 deacetylase participates in epigenetic reprogramming during endotoxin tolerance. J. Biol. Chem. 286, 9856–9864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scarpa M., and Stylianou E. (2012) Epigenetics: concepts and relevance to IBD pathogenesis. Inflamm. Bowel. Dis. 18, 1982–1996 [DOI] [PubMed] [Google Scholar]
- 37. Ha S. D., Han C. Y., Reid C., and Kim S. O. (2014) HDAC8-mediated epigenetic reprogramming plays a key role in resistance to anthrax lethal toxin-induced pyroptosis in macrophages. J. Immunol. 193, 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu E., Chen Z., Fredrickson T., Zhu Y., Kirkpatrick R., Zhang G. F., Johanson K., Sung C. M., Liu R., and Winkler J. (2000) Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J. Biol. Chem. 275, 15254–15264 [DOI] [PubMed] [Google Scholar]
- 39. Buggy J. J., Sideris M. L., Mak P., Lorimer D. D., McIntosh B., and Clark J. M. (2000) Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem. J. 350, 199–205 [PMC free article] [PubMed] [Google Scholar]
- 40. Gregoretti I. V., Lee Y. M., and Goodson H. V. (2004) Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338, 17–31 [DOI] [PubMed] [Google Scholar]
- 41. Wolfson N. A., Pitcairn C. A., and Fierke C. A. (2013) HDAC8 substrates: histones and beyond. Biopolymers 99, 112–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith E., and Shilatifard A. (2014) Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 21, 210–219 [DOI] [PubMed] [Google Scholar]
- 43. Lam M. T., Li W., Rosenfeld M. G., and Glass C. K. (2014) Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 39, 170–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., Wu J., Harmin D. A., Laptewicz M., Barbara-Haley K., Kuersten S., Markenscoff-Papadimitriou E., Kuhl D., Bito H., Worley P. F., Kreiman G., and Greenberg M. E. (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Natoli G., and Andrau J. C. (2012) Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 46, 1–19 [DOI] [PubMed] [Google Scholar]
- 46. Ørom U. A., and Shiekhattar R. (2013) Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154, 1190–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B. K., Muller H., Ragoussis J., Wei C. L., and Natoli G. (2010) A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLos Biol. 8, e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim T. K., Hemberg M., and Gray J. M. (2015) Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 7, a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaikkonen M. U., Spann N. J., Heinz S., Romanoski C. E., Allison K. A., Stender J. D., Chun H. B., Tough D. F., Prinjha R. K., Benner C., and Glass C. K. (2013) Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A., Boyer L. A., Young R. A., and Jaenisch R. (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., and Wysocka J. (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zentner G. E., Tesar P. J., and Scacheri P. C. (2011) Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 21, 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ha S. D., Park S., Han C. Y., Nguyen M. L., and Kim S. O. (2012) Cellular adaptation to anthrax lethal toxin-induced mitochondrial cholesterol enrichment, hyperpolarization, and reactive oxygen species generation through downregulating MLN64 in macrophages. Mol. Cell. Biol. 32, 4846–4860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Warren M. K., and Vogel S. N. (1985) Bone marrow-derived macrophages: development and regulation of differentiation markers by colony-stimulating factor and interferons. J. Immunol. 134, 982–989 [PubMed] [Google Scholar]
- 55. Park S., Ha S. D., Coleman M., Meshkibaf S., and Kim S. O. (2013) p62/SQSTM1 enhances NOD2-mediated signaling and cytokine production through stabilizing NOD2 oligomerization. PLoS ONE 8, e57138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Murko C., Lagger S., Steiner M., Seiser C., Schoefer C., and Pusch O. (2010) Expression of class I histone deacetylases during chick and mouse development. Int. J. Dev. Biol. 54, 1527–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lam M. T., Cho H., Lesch H. P., Gosselin D., Heinz S., Tanaka-Oishi Y., Benner C., Kaikkonen M. U., Kim A. S., Kosaka M., Lee C. Y., Watt A., Grossman T. R., Rosenfeld M. G., Evans R. M., and Glass C. K. (2013) Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Medzhitov R., and Horng T. (2009) Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9, 692–703 [DOI] [PubMed] [Google Scholar]
- 59. Murray P. J., and Smale S. T. (2012) Restraint of inflammatory signaling by interdependent strata of negative regulatory pathways. Nat. Immunol. 13, 916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. IIott N. E., Heward J. A., Roux B., Tsitsiou E., Fenwick P. S., Lenzi L., Goodhead I., Hertz-Fowler C., Heger A., Hall N., Donnelly L. E., Sims D., and Lindsay M. A. (2014) Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 5, 3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A. Y., Merkurjev D., Zhang J., Ohgi K., Song X., Oh S., Kim H. S., Glass C. K., and Rosenfeld M. G. (2013) Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Melo C. A., Drost J., Wijchers P. J., van de Werken H., de Wit E., Oude Vrielink J. A., Elkon R., Melo S. A., Léveillé N., Kalluri R., de Laat W., and Agami R. (2013) eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 49, 524–535 [DOI] [PubMed] [Google Scholar]
- 63. Heward J. A., Roux B. T., and Lindsay M. A. (2015) Divergent signalling pathways regulate lipopolysaccharide-induced eRNA expression in human monocytic THP1 cells. FEBS Lett. 589, 396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang X. J., and Seto E. (2003) Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr. Opin. Genet. Dev. 13, 143–153 [DOI] [PubMed] [Google Scholar]
- 65. Waltregny D., Glénisson W., Tran S. L., North B. J., Verdin E., Colige A., and Castronovo V. (2005) Histone deacetylase HDAC8 associates with smooth muscle α-actin and is essential for smooth muscle cell contractility. FASEB J. 19, 966–968 [DOI] [PubMed] [Google Scholar]
- 66. Bjerling P., Silverstein R. A., Thon G., Caudy A., Grewal S., and Ekwall K. (2002) Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 22, 2170–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Balasubramanian S., Ramos J., Luo W., Sirisawad M., Verner E., and Buggy J. J. (2008) A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 22, 1026–1034 [DOI] [PubMed] [Google Scholar]
- 68. Saha A., Pandian G. N., Sato S., Taniguchi J., Hashiya K., Bando T., and Sugiyama H. (2013) Synthesis and biological evaluation of a targeted DNA-binding transcriptional activator with HDAC8 inhibitory activity. Bioorg. Med. Chem. 21, 4201–4209 [DOI] [PubMed] [Google Scholar]
- 69. Zhao B., and Ricciardi R. P. (2006) E1A is the component of the MHC class I enhancer complex that mediates HDAC chromatin repression in adenovirus-12 tumorigenic cells. Virology 352, 338–344 [DOI] [PubMed] [Google Scholar]
- 70. Durst K. L., Lutterbach B., Kummalue T., Friedman A. D., and Hiebert S. W. (2003) The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol. Cell. Biol. 23, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wilson B. J., Tremblay A. M., Deblois G., Sylvain-Drolet G., and Giguère V. (2010) An acetylation switch modulates the transcriptional activity of estrogen-related receptor α. Mol. Endocrinol. 24, 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Olson D. E., Udeshi N. D., Wolfson N. A., Pitcairn C. A., Sullivan E. D., Jaffe J. D., Svinkina T., Natoli T., Lu X., Paulk J., McCarren P., Wagner F. F., Barker D., Howe E., Lazzaro F., et al. (2014) An unbiased approach to identify endogenous substrates of “histone” deacetylase 8. ACS Chem. Biol. 9, 2210–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Canettieri G., Morantte I., Guzmán E., Asahara H., Herzig S., Anderson S. D., Yates J. R. 3rd, and Montminy M. (2003) Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat. Struct. Biol. 10, 175–181 [DOI] [PubMed] [Google Scholar]
- 74. Karolczak-Bayatti M., Sweeney M., Cheng J., Edey L., Robson S. C., Ulrich S. M., Treumann A., Taggart M. J., and Europe-Finner G. N. (2011) Acetylation of heat shock protein 20 (Hsp20) regulates human myometrial activity. J. Biol. Chem. 286, 34346–34355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee H., Sengupta N., Villagra A., Rezai-Zadeh N., and Seto E. (2006) Histone deacetylase 8 safeguards the human ever-shorter telomeres 1B (hEST1B) protein from ubiquitin-mediated degradation. Mol. Cell. Biol. 26, 5259–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gao J., Siddoway B., Huang Q., and Xia H. (2009) Inactivation of CREB mediated gene transcription by HDAC8 bound protein phosphatase. Biochem. Biophys. Res. Commun. 379, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang P., Lin C., Smith E. R., Guo H., Sanderson B. W., Wu M., Gogol M., Alexander T., Seidel C., Wiedemann L. M., Ge K., Krumlauf R., and Shilatifard A. (2009) Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29, 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., and Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 79. Herz H. M., Hu D., and Shilatifard A. (2014) Enhancer malfunction in cancer. Mol. Cell 53, 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hu D., Gao X., Morgan M. A., Herz H. M., Smith E. R., and Shilatifard A. (2013) The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y., and Ge K. (2011) Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang S. A., Hung C. Y., Chuang J. Y., Chang W. C., Hsu T. I., and Hung J. J. (2014) Phosphorylation of p300 increases its protein degradation to enhance the lung cancer progression. Biochim. Biophys. Acta 1843, 1135–1149 [DOI] [PubMed] [Google Scholar]
- 83. Nusinzon I., and Horvath C. M. (2006) Positive and negative regulation of the innate antiviral response and β interferon gene expression by deacetylation. Mol. Cell. Biol. 26, 3106–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li S., Fossati G., Marchetti C., Modena D., Pozzi P., Reznikov L. L., Moras M. L., Azam T., Abbate A., Mascagni P., and Dinarello C. A. (2015) Specific inhibition of histone deacetylase 8 reduces gene expression and production of proinflammatory cytokines in vitro and in vivo. J. Biol. Chem. 290, 2368–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cote C. K., and Welkos S. L. (2015) Anthrax toxins in context of Bacillus anthracis spores and spore germination. Toxins 7, 3167–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Boyer A. E., Quinn C. P., Hoffmaster A. R., Kozel T. R., Saile E., Marston C. K., Percival A., Plikaytis B. D., Woolfitt A. R., Gallegos M., Sabourin P., McWilliams L. G., Pirkle J. L., and Barr J. R. (2009) Kinetics of lethal factor and poly-d-glutamic acid antigenemia during inhalation anthrax in rhesus macaques. Infect. Immun. 77, 3432–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Saccani S., Pantano S., and Natoli G. (2002) p38-dependent marking of inflammatory genes for increased NF-κB recruitment. Nat. Immunol. 3, 69–75 [DOI] [PubMed] [Google Scholar]
- 88. Arbibe L., Kim D. W., Batsche E., Pedron T., Mateescu B., Muchardt C., Parsot C., and Sansonetti P. J. (2007) An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8, 47–56 [DOI] [PubMed] [Google Scholar]
- 89. Hamon M. A., Batsché E., Régnault B., Tham T. N., Seveau S., Muchardt C., and Cossart P. (2007) Histone modifications induced by a family of bacterial toxins. Proc. Natl. Acad. Sci. U.S.A. 104, 13467–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]