Abstract
Most proinflammatory actions of C-reactive protein (CRP) are only expressed following dissociation of its native pentameric assembly into monomeric form (mCRP). However, little is known about what underlies the greatly enhanced activities of mCRP. Here we show that a single sequence motif, i.e. cholesterol binding sequence (CBS; a.a. 35–47), is responsible for mediating the interactions of mCRP with diverse ligands. The binding of mCRP to lipoprotein component ApoB, to complement component C1q, to extracellular matrix components fibronectin and collagen, to blood coagulation component fibrinogen, and to membrane lipid component cholesterol, are all found to be markedly inhibited by the synthetic CBS peptide but not by other CRP sequences tested. Likewise, mutating CBS in mCRP also greatly impairs these interactions. Functional experiments further reveal that CBS peptide significantly reduces the effects of mCRP on activation of endothelial cells in vitro and on acute induction of IL-6 in mice. The potency and specificity of CBS are critically determined by the N-terminal residues Cys-36, Leu-37, and His-38; while the versatility of CBS appears to originate from its intrinsically disordered conformation polymorphism. Together, these data unexpectedly identify CBS as the major recognition site of mCRP and suggest that this motif may be exploited to tune the proinflammatory actions of mCRP.
Keywords: atherosclerosis, endothelial dysfunction, extracellular matrix, inflammation, protein motif, C-reactive protein, intrinsically disordered region
Introduction
C-reactive protein (CRP)3 is a major human acute phase reactant composed of five identical subunits (1, 2). Accumulating evidence demonstrates that the actions of CRP depend on conformation and localization (3–5). CRP is primarily produced by the liver and circulates in the blood as a pentamer but is induced to dissociate into subunits (monomeric CRP, mCRP) upon interaction with the microenvironment at inflammatory loci (6–18). mCRP exhibits markedly enhanced activities and recognizes an expanded list of binding partners. Following reduction of the intra-subunit disulfide bond, mCRP can be further activated (19). The degradation of mCRP, on the other hand, would generate bioactive fragments showing anti-inflammatory actions (20–23). The differential contributions of these CRP conformations at distinct locations may therefore account for the intense controversies regarding the function of CRP in animal models and in clinical studies (3–5).
mCRP appears to be the major conformation of CRP that regulates local inflammation (3–5), yet how it acts remains incompletely understood. In particular, though the markedly enhanced binding capability of mCRP underlies its actions, little is known through which sites mCRP recognizes diverse ligands. Consequently, no means is available to specifically modulate the actions of mCRP, which is necessary for clarifying the exact contributions of different CRP conformations in vitro and in vivo. The current study was designed to identify the recognition site of mCRP for ligand binding. The results unexpectedly demonstrated cholesterol binding sequence (CBS) as a versatile motif that mediates the interactions of mCRP with different types of ligands, including two newly identified herein. We further showed that synthetic CBS peptide was able to inhibit the proinflammatory actions of mCRP both in vitro and in vivo. Hence, optimized CBS peptide may be developed as a potential inhibitor of mCRP.
Experimental Procedures
Reagents
Human native CRP (purity > 99%) purified from ascites was purchased from the BindingSite (Birmingham, UK; catalogue number: BP300.X). mCRP and acylated Cys-mutated mCRP was prepared as described (19, 24). Proteins were dialyzed to remove NaN3, and passed through Detoxi-Gel Columns (Thermo Fisher Scientific, Rockford, IL; catalogue number: 20344) to remove endotoxin when necessary. CRP peptides (purity > 98%) were synthesized by Science Peptide Biological Technology (Shanghai, China). Lyophilized peptides were reconstituted aseptically with DMSO at 40 mg/ml and stored at −20 °C in aliquots or kept at 4 °C for a maximum of 1 week. Mouse anti-human mCRP mAbs were generated as described (25).
Ligands examined in the present study include: C1q (Calbiochem; catalogue number: 204876), fibronectin (BD Biocoat, San Jose, CA; catalogue number: 354008), ApoB (Biodesign, Memphis, TN; catalogue number: A50220H), collagen IV (Sigma, catalogue number: C7521), fibrinogen (Merck, Kenilworth, NJ; catalogue number: 341576) and cholesterol (Avanti, Alabaster, AL; catalogue number: 700000). Other reagents were from Sigma unless otherwise stated.
Purification of mCRP Mutants
Expression vectors of His-tagged wild type and mutant mCRP were induced in Escherichia coli. with 0.5 mm IPTG for 4 h at 37 °C. Inclusion bodies were collected, washed, and solubilized overnight with 6 m GuHCl at 4 °C. The supernatants were filtered twice with 0.22 μm membranes and loaded onto Ni-NTA His Bind columns. The bound proteins were washed with PBS containing 40 mm imidazole, 6 m GuHCl, followed by elution with PBS containing 500 mm imidazole, 6 m GuHCl. The eluted proteins were regenerated, citraconylated, and kept in buffers of 10 mm Tris, 15 mm NaCl, pH 7.4.
Ligand Binding
The interaction of mCRP with its ligands were determined with ELISA (19, 26). Briefly, microtiter wells were coated with the indicated ligand overnight at 4 °C. All the following steps were performed at 37 °C. Wells were washed three times with TBS (10 mm Tris, 140 mm NaCl, pH 7.4) containing 0.02% Nonidet P-40 and blocked with 1% BSA, TBS. mCRP with or without indicated synthetic peptide was then added for 1 h. mCRP binding was detected with 3H12 mAb and a HRP-conjugated secondary antibody. Wells were developed with TMB and stopped with 1 m H2SO4.
Membrane Insertion
The interactions of synthetic peptides with lipid membrane were examined with μTrough-S microbalance (Kibron, Finland) (27, 28). The trough was filled with TBS followed by spreading of lipid onto the buffer surface. Peptides were then injected into the subphase to a final concentration of 600 nm through the side hole. The surface pressure of monolayer was recorded at an interval of 1 s for about 6000 s. All experiments were performed at 23.0 ± 0.5 °C. An increase of surface pressure reflects the insertion of peptide into the hydrophobic core of lipid monolayer.
Monocyte Adhesion to Endothelial Cells
Primary human aortic endothelial cells (HAEC, Lifeline Cell Technology, Frederick, MD; catalogue number: FC-0027; lot number: 01381) were cultured as described (19, 29). Confluent HAEC monolayers (passages 3–6) in 24-well plates or on coverslips were rinsed twice with PBS containing 1 mm CaCl2 and 1 mm MgCl2 and labeled with Calcein AM for 15 min at 37 °C. Human monocytic cell line U937 were labeled with Hoechst 33342 for 10 min at 37 °C. HAEC monolayers were pre-incubated with 2.5 mg/ml peptides for 30 min. After gentle rinsing, 2 × 105 U937 cells were added together with 50 μg/ml mCRP for 60 min at 37 °C. Non-adherent monocytes were removed by washing and adherent cells were counted by confocal microscopy.
Induction of Inflammatory Response in Mice
Male Kunming mice (25 ± 2g) were intraperitoneal injected with PBS or mCRP (2.5 mg/kg) with or without the indicated peptide (14 mg/kg). Peritoneal fluid was collected 2 h later and assayed for IL-6 content by specific ELISA kit. The experiments conformed to the Guide for the Care and Use of Laboratory Animals published by NIH, and were conducted according to the protocols approved by the Ethics Committee of Animal Experiments of Xi'an Jiaotong University.
Molecular Dynamics (MD) Simulation
The initial structures of CRP peptides were taken from the crystal structure (PDB 1B09) (30). CBS and control CRP peptides were immersed together into the explicit solvate box with the size of 57 × 50 × 52 Å3 using TIP3P water model with infinite periodic boundary conditions (31). The minimization protocol used the conjugate gradient method. The nonbonded cutoff takes 12 Å for the calculation of electrostatics potential according to particle-mesh Ewald (PME) method. MD simulations were performed by NAMD 2.9 with CHARMM27 force field (32) under constant pressure (1 bar) and temperature (300 K) employing the langevin thermostat and langevin barostat method, respectively. The entire system was minimized for 2 ps, equilibrated for 1000 ps, and finally simulated for 100 ns with a 2-fs step. The results were visualized and analyzed with VMD software (33).
Statistical Analysis
Data were presented as mean ± S.E. Statistical analysis was performed by the two-tailed Student's t test, one-way ANOVA with Tukey post hoc or K-S tests as appropriate. Values of p < 0.05 were considered significant.
Results
Cholesterol Binding Sequence Inhibits mCRP Binding to Diverse Ligands
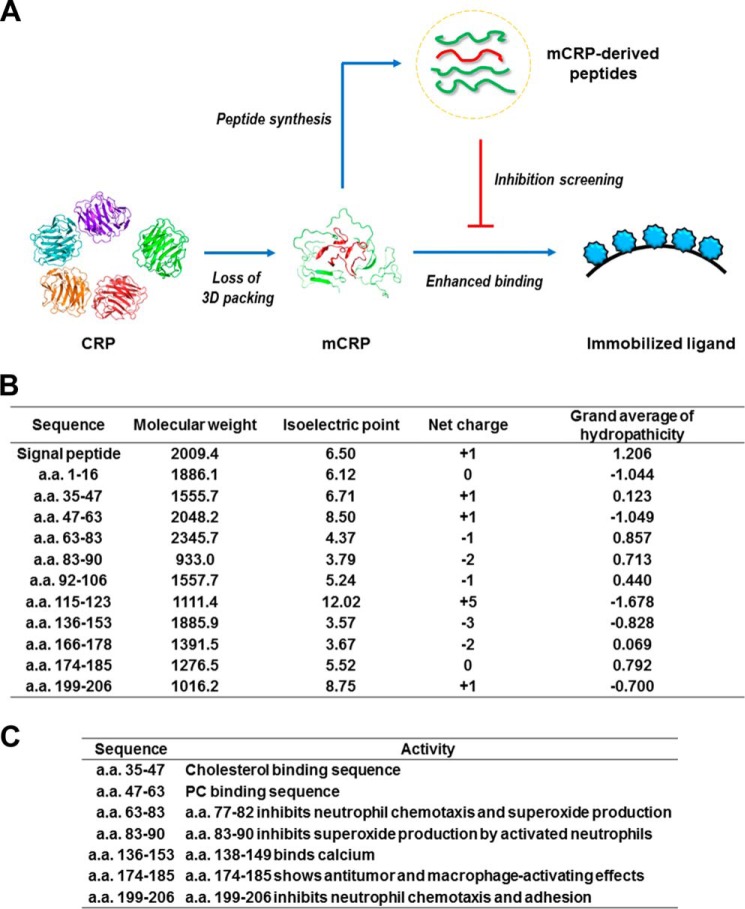
As the generation of mCRP is associated with a dramatic loss of tertiary packing (6, 19, 34), we reasoned that linear sequence motifs may play a predominant role in mediating the interactions of mCRP with its ligands. We thus synthesized a panel of peptides derived from CRP sequence, and examined whether they were inhibitory on ligand binding of mCRP (Fig. 1A). These peptides are distributed across the entire length of CRP sequence and covered the major secondary structure elements, showing variations in their physical properties such as molecular weight, isoelectric point, net charge and hydrophobicity (Fig. 1B). Moreover, most of the peptides have also been reported to be biological active (Fig. 1C) (20–23, 35–41).
FIGURE 1.
An overview of examined CRP peptides. A, experiment design. The physical properties of synthesized peptides computed by ProtParam online tool (B) and their reported bioactivities (C) are also indicated.
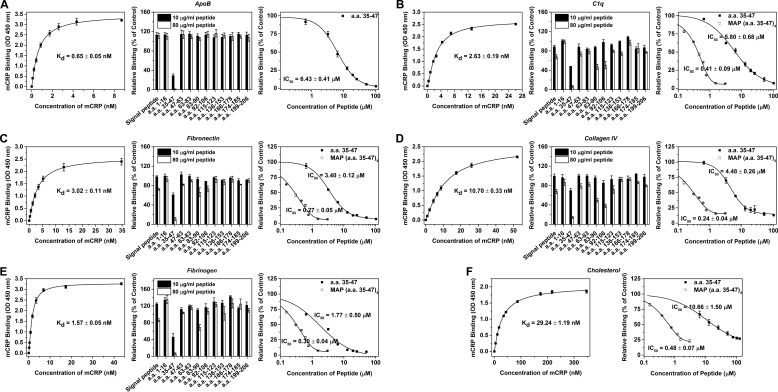
ApoB is a known ligand for mCRP to regulate metabolism of low density lipoprotein (Fig. 2A and ref. 27). Competition assays revealed that a.a. 35–47, i.e. the CBS peptide, was able to abrogate the high affinity binding of fluid phase mCRP to immobilized ApoB; while no inhibition was observed with other peptides (Fig. 2A). When competition assays were conducted in a reverse setting of fluid phase ApoB and immobilized mCRP, comparable results were obtained (data not shown). Though reduced and Cys-mutated mCRP are stronger binder than (non-reduced) mCRP (19), their binding to ApoB could still be abrogated by CBS peptide (data not shown).
FIGURE 2.
CBS peptide inhibits the binding of mCRP to various types of ligands. ApoB (A), C1q (B), fibronectin (C), collagen IV (D), fibrinogen (E) in TBS or cholesterol (F) in ethanol was immobilized onto microtiter wells (n = 3–4). mCRP at different concentrations was added and binding was evaluated using mCRP-specific mAb 3H12 (left panel). The half-saturated binding concentration of mCRP was selected to test the inhibitory effects of co-added peptides (middle panel). The peptide of CBS (a.a. 35–47) emerged as the most potent inhibitor against all interactions. The IC50 of CBS or CBS in multiple antigenic peptide (MAP) format with 4 branches was further determined (right panel).
C1q is a known ligand for mCRP to activate the classical pathway of complement (Fig. 2B and Ref. 26). Because mCRP is predominantly tissue-associated and may contribute to thrombosis (3–5), we also tested whether extracellular matrix (fibronectin and collagen I and IV) and blood coagulation (fibrinogen) proteins were mCRP ligands in the present study. Avid binding of mCRP to these proteins was observed (Fig. 2, C–E), supporting a role of mCRP in tissue localized, extracellular matrix relevant processes. Surprisingly, CBS peptide was again most potent in inhibiting these interactions (Fig. 2, B–E). Amino acid sequences 83–90 and 92–106 also showed some extent of inhibition albeit the potency was much lower, indicating an auxiliary contribution at best.
CBS has been shown to mediate the interaction of mCRP with cholesterol-enriched lipid rafts in cell membranes (41). Accordingly, we found CBS peptide was inhibitory in assays evaluating the binding of mCRP to cholesterol (Fig. 2F). Additional experiments confirmed the direct binding of CBS, with a C-terminal labeled biotin, to all the 6 ligands (data not shown). These suggest that CBS may form the major recognition site of mCRP with sufficient plasticity to accommodate ligands with widely varied topologies, ranging from protein to lipid. Of note, the tendency of mCRP to form high order assemblies (i.e. aggregates) results in the presentation of multivalent binding clusters, which could explain the relative low IC50 of CBS peptide. Accordingly, CBS synthesized as the multiple antigenic peptide with 4 branches improved the IC50 by over an order (Fig. 2).
CBS Exhibits Intrinsically Disordered Region-like Characteristics
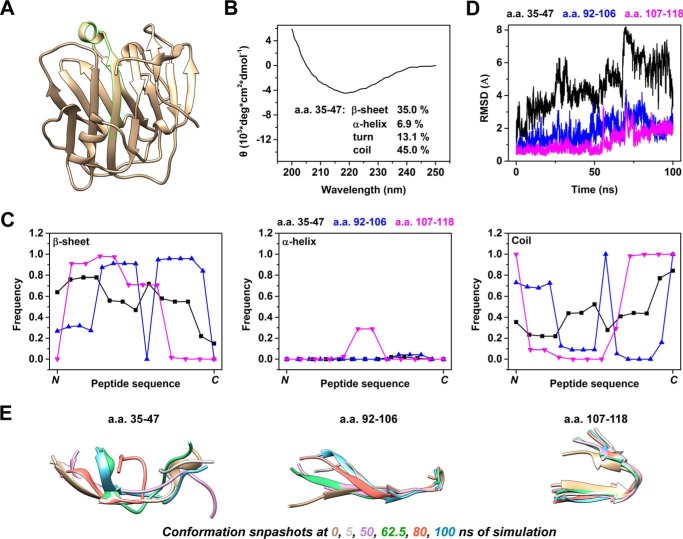
To understand the mechanism of binding, we first characterized the structural features of CBS. Within the crystal structure of CRP subunit as resolved in its pentameric conformation, CBS exhibits β-strand and primarily loop conformation on the N- and C-terminal halves, respectively (Fig. 3A). By using circular dichroism, the secondary structure of synthetic CBS peptide was also determined to be mainly composed of β-sheet (∼35%) and unstructured elements (∼13% turn, ∼45% coil) (Fig. 3B). These features were largely preserved during the 100-ns molecular dynamics simulation, in which the N-terminal portion of CBS peptide was more likely to be β-strand while the C-terminal portion tended to be unstructured (Fig. 3C).
FIGURE 3.
CBS exhibits intrinsically disordered region-like conformation features. A, crystal structure of subunit A of pentameric CRP (PDB 1B09) in which a.a. 35–47 is highlighted. B, circular dichroism spectra of CBS peptide. Estimated contents of secondary structure elements are indicated. C–E, structures of CBS, a.a. 92–106 and a.a. 107–118 were taken from the crystal structure (PDB 1B09) and subjected to a 100-ns molecular dynamics simulation. C, frequency of each residue in the indicated peptide exists as β-sheet, α-helix or coil conformations across the 100-ns simulation. D, backbone root-mean-squared deviation (RMSD) from the initial crystal structure was plotted with time. E, conformation snapshots at 0, 5, 50, 62.5, 80, and 100 ns of simulation. CBS fluctuated through a range of conformations with preserved N-terminal β-strand and C-terminal loop. These were in contrast to the rather stable conformations of control CRP peptides during the simulation.
Nevertheless, CBS peptide also exhibited continuous and prominent conformation fluctuations throughout the simulation. At the residue level, spontaneous and reversible conversion between β-sheet and coil conformations was evident though the probability of conversion was non-uniform along the length of CBS (Fig. 3C). The most striking changes, however, occurred at the level of the overall configuration as evidenced by the up to 8 Å backbone root-mean-squared deviation (Fig. 3D). Such changes were due to large variations in the relative spatial positioning between the N and C termini of CBS peptide (Fig. 3E). By contrast, two control CRP peptides, i.e. a.a. 92–106 (with structural features similar to CBS) and a.a. 107–118, kept their conformations stable during the same simulation.
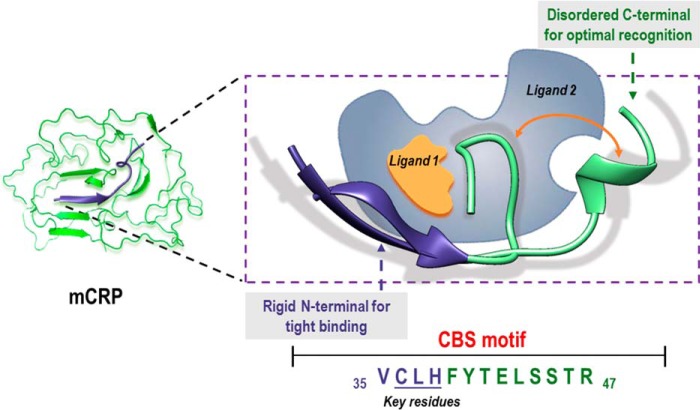
The frequent oscillation to and from coil conformation, the dynamic fluctuation though a range of overall configurations and the binding versatility of CBS closely resemble the structural and functional features of intrinsically disordered region (42). Indeed, most part of CBS except for a.a. 36–39 was predicted to be disordered using the Protein Disorder Prediction Server (43) at a 3% false positive rate cut-off. It has been shown that structural polymorphism of intrinsically disordered region allows dynamic sampling of configurations suitable for recognizing different targets, with the binding process being facilitated by pre-formed secondary structural elements (42). Therefore, we reasoned that the N-terminal residues, in particular a.a. 35–38, with high tendency to form rigid β-sheet (Fig. 3C), provide the scaffold for tight interaction, while the disordered C-terminal portion might confer the required plasticity to accommodate distinct ligands.
N-terminal Residues Determine the Performance of CBS
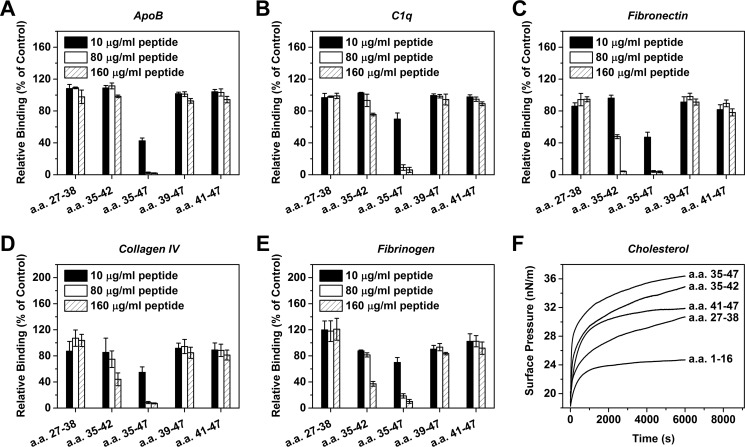
With above hints, we synthesized a series of CBS truncation mutants and assessed their inhibitory effects. The absence of N-terminal residues (i.e. peptides of a.a. 41–47 and a.a. 39–47) abrogated, whereas the absence of C-terminal residues (i.e. peptide of a.a. 35–42) markedly impaired but did not abrogate the capacity of CBS mutants to inhibit ligand binding of mCRP (Fig. 4). These results are consistent with the aforementioned interpretation that the 4 N-terminal residues (i.e. a.a. 35–38) are essential for binding with the C-terminal residues necessary for optimized recognition. The necessity of the latter was further exemplified by the ineffectiveness of a.a. 27–38 in inhibiting mCRP-ligand interactions (Fig. 4).
FIGURE 4.
The N- and C-terminal contribute differentially to the inhibitory capacity of CBS peptide. CBS mutants with truncations in its N or C terminus were tested for their effects on mCRP binding to ApoB (A), C1q (B), fibronectin (C), collagen IV (D), fibrinogen (E), and insertion into the cholesterol-enriched monolayers (F) (n = 3). In A–E, assays were performed as described in Fig. 2. In F, monolayer technique (see “Experimental Procedures”) was used to more appropriately model the interaction of mCRP with cholesterol in a membraneous environment. In most cases, the inhibitory effects of CBS were completely lost following N-terminal truncation, but was not abrogated by C-terminal truncation.
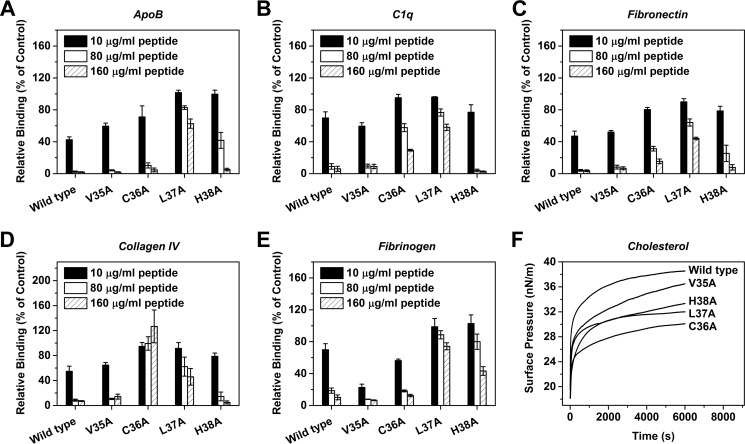
Alanine scanning was then performed to delineate the respective contributions of the 4 N-terminal residues (Fig. 5). The results clearly indicated that the recognition of different ligands by CBS depended on distinct combinations of individual residues. Of the 4 residues, Leu-37 was the most essential one being the common requirement for binding to each ligand. By contrast, Val-35 appeared to be marginally involved. His-38 was also infrequently required, suggesting that CBS binds ligands mainly via hydrophobic interactions. The frequent involvement of Cys-36, on the other hand, highlights the importance of the redox regulation of the intra-subunit disulfide bond on the actions of CBS.
FIGURE 5.
The 3 N-terminal residues determine the ligand specificity of CBS. CBS mutants with point mutations at the indicated N-terminal residues were tested for their effects on mCRP binding to ApoB (A), C1q (B), fibronectin (C), collagen IV (D), fibrinogen (E), and insertion into the cholesterol-enriched monolayers (F) (n = 3). The inhibition of mCRP binding to distinct ligands depend on different combinations of N-terminal residues.
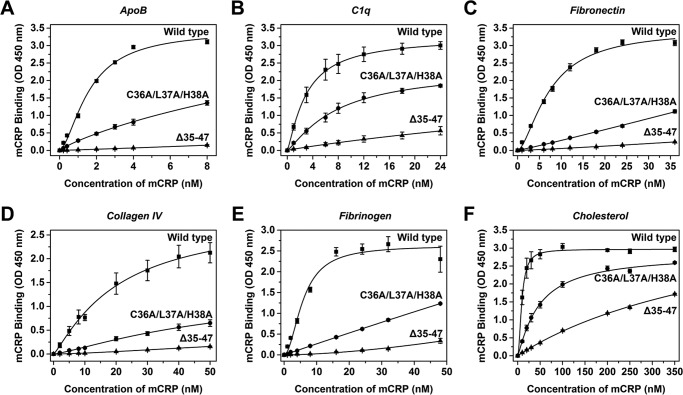
To affirm the role of CBS and its N-terminal residues in mediating the ligand interactions of mCRP, wild type, and CBS-mutated mCRP were expressed in E. coli and purified to homogeneity. Recombinant wild type mCRP showed comparable binding capacity as that prepared by urea-denaturation of purified serum CRP (Fig. 6). Mutating the 3 N-terminal residues in CBS of recombinant full-length mCRP (C36A/L37A/H38A) greatly, albeit differentially, impaired the interaction of recombinant mCRP with ligands. As comparison, deleting the C-terminal residues of CBS (Δ39–47) had only marginal effects on maximal binding of mCRP but showed 5–20 times reduced affinities (data not shown). Complete deletion of CBS (Δ35–47), however, nearly abrogated the binding capacity of mCRP. Together, the above results demonstrate that the specificity and potency of CBS are critically determined by Cys-36, Leu-37, and His-38 with the plasticity of the binding site being conferred by the disordered C-terminal residues (see also Fig. 9).
FIGURE 6.
Mutating CBS in mCRP impairs ligand binding. mCRP wild type or mutants with altered CBS were expressed in E. coli and purified to test their binding to ApoB (A), C1q (B), fibronectin (C), collagen IV (D), fibrinogen (E), and cholesterol (F) (n = 4). The binding of mCRP mutants was greatly impaired or nearly abrogated by mutating the 3 N-terminal residues (C36A/L37A/H38A) or deleting CBS (Δ35–47).
FIGURE 9.
Schematic illustration of CBS-mediated ligand binding of mCRP. The N-terminal residues in β conformation likely constitute the major interface for ligand binding, whereas the intrinsically disordered C-terminal residues may contribute to the optimal recognition of ligands with distinct topologies by dynamical sampling of appropriate spatial configurations.
CBS Inhibits Proinflammatory Actions of mCRP in Vitro and in Vivo
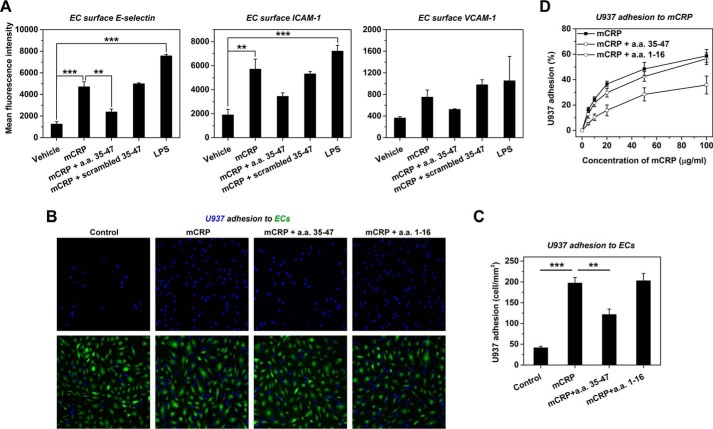
We next examined whether CBS peptide had functional bioactivities in inhibiting the proinflammatory actions of mCRP. mCRP is a potent activator of endothelial cells (ECs) through interaction with cholesterol-enriched lipid rafts when applied apically (29, 44) and EC activation is an early event in atherosclerosis, leading to the adhesion and entry of monocytes into vessel walls (45). Apical stimulation of HAEC monolayers with mCRP strongly up-regulated surface expression of adhesion molecules (Fig. 7A) leading to a 5-fold increase of U937 monocyte adhesion (Fig. 7, B and C). Tissue-associated mCRP may also contribute to the retention of monocytes via interacting with integrins (46). Indeed, mCRP coating greatly enhanced the adhesion of U937 monocytes (Fig. 7D). Notably, these actions of mCRP were all significantly reversed by pre-treatment with CBS but not by scrambled CBS or a control peptide of a.a. 1–16 (Fig. 7).
FIGURE 7.
CBS inhibits mCRP-induced cell adhesion. A, confluent HAEC monolayers were pre-incubated with 0.5 mg/ml CBS or scrambled CBS followed by stimulation with 50 μg/ml Cys-mutated mCRP. Treatment with 1 μg/ml LPS was used as the positive control. Adhesion molecules on the surfaces of HAECs were evaluated by flow cytometry using PE-labeled anti-E-selectin, APC-labeled anti-ICAM-1, and FITC-labeled anti-VCAM-1 mAbs (n = 3). B, confluent monolayers of Calcein AM-labeled HAECs were pre-incubated with 2.5 mg/ml CBS or control peptide (a.a. 1–16) for 30 min. Following gentle washing, Hoechst 33342-labeled U937 cells were added together with 50 μg/ml Cys-mutated mCRP for 60 min. U397 adhesion was visualized by confocal microscopy. In control experiments, peptide pre-treatment and mCRP stimulation were omitted. C, quantification of experiments in B (n = 9). ***, p < 0.001; **, p < 0.01. D, mCRP at the indicated concentrations was immobilized onto microtiter wells overnight at 4 °C. After blocking with 0.1% BSA, U937 cells were added with 2.5 mg/ml CBS or control peptide (a.a. 1–16) for 60 min followed by gentle rinsing and fixation. U937 adhesion was quantified by crystal violet staining (n = 3–4).
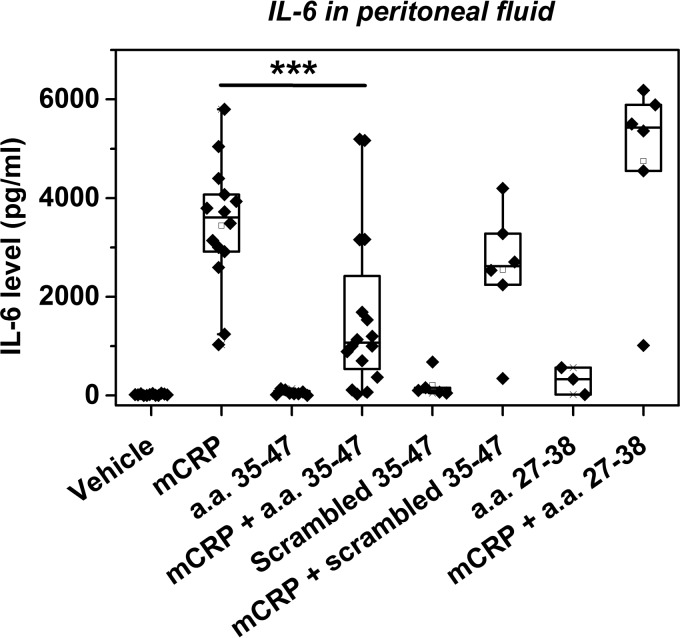
Intraperitoneal injection of mCRP into mice has been shown to induce an acute increase of IL-6 in peritoneal fluid (19). As the majority of peritoneal cells are macrophages (47), they are likely to be the main producers of IL-6 in response to mCRP. Of note, the interactions of mCRP with monocytes (7, 48) and macrophages (27) have both been shown to also depend on lipid rafts. Accordingly, co-injection with CBS peptide markedly inhibited mCRP-induced IL-6 release (Fig. 8). By contrast, neither scrambled CBS nor a control peptide of a.a. 27–38 showed inhibition. Injection of CBS or the control peptides alone did not affect the basal levels of IL-6. These results demonstrate CBS peptide as a functional inhibitor of mCRP.
FIGURE 8.
CBS inhibits mCRP-induced peritoneal IL-6 release. Male Kunming mice were intraperitoneally injected with PBS or Cys-mutated mCRP (2.5 mg/kg) with vehicle, CBS, scrambled CBS or a control peptide (a.a. 27–38) (14 mg/kg). Peritoneal fluid was collected 2 h later and assayed for IL-6. ***, p < 0.001.
Discussion
The major hurdle for defining the pathophysiological role of CRP, particularly in complex or chronic diseases, is the conditional expression of distinct or even contrasting actions by different CRP conformations at systemic or local levels (3–5). Conformation-specific modulation is therefore mandatory to clear this hurdle. With the aid of crystal structure, a specific inhibitor blocking the binding of pentameric CRP to phosphorylcholine has been developed showing beneficial effects in a rat model undergoing acute myocardial infarction (49). However, given the putative function of CRP in host defense (50) and its (tonic and global) anti-inflammatory activities (4, 51), the systemic long-term inhibition appears to be problematic.
mCRP may be the functionally dominant conformation of CRP active in inflammatory loci (3–5). It could be formed by localized dissociation of the hepatically produced CRP, or may be produced locally by stimulated monocytes (3–5). The recurrent promoter mutation of CRP identified in human cancers strongly supports the importance of locally produced CRP and its conversion to mCRP in promoting chronic inflammation underlying tumorigenesis (52, 53). Targeting the local proinflammatory actions of mCRP thus represents not only a valuable research tool but a potential therapy against certain diseases. The lack of high resolution structure of mCRP, however, impedes the rational design of its inhibitor. Nevertheless, the prominent loss of tertiary packing in mCRP (6, 19, 34) hints the contributions of sequence motifs that are amenable to identification by peptide screening.
One striking result of the screening is that a single peptide, corresponding to the CBS motif, abrogates the binding of mCRP to various types of ligands (Fig. 2) and inhibits its proinflammatory actions on endothelial cells and monocytes/macrophages (Figs. 7 and 8). This strongly argues that CBS forms the major recognition site of mCRP. Such an unexpected versatility of CBS is not an artifact because (i) no other tested peptides show comparable behavior (Fig. 2) though CBS appears not to process extraordinary physical properties (Fig. 1), (ii) the features of inhibition by CBS differ with ligands (Fig. 2), and (iii) distinct sets of N-terminal residues of CBS determine its specificity and potency toward different ligand interactions (Fig. 5).
Consistent with the functional importance, CBS, in particular its N-terminal portion, exhibits stronger evolutionary conservation over the full-length sequence (data not shown). Of the N-terminal residues, Cys-36 is invariable during the evolution and is frequently involved in ligand binding (Fig. 5). As Cys-36 normally forms an intra-subunit disulfide bond with Cys-97, this highlights the importance of redox status of Cys-36 in tuning the activities of CBS (19). Nonetheless, such tuning would be expected not to have an on/off consequence for most ligands since Cys-36 is decisive only in interactions with collagen and cholesterol. Rather, the impact would be on the extent of accessibility with a moderate effect on the binding capacity of CBS.
Leu-37 is required for CBS binding to all the tested ligands (Fig. 5). Though less conserved, variants to Leu-37 tend to be hydrophobic. His-38, by contrast, swings between hydrophilic His/Arg/Gln and hydrophobic Tyr/Trp during the evolution (data not shown). Considering the minor contribution of His-38 in CBS recognition, it is plausible that hydrophobic interactions dominate the actions of CBS. On the other hand, Val-35 is not required for ligand binding but is nonetheless highly conserved, suggesting a role of this residue in the structural aspect of CRP. Such information thus provides the basis for rational optimization of CBS peptide to further enhance its performance as an inhibitor of mCRP (Fig. 9).
The relatively low IC50 of CBS peptide determined in in vitro inhibition assays may be the major concern for its future application. Another concern is likely related to the versatility of CBS peptide, because not all ligand interactions lead to a detrimental outcome. These concerns could in principle be addressed by adjusting the key contributing residues in CBS peptide and/or its multivalency to improve potency and/or selectivity. Moreover, the performance of CBS peptide in vivo (Fig. 8) is very much better than that in vitro (Fig. 2), indicating that the experimental setting of the latter might underestimate the actual performance. Therefore, the optimized CBS peptide represents a promising option as a research tool and also a potential therapy.
Author Contributions
H. Y. L., Y. W., and S. R. J. designed the research. H. Y. L., J. W., F. M., Z. K. J., Y. S., Q. F. B., L. L. L., and F. R. M. performed the research. Y. W., S. R. J., H. Y. L., L. A. P., and Y. B. Y. analyzed the data and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Li-Ping Guan and Liang Peng for their excellent technical assistance, and thank the support of computing facilities of Extremadura Research Centre for Advanced Technologies (CETA-CIEMAT).
This work was supported by grants from the National Natural Science Foundation of China (Grant Numbers, 31270813, 31222015, 30930024) and the China Postdoctoral Science Foundation (Grant Numbers 2014M560075). The authors declare that they have no conflicts of interest with the contents of this article.
- CRP
- C-reactive protein
- mCRP
- monomeric C-reactive protein
- CBS
- cholesterol binding sequence
- MD
- molecular dynamics.
References
- 1. Pepys M. B., and Hirschfield G. M. (2003) C-reactive protein: a critical update. J. Clin. Invest. 111, 1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Singh S. K., Suresh M. V., Voleti B., and Agrawal A. (2008) The connection between C-reactive protein and atherosclerosis. Ann. Med. 40, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma X., Ji S. R., and Wu Y. (2013) Regulated conformation changes in C-reactive protein orchestrate its role in atherogenesis. Chinese Sci. Bull. 58, 1642–1649 [Google Scholar]
- 4. Wu Y., Potempa L. A., El Kebir D., and Filep J. G. (2015) C-reactive protein and inflammation: conformational changes affect function. Biol. Chem. 396, 1181–1197 [DOI] [PubMed] [Google Scholar]
- 5. Thiele J. R., Zeller J., Bannasch H., Stark G. B., Peter K., and Eisenhardt S. U. (2015) Targeting C-Reactive protein in inflammatory disease by preventing conformational changes. Mediators Inflamm. 2015, 372432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ji S. R., Wu Y., Zhu L., Potempa L. A., Sheng F. L., Lu W., and Zhao J. (2007) Cell membranes and liposomes dissociate C-reactive protein (CRP) to form a new, biologically active structural intermediate: mCRP(m). FASEB J. 21, 284–294 [DOI] [PubMed] [Google Scholar]
- 7. Eisenhardt S. U., Habersberger J., Murphy A., Chen Y. C., Woollard K. J., Bassler N., Qian H., von Zur Muhlen C., Hagemeyer C. E., Ahrens I., Chin-Dusting J., Bobik A., and Peter K. (2009) Dissociation of pentameric to monomeric C-reactive protein on activated platelets localizes inflammation to atherosclerotic plaques. Circ. Res. 105, 128–137 [DOI] [PubMed] [Google Scholar]
- 8. Molins B., Peña E., de la Torre R., and Badimon L. (2011) Monomeric C-reactive protein is prothrombotic and dissociates from circulating pentameric C-reactive protein on adhered activated platelets under flow. Cardiovasc. Res. 92, 328–337 [DOI] [PubMed] [Google Scholar]
- 9. de la Torre R., Peña E., Vilahur G., Slevin M., and Badimon L. (2013) Monomerization of C-reactive protein requires glycoprotein IIb-IIIa activation: pentraxins and platelet deposition. J. Thromb. Haemost. 11, 2048–2058 [DOI] [PubMed] [Google Scholar]
- 10. Xu P. C., Lin S., Yang X. W., Gu D. M., Yan T. K., Wei L., and Wang B. L. (2015) C-reactive protein enhances activation of coagulation system and inflammatory response through dissociating into monomeric form in antineutrophil cytoplasmic antibody-associated vasculitis. BMC Immunol. 16, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mihlan M., Blom A. M., Kupreishvili K., Lauer N., Stelzner K., Bergström F., Niessen H. W., and Zipfel P. F. (2011) Monomeric C-reactive protein modulates classic complement activation on necrotic cells. FASEB J. 25, 4198–4210 [DOI] [PubMed] [Google Scholar]
- 12. Lauer N., Mihlan M., Hartmann A., Schlötzer-Schrehardt U., Keilhauer C., Scholl H. P., Charbel Issa P., Holz F., Weber B. H., Skerka C., and Zipfel P. F. (2011) Complement regulation at necrotic cell lesions is impaired by the age-related macular degeneration-associated factor-H His402 risk variant. J. Immunol. 187, 4374–4383 [DOI] [PubMed] [Google Scholar]
- 13. Thiele J. R., Habersberger J., Braig D., Schmidt Y., Goerendt K., Maurer V., Bannasch H., Scheichl A., Woollard K. J., von Dobschütz E., Kolodgie F., Virmani R., Stark G. B., Peter K., and Eisenhardt S. U. (2014) Dissociation of pentameric to monomeric C-reactive protein localizes and aggravates inflammation: in vivo proof of a powerful proinflammatory mechanism and a new anti-inflammatory strategy. Circulation 130, 35–50 [DOI] [PubMed] [Google Scholar]
- 14. Braig D., Kaiser B., Thiele J. R., Bannasch H., Peter K., Stark G. B., Koch H. G., and Eisenhardt S. U. (2014) A conformational change of C-reactive protein in burn wounds unmasks its proinflammatory properties. Int. Immunol. 26, 467–478 [DOI] [PubMed] [Google Scholar]
- 15. Habersberger J., Strang F., Scheichl A., Htun N., Bassler N., Merivirta R. M., Diehl P., Krippner G., Meikle P., Eisenhardt S. U., Meredith I., and Peter K. (2012) Circulating microparticles generate and transport monomeric C-reactive protein in patients with myocardial infarction. Cardiovasc Res. 96, 64–72 [DOI] [PubMed] [Google Scholar]
- 16. Wang M. S., Messersmith R. E., and Reed S. M. (2012) Membrane curvature recognition by C-reactive protein using lipoprotein mimics. Soft Matter 8, 7909–7918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strang F., Scheichl A., Chen Y. C., Wang X., Htun N. M., Bassler N., Eisenhardt S. U., Habersberger J., and Peter K. (2012) Amyloid plaques dissociate pentameric to monomeric C-reactive protein: a novel pathomechanism driving cortical inflammation in Alzheimer's disease? Brain Pathol. 22, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hammond D. J. Jr., Singh S. K., Thompson J. A., Beeler B. W., Rusiñol A. E., Pangburn M. K., Potempa L. A., and Agrawal A. (2010) Identification of acidic pH-dependent ligands of pentameric C-reactive protein. J. Biol. Chem. 285, 36235–36244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang M. Y., Ji S. R., Bai C. J., El Kebir D., Li H. Y., Shi J. M., Zhu W., Costantino S., Zhou H. H., Potempa L. A., Zhao J., Filep J. G., and Wu Y. (2011) A redox switch in C-reactive protein modulates activation of endothelial cells. FASEB J. 25, 3186–3196 [DOI] [PubMed] [Google Scholar]
- 20. Heuertz R. M., Ahmed N., and Webster R. O. (1996) Peptides derived from C-reactive protein inhibit neutrophil alveolitis. J. Immunol. 156, 3412–3417 [PubMed] [Google Scholar]
- 21. Zouki C., Beauchamp M., Baron C., and Filep J. G. (1997) Prevention of in vitro neutrophil adhesion to endothelial cells through shedding of L-selectin by C-reactive protein and peptides derived from C-reactive protein. J. Clin. Invest. 100, 522–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. El Kebir D., Zhang Y., Potempa L. A., Wu Y., Fournier A., and Filep J. G. (2011) C-reactive protein-derived peptide 201–206 inhibits neutrophil adhesion to endothelial cells and platelets through CD32. J. Leukoc. Biol. 90, 1167–1175 [DOI] [PubMed] [Google Scholar]
- 23. Shephard E. G., Anderson R., Rosen O., Myer M. S., Fridkin M., Strachan A. F., and De Beer F. C. (1990) Peptides generated from C-reactive protein by a neutrophil membrane protease. Amino acid sequence and effects of peptides on neutrophil oxidative metabolism and chemotaxis. J. Immunol. 145, 1469–1476 [PubMed] [Google Scholar]
- 24. Potempa L. A., Yao Z.-Y., Ji S.-R., Filep J. G., and Wu Y. (2015) Solubilization and purification of recombinant modified C-reactive protein from inclusion bodies using reversible anhydride modification. Biophys. Rep. 1, 18–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ying S. C., Gewurz H., Kinoshita C. M., Potempa L. A., and Siegel J. N. (1989) Identification and partial characterization of multiple native and neoantigenic epitopes of human C-reactive protein by using monoclonal antibodies. J. Immunol. 143, 221–228 [PubMed] [Google Scholar]
- 26. Ji S. R., Wu Y., Potempa L. A., Liang Y. H., and Zhao J. (2006) Effect of modified C-reactive protein on complement activation: a possible complement regulatory role of modified or monomeric C-reactive protein in atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 26, 935–941 [DOI] [PubMed] [Google Scholar]
- 27. Ji S. R., Wu Y., Potempa L. A., Qiu Q., and Zhao J. (2006) Interactions of C-reactive protein with low-density lipoproteins: implications for an active role of modified C-reactive protein in atherosclerosis. Int. J. Biochem. Cell Biol. 38, 648–661 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y. J., Shi J. M., Bai C. J., Wang H., Li H. Y., Wu Y., and Ji S. R. (2012) Intra-membrane oligomerization and extra-membrane oligomerization of amyloid-beta peptide are competing processes as a result of distinct patterns of motif interplay. J. Biol. Chem. 287, 748–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H. Y., Wang J., Wu Y. X., Zhang L., Liu Z. P., Filep J. G., Potempa L. A., Wu Y., and Ji S. R. (2014) Topological localization of monomeric C-reactive protein determines proinflammatory endothelial cell responses. J. Biol. Chem. 289, 14283–14290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thompson D., Pepys M. B., and Wood S. P. (1999) The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure 7, 169–177 [DOI] [PubMed] [Google Scholar]
- 31. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 32. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33- 38, 27–38 [DOI] [PubMed] [Google Scholar]
- 34. Potempa L. A., Maldonado B. A., Laurent P., Zemel E. S., and Gewurz H. (1983) Antigenic, electrophoretic and binding alterations of human C-reactive protein modified selectively in the absence of calcium. Mol. Immunol. 20, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 35. Shephard E. G., Anderson R., Rosen O., and Fridkin M. (1992) C-reactive protein (CRP) peptides inactivate enolase in human neutrophils leading to depletion of intracellular ATP and inhibition of superoxide generation. Immunology 76, 79–85 [PMC free article] [PubMed] [Google Scholar]
- 36. Swanson S. J., and Mortensen R. F. (1990) Binding and immunological properties of a synthetic peptide corresponding to the phosphorylcholine-binding region of C-reactive protein. Mol. Immunol. 27, 679–687 [DOI] [PubMed] [Google Scholar]
- 37. Kinoshita C. M., Ying S. C., Hugli T. E., Siegel J. N., Potempa L. A., Jiang H., Houghten R. A., and Gewurz H. (1989) Elucidation of a protease-sensitive site involved in the binding of calcium to C-reactive protein. Biochemistry 28, 9840–9848 [DOI] [PubMed] [Google Scholar]
- 38. Thomassen M. J., Meeker D. P., Deodhar S. D., Wiedemann H. P., and Barna B. P. (1993) Activation of human monocytes and alveolar macrophages by a synthetic peptide of C-reactive protein. J Immunother. Emphasis Tumor Immunol. 13, 1–6 [DOI] [PubMed] [Google Scholar]
- 39. Barna B. P., Eppstein D. A., Thomassen M. J., Nestor J. J. Jr., Ho T., Medendorp S. V., and Deodhar S. D. (1993) Therapeutic effects of a synthetic peptide of C-reactive protein in pre-clinical tumor models. Cancer Immunol. Immunother. 36, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhong W., Zen Q., Tebo J., Schlottmann K., Coggeshall M., and Mortensen R. F. (1998) Effect of human C-reactive protein on chemokine and chemotactic factor-induced neutrophil chemotaxis and signaling. J. Immunol. 161, 2533–2540 [PubMed] [Google Scholar]
- 41. Ji S. R., Ma L., Bai C. J., Shi J. M., Li H. Y., Potempa L. A., Filep J. G., Zhao J., and Wu Y. (2009) Monomeric C-reactive protein activates endothelial cells via interaction with lipid raft microdomains. FASEB J. 23, 1806–1816 [DOI] [PubMed] [Google Scholar]
- 42. Wright P. E., and Dyson H. J. (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ishida T., and Kinoshita K. (2007) PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khreiss T., József L., Potempa L. A., and Filep J. G. (2004) Conformational rearrangement in C-reactive protein is required for proinflammatory actions on human endothelial cells. Circulation 109, 2016–2022 [DOI] [PubMed] [Google Scholar]
- 45. Ross R. (1999) Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 340, 115–126 [DOI] [PubMed] [Google Scholar]
- 46. Fujita M., Takada Y. K., Izumiya Y., and Takada Y. (2014) The binding of monomeric C-reactive protein (mCRP) to Integrins αvβ3 and α4β1 is related to its pro-inflammatory action. PLoS ONE 9, e93738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eischen A., Duclos B., Schmitt-Goguel M., Rouyer N., Bergerat J. P., Hummel M., Oskam R., and Oberling F. (1994) Human resident peritoneal macrophages: phenotype and biology. Br J. Haematol. 88, 712–722 [DOI] [PubMed] [Google Scholar]
- 48. Zhao J., and Shi X. H. (2010) Study of the interaction of the C-reactive protein monomer with the U937 monocyte. Cell Mol. Biol. Lett. 15, 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pepys M. B., Hirschfield G. M., Tennent G. A., Gallimore J. R., Kahan M. C., Bellotti V., Hawkins P. N., Myers R. M., Smith M. D., Polara A., Cobb A. J., Ley S. V., Aquilina J. A., Robinson C. V., Sharif I., Gray G. A., Sabin C. A., Jenvey M. C., Kolstoe S. E., Thompson D., and Wood S. P. (2006) Targeting C-reactive protein for the treatment of cardiovascular disease. Nature 440, 1217–1221 [DOI] [PubMed] [Google Scholar]
- 50. Du Clos T. W. (2013) Pentraxins: structure, function, and role in inflammation. ISRN Inflamm. 2013, 379040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang L., Liu S. H., Wright T. T., Shen Z. Y., Li H. Y., Zhu W., Potempa L. A., Ji S. R., Szalai A. J., and Wu Y. (2015) C-reactive protein directly suppresses Th1 cell differentiation and alleviates experimental autoimmune encephalomyelitis. J. Immunol. 194, 5243–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang M. Y., Zhou H. H., Zhang S. C., Hui F., Zhu W., Su H. X., Guo H. Y., Li X. W., Ji S. R., and Wu Y. (2014) Recurrent mutations at C-reactive protein gene promoter SNP position −286 in human cancers. Cell Res. 24, 505–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Su H. X., Zhou H. H., Wang M. Y., Cheng J., Zhang S. C., Hui F., Chen X. Z., Liu S. H., Liu Q. J., Zhu Z. J., Hu Q. R., Wu Y., and Ji S. R. (2014) Mutations of C-reactive protein (CRP) −286 SNP, APC and p53 in colorectal cancer: implication for a CRP-Wnt crosstalk. PLoS ONE 9, e102418. [DOI] [PMC free article] [PubMed] [Google Scholar]