Abstract
The supply of MHC class I-restricted peptides is primarily ensured by the degradation of intracellular proteins via the ubiquitin-proteasome system. Depending on the target and the enzymes involved, ubiquitination is a process that may dramatically vary in terms of linkages, length, and attachment sites. Here we identified the unique lysine residue at position 124 of the NY-ESO-1 cancer/testis antigen as the acceptor site for the formation of canonical Lys-48-linkages. Interestingly, a lysine-less form of NY-ESO-1 was as efficient as its wild-type counterpart in supplying the HLA-A*0201-restricted NY-ESO-1157–165 antigenic peptide. In fact, we show that the regulation of NY-ESO-1 processing by the ubiquitin receptors Rpn10 and Rpn13 as a well as by the standard and immunoproteasome is governed by non-canonical ubiquitination on non-lysine sites. In summary, our data underscore the significance of atypical ubiquitination in the modulation of MHC class I antigen processing.
Keywords: antigen processing, proteasome, protein degradation, tumor immunology, ubiquitin
Introduction
Most of the MHC class I-restricted antigenic peptides are generated through the degradation of intracellular proteins by proteasomes (1, 2). Proteasomes are multicatalytic assemblies that are primarily found as 20S and/or 26S complexes in the cytoplasm and nuclei of all eukaryotic cells. The 20S particle is defined as the latent and free form of proteasomes and consists of two copies of seven different α and seven different β subunits with two α rings surrounding two β rings (3, 4). The 26S complex is made up of one 20S catalytic core that is bound to each end by one 19S regulatory complex (5). The peptidase activity of both 20S and 26S complexes is ensured by the incorporation of the β5, β1, and β2 catalytic subunits. Upon exposure to IFN-α/β and/or IFN-γ, three additional non-essential catalytic β subunits (β5i, β1i, and β2i) are up-regulated and incorporated in place of the β5, β1, and β2 subunits during de novo assembly, ensuring a shift from the standard proteasome configuration to an immunoproteasome (IP),2 which has been shown to be more effective at the removal of damaged proteins (6–8). Under physiological conditions, the majority of proteasomes exist in the form of 26S complexes, which is the configuration responsible for ATP- and ubiquitin (Ub)-dependent degradation (9).
In this pathway, substrates destined to be broken down must undergo a so-called ubiquitination process (sometimes also referred as to ubiquitylation) that relies on a three-step enzymatic cascade involving E1, E2, and E3 enzymes that allows the covalent transfer of cellular Ub to target proteins. Ubiquitination primarily occurs via isopeptide bonds to lysine residues of target proteins (10). Meanwhile, a number of protein substrates have been shown to be conjugated on threonine, serine, and cysteine residues as well as at the N terminus (11–13). The Ub molecule itself possesses seven lysine residues (Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, and Lys-63) as well as an NH2-terminal methionine (referred as to M1), each of which serves as a potential acceptor site for the formation of poly-Ub chains (14). This allows the assembly of at least eight types of poly-Ub chains with distinct topologies and outcomes for the modified protein. Importantly, ubiquitination is thought to operate at least at two distinct levels in vivo. It may be initiated on fully translated mature proteins or during protein synthesis. The latter is part of a quality control process that allows the rapid ubiquitination and subsequent proteasome-mediated degradation of newly synthetized proteins that have accumulated errors and/or damage during translation (i.e. defective ribosomal products or DRiPs) (15, 16). Both mature proteins and DRiPs have been shown to be sources for MHC class I antigen presentation (17, 18).
Depending on the number and/or nature of acceptor sites, mature proteins or DRiPs may therefore be subjected to a considerable variety of ubiquitination combinations. Whether these ubiquitination sites and/or linkage types directly influence MHC class I antigen presentation is unknown. Here we set out to address this issue by analyzing the ubiquitination profile of the NY-ESO-1 cancer/testis antigen, which presents the unique advantage of containing only one lysine residue at position 124. In this work, we show that the mature NY-ESO-1 form devoid of Lys-124 (NY-ESO-1K0) greatly differs from its wild-type counterpart in terms of attachment sites and linkages. However, despite those differences, the presentation of the HLA-A*0201-restricted NY-ESO-1157–165 peptide emerging from NY-ESO-1K0 was not altered. Our data suggest that atypical linkages and/or sites actively participate in the regulation of MHC class I antigen processing.
Experimental Procedures
Reagents
The proteasome inhibitors MG-132 and epoxomicin were purchased from Calbiochem. N-ethylmaleimide and cycloheximide were obtained from Sigma. The PR-893-specific inhibitor for the β5 standard subunit was provided by Onyx Pharmaceuticals, Inc. (an Amgen subsidiary, South San Francisco, CA) and the YU-102 β1/LMP2-specific inhibitor was a gift from Dr. Kyung-Bo Kim (Department of Pharmaceutical Sciences, University of Kentucky, Lexington, KY).
Antibodies
The mouse monoclonal antibodies against NY-ESO-1 (clone E978), LMP7 (clone A-12), c-myc (clone 9E10), Rpn13 (clone Q31), cathepsin D (sc-6486), and β-actin (clone C4) were obtained from Santa Cruz Biotechnology, Inc. Antibodies directed against ubiquitin (clone FK2), β1 (clone MCP421), β2 (clone MCP165), Rpn10 (clone S5a-18), and α6 (clone MCP20) were purchased from Enzo Life Sciences. The goat anti-MECL1 polyclonal antibody (PA5-19146) was obtained from Thermo Scientific. Rabbit anti-β5 (ab3330) and anti-LMP2 (ab3328) antibodies were purchased from Abcam. Other antibodies used in this work included HA.11 (clone 16B12, Covance), GFP (clone JL-8, TaKaRa, Clontech), LC3B (2775, Cell Signaling Technology), and ubiquitin (Dako). The secondary goat anti-rabbit or anti-mouse antibodies conjugated with peroxidase were obtained from Calbiochem and used at a 1:5000 dilution.
Cells and Culture Conditions
HeLa cells were cultivated in Iscove's medium supplemented with 10% FCS, 1% l-glutamine, and 1% penicillin/streptomycin (all purchased from Biochrom AG). The 33/2 cell line is a derivative of HeLa cells and has been described previously (19). The CTL clone RG39 specific for the HLA-A*0201-restricted NY-ESO-1157–165 peptide was prepared as described previously (19).
Plasmid Construction and Transfection
The cDNA for CTAG1b encoding the full-length NY-ESO-1 protein was amplified by RT-PCR from RNA of HT1080 cells and cloned into pcDNA3.1/myc-HIS version B (Invitrogen) to construct a C-terminally myc-HIS-tagged NY-ESO-1. A single amino acid substitution (Lys-3→Arg-3) of the EQKLISEEDL myc tag sequence was introduced by site-directed mutagenesis using primer pairs containing the appropriate base changes to generate a pcDNA3.1/NY-ESO-1/myc*-HIS with a modified EQRLISEEDL myc* tag devoid of lysine residues. The pcDNA3.1/NY-ESO-1/myc*-HIS was used again as a template for site-directed mutagenesis to substitute the unique lysine residue of NY-ESO-1 at position 124 into an arginine (K124R) and to generate a pcDNA3.1/NY-ESO-1K124R/myc*-HIS construct (referred as to NY-ESO-1K0 in this paper). For N-terminal tagging of NY-ESO-1 and NY-ESO-1K0, oligonucleotides encoding a short 9-mer V5 sequence (IPNPLLGLD) fused to a tobacco etch virus (TEV) protease-sensitive site (ENLYFQG) were constructed, annealed, and inserted in-frame into the pcDNA3.1/NY-ESO-1/myc*-HIS and pcDNA3.1/NY-ESO-1K0/myc*-HIS plasmids to generate V5-TEV-NY-ESO-1-myc*/HIS and V5-TEV-NY-ESO-1K0-myc*/HIS constructs. Both of these constructs were further used as templates for site-directed mutagenesis to substitute the NY-ESO-1 methionine initiation codon at position 20 into a valine one and to generate the V5-TEV-NY-ESO-1M20V-myc*/HIS, V5-TEV-NY-ESO-1M20V/K0-myc*/HIS variants. The HA-Ub-GFP construct has been described previously (19). Multiple lysine mutations of HA-Ub-GFP were generated by site-directed mutagenesis to generate HA-UbK6-only-GFP, HA-UbK11-only-GFP, HA-UbK27-only-GFP, HA-UbK29-only-GFP, HA-UbK33-only-GFP, HA-UbK48-only-GFP, and HA-UbK63-only-GFP. Mammalian cells were transfected using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer.
NY-ESO-1 Pulldown
Subconfluent HeLa or 33/2 cells grown in 10-cm tissue culture dishes were subjected to a first transfection with the wild-type HA-Ub-GFP plasmid or one of the HA-Ub-GFP mutant constructs for 24 h prior to another subsequent transfection with either NY-ESO-1 or NY-ESO-1K0. Sixteen hours after the second transfection, cells were incubated with 10 μm MG-132 for 6 h before collection. Cells expressing an NY-ESO-1 antigen form or not (as negative control) were washed twice with PBS and lysed in lysis buffer containing 50 mm Tris HCl (pH 8.0), 150 mm NaCl, 1% Triton X-100, as well as 10 μm MG-132 and 50 mm N-ethylmaleimide. Whole cell extracts were incubated for 30 min at 4 °C and subsequently centrifuged at 10,000 rpm for 10 min at 4 °C. Supernatants were then collected and used immediately for the pulldown of myc-tagged NY-ESO-1 proteins using magnetic beads conjugated with μMACS anti-myc antibodies (Miltenyi Biotec). Briefly, 5 mg of protein lysate was supplemented with 50 μl of μMACS anti-myc beads for 30 min at 4 °C. Subsequently, labeled cell lysates were applied to MACS columns in a separator (Miltenyi Biotec) and washed five times with a first buffer containing 50 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS. In some experiments, the NaCl concentration in this wash buffer was increased up to 3 m. The second wash was carried out using a buffer consisting of 20 mm Tris HCl (pH 7.5). After the last washing step, proteins were eluted in 85 μl of elution buffer (50 mm Tris HCl (pH 6.8), 50 mm DTT, 1 mm EDTA, 0.005% bromphenol blue, and 10% glycerol). The precipitates were finally subjected to SDS-PAGE, followed by Western blotting using specific antibodies.
TEV Protease On-bead Digestion Assay
HeLa cells were transfected with HA-Ub-GFP together with V5-TEV-NY-ESO-1M20V-myc*/HIS, V5-TEV-NY-ESO-1M20V/K0-myc*/HIS, or MART-1-myc/HIS (as a control). One day after transfection, cells were exposed to a 6-h treatment with 10 μm MG-132 prior to a NY-ESO-1 pulldown as described above. Digestion of V5-TEV-NY-ESO-1M20V-myc*/HIS and V5-TEV-NY-ESO-1M20V/K0-myc*/HIS was carried out while the proteins were still bound to μMACS anti-myc beads by adding 20 units of TEV protease for 150 min at 22 °C. Subsequently, the beads were washed several times, as described above, and the eluted material was further assessed by Western blotting.
siRNA Transfection
ON-TARGET-plus SMARTpool siRNAduplexes (Dharmacon) against Rpn10 (PSMD4, L-011365-00-0005), Rpn13 (ADMR1, L-01234-00-0005), LMP7 (PSMB8, L-006022-00-0005), and LMP2 (PSMB9, L-006023-00-0005) were used at 50 nm final concentration. Approximately 2 × 105 cells were seeded into one well of a 6-well tissue culture plate the day before siRNA transfection. All transfections were done using X-tremeGENE siRNA reagent (Roche). The siRNA efficiency in this work was validated by monitoring the siRNA-induced knockdown by Western blotting.
RNA Isolation, Reverse Transcription, and PCR Analysis
Total RNA was isolated using the kit from Roche according to the instructions of the manufacturer. RT-PCR was then performed using 1 μg of total RNA with a primer specific for NY-ESO-1 and GAPDH (loading control).
Antigen Presentation Assays
HeLa cells were transiently transfected with HLA-A*0201 together with either NY-ESO-1 or NY-ESO-1K0 and used as targets for their potential to activate the production of IFN-γ by the CTL clone RG39 recognizing the NY-ESO-1157–165 peptide. Following a 24-h transfection, target cells were serially diluted and then co-cultured with a fixed amount of T cells, resulting in graded effector:target ratio in a final volume of 100 μl on 96-well plates. After 16-h incubation, the supernatants were collected, and the IFN-γ content was determined using a commercially available human ELISA kit (BD Biosciences) according to the instructions of the manufacturer. The data in the figures refer to the mean of two replicates. The standard deviation was below 5% of the mean.
Statistics
Student's t test (one-tailed) was used for data analysis when appropriate.
Results
The Removal of the Unique Lysine Residue of the NY-ESO-1 Antigen Does Not Affect the Presentation of the NY-ESO-1157–165 Antigenic Peptide
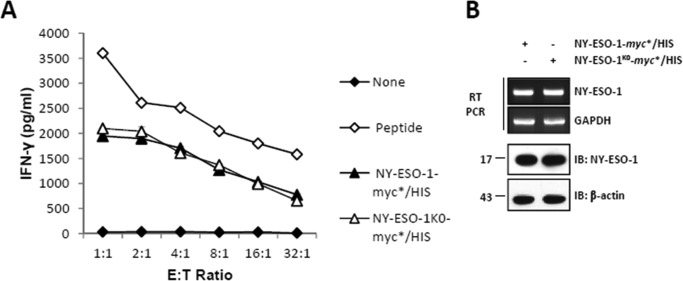
To determine whether the unique lysine residue of NY-ESO-1 is an essential target for ubiquitination, allowing degradation and subsequent MHC class I antigen presentation, we generated a lysine-free form of NY-ESO-1 (i.e. NY-ESO-1K0) by introducing a single Lys-to-Arg change into a C-terminal myc*/HIS-tagged version of NY-ESO-1 protein at position 124. As shown in Fig. 1A, NY-ESO-1 and NY-ESO-1K0 yielded comparable NY-ESO-1157–165CTL responses when expressed in HeLa cells, as evidenced by similar IFN-γ levels produced by our RG39 clone. Likewise, the Lys-to-Arg mutation had no substantial effect on the transcription and/or steady-state expression level of NY-ESO-1 and/or NY-ESO-1K0, as determined by RT-PCR and Western blotting, respectively (Fig. 1B). Together, these data demonstrate that Lys-124, as the unique canonical ubiquitination site of the NY-ESO-1 antigen, is not essential for the presentation of NY-ESO-1157–165 peptide.
FIGURE 1.
The unique lysine residue of NY-ESO-1 at position 124 is not essential for NY-ESO-1157–165 presentation. A, HeLa cells were subjected to 24-h transfection with HLA-A*0201 together with either NY-ESO-1 or NY-ESO-1K0. The NY-ESO-1157–165 CTL response was assessed using the CTL clone RG39 specific for NY-ESO-1157–165 at various effector:target ratios as indicated. Internal controls in this experiment consisted of HLA-A*0201-expressing HeLa cells or cells loaded with 10 μm of the 9-mer SLLMWITQV synthetic peptide. After 16 h of co-culture, supernatants were tested for their IFN-γ content by ELISA. The results are expressed as mean ± S.D. of duplicate values. Shown is one representative experiment of three. B, NY-ESO-1 expression was analyzed by RT-PCR (top panel) and Western blotting (bottom panel) as indicated. IB, immunoblot.
NY-ESO-1157–165 Presentation Relies on Rpn10 and, to a Lesser Extent, on Rpn13
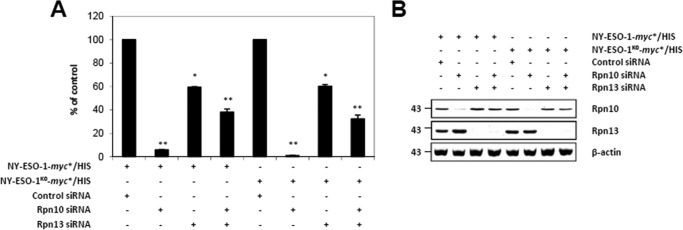
We next determined whether our two NY-ESO-1 antigen sources differ in their capacity to bind the Ub receptors Rpn10 (20, 21) and/or Rpn13 (22). Consistent with its role as the major Ub receptor, Rpn10 knockdown led to a nearly complete inhibition of NY-ESO-1157–165 presentation regardless of the NY-ESO-1 form used (Fig. 2A). Interestingly, a much lower inhibition of the NY-ESO-1157–165 CTL response was observed in cells with an individual knockdown of Rpn13 or a combined knockdown of Rpn10 and Rpn13 for both NY-ESO-1 and NY-ESO-1K0. The observation that Rpn10/Rpn13 double knockdown was less efficient than the Rpn10 single one in impeding the NY-ESO-1157–165 CTL response can likely be attributed to the fact that cells exposed to both Rpn10 and Rpn13 siRNA achieved a less effective down-regulation of Rpn10 compared with cells exposed to Rpn10 siRNA alone (Fig. 2B). The exact similarity in the levels of NY-ESO-1157–165 CTL response between NY-ESO-1 and NY-ESO-1K0 following depletion of Rpn10 and/or Rpn13 suggests that both of these antigens are equally well recognized by 26 proteasomes.
FIGURE 2.
The Rpn10 and/or Rpn13 dependences do not vary between NY-ESO-1 and NY-ESO-1K0 for presentation of the NY-ESO-1157–165 antigenic peptide. A, HeLa cells were exposed to control siRNA or siRNA specific against Rpn10 and/or Rpn13 for 3 days prior to subsequent transfection with either NY-ESO-1 or NY-ESO-1K0, as indicated. The NY-ESO-1157–165 CTL response arising from NY-ESO-1 or NY-ESO-1K0 in cells exposed to Rpn10 and/or Rpn13 siRNA was assessed by culturing these cells in the presence of the RG39 CTL clone at a ratio of 1:1. After 16 h of co-culture, supernatants were tested for their IFN-γ content by ELISA. Data are expressed as the percentage relative to cells exposed to control siRNA. Shown is one representative experiment of three. *, p < 0.05; **, p < 0.01 versus control siRNA (Student's t test). B, the knockdown efficiency of Rpn10 and/or Rpn13 in these cells was evaluated by monitoring the steady-state levels of both of these proteins by Western blotting with antibodies specific for Rpn10 and Rpn13.
NY-ESO-1 Critically Requires Proteasomal β2 Catalytic Activity for Generation of the NY-ESO-1157–165 Antigenic Peptide
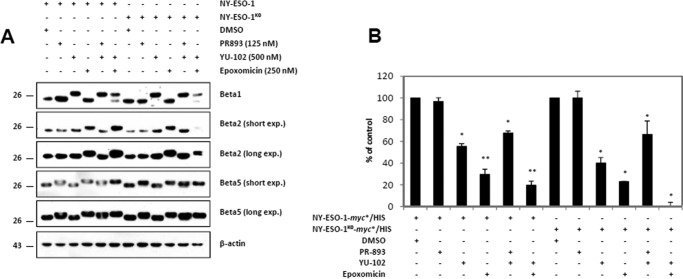
To unveil a possible relationship between ubiquitination and proteasomal processing, we next profiled the contribution of the three catalytic activities of the β5, β1, and β2 standard subunits to the generation of the NY-ESO-1157–165 peptide. To this end, HeLa cells were engineered to express either NY-ESO-1 or NY-ESO-1K0 before being treated with the PR-893, YU-102, and/or epoxomicin inhibitors. PR-893 is the human homolog of the PR-825 inhibitor specific for the β5 subunit, whereas YU-102 is an inhibitor known for its capacity to block β1-activity without affecting β5 and β2 activities (23). Epoxomicin is an irreversible proteasome inhibitor that blocks β5 at low concentrations and both β5 and β2 when incubated with cells at higher concentrations (24). The specificity of these inhibitors in our cell system was controlled by monitoring their potential binding to any of the β1, β2, and β5 subunits by Western blotting. As expected, β5 and β1 shifted up in the presence of PR-893 and YU-102, respectively, confirming subunit inhibition specificity in this assay (Fig. 3A). Exposure of the cells to 250 nm epoxomicin resulted in an evident upward shift of the β5 and β2 bands.
FIGURE 3.
The generation of the NY-ESO-1157–165 peptide from NY-ESO-1 or NY-ESO-1K0 is mainly driven by the β2 catalytic proteasomal subunit. A, HeLa cells expressing HLA-A*0201 together with either NY-ESO-1 or NY-ESO-1K0 were treated for 2 h with PR-893 (125 nm), YU-102 (500 nm), and epoxomicin (250 nm) individually or in combination, as indicated. Covalent binding of the inhibitors to any of the three standard subunits in cells expressing HLA-A*0201/NY-ESO-1 or HLA-A*0201/NY-ESO-1K0 was monitored by Western blotting using antibodies specific for β1, β2, β5, and β-actin (loading control). DMSO, dimethyl sulfoxide. B, the effects of the PR-893, YU-102, and/or epoxomicin on the NY-ESO-1157–165 CTL responses emerging from NY-ESO-1 or NY-ESO-1K0 were examined in a 16-h CTL assay using RG39 CTL. IFN-γ was measured by ELISA. The results are expressed as percentage relative to cells exposed to dimethyl sulfoxide. Shown is one representative experiment of three. *, p < 0.05; **, p < 0.01 versus dimethyl sulfoxide (Student's t test).
Treatment of cells with epoxomicin together with YU-102 resulted in the covalent modification of all three subunits (Fig. 3A) and was accompanied by a significant drop in NY-ESO-1157–165 presentation regardless of the NY-ESO-1 antigen form used (Fig. 3B). These data unambiguously indicate that the generation of the NY-ESO-1157–165 peptide is proteasome-dependent. Unexpectedly, PR-893 alone failed to substantially influence the NY-ESO-1157–165 CTL response emerging from NY-ESO-1 or NY-ESO-1K0. By contrast, a substantial reduction in NY-ESO-1157–165 presentation could be observed following exposure of the cells to the YU-102 inhibitor. Of note, PR-893 showed no substantial additive effect on the YU-102-induced down-regulation of the NY-ESO-1157–165 presentation, confirming that the activity of the β5 subunits was dispensable in this process. Because epoxomicin, which blocks both β5 and β2, dramatically affects presentation and given that β5 was not required, we reasoned that generation of NY-ESO-1157–165 is mainly driven by the β2 subunit.
The NY-ESO-1157–165 CTL Response Is Affected by IP
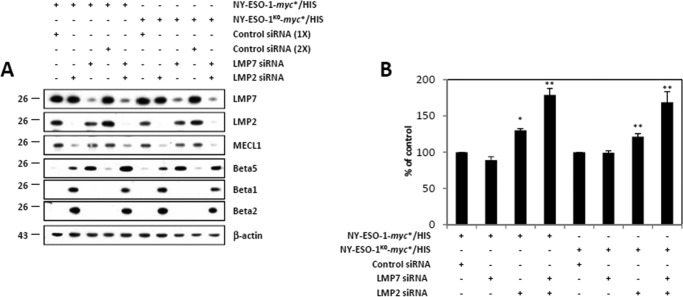
We next compared the abilities of NY-ESO-1 and NY-ESO-1K0 to generate the NY-ESO-1157–165 peptide in the presence of immunoproteasomes. To this end, 33/2 cells that stably expressed LMP7, LMP2, and MECL1 (19) were treated with siRNA directed against each of the inducible subunit prior to a 24-h transfection with plasmids encoding either NY-ESO-1 or NY-ESO-1K0. As expected the down-regulation of LMP7 was accompanied by a parallel rise of its β5 standard counterpart subunit (Fig. 4A). Similarly, the suppression of LMP2 and MECL1 by LMP2 siRNA resulted in an increased expression of both of the β1 and β2 standard subunits. As shown in Fig. 4B, the NY-ESO-1157–165 CTL response was not affected by LMP7 depletion in cells expressing either NY-ESO-1 or NY-ESO-1K0. However, the NY-ESO-1157–165 presentation was improved in 33/2 cells expressing either NY-ESO-1 or NY-ESO-1K0 following exposure to LMP2 siRNA. Likewise, 33/2 cells with an LMP7/LMP2 double knockdown and expressing either NY-ESO-1 or NY-ESO-1K0 exhibited a typical standard proteasome composition that was by far much more efficient than the IP of 33/2 cells exposed to control siRNA in generating the NY-ESO-1157–165 antigenic peptide (Fig. 4B). These data show that standard proteasomes process the NY-ESO-1157–165 peptide better than IPs.
FIGURE 4.
Both NY-ESO-1 and NY-ESO-1K0 are more effective at producing the NY-ESO-1157–165 antigenic peptide in cells lacking the IP-inducible subunits. A, 33/2 cells were exposed to control (once or twice), LMP7, and/or LMP2 siRNA for 2 days before being subjected to a subsequent transfection with NY-ESO-1 or NY-ESO-1K0. Cell extracts were analyzed by Western blotting using antibodies specific for LMP7, LMP2, MECL1, β5, β1, β2, and β-actin (loading control). B, 33/2 cells with a knockdown of LMP7 and/or LMP2 and expressing either NY-ESO-1 or NY-ESO-1K0 were evaluated for their capacity to present the NY-ESO-1157–165 peptide by exposing them to our CTL clone RG39 for 16 h at a effector:target ratio of 8:1. IFN-γ was measured by ELISA. The data are expressed as percentage relative to cells exposed to control siRNA. Shown is one representative experiment of three. *, p < 0.05; **, p < 0.001 versus control siRNA (Student's t test).
Lys-124 Removal Affects the Ubiquitination Profile of the NY-ESO-1 Mature Protein
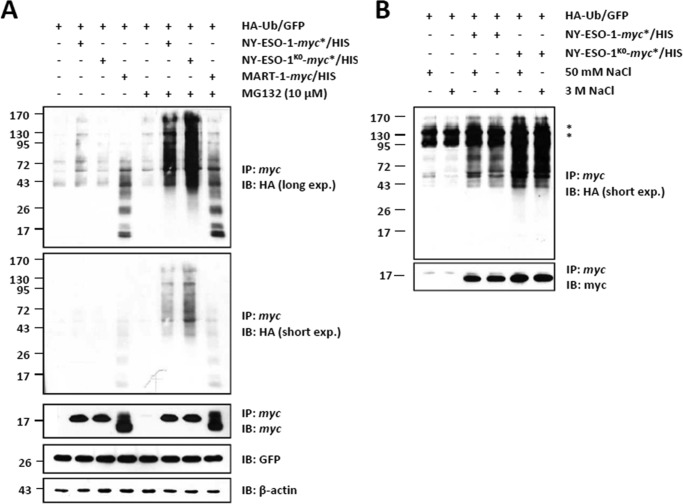
To determine whether the K124R mutation affects NY-ESO-1 ubiquitination, HeLa cells were transiently transfected with a C-terminal myc*/HIS-tagged version of NY-ESO-1 or NY-ESO-1K0 or the MART-1 melanoma antigen (as a control) in combination with an HA-tagged Ub-GFP (HA-Ub-GFP) construct. As shown in Fig. 5A, significant amounts of slowly migrating forms of NY-ESO-1 were detected with both NY-ESO-1 and NY-ESO-1K0 precipitates using the HA antibody. Importantly, such HA-protein conjugates were only detectable in the presence of MG-132, indicating that they serve as a target signal for proteasome-mediated degradation. Noteworthy, apart from the fact that the NY-ESO-1 pulldown was conducted utilizing ionic detergent-based buffers, the exposure of the NY-ESO-1 bead-bound immune complexes to very high salt concentrations failed to alter the HA smears of NY-ESO-1 and NY-ESO-1K0 (Fig. 5B). This indicates that the binding of HA-Ub to NY-ESO-1 or NY-ESO-1K0 is indeed of a covalent nature. Surprisingly, the conjugation of NY-ESO-1K0 to the HA-Ub construct was even superior to that observed with NY-ESO-1. Because a C terminus-mediated sample enrichment exclusively provides full-length (i.e. fully translated) products, these data suggest that Lys-124 removal increases ubiquitination arising from the mature NY-ESO-1 protein.
FIGURE 5.
The unique lysine residue of NY-ESO-1 at position 124 is not required for NY-ESO-1 ubiquitination. A, whole cell extracts were prepared from HeLa cells co-expressing HA-Ub-GFP together with NY-ESO-1, NY-ESO-1K0, or the MART-1 melanoma antigen (as a control) in the presence or absence of a 6-h treatment with 10 μm MG-132. In vivo ubiquitination of NY-ESO-1 or NY-ESO-1K0 was visualized by pulling down NY-ESO-1 with anti-myc beads, followed by Western blotting using antibodies specific for HA and NY-ESO-1 (which served as a loading control for the pulldown). Cell extracts were subjected to Western blotting analysis using antibodies specific for HA, myc, GFP, and β-actin (loading control) as indicated. IP, immunoprecipitation; IB, immunoblot. B, in vivo ubiquitination of NY-ESO-1 or NY-ESO-1K0 was visualized by precipitating NY-ESO-1 with anti-myc beads prior to five stringent washes with either 50 mm or 3 m NaCl as indicated. After a final wash with 20 mm Tris, the anti-myc-bound material was eluted, and the precipitates were analyzed by Western blotting using antibodies specific for HA and myc. The asterisks represent nonspecific bands. Shown is one representative experiment of two.
NY-ESO-1 and NY-ESO-1K0 Use Different Acceptor Sitesfor Ubiquitination
Having demonstrated that Lys-124 of NY-ESO-1 is not critical for either ubiquitination and/or presentation of the NY-ESO-1157–165 peptide, we next sought to identify other sites responsible for NY-ESO-1 ubiquitination. To determine whether NY-ESO-1 undergoes ubiquitination via Ser and/or Thr residues, HeLa cells were first engineered to express HA-Ub-GFP in combination with NY-ESO-1, NY-ESO-1K0, or MART-1 (as a control) prior to a 6-h treatment with 10 μm MG-132. Following NY-ESO-1 pulldown, the precipitates were left untreated or treated with 50 mm NaOH for 25 min at 32 °C. Fig. 6 shows that the ubiquitination of NY-ESO-1 and NY-ESO-1K0 was reduced following alkaline treatment. Importantly, peptide bonds were not hydrolyzed under these conditions, as evidenced by the levels of eluted NY-ESO-1, which remained unchanged following sodium hydroxide treatment. This indicates that both NY-ESO-1 and NY-ESO-1K0 are modified on Ser and/or Thr residues in these cells. Noteworthy, the use of the anti-Ub antibody (FK2) revealed the presence of Lys-29, Lys-48, and/or Lys-63 linkages on NY-ESO-1K0 that were resistant to NaOH treatment (Fig. 6).
FIGURE 6.
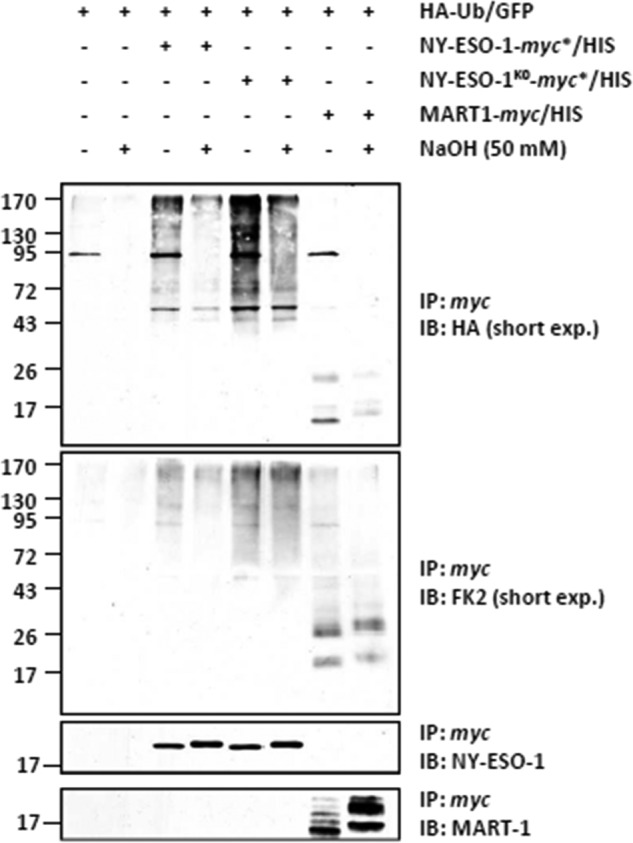
Ubiquitination of the NY-ESO-1 and NY-ESO-1K0 tumor antigens occurs on serine and/or threonine residues in HeLa cells. HeLa cells expressing HA-Ub-GFP in combination with NY-ESO-1, NY-ESO-1K0, or the MART-1 melanoma antigen (as a control) were subjected to protein extraction. NY-ESO-1 and/or NY-ESO-1K0 were pulled down by anti-myc beads, and the precipitates were treated with 50 mm NaOH for 25 min at 32 °C or left untreated, as indicated. The negative control for the immunoprecipitation (IP) in this experiment consisted in HeLa cells only transfected with the HA-Ub-GFP construct (first and second lanes). The effect of the alkaline treatment on the ubiquitination of NY-ESO-1, NY-ESO-1K0, and MART-1 was assessed by Western blotting using antibodies specific for HA, ubiquitin, and NY-ESO-1 (loading control). IB, immunoblot.
NY-ESO-1 and NY-ESO-1K0 Do Not Use Their Respective N Termini for Ubiquitination
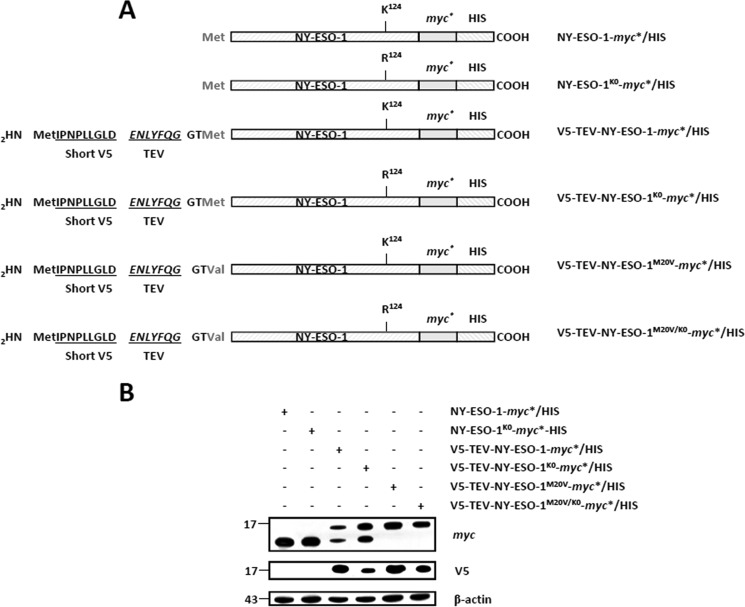
We next asked whether the N terminus of the NY-ESO-1 tumor antigen may act as a site for ubiquitination. To address this question, we introduced a short V5 (IPNPLLGLD) epitope at the N terminus of both of the NY-ESO-1-myc*/HIS and NY-ESO-1K0-myc*/HIS constructs, which was immediately followed by a TEV protease recognition site (ENLYFQG) (Fig. 7A). To avoid the expression of untagged NY-ESO-1 proteins deprived of the N-terminal V5 epitope, the original NY-ESO-1 methionine initiator codon at position 20 of both of the V5-TEV-NY-ESO-1-myc*/HIS and V5-TEV-NY-ESO-1K0-myc*/HIS constructs was changed into a valine (M20V), as depicted in Fig. 7, A and B. If the ubiquitination of NY-ESO-1 and/or NY-ESO-1K0 indeed partially relies on their N termini, it should be affected upon treatment with the TEV protease (Fig. 8A). Remarkably, on-bead digestion of the cell lysates with TEV protease resulted in loss of the V5 epitope as well as in a decrease in size of the eluted NY-ESO-1 proteins, as determined by Western blotting (Fig. 8B). This unambiguously indicates that the N-terminal V5 tag was efficiently cleaved off and stripped away from both of the NY-ESO-1 full-length proteins. Interestingly, despite being devoid of their respective N termini, both NY-ESO-1 and NY-ESO-1K0 exhibited an HA staining that was comparable with that observed with the untreated NY-ESO-1 proteins (Fig. 8B). These data demonstrate that the free N terminus of either NY-ESO-1 or NY-ESO-1K0 is not essentially involved in the ubiquitination process under these conditions.
FIGURE 7.
The expression of an N-terminal V5-tagged version of NY-ESO-1 and/or NY-ESO-1K0 requires the removal of the original NY-ESO-1 start codon. A, sequences of the NY-ESO-1 constructs used in this study. All NY-ESO-1 constructs were C-terminally tagged with a double myc*/HIS epitope. In some experiments, NY-ESO-1-myc*/HIS and NY-ESO-1K0-myc*/HIS were N-terminally tagged with a short 9-mer V5 epitope (IPNPLLGLD) directly upstream of a TEV protease-sensitive site (ENLYFQG). Both V5-TEV-NY-ESO-1-myc*/HIS and V5-TEV-NY-ESO-1K0-myc*/HIS were finally subjected to M20V site-directed mutagenesis to remove the NY-ESO-1 original methionine initiator codon. B, HeLa cells were transfected with each of the six NY-ESO-1 variant for 24 h prior to protein extraction. Whole cell extracts were resolved on 15% SDS-PAGE and subsequently analyzed by Western blotting using anti-myc, anti-V5, and anti-β-actin (loading control) antibodies as indicated. The M20V in both of V5-TEV-NY-ESO-1-myc*/HIS and V5-TEV-NY-ESO-1K0-myc*/HIS prevents the expression of N-terminally untagged NY-ESO-1-myc*/HIS and NY-ESO-1-myc*/HIS, respectively.
FIGURE 8.
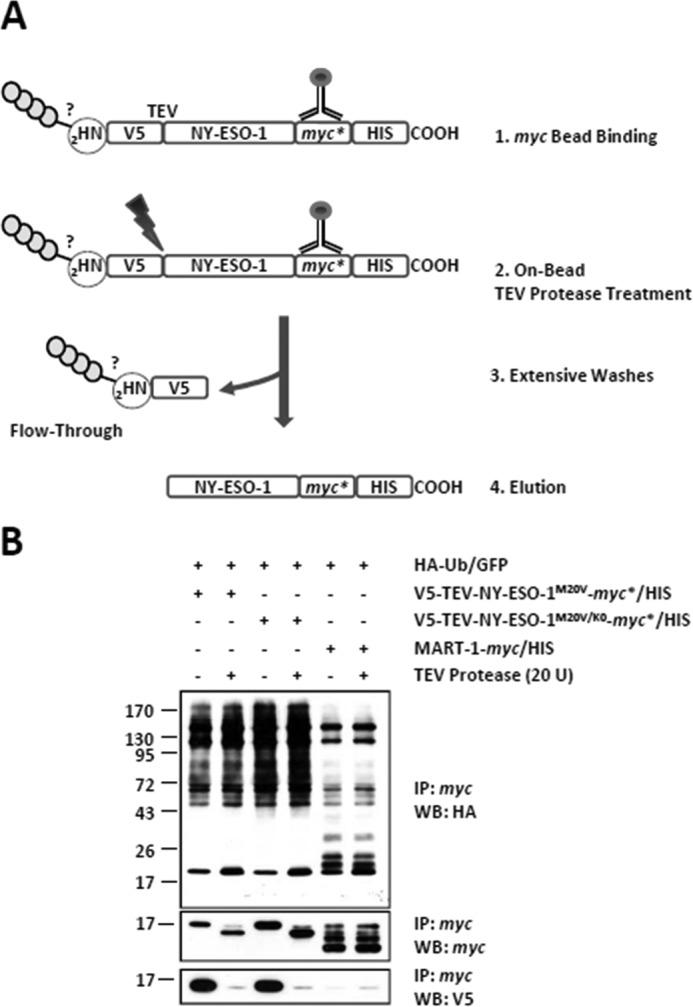
The N-terminal −NH2 group of either NY-ESO-1 or NY-ESO-1K0 is not a site of conjugation. A, HeLa cells expressing HA-Ub-GFP in combination with V5-TEV-NY-ESO-1M20V-myc*/HIS or V5-TEV-NY-ESO-1M20V/K0-myc*/HIS were subjected to protein extraction and incubated with μMACS anti-myc beads that were subsequently left untreated or treated with 20 U TEV protease for 150 min at 22 °C as indicated. Extensive washing of the μMACS anti-myc bead-bound material resulted in the elimination of the N terminus and its potential anchored Ub chains in the flow-through fraction. After five washes, the material bound to the μMACS anti-myc beads was eluted. B, the effect of the removal of the V5 N terminus on the ubiquitination of NY-ESO-1 and NY-ESO-1K0 was assessed by Western blotting (WB) using antibodies specific for HA, ubiquitin, V5, and myc (loading control). The negative control for the immunoprecipitation (IP) in this experiment consisted of HeLa cells transfected with the HA-Ub-GFP construct in combination with an expression vector encoding the MART-1 melanoma antigen (fifth and sixth lanes). Shown is one representative experiment of three.
Removal of Lys-124 of NY-ESO-1 Results in the Loss of Lys-48-linked Ub Chains
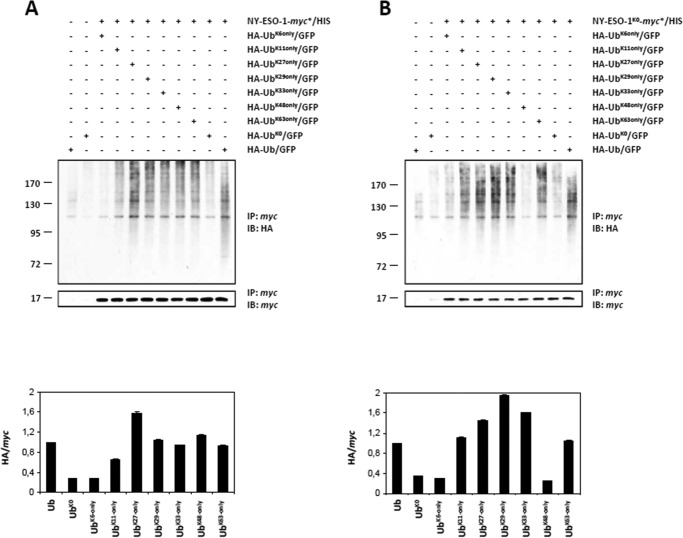
We next set out to identify the linkages of the poly-Ub chains assembled on both NY-ESO-1 and NY-ESO-1K0. To this end, both of these proteins were expressed together with HA-tagged Lys-6-only, Lys-11-only, Lys-27-only, Lys-29-only, Lys-33-only, Lys-48-only, and Lys-63-only Ub constructs in which all lysine residues were altered to arginine except for lysines 6, 11, 27, 29, 33, 48, and 63, respectively. Internal controls in this experiment consisted of the use of a wild-type HA-Ub construct and an HA-tagged K0 Ub mutant (HA-UbK0) devoid of all of its lysine residues. NY-ESO-1 poly-ubiquitination was supported by wild-type, Lys-11-only, Lys-27-only, Lys-29-only, Lys-33-only, Lys-48-only, and Lys-63-only Ub chains but not by the Lys-6-only mutant (Fig. 9A). NY-ESO-1K0 showed a ladder of anti-HA reactivity when cells were engineered to express wild-type Ub as well as Lys-11-only, Lys-27-only, Lys-29-only, Lys-33-only, or Lys-63-only Ub mutants, suggesting efficient poly-ubiquitination of NY-ESO-1K0 by these Ub species (Fig. 9B). Strikingly, co-expression of NY-ESO-1K0 with either Lys-6-only or Lys-48-only mutants did not result in the formation of HA-modified protein conjugates, indicating that both of these mutants were not used for conjugation and that such linkages were therefore dispensable for poly-Ub chain formation. Besides the loss of Lys-48 chains, our densitometric analysis demonstrated an increased formation of Lys-11, Lys-29 and Lys-33 linkages following the Lys-to-Arg mutation, whereas the levels of Lys-27 and Lys-63 linkages remained unchanged.
FIGURE 9.
NY-ESO-1 and NY-ESO-1K0 distinguish themselves in terms of poly-Ub chains. A and B, ubiquitination of NY-ESO-1 (A) and NY-ESO-1K0 (B) was examined following overexpression of a HA-tagged wild-type Ub, a Lys-0 HA-tagged Ub (HA-UbK0) devoid of all its seven lysine residues, as well as HA-tagged Lys-6-only, Lys-11-only, Lys-27-only, Lys-29-only, Lys-33-only, Lys-48-only, or Lys-63-only Ub mutants in which all lysine residues were altered to arginine except for lysines 6, 11, 27, 29, 33, 48, and 63, respectively, and as indicated. Ubiquitination of NY-ESO-1 and/or NY-ESO-1K0 was visualized by pulling down NY-ESO-1 with anti-myc beads, followed by Western blotting using antibodies specific for HA and NY-ESO-1 (loading control). Pulldowns with cells overexpressing HA-Ub or HA-UbK0 (first and second lanes) but lacking NY-ESO-1 (A) or NY-ESO-1K0 (B) demonstrate the specificity of the assay. A densitometric analysis of ubiquitinated NY-ESO-1 and NY-ESO-1K0 normalized to pulled-down NY-ESO-1 and NY-ESO-1K0 is plotted (mean ± S.D., n = 2). For both of the ubiquitinated species of NY-ESO-1 and NY-ESO-1K0, the mean of the signals obtained with the wild-type HA-Ub-GFP construct was set at 1. IP, immunoprecipitation; IB, immunoblot.
Discussion
In this paper, we identified Lys-124 of the NY-ESO-1 cancer/testis antigen as the major site for the formation of canonical Lys-48-linked poly-Ub chains (Fig. 8, A and B). Unexpectedly, abrogation of this site did not prevent the formation of NY-ESO-1 Ub-conjugates or the presentation of the NY-ESO-1157–165antigenic peptide (Figs. 1A and 5). This suggests that the MHC class I processing of the NY-ESO-1 antigen is driven by non-lysine residues and non-canonical poly-Ub chains. Consistently, our investigations show that NY-ESO-1 uses the hydroxyl groups of its Ser and/or Thr residues to form ester bonds with Ub, which eventually results in the formation of atypical chains containing Lys-11, Lys-27, Lys-29, Lys-33, and Lys-63 linkages (Figs. 6 and 9, A and B). Noteworthy is that such NY-ESO-1 Ub species were detected in the presence of MG-132, confirming their potency as signals for targeting NY-ESO-1 to proteasome-mediated degradation. The notion that proteasomes do not necessarily recognize canonical linkages is in agreement with previous studies showing that atypical poly-Ub may actively participate in the regulation of protein breakdown (25–27). Importantly, our analysis of the NY-ESO-1 ubiquitination profile was carried out following enrichment of the full-length protein using a C-terminal epitope tagging strategy. Such an experimental design does not preclude the pulldown of incompletely synthetized NY-ESO-1 polypeptides, including those resulting from premature translation termination, which are believed to represent a form of DRiPs (15–17). This point is of great importance because DRiPs have been shown to largely contribute to the supply of the pool of antigenic peptides (15, 16). One should, however, emphasize that the HLA-A*0201-restricted NY-ESO-1157–165 epitope lies very close to the extreme C terminus of the 180-amino acid-long NY-ESO-1 mature protein. It is therefore highly unlikely that NY-ESO-1157–165 presentation arises from ubiquitination of incomplete NY-ESO-1 polypeptides during protein biosynthesis. Nonetheless, this does not necessarily exclude DRiPs as the relevant antigen source for presentation of the NY-ESO-1157–165peptide. It is indeed conceivable that the 9-mer peptide is generated from co-translation ubiquitination of full-length misfolded NY-ESO-1 proteins. The observation that the Lys-124 removal is accompanied by increased ubiquitination, whereas the steady-state level of the mature protein does not change, strongly suggests the generation of NY-ESO-1 DRiPs in the form of improperly folded full-length proteins (Fig. 5).
Remarkably, the NY-ESO-1157–165 CTL response was entirely dependent on Rpn10 (Fig. 2A). These findings show that the recognition of Ub-modified proteins by Rpn10 and Rpn13 is not restricted to those bearing Lys-48-linked poly-Ub chains. These data also suggest that Rpn10 is a unique receptor for the recognition and subsequent degradation of both NY-ESO-1 and NY-ESO-1K0. However, this assumption is somehow in conflict with the observation that the NY-ESO-1157–165 presentation was also affected in cells with an individual knockdown of Rpn13, albeit to a much lesser extent (∼50%). One possible explanation for these apparent discrepancies might be that Rpn13 requires the presence of Rpn10 in 26S complexes to exert its function as Ub receptor. As such, any individual down-regulation of Rpn10 would inactivate Rpn13 and result in a nearly complete loss of capacity for 26S complexes to cope with ubiquitinated proteins. In agreement with previous reports, our findings point to a structure-function relationship between Rpn10 and Rpn13 that regulates the access of Ub-modified proteins to 26S proteasome complexes (28).
There is disagreement in the field with respect to the involvement of 26S complexes in the generation of the NY-ESO-1157–1659-mer antigenic peptide. Indeed, early reports argued that the NY-ESO-1157–165 presentation was insensitive to proteasome inhibitors (29), suggesting that the NY-ESO-1 protein may be degraded by an alternative route. However, it was later shown that dendritic cells were capable of stabilizing NY-ESO-1 when treated with the N-acetyl-l-leucyl-l-leucyl-l-norleucinal (LLnL) proteasome inhibitor (30). In our hands, the NY-ESO-1157–165 presentation arising from either NY-ESO-1 or NY-ESO-1K0 in HeLa cells fully relied on proteasomal activity (Fig. 3B). Surprisingly, the generation of the NY-ESO-1157–165 antigenic peptide was not affected when the β5 catalytic subunit was inhibited, suggesting that the β5 activity in this situation was either dispensable or could be compensated for by the other β1 and/or β2 catalytic subunits. Of note, epoxomicin, which blocks both β5 and β2, was accompanied by a significant decrease in NY-ESO-1157–165 CTL response. Given that β5 is not important, we deduced from this experiment that the major driving force for the generation of the NY-ESO-1157–165 peptide is the β2 catalytic subunit.
The role of the inducible subunits in this process was addressed by using the 33/2 cell line. The siRNA-mediated down-regulation of LMP7 had no substantial influence on the generation of the NY-ESO-1157–165 antigenic peptide, reinforcing the notion that the β5/LMP7 chymotryptic-like activity is not relevant in this process. Combined down-regulation of LMP7 and LMP2 resulted in a substantial increase in the production of the NY-ESO-1157–165 peptide (Fig. 4B). Our data therefore identify NY-ESO-1157–165 as a new member the growing family of peptides such as MART-126–35 (31) and gp100209–217(32) whose presentation has been reported to be impaired by IPs. Importantly, the preferential processing of NY-ESO-1157–165by standard proteasomes occurred independently of the type of ubiquitination harbored by NY-ESO-1.
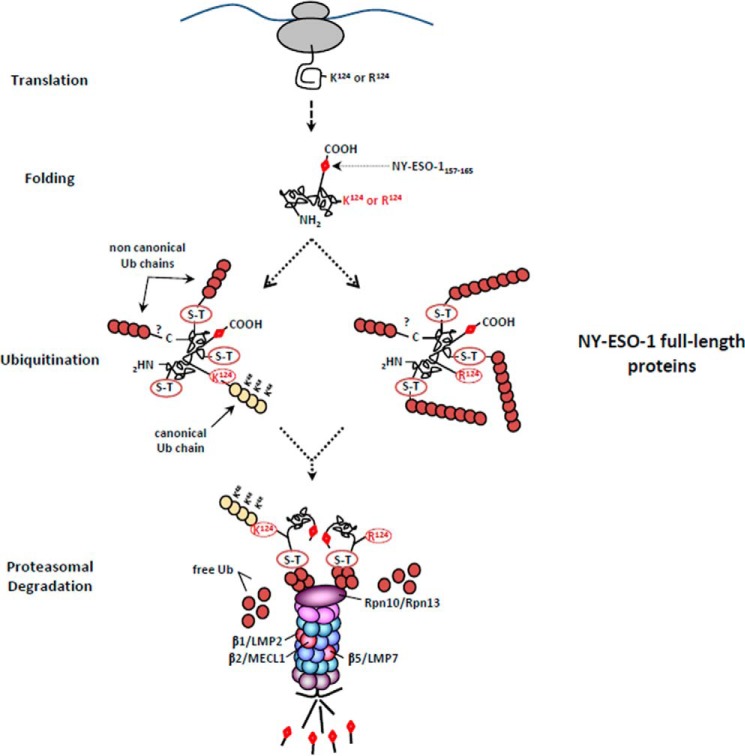
Overall, using NY-ESO-1 as a model substrate, we show that neither lysine residues nor canonical Lys-48 linkages represent a prerequisite for MHC class I antigen processing and presentation. As illustrated in Fig. 10, although the lysine-less and wild-type forms of NY-ESO-1 exhibit ubiquitination patterns that are quantitatively and qualitatively different, they both enter the same proteasome-dependent MHC class I pathway, which is regulated by Rpn10, Rpn13, and the standard β2 subunit. Together, these data point to a critical contribution of atypical ubiquitination on non-lysine residues in MHC class I antigen processing that may be much more important than previously assumed.
FIGURE 10.
The processing of NY-ESO-1 relies on atypical ubiquitination occurring on non-lysine sites. The presence of the NY-ESO-1157–165 antigenic peptide at the C terminus of the 180-amino acid-long NY-ESO-1 full-length protein required the protein to be fully synthetized before ubiquitination. Following translation, the full-length NY-ESO-1 protein was subjected to both typical and atypical ubiquitination on Lys-124 and on non-canonical sites (Ser-Thr), respectively. The removal of Lys-124 abrogated the formation of Lys-48-linked poly-Ub chains, which was also accompanied by increased generation of atypical chains. Despite these qualitative and quantities differences, both the wild-type and lysine-less forms of NY-ESO-1 converged on the same regulatory proteasome-mediated MHC class I processing pathway involving the Rpn10 and Rpn13 19S subunits as well as the trypsin-like activity of the β2 standard subunit. This eventually leads to similar production of antigenic peptides, which are equally presented on HLA-A*0201 molecules.
Author Contributions
R. G. and A. L. performed the research and analyzed the data. P. M. K. designed the research and analyzed the data. F. E. designed the research, analyzed the data, and wrote the manuscript.
Acknowledgments
We thank Ellen Hilgenberg (Deutsches Rheuma-Forschungszentrum Berlin) for help with the irradiation of peripheral blood mononuclear cells and EBV-transformed B cells.
This work was supported by grants from the Berlin Institute of Health (CRG Project) (to P. M. K.) and from the Einstein Foundation Berlin (to P. M. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- IP
- immunoproteasome
- Ub
- ubiquitin
- DRiP
- defective ribosomal product
- CTL
- cytotoxic T lymphocyte
- TEV
- tobacco etch virus
- NY-ESO-1
- New York esophageal squamous cell carcinoma 1.
References
- 1. Sijts E. J., and Kloetzel P. M. (2011) The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell. Mol. Life Sci. 68, 1491–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rock K. L., York I. A., Saric T., and Goldberg A. L. (2002) Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 80, 1–70 [DOI] [PubMed] [Google Scholar]
- 3. Murata S., Yashiroda H., and Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 4. Ciechanover A., and Stanhill A. (2014) The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta 1843, 86–96 [DOI] [PubMed] [Google Scholar]
- 5. DeMartino G. N., and Slaughter C. A. (1999) The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 274, 22123–22126 [DOI] [PubMed] [Google Scholar]
- 6. Seifert U., Bialy L. P., Ebstein F., Bech-Otschir D., Voigt A., Schröter F., Prozorovski T., Lange N., Steffen J., Rieger M., Kuckelkorn U., Aktas O., Kloetzel P. M., and Krüger E. (2010) Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 142, 613–624 [DOI] [PubMed] [Google Scholar]
- 7. Strehl B., Seifert U., Krüger E., Heink S., Kuckelkorn U., and Kloetzel P. M. (2005) Interferon-γ, the functional plasticity of the ubiquitin-proteasome system, and MHC class I antigen processing. Immunol. Rev. 207, 19–30 [DOI] [PubMed] [Google Scholar]
- 8. Shin E. C., Seifert U., Kato T., Rice C. M., Feinstone S. M., Kloetzel P. M., and Rehermann B. (2006) Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. J. Clin. Invest. 116, 3006–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Livnat-Levanon N., Kevei É., Kleifeld O., Krutauz D., Segref A., Rinaldi T., Erpapazoglou Z., Cohen M., Reis N., Hoppe T., and Glickman M. H. (2014) Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 7, 1371–1380 [DOI] [PubMed] [Google Scholar]
- 10. Breitschopf K., Bengal E., Ziv T., Admon A., and Ciechanover A. (1998) A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17, 5964–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McDowell G. S., Kucerova R., and Philpott A. (2010) Non-canonical ubiquitylation of the proneural protein Ngn2 occurs in both Xenopus embryos and mammalian cells. Biochem. Biophys. Res. Commun. 400, 655–660 [DOI] [PubMed] [Google Scholar]
- 12. Tait S. W., de Vries E., Maas C., Keller A. M., D'Santos C. S., and Borst J. (2007) Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J. Cell Biol. 179, 1453–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vosper J. M., McDowell G. S., Hindley C. J., Fiore-Heriche C. S., Kucerova R., Horan I., and Philpott A. (2009) Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J. Biol. Chem. 284, 15458–15468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 15. Yewdell J. W., and Nicchitta C. V. (2006) The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 27, 368–373 [DOI] [PubMed] [Google Scholar]
- 16. Dolan B. P., Bennink J. R., and Yewdell J. W. (2011) Translating DRiPs: progress in understanding viral and cellular sources of MHC class I peptide ligands. Cell. Mol. Life Sci. 68, 1481–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rock K. L., Farfán-Arribas D. J., Colbert J. D., and Goldberg A. L. (2014) Re-examining class-I presentation and the DRiP hypothesis. Trends Immunol. 35, 144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., and Bennink J. R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 [DOI] [PubMed] [Google Scholar]
- 19. Ebstein F., Lehmann A., and Kloetzel P. M. (2012) The FAT10- and ubiquitin-dependent degradation machineries exhibit common and distinct requirements for MHC class I antigen presentation. Cell. Mol. Life Sci. 69, 2443–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deveraux Q., van Nocker S., Mahaffey D., Vierstra R., and Rechsteiner M. (1995) Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26S protease subunit S5a. J. Biol. Chem. 270, 29660–29663 [DOI] [PubMed] [Google Scholar]
- 21. van Nocker S., Deveraux Q., Rechsteiner M., and Vierstra R. D. (1996) Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 93, 856–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., and Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Myung J., Kim K. B., Lindsten K., Dantuma N. P., and Crews C. M. (2001) Lack of proteasome active site allostery as revealed by subunit-specific inhibitors. Mol. Cell 7, 411–420 [DOI] [PubMed] [Google Scholar]
- 24. Kisselev A. F., Callard A., and Goldberg A. L. (2006) Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 281, 8582–8590 [DOI] [PubMed] [Google Scholar]
- 25. Dammer E. B., Na C. H., Xu P., Seyfried N. T., Duong D. M., Cheng D., Gearing M., Rees H., Lah J. J., Levey A. I., Rush J., and Peng J. (2011) Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 286, 10457–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bedford L., Layfield R., Mayer R. J., Peng J., and Xu P. (2011) Diverse polyubiquitin chains accumulate following 26S proteasomal dysfunction in mammalian neurons. Neurosci. Lett. 491, 44–47 [DOI] [PubMed] [Google Scholar]
- 27. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., and Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Besche H. C., Sha Z., Kukushkin N. V., Peth A., Hock E. M., Kim W., Gygi S., Gutierrez J. A., Liao H., Dick L., and Goldberg A. L. (2014) Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 33, 1159–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen J. L., Dunbar P. R., Gileadi U., Jäger E., Gnjatic S., Nagata Y., Stockert E., Panicali D. L., Chen Y. T., Knuth A., Old L. J., and Cerundolo V. (2000) Identification of NY-ESO-1 peptide analogues capable of improved stimulation of tumor-reactive CTL. J. Immunol. 165, 948–955 [DOI] [PubMed] [Google Scholar]
- 30. Batchu R. B., Moreno A. M., Szmania S. M., Bennett G., Spagnoli G. C., Ponnazhagan S., Barlogie B., Tricot G., and van Rhee F. (2005) Protein transduction of dendritic cells for NY-ESO-1-based immunotherapy of myeloma. Cancer Res. 65, 10041–10049 [DOI] [PubMed] [Google Scholar]
- 31. Morel S., Lévy F., Burlet-Schiltz O., Brasseur F., Probst-Kepper M., Peitrequin A. L., Monsarrat B., Van Velthoven R., Cerottini J. C., Boon T., Gairin J. E., and Van den Eynde B. J. (2000) Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity 12, 107–117 [DOI] [PubMed] [Google Scholar]
- 32. Chapiro J., Claverol S., Piette F., Ma W., Stroobant V., Guillaume B., Gairin J. E., Morel S., Burlet-Schiltz O., Monsarrat B., Boon T., and Van den Eynde B. J. (2006) Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J. Immunol. 176, 1053–1061 [DOI] [PubMed] [Google Scholar]