Abstract
Poly(ADP-ribose) polymerases (PARPs) synthesize and bind branched polymers of ADP-ribose to acceptor proteins using NAD as a substrate and participate in the control of gene transcription and DNA repair. PARP1, the most abundant isoform, regulates the expression of proinflammatory mediator cytokines, chemokines, and adhesion molecules, and inhibition of PARP1 enzymatic activity reduced or ameliorated autoimmune diseases in several experimental models, including colitis. However, the mechanism(s) underlying the protective effects of PARP1 inhibition in colitis and the cell types in which Parp1 deletion has the most significant impact are unknown. The objective of the current study was to determine the impact of Parp1 deletion on the innate immune response to mucosal injury and on the gut microbiome composition. Parp1 deficiency was evaluated in DSS-induced colitis in WT, Parp1−/−, Rag2−/−, and Rag2−/−×Parp1−/− double knock-out mice. Genome-wide analysis of the colonic transcriptome and fecal 16S amplicon profiling was performed. Compared with WT, we demonstrated that Parp1−/− were protected from dextran-sulfate sodium-induced colitis and that this protection was associated with a dramatic transcriptional reprogramming in the colon. PARP1 deficiency was also associated with a modulation of the colonic microbiota (increases relative abundance of Clostridia clusters IV and XIVa) and a concomitant increase in the frequency of mucosal CD4+CD25+ Foxp3+ regulatory T cells. The protective effects conferred by Parp1 deletion were lost in Rag2−/− × Parp1−/− mice, highlighting the role of the adaptive immune system for full protection.
Keywords: colitis, gene expression, gene knockout, innate immunity, microarray, Poly (ADP-ribose) polymerase, dextran sulfate, gut microbiota, regulatory T cells
Introduction
Poly(ADP-ribose) polymerases (PARPs)4 represent a family of 18 cell-signaling enzymes involved in the regulation of multiple nuclear proteins and participate in the complex regulatory networks involved in mucosal immunity. PARP-mediated post-translational modifications include the transfer of polymers of ADP-ribose (parylation) from NAD+ donor molecules to glutamic acid, aspartic acid, or lysine on target proteins (1). Among PARPs, PARP1 plays essential roles in the maintenance of genomic integrity (DNA-damage sensing), facilitation of cell survival, and the relaxation of chromatin structure, which fosters protein/protein interactions and protein-DNA binding (2). PARP1 targets include core histones and diverse transcriptional factors (3). PARP1 activity can be regulated by several endogenous and exogenous factors, such as estrogen and 1,25(OH)2 vitamin D3 (4).
PARP1 inhibition mitigates morbidity and mortality in multiple inflammatory and autoimmune diseases such as diabetes mellitus, rheumatoid arthritis, septic shock, ischemic stroke, acute pancreatitis, asthma, and inflammatory bowel disease-like colitis (5). Genetic ablation or pharmacological inhibition of PARP1 reduces the biological and physical manifestations of experimental colitis; Il-10−/− mice treated with a PARP1 inhibitor (3-aminobenzamide) show a significant reduction in proinflammatory cytokine production associated with reduced intestinal permeability (6). Other PARP1 inhibitors have also been used successfully to prevent hapten-induced colitis in rats (7–11) and in mice (12, 13). However, the mechanism(s) underlying PARP-mediated promotion of immune dysregulation in colitis remains unknown, although depletion of cellular energetic pools, which could culminate in cell dysfunction and necrosis as well as the promotion of the transcription of proinflammatory genes, has been postulated (2).
PARP1 has been shown to enhance inflammatory responses by promoting the expression of inflammation-associated genes, such as proinflammatory cytokines, inducible nitric-oxide synthase, intracellular adhesion molecule 1 (ICAM-1), cyclooxygenase 2 (COX-2), and NADPH oxidase as well as major histocompatibility complex class II (MHC-II). PARP1 potentiates NF-κB activity and AP-1 expression, fostering the expression of major NF-κB and AP-1-depandant proinflammatory mediators (12). In intestinal epithelial cells in vitro, inhibition of PARP-1 with PJ-34 is protective during invasive Salmonella spp. infection by up-regulating IL-6 production through ERK and NF-κB signaling pathways (14), suggesting an enhanced innate immune response to pathogenic bacteria. It is, therefore, plausible that PARP1 inhibition in vivo could modulate the inflammatory tone in the colonic mucosa, which might prove protective in the face of epithelial barrier breach and translocation of commensal bacteria. To test this, we utilized a model of dextran-sulfate sodium (DSS)-mediated mucosal injury, which leads to bacterial translocation and transient ulcerative colitis-like inflammation in Parp1−/− mice and their wild-type littermates. We performed genome-wide analysis of the colonic transcriptome profile in combination with 16S amplicon library profiling to test the potential effect of PARP1 deficiency on the colonic microbiome. We also tested the effects of Parp1 deletion in T-and B-cell-deficient Rag2−/− mice in the DSS model of colitis. We show that Parp1 deficiency offers significant protection from mucosal injury in Parp1−/− mice and that deletion of the Parp1 gene leads to dramatic transcriptional reprogramming in the colonic mucosa. This effect is accompanied by modulation of the composition of colonic microbiota and an increased relative abundance of Firmicutes, including Clostridia clusters IV and XIVa. Consistent with this, colonic lamina propria CD4+CD25+Foxp3+ iTreg were present at a higher frequency in Parp1−/− mice. Protective effects related to Parp1 deletion were lost in DSS-treated Parp1/Rag1 double knock-out mice, thus suggesting the requirement for elements of the adaptive immune system for full protection.
Experimental Procedures
Experimental Animals
Specific pathogen-free wild-type (WT) 129/SvEv mice, Parp1−/− and Rag2−/− mice on the same genetic background, were originally obtained from Taconic (Germantown, NY). Rag2−/− were crossed with Parp1−/− mice to develop double knockouts (DKOs). All mice were maintained in a conventional animal facility at the University of Arizona Health Sciences Center. Sentinel mice were routinely monitored and determined as free from common murine pathogens (MHV, MPV, MVM, TMEV, Mycoplasma pulmonis, Sendai, EDIM, MNV, and ecto- and endoparasites). All animal protocols and procedures were approved by the University of Arizona Animal Care and Use Committee.
Experimental Model of Colitis
Colitis was induced by DSS treatment (Affymetrix, Santa Clara, CA), administrated in drinking water. 6–8-Week-old Parp1−/− mice or their genetically matched WT mice were left untreated or given drinking water with DSS for up to 7 days. Mortality and body weight were monitored daily. In the initial experiment, 4% DSS was used for 7 days followed by H2O for another week. The DSS lot used was determined to be too toxic at this concentration, as it led to 70% mortality during the recovery period (Fig. 1A). In all subsequent experiments, 3% DSS was used for 7 days only to determine the effects of Parp1 status on acute mucosal injury and immune response. A separate study aimed at microarray analysis of colonic gene expression was performed with a new lot of DSS, empirically determined to result in no mortality, at a dose of 4% in drinking water for 7 days.
FIGURE 1.
Reduced mortality and mucosal inflammation in DSS-treated Parp1−/− mice. A, no mortality was observed in Parp1−/− mice treated with 4% DSS for 7 days. B, representative H&E staining of the proximal and distal colon of WT and Parp1−/− mice treated with 3% DSS for 7 days. C, colonic mucosal cytokine mRNA expression in WT and Parp1−/− mice treated with 3% DSS for 7 days evaluated by real-time RT-PCR (n = 3–7; asterisks indicate statistical significance at p < 0.05 between DSS-treated WT and WT and Parp1−/− mice; ANOVA followed by Fisher protected least significant difference post hoc test). D, secretion of IFNγ and TNFα in colonic explant culture from 3% DSS-treated WT and Parp1−/− mice.
Colonic Histology
Proximal and distal colons were harvested and fixed in 10% neutral buffered formalin (Fisher Scientific, Tustin, CA). Fixed tissues were then embedded in paraffin, and 5-μm-thick tissue cuts were stained with hematoxylin and eosin (H&E) for light microscopic examination.
Colonic Explant Culture, ELISA, and xMAP Assays
Colon segments were flushed with phosphate-buffered saline (PBS) to remove fecal contents, opened lengthwise, and shaken vigorously for 10 min in PBS. Tissue was then apportioned to the wells (50–100 mg of tissue per well) of a 24-well tissue culture plate (Corning Costar, Lowell, MA) and cultured in 1 ml of complete RPMI 1640 medium containing 5% heat-inactivated fetal bovine serum, penicillin, streptomycin, and amphotericin B (all from Invitrogen). Tissues were incubated at 37 °C for 18 h, and supernatants were collected and stored at −80 °C until being assayed. Selected cytokine concentrations were evaluated by an xMAP assay (Millipore, Billerica, MA) and Luminex 100 platform (Millipore, Danvers, MA).
Real-time RT-PCR
Real-time RT-PCR was used to evaluate mucosal expression of IL-17, IL-12p40, TNFα, IL-1β, IFNγ, and MMP-8 mRNA (the latter as a surrogate marker of neutrophil infiltration). Total RNA was isolated from mouse proximal and distal colon using the Qiagen RNAeasy kit (Qiagen, Valencia, CA). 250 ng of total RNA was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad). Subsequently, 20 μl of the PCR reactions were set up in 96-well plates containing 10 μl of 2× IQ Supermix (Bio-Rad), 1 μl of TaqMan® primer/probe set (ABI, Foster City, CA), 2 μl of the cDNA synthesis reaction (10% of the reverse transcription reaction), and 7 μl of nuclease-free water. Reactions were run and analyzed on a Bio-Rad CFX real-time PCR detection system. Data were analyzed by using the comparative Ct method as the means of relative quantification, normalized to an endogenous reference (TATA box-bonding protein (TBP) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH)) and relative to a calibrator (normalized Ct value obtained from control mice) and expressed as 2−ΔΔCt (Applied Biosystems User Bulletin #2: Rev B “Relative Quantification of Gene Expression”).
Microarray Analysis of Colonic Gene Expression Profile
Amplified and biotinylated sense-stranded DNA targets were generated from total RNA isolated from WT and Parp1−/− mice treated with water or DSS (n = 3 in each genotype/treatment group) using GeneChip® WT PLUS Reagent kit (Affymetrix) and hybridized to GeneChip® Mouse Gene 2.0 ST Arrays (Affymetrix). Gene expression analysis was performed using GeneSpring 13.0 software (Agilent Technologies, Santa Clara, CA). Data were processed using the RMA16 summarization algorithm and normalized against the median of all samples or, in some instances, to the median of control samples (WT + H2O). Statistical analysis was performed using built-in tools, including normalized t test or two-way ANOVA, in both cases with Benjamini-Hochberg multiple testing correction. Gene ontology (GO) functional annotation analysis was performed either with GeneSpring or using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.7 online tool (15). Single-site analysis to detect over-represented conserved transcription factor binding sites in a set of genes regulated in Parp1 deficiency was performed using the oPOSSUM 3.0 online tool (16). More detailed results of the analyses, including raw and normalized expression values, can be viewed at the National Center for Biotechnology Information Gene Expression Omnibus microarray depository web site (www.ncbi.nlm.nih.gov; GEO accession no. GSE76658).
Preparation of Single Cell Suspension from Lamina Propria
WT and Parp1−/− mouse colons were harvested and cleaned of luminal contents using 1× PBS. The colons were cut open longitudinally and into ∼5-mm pieces. Colonic pieces were washed extensively in Ca2+/Mg2+-free Hanks' balanced salt solution containing 5% FBS and 10 mm HEPES (HHF) (Life Technologies) and incubated for 20 min at 37 °C in the pre-digestion solution: 5 mm EDTA and 1 mm DTT in HHF to remove epithelial cells. Free of intestinal epithelial cells, colonic pieces were further digested with a solution containing 100 units/ml collagenase (Worthington, NJ) and 40 μg/ml DNase (Roche Diagnostics) in HHF at 37 °C for two consecutive 15-min incubations. After each incubation, cells passing through a 100-μm strainer were collected in RPMI with 10% FBS. After centrifugation at 400 × g at 4 °C, cells were resuspended in 5 ml of RPMI 1640 medium and counted with a Beckman Coulter viability counter (Vi-Cell XR).
Flow Cytometry
Colonic lamina propria single cell suspensions were stained for Live-Dead cells (Zombie Aqua, Biolegend, San Diego, CA) followed by anti-mouse CD16/CD32 blocking for 15 min (2.4G2 antibody). Cells were labeled with different conjugated antibodies in the dark for 30 min at 4 °C: anti-mouse CD4-FITC, CD25-Brilliant Violet 421, FoxP3-APC, and CCR9-PE (BD Biosciences or eBioscience Inc., San Diego, CA). After washing in FACS stain buffer (BD Biosciences), cells were fixed and permeabilized at 4 °C overnight using fixation/permeabilization buffer following the manufacturer's instructions (eBioscience). Intracellular staining for Foxp3 was performed following the manufacturer's recommendation for the Treg staining kit (ebioscience). Data were acquired using an LSR Fortessa flow cytometer (Becton Dickinson) and analyzed using FlowJo Software (Tree Star Inc, Ashland, OR).
Microbiome Analysis
Fecal DNA Extraction and Quantification of Bacterial DNA
Fecal pellets were collected from mice and stored at −80 °C. Fecal DNA was extracted using the FAST DNA SPIN Kit (MP Biomedicals, Santa Ana, CA) according to the manufacturer's instructions. Purified DNA was resuspended in 200 μl of Tris-EDTA buffer and quantified on a Nanodrop ND-1000. DNA samples were stored at −80 °C.
Analysis of Bacterial 16S Genes and Gut Community Profiling
The hypervariable region V4 of the 16S rRNA gene was amplified from each sample using barcoded 806R primers and 515F primer (17) and 5 Prime Hot MasterMix (5 Prime, Hilden, Germany) in triplicate. The quality of the amplicons and potential contaminants was checked on a 1.5% agarose gel. Replicates were pooled and quantified using Picogreen (Invitrogen) according to the manufacturer's protocol. Equal amounts of 240 ng of DNA from each sample were pooled into one multiplexed library and cleaned using an UltraClean PCR Clean-Up kit (MoBio). Pooled amplicons were diluted, denatured (0.2 n NaOH), and sequenced on the MiSeq platform (Illumina) using custom primers (17). Due to the limited sequence diversity among 16S rRNA amplicons, 10% of the PhiX control library (Illumina) made from phiX174 was added to the run. At a final concentration of 6.75 pm, the pooled 16S rRNA library was subjected to paired-end sequencing using the 2 × 150bp MiSeq Reagent Kit V2 (Illumina). Sequencing was performed in our laboratories on the Illumina MiSeq (serial #M03190, with the MiSeq Control Software v 2.5.0.5). The run of 105 pooled samples generated 12,491,854 sequences. After de-multiplexing and quality filtering 11,371,676 reads remained. Of 105 samples, only 20 pertained to the experiment with a total number of 4,227,058 reads. The reads had a median length of 253 bases.
De-multiplexing and filtering were done using the QIIME 1.9.1 software package (18). Sequences were assigned to operational taxonomic units (OTU) with a 97% similarity threshold using QIIME's uclust-based open-reference OTU picking protocol against the SILVA reference database (release 119). The average number of sequences per sample was 203,918.6 ± 21,727.4 (mean ± S.D.). The threshold for the minimum number of sequences in a given sample was set to 24,000. The minimum number of reads per sample in this experiment was 173,086; therefore, all samples were included in our analysis.
Clostridia clusters IV and XIVa were amplified from 10 ng of fecal DNA using PerfeCTa SYBR Green Fastmix (Quanta Biosciences) at primer annealing temperatures optimized by gradient PCR and melting curve analysis. Quantification cycle (Cq) values were used to calculate relative abundance. All samples were normalized to all bacteria/archaea detectable with universal 16S primers 926F and 1062R. The following PCR protocol was run: 95 °C (30 s), then 95 °C (5 s), variable annealing temperature (15 s), and 72 °C (10 s) for 30 cycles. Each sample was run in duplicate, and the mean Ct value was used to calculate 2−ΔΔCt values. p values were calculated using the two-tailed Student's t test. Selected primers (19–21), their sequences, and annealing temperatures can be found in Table 1.
TABLE 1.
| Target group | Primer name | Forward primer | Annealing temperature |
|---|---|---|---|
| °C | |||
| Universal | 926F | AAACTCAAAKGAATTGACGG | 59 |
| 1062R | CTCACRRCACGAGCTGAC | ||
| C. leptum (IV) | CL-IV-F | CCTTCCGTGCCGSAGTTA | 52.8 |
| CL-IV-R | GAATTAAACCACATACTCCACTGCTT | ||
| C. coccoides (XIVa) | CL-XIVa-F | AAATGACGGTACCTGACTAA | 57.0 |
| CL-XIVa-R | CTTTGAGTTTCATTCTTGCGAA |
Statistical Analysis
Statistical significance of non-array data was determined by ANOVA followed by Fisher protected least significant difference post hoc test or by unpaired two-tailed Student's t test, as appropriate, using the StatView software package v.4.53 (SAS Institute, Cary, NC). Data are expressed as the means ± S.E. of mean.
Results
PARP1 Deficiency Protects against DSS-induced Colitis
At baseline, Parp1−/− mice were phenotypically indistinguishable from their WT littermates, with normal colonic histology and no evidence of mucosal inflammation (not shown). To determine the influence of Parp1 in the modulation of the mucosal immune response during epithelial injury (acute colitis), colitis was initially induced in WT or Parp1−/− mice with 4% DSS in drinking water for 7 days followed by 7 days of H2O alone. In this initial study, the DSS lot used resulted in 70% mortality during the recovery period. However, Parp1−/− mice had a 100% survival rate (Fig. 1A). All subsequent experiments focused on the development of acute colitis 7 days into the administration of reduced-dose DSS (3%). WT mice had bloody stools and extensive changes primarily in distal colonic tissue architecture with hyperplasia, immune cell infiltration, loss of goblet cells, extensive mucosal destruction, and loss of the surface epithelium (Fig. 1B). In contrast, Parp1−/− mice displayed no overt pathology (no blood in stools or diarrhea) and showed significantly milder histopathologic changes (Fig. 1B). In the colonic mucosa, TNFα and IL-17 mRNA and protein expression were significantly lower in DSS-treated Parp1−/− mice compared with DSS-treated WT mice (Fig. 1, C and D). Potential anti-inflammatory effects of IL-10 and IL-22, both protective cytokines in the DSS model, were unlikely to account for colitis resistance in Parp1−/− mice as their mucosal expression pattern closely followed that of other cytokines, i.e. no significant difference between genotypes at baseline and blunted response to DSS in Parp1−/− mice (supplemental Fig. S1). In DSS-treated Parp1−/− mice, although not statistically significant, a clear trend toward decreased IFNγ mRNA, IFNγ protein, and IL12p40 mRNA production in colonic explant culture was observed (Fig. 1D).
Transcriptional Reprogramming in the Colon of Parp1−/− Mice
To better understand the PARP1-mediated effects on inflammation, we performed a comprehensive microarray analysis of gene expression patterns in WT and Parp1−/− mice with and without DSS treatment. Hierarchical clustering of all samples with Euclidean similarity measure indicated that the gene expression pattern in Parp1−/− mice was dramatically different from WT mice at baseline. Limited changes in the gene expression profiles of Parp1−/− mice treated with water or DSS were reflected by close clustering of those two experimental groups (Fig. 2). Interestingly, Parp1−/− status, irrespective of treatment, was closely related to DSS-treated WT mice (Fig. 2). The same clustering algorithm was applied to non-averaged data to demonstrate uniformity of gene expression within each experimental group (supplemental Fig. S2).
FIGURE 2.
Gene expression patterns in control and 3% DSS-treated WT and Parp1−/− mice; hierarchical condition tree. A, data were normalized to the median of all samples, filtered on expression levels (raw data >20.0 in at least one of 12 analyzed samples) and without statistical analysis or pre-selection selection, were subjected to hierarchical clustering. B, analogous clustering analysis but with a preselected gene list from 2-way ANOVA analysis with the corrected p value for genotype-treatment of <0.05, with Benjamini-Hochberg test used as the multiple testing correction.
Dramatic Difference in Gene Expression Pattern in Parp1−/− at Baseline
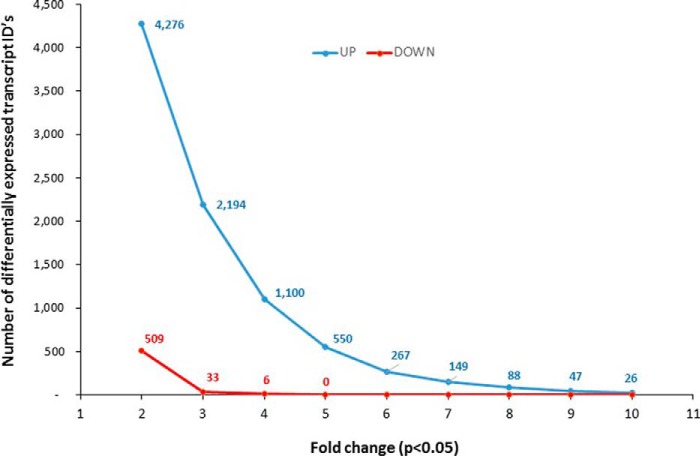
When WT and Parp1−/− colonic transcriptomes were compared at baseline (no DSS), 10,787 transcript cluster IDs were identified as significantly different (p < 0.05, moderated t test with Benjamini-Hochberg multiple testing correction), of which 4,785 transcripts were affected 2-fold or more. The magnitude of this difference is depicted in Fig. 3 as subsets of this list with increasing -fold change (up- or down-regulated transcript cluster IDs) between 2- and 10-fold. The emerging pattern, which favored up-regulated genes, strongly implied a transcriptional de-repression in Parp1−/− mice and suggests that the primary role of PARP1 is to suppress the transcription of a significant proportion of the genes. We further performed gene ontology analysis to identify the most overrepresented categories (biological process) among genes differentially expressed between the genotypes at baseline. The 4785 transcript cluster IDs were converted into 3587 unique annotated genes that were analyzed with the DAVID functional annotation tool under high stringency (minimum 10 genes per category, EASE score <0.01). As shown in Fig. 4 (limited to categories with >100 genes each), the Parp1-regulated genes were greatly enriched in the categories related to proteolysis, protein localization, metabolic processes, cell cycle and cell death, and RNA metabolism and translation.
FIGURE 3.
The magnitude of transcriptional reprogramming in the colon of Parp1−/− mice at baseline. Data were normalized and filtered as in Fig. 2 and statistically analyzed for transcript IDs that statistically differed between WT and Parp1−/− mice at baseline (without DSS) (moderated t test with Benjamini-Hochberg test used as the multiple testing correction). The transcript IDs identified as significantly different (p < 0.05) were plotted as the number of up-regulated (blue line) or down-regulated (red line) transcripts with -fold change increasing from 2- to 10-fold over WT controls.
FIGURE 4.
Gene ontology (GO) analysis of genes differentially expressed in WT and Parp1−/− mice at baseline. 4,785 transcript cluster IDs (WT versus Parp1−/−; -fold change ≥2, p < 0.05) were converted into 3,587 uniquely annotated proteins and analyzed with the DAVID functional annotation tool with the following options: minimum 10 per category, Ease score <0.01. To reduce the results for presentation, biological process categories with ≥100 genes/proteins in each were selected. Black bars represent the number of genes in a given category (upper horizontal axis), and orange dots represent EASE score for each category (lower horizontal axis).
Prediction of Transcription Factors Affected by Parp1 Deficiency in the Mouse Colon
To gain insight into putative colonic transcriptional regulators affected by Parp1 deletion, we performed the single-site analysis of over- and under-represented conserved transcription factor (TF) binding sites in a defined subset of the genes differentially expressed between the two genotypes. We used oPOSSUM v.3.0 online tool, which compares an a priori defined gene list against a database of 29,347 mouse genes, 5,000 bp upstream and 5,000 bp downstream of the transcription start site. Because this tool limits the query list to 2,000 genes, we used a sliding scale of moderated t test p value to reduce the number of transcript cluster IDs to this number and arrived at p ≤ 0.00365. From this set, oPOSSUM algorithm selected 1600 known genes for analysis. 53 TFs were found as overrepresented (Z-score >1.0), and 53 were underrepresented (Z score <−1.0). When these two lists was grouped into TF classes, the emerging picture indicated that the majority of overrepresented TF binding sites belonged to helix-turn-helix and other α-helix classes, whereas underrepresented cis elements were dominated by binding sites for zinc-coordinating and Ig-fold TFs (Fig. 5). The latter is particularly interesting, as it implies that normally, PARP1 positively regulates the Ig-fold TF class, which includes several key transcriptional regulators of the inflammatory response, such as NF κ-B family members and STATs.
FIGURE 5.
oPOSSUM analysis of transcription factors binding sites over-represented or under-represented in genes regulated in the colon of Parp1−/− mice at baseline. To limit the query to the allowed list to 2,000 genes, we used a sliding scale of moderated t test p value (untreated WT versus Parp1−/− mice) and arrived at p ≤ 0.00365. From this set, oPOSSUM algorithm selected 1600 known genes for analysis. 53 TFs were found as overrepresented (Z-score >1.0; range 33.54 to 1.12), and 53 were underrepresented (Z score < −1.0;l range −35.92 to −1.21). The relative contribution of the under- and over-represented cis elements grouped into respective transcription factor classes is depicted.
Effects of DSS on Colonic Transcriptome in WT and Parp1−/− Mice
In WT mice, stringent statistical analysis resulted in identification of 755 transcript cluster IDs significantly altered (down- or up-regulated) by DSS treatment (p < 0.05, moderated t test with Benjamini-Hochberg multiple testing correction, ≥2-fold change). The same comparison applied to Parp1−/− mice yielded only three transcript cluster IDs. These included 8.3-fold up-regulation of TNF-inducible metalloreductase Steap4, 6.2-fold up-regulation of tissue plasminogen activator Plat, and 3.4-fold reduction of nuclear- encoded rRNA n-RSs176. The difference in transcriptional response to DSS between WT and Parp1−/− mice is shown in Fig. 6, which depicts the 755 transcript cluster IDs altered in WT mice (panel A) next to the same IDs plotted for Parp1−/− mice (panel B). Interestingly, the vast majority of transcript cluster IDs that were up-regulated or down-regulated by DSS treatment in WT mice were already up- or down-regulated in Parp1−/− mice, in whom DSS treatment did not result in further changes (supplemental Fig. S3). When all genotype/treatment groups were compared by 2-way ANOVA, 959 differentially expressed transcript cluster IDs mapped primarily to metabolic and not immune pathways (supplemental Fig. S4A) with the exception of the Jak-Stat cascade, represented by JAK2, Nemo-like kinase Nlk, protein inhibitor of activated Stat1 Pias, and interleukin 23 receptor IL23R (supplemental Fig. S4B). To increase the power of analysis, we performed gene set enrichment analysis, where a predefined GO gene set of 316 entities described as Immune_System_Process was selected and analyzed by 2-way ANOVA. The analysis with genotype as the variable resulted in identification of 54 unique genes (supplemental Table S1 and Fig. S5) involved in cytokine and chemokine signaling, T- and B-cell receptor signaling, and MAPK signaling pathways, among other functions.
FIGURE 6.
Differential effect of genotype (WT versus Parp1−/−) and treatment (H2O) versus 3% DSS on colonic gene expression. A, data were normalized and filtered as in Fig. 2 and statistically analyzed for transcript IDs that statistically differed between H2O- and DSS-treated WT mice (moderated t test p ≤ 0.05, -fold change cutoff of ≥2.0). 755 transcript IDs identified in WT mice are plotted. B, the same 755 transcript IDs were selected and plotted with respective normalized values derived from H2O- and DSS-treated Parp1−/− mice.
Colonic Microbiome Composition Is Altered in Parp1-deficient Mice
In light of the dramatic differences in gene expression patterns observed between WT and Parp1−/− mice under steady-state conditions, we hypothesized this may translate into baseline differences in the composition of the gut microbiota. We observed a trend toward increased microbial alpha diversity in Parp1−/− mouse fecal samples, although this did not reach statistical significance (data not shown). Taxonomic analysis at the phylum level also did not indicate significant differences (not shown). To look for more subtle changes, we compared relative microbial abundance at the order level, where we observed significant reduction of Lactobacillales in Parp1−/− mice (3.9% versus 12.51% in WT mice, p < 0.05; Fig. 7A). This decrease was compensated by the relative expansion of the order Clostridiales (32.01% in WT and 45.27% in Parp1−/− mice), although this was not statistically significant. Breakdown of Clostridiales at the family level showed a similar pattern in both mouse strains, thus suggesting that the expansion of this order is not specific to any particular family. Because butyrate-producing Clostridium clusters IV and XIVa (Clostridium leptum and Clostridium coccoides groups, respectively) represent dominant Firmicutes spanning multiple families and are known for their beneficial effects on epithelial integrity and the regulatory T cell compartment (22), we analyzed their abundance in WT and Parp1−/− mice by qPCR. Relative abundance of both clostridial clusters were significantly increased in Parp1−/− mice compared with WT (Fig. 7B). Members of these two clusters have been shown to provide antigens and a local microenvironment facilitating expansion and differentiation of regulatory T cells (Treg; Ref. 23). Consistent with this, Parp1−/− mice showed increased frequencies of CD4+CD25+FoxP3+ Tregs in the colonic lamina propria (Fig. 7C).
FIGURE 7.
Colonic microbial community in Parp1−/− mice. A, next-generation sequencing analysis of the contributions of the major bacterial orders in the colonic contents of WT and Parp1−/− mice. Increase in the order Clostridiales in PARP1-deficient mice was further analyzed at the family level. Statistically significant differences are indicated with an asterisk (p = 0.028). B, real-time PCR analysis of the relative abundance of Clostridia clusters IX and XIVa in WT and Parp1−/− mice at baseline. Data were normalized to all bacteria/archaea detected with universal 16S primers depicted in Table 1. C, flow cytometry analysis of CD4+CD25+FoxP3+ Tregs in the colonic lamina propria of WT and Parp1−/− mice. Representative dot plots (left panels) and summary graph (right panel) are shown; p value from unpaired two-tail t test is indicated.
Protective Effects of Parp1 Deficiency in DSS-mediated Mucosal Injury Model Require the Adaptive Immune System
To address the role of the adaptive immune system cells, including Tregs, in the protective effects of Parp1 deficiency, we crossed Parp1−/− mice with Rag2−/− mice on the same 129/SvEv background. Parp1−/− × Rag2−/− DKO mice were viable, bred well as homozygotes, and showed no symptoms. Considering the observed increase in mucosal Tregs in Parp1−/− mice and the protective role of these cells in the DSS model (24), we compared the response to DSS in Rag2−/− and DKO mice. Initial experiments with 4% DSS resulted in similar body weight loss in the two strains and a trend to decreased survival in DKO mice (Fig. 8A), thus suggesting a lack of protective effects of Parp1 deletion in the latter strain. As with the WT and Parp1−/− mice, we altered the protocol for the subsequent studies to lower the concentration of DSS to 3%. We did not observe any difference between DSS-treated Rag2−/− and DKO mice in terms of mortality or body weight loss (data not shown). Contrary to T- and B-cell-competent mice, Parp1 deletion in Rag2−/− mice was not associated with significant protection against 3% DSS-induced colitis, as indicated by proximal or distal colonic histology (representative H&E sections are depicted in Fig. 8B) or with statistically significant differences in mucosal expression of the key inflammatory mediators TNFα and IFNγ (Fig. 8C). Collectively, these observations suggest that the protective effects related to Parp1 deletion require the presence of the cells of the adaptive immune system, with Tregs representing plausible candidates to mediate this protection.
FIGURE 8.
Loss of protective effects of Parp1 deficiency in Parp1−/−x Rag2−/− DKO mice. A, body weight loss and mortality in response to treatment with 4% DSS for 7 days. B, representative H&E histology staining of the proximal and distal colon of Rag2−/− and DKO mice treated with H2O or 3% DSS for 7 days. C, mucosal TNFα and IFNγ expression in the proximal and distal colon of Rag2−/− and DKO mice treated with 3% DSS for 7 days (N.S., not statistically significant).
Discussion
PARP1 is the founding member of the family of enzymes capable of transferring ADP-ribose from NAD+ with the ability to synthesize long polymers of ADP-ribose (PAR) on itself and other target proteins. Poly-ADP-ribosylation itself is an evolutionarily highly conserved protein modification known to modulate chromatin structure, DNA metabolism, transcription factor activity, gene transcription, and overall cellular metabolism. Both extracellular and intracellular NAD+-dependent enzymes have been shown to modulate immune processes (25, 26). Two examples of extracellular ADP-ribosyltransferases (ARTS) involved in the function of the immune system are ART1 and ART2, expressed primarily in neutrophils and T cells, respectively. These extracellular ADP-ribosyl-transferases are thought to act primarily as danger sensors, which react to elevated NAD+ concentrations resulting from tissue injury and cell death during acute inflammation; ART1 to down-modulate the cytotoxic effects of defensin 1α (27) and ART2 to increase T cell sensitivity to NAD+-induced cell death by increasing the activity of the P2X7 calcium channel (28). Intracellular (cytoplasmic and nuclear) PARP1 is known for protection against genomic instability but also controls several forms of cell death, the effect attributed to energetic failure due to NAD+ depletion (29), activation of mitochondrial apoptosis inducible factor via its PAR-binding domain (30), or through activation of redox-sensitive cation channel TRPM2 (31) among other mechanisms. Therefore, the protection we observed in Parp1−/− mice from DSS-mediated colitis, a model reliant on a direct toxic and pro-apoptotic effect on colonic epithelial cells (32), could be attributed to the resistance of the colonocytes to DSS-related cytotoxicity. However, the observed lack of protection in B- and T-cell-deficient Parp1−/− mice (Rag2−/−× Parp1−/− DKO) suggests a much more complex mechanism of protection going beyond the innate defenses against epithelial injury and bacterial translocation. Forsythe et al. (33) demonstrated that inhibition of PARP1 reduced epithelial cell permeability and bacterial translocation in LPS/NOx-treated epithelial cells in vitro. Similar reduction of epithelial permeability was observed in IL10−/− mice treated with PARP1 inhibitor, 3-aminobenzamide, although this report did not investigate the effects of PARP1 inhibition in healthy mice (6). Cuzzocrea et al. (34) showed that in zymosan-treated mice, increased intestinal permeability was significantly reduced with a specific PARP1 inhibitor. However, baseline permeability was not affected by PARP1 inhibition. These limited published data indicate that PARP1 may modulate permeability indirectly via inhibition of inflammatory response rather then by directly affecting the epithelial apical junction complexes. Future studies with epithelial cell-specific conditional knock-out of PARP1 may help elucidate the role of PARP1 in the colonic epithelial barrier function.
Microarray analysis of colonic gene expression at baseline and after DSS challenge revealed a stunning degree of transcriptional reprogramming in Parp1−/− mice, with 4785 transcripts significantly up- or down-regulated ≥2-fold. This is consistent with the role of PARP1 as a modulator of chromatin structure, modulator of enhancers and transcriptional coregulators, and of its insulator function (35). Functional classification analysis of the differentially expressed genes in Parp1−/− mice at baseline identified overrepresentation of genes involved in proteolysis, protein localization, metabolic processes, cell cycle and cell death, and RNA metabolism and translation and to a much lesser extent related to inflammation. Moreover, this skewed transcriptional profile in Parp1−/− mice strongly favored up-regulation of transcription, implying a transcriptional de-repression, and suggests that the primary role of PARP1 is to suppress the transcription of a significant proportion of the genes in the colon. Analysis of over- or underrepresentation of transcription factor binding sites in the regulatory regions of the genes affected by PARP1 deficiency using the oPOSSUM algorithm also identified a skewed utilization of transcription factors of various classes, potentially indicating a preferential role of PARP1 in modulating the function of specific transcriptional regulators. Of importance, binding sites for the Ig-fold transcription factors, including key transcriptional regulators of the inflammatory response, such as NFκ-B family members and STATs, were underrepresented in genes regulated in Parp1−/− mice. This finding implies that normally PARP1 serves as a positive regulator of these transcription factors. Indeed, activation of NFκ-B by PARP1 has been reported by our group and others (36–38). A comparison of the response to DSS challenge in WT and Parp1−/− mice yielded a fascinating observation: the vast majority of transcripts that were up-regulated or down-regulated by DSS treatment in WT mice were already up- or down-regulated in Parp1−/− mice and did not further change with DSS treatment. This pattern was not reflected by baseline intestinal inflammation in Parp1−/− mice, which were phenotypically indistinguishable from healthy WT mice, but rather suggested a new equilibrium resulting in more resilient mucosal function.
This new transcriptional equilibrium was accompanied by increased frequencies of mucosal CD4+CD25+FoxP3+ Tregs, known for limiting and resolving mucosal inflammation in human inflammatory bowel disease and animal models of colitis, including DSS injury (24). This increase paralleled the relative expansion of colonic butyrate-producing Clostridia clusters IV and XIVa, recently identified as creating a microenvironment that facilitates expansion and differentiation of Tregs (23) and that decreases in prevalence in fecal samples from inflammatory bowel disease patients (39, 40). Although metabolic changes in the host may be responsible for the expansion of these protective microbes and the resulting Treg expansion, Treg-intrinsic effects of Parp1 deletion cannot be ruled out. Nasta et al. (41) recently demonstrated an increased Treg frequency in the peripheral lymph nodes of Parp1−/− mice, and PARP1 has been shown to reduce the immunosuppressive function of regulatory T cells by destabilizing Foxp3 binding to CNS2 (conserved non-coding DNA sequence 2) (42). Our own experiments not included in this manuscript showed that although the differentiation of iTreg from naïve T cells isolated from Parp1−/− mice is increased and the obtained cells showed higher expression of FoxP3 mRNA, the phenotype and in vivo immunosuppressive functions of primary Parp1−/− Tregs were similar to those of WT iTreg cells (data not shown). Thus, the change in the frequency of colonic lamina propria Tregs in Parp1−/− mice may be of more importance for mucosal protection than their altered function. Indeed, lack of protection in DSS-treated Rag2−/− × Parp1−/− DKO mice may provide support for the contribution of Tregs to this observed phenomenon. With the understanding of the limitations of Rag2−/− mice in mind, it is important to note that directly testing the role of CD4+CD25+FoxP3+ Tregs in the protection against mucosal injury observed in DSS-treated Parp1−/− mice is not straightforward. Depletion of Tregs leads to spontaneous wasting disease and very high mortality in response to DSS challenge (24), which would confound the effects of Parp1 status and yield results that are difficult to interpret. Rag-deficient mice, on the other hand, although lacking the optimal resolution of Treg-deficient mice, develop comparable colitis to WT mice after high-dose DSS (5%) and show mild resistance to low dose DSS (1.5%) (43). Therefore, we propose that a cooperative mechanism between PARP1-deficient innate immune cells (macrophages or intestinal epithelial cells) and regulatory T cells is required to mediate the full protective effect observed in DSS-induced colitis and that further studies are required to fully understand the role of PARP1 and its inhibition in the pathogenesis and potential treatment of inflammatory bowel disease.
Author Contributions
C. B. L. obtained the funding and provided the study concept and design, data acquisition, data analysis and interpretation, statistical analysis and drafting, and revision of the manuscript. K. W. S. acquired the data and provided data analysis and interpretation, statistical analysis, and drafting, and revision of the manuscript. D. L. provided technical support, data acquisition and analysis, and interpretation of data. D. R. J. provided technical support, data acquisition and analysis, and interpretation of data. P. R. K. obtained funding, provided the study concept and design of the experiments, statistical analysis, drafting of the manuscript, critical revision of the manuscript, and study supervision. F. K. G. obtained funding, study supervision, and critical revision of the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Candice J. Mason and Dr. George Watts for assistance with sample quality control and microarray hybridization at the Genomics Shared Service, University of Arizona Cancer Center.
This work was supported by National Institutes of Health Grants R37DK033209 (to F. K. G.) and 1K01DK099268 (to C. B. L.) through the NIDDK. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1 and Figs. S1–S5.
- PARP
- poly(ADP-ribose) polymerase
- DSS
- dextran-sulfate sodium
- DKO
- double knockout
- ANOVA
- analysis of variance
- TF
- transcription factor.
References
- 1. Suzuki H., Quesada P., Farina B., and Leone E. (1986) In vitro poly(ADP-ribosyl)ation of seminal ribonuclease. J. Biol. Chem. 261, 6048–6055 [PubMed] [Google Scholar]
- 2. Sodhi R. K., Singh N., and Jaggi A. S. (2010) Poly(ADP-ribose) polymerase-1 (PARP-1) and its therapeutic implications. Vascul. Pharmacol. 53, 77–87 [DOI] [PubMed] [Google Scholar]
- 3. Kim M. Y., Mauro S., Gévry N., Lis J. T., and Kraus W. L. (2004) NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119, 803–814 [DOI] [PubMed] [Google Scholar]
- 4. Szabo C., Pacher P., and Swanson R. A. (2006) Novel modulators of poly(ADP-ribose) polymerase. Trends Pharmacol. Sci. 27, 626–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peralta-Leal A., Rodríguez-Vargas J. M., Aguilar-Quesada R., Rodríguez M. I., Linares J. L., de Almodóvar M. R., and Oliver F. J. (2009) PARP inhibitors: new partners in the therapy of cancer and inflammatory diseases. Free Radic. Biol. Med. 47, 13–26 [DOI] [PubMed] [Google Scholar]
- 6. Jijon H. B., Churchill T., Malfair D., Wessler A., Jewell L. D., Parsons H. G., and Madsen K. L. (2000) Inhibition of poly(ADP-ribose) polymerase attenuates inflammation in a model of chronic colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 279, G641–G651 [DOI] [PubMed] [Google Scholar]
- 7. Di Paola R., Mazzon E., Xu W., Genovese T., Ferrraris D., Muià C., Crisafulli C., Zhang J., and Cuzzocrea S. (2005) Treatment with PARP-1 inhibitors, GPI 15427 or GPI 16539, ameliorates intestinal damage in rat models of colitis and shock. Eur. J. Pharmacol. 527, 163–171 [DOI] [PubMed] [Google Scholar]
- 8. Mabley J. G., Jagtap P., Perretti M., Getting S. J., Salzman A. L., Virág L., Szabó E., Soriano F. G., Liaudet L., Abdelkarim G. E., Haskó G., Marton A., Southan G. J., and Szabó C. (2001) Anti-inflammatory effects of a novel, potent inhibitor of poly (ADP-ribose) polymerase. Inflamm. Res. 50, 561–569 [DOI] [PubMed] [Google Scholar]
- 9. Mazzon E., Dugo L., Li J. H., Di Paola R., Genovese T., Caputi A. P., Zhang J., and Cuzzocrea S. (2002) GPI 6150, a PARP inhibitor, reduces the colon injury caused by dinitrobenzene sulfonic acid in the rat. Biochem. Pharmacol. 64, 327–337 [DOI] [PubMed] [Google Scholar]
- 10. Sánchez-Fidalgo S., Villegas I., Martín A., Sánchez-Hidalgo M., and Alarcón de la Lastra C. (2007) PARP inhibition reduces acute colonic inflammation in rats. Eur. J. Pharmacol. 563, 216–223 [DOI] [PubMed] [Google Scholar]
- 11. Zingarelli B., O'Connor M., and Hake P. W. (2003) Inhibitors of poly (ADP-ribose) polymerase modulate signal transduction pathways in colitis. Eur. J. Pharmacol. 469, 183–194 [DOI] [PubMed] [Google Scholar]
- 12. Zingarelli B., Hake P. W., Burroughs T. J., Piraino G., O'connor M., and Denenberg A. (2004) Activator protein-1 signalling pathway and apoptosis are modulated by poly(ADP-ribose) polymerase-1 in experimental colitis. Immunology 113, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zingarelli B., Szabó C., and Salzman A. L. (1999) Blockade of Poly(ADP-ribose) synthetase inhibits neutrophil recruitment, oxidant generation, and mucosal injury in murine colitis. Gastroenterology 116, 335–345 [DOI] [PubMed] [Google Scholar]
- 14. Huang F. C. (2009) Upregulation of Salmonella-induced IL-6 production in Caco-2 cells by PJ-34, PARP-1 inhibitor: involvement of PI3K, p38 MAPK, ERK, JNK, and NF-κB. Mediators Inflamm. 2009, 103890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 16. Kwon A. T., Arenillas D. J., Worsley Hunt R., and Wasserman W. W. (2012) oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 2, 987–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J. A., Smith G., and Knight R. (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bacchetti De Gregoris T., Aldred N., Clare A. S., and Burgess J. G. (2011) Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J. Microbiol. Methods 86, 351–356 [DOI] [PubMed] [Google Scholar]
- 20. Furet J. P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., Doré J., and Corthier G. (2009) Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 68, 351–362 [DOI] [PubMed] [Google Scholar]
- 21. Matsuki T., Watanabe K., Fujimoto J., Miyamoto Y., Takada T., Matsumoto K., Oyaizu H., and Tanaka R. (2002) Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68, 5445–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lopetuso L. R., Scaldaferri F., Petito V., and Gasbarrini A. (2013) Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 5, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., Kim S., Fritz J. V., Wilmes P., Ueha S., Matsushima K., Ohno H., Olle B., Sakaguchi S., Taniguchi T., Morita H., Hattori M., and Honda K. (2013) Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 [DOI] [PubMed] [Google Scholar]
- 24. Boehm F., Martin M., Kesselring R., Schiechl G., Geissler E. K., Schlitt H. J., and Fichtner-Feigl S. (2012) Deletion of Foxp3+ regulatory T cells in genetically targeted mice supports development of intestinal inflammation. BMC Gastroenterol. 12, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosado M. M., Bennici E., Novelli F., and Pioli C. (2013) Beyond DNA repair, the immunological role of PARP-1 and its siblings. Immunology 139, 428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Welsby I., Hutin D., and Leo O. (2012) Complex roles of members of the ADP-ribosyl transferase super family in immune defences: looking beyond PARP1. Biochem. Pharmacol. 84, 11–20 [DOI] [PubMed] [Google Scholar]
- 27. Paone G., Stevens L. A., Levine R. L., Bourgeois C., Steagall W. K., Gochuico B. R., and Moss J. (2006) ADP-ribosyltransferase-specific modification of human neutrophil peptide-1. J. Biol. Chem. 281, 17054–17060 [DOI] [PubMed] [Google Scholar]
- 28. Seman M., Adriouch S., Scheuplein F., Krebs C., Freese D., Glowacki G., Deterre P., Haag F., and Koch-Nolte F. (2003) NAD-induced T cell death: ADP-ribosylation of cell surface proteins by ART2 activates the cytolytic P2X7 purinoceptor. Immunity 19, 571–582 [DOI] [PubMed] [Google Scholar]
- 29. Alano C. C., Garnier P., Ying W., Higashi Y., Kauppinen T. M., and Swanson R. A. (2010) NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. J. Neurosci. 30, 2967–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y., Kim N. S., Haince J. F., Kang H. C., David K. K., Andrabi S. A., Poirier G. G., Dawson V. L., and Dawson T. M. (2011) Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci. Signal 4, ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perraud A. L., Fleig A., Dunn C. A., Bagley L. A., Launay P., Schmitz C., Stokes A. J., Zhu Q., Bessman M. J., Penner R., Kinet J. P., and Scharenberg A. M. (2001) ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature 411, 595–599 [DOI] [PubMed] [Google Scholar]
- 32. Qiu W., Wu B., Wang X., Buchanan M. E., Regueiro M. D., Hartman D. J., Schoen R. E., Yu J., and Zhang L. (2011) PUMA-mediated intestinal epithelial apoptosis contributes to ulcerative colitis in humans and mice. J. Clin. Invest. 121, 1722–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Forsythe R. M., Xu D. Z., Lu Q., and Deitch E. A. (2002) Lipopolysaccharide-induced enterocyte-derived nitric oxide induces intestinal monolayer permeability in an autocrine fashion. Shock 17, 180–184 [DOI] [PubMed] [Google Scholar]
- 34. Cuzzocrea S., Mazzon E., De Sarro A., and Caputi A. P. (2000) Role of free radicals and poly(ADP-ribose) synthetase in intestinal tight junction permeability. Mol. Med. 6, 766–778 [PMC free article] [PubMed] [Google Scholar]
- 35. Kraus W. L. (2008) Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakajima H., Nagaso H., Kakui N., Ishikawa M., Hiranuma T., and Hoshiko S. (2004) Critical role of the automodification of poly(ADP-ribose) polymerase-1 in nuclear factor-κB-dependent gene expression in primary cultured mouse glial cells. J. Biol. Chem. 279, 42774–42786 [DOI] [PubMed] [Google Scholar]
- 37. Hassa P. O., Covic M., Hasan S., Imhof R., and Hottiger M. O. (2001) The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. J. Biol. Chem. 276, 45588–45597 [DOI] [PubMed] [Google Scholar]
- 38. Majewski P. M., Thurston R. D., Ramalingam R., Kiela P. R., and Ghishan F. K. (2010) Cooperative role of NF-κB and poly(ADP-ribose) polymerase 1 (PARP-1) in the TNF-induced inhibition of PHEX expression in osteoblasts. J. Biol. Chem. 285, 34828–34838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., and Pace N. R. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., Roca J., and Dore J. (2006) Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nasta F., Laudisi F., Sambucci M., Rosado M. M., and Pioli C. (2010) Increased Foxp3+ regulatory T cells in poly(ADP-Ribose) polymerase-1 deficiency. J. Immunol. 184, 3470–3477 [DOI] [PubMed] [Google Scholar]
- 42. Zhang P., Maruyama T., Konkel J. E., Abbatiello B., Zamarron B., Wang Z. Q., and Chen W. (2013) PARP-1 controls immunosuppressive function of regulatory T cells by destabilizing Foxp3. PloS ONE 8, e71590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim T. W., Seo J. N., Suh Y. H., Park H. J., Kim J. H., Kim J. Y., and Oh K. I. (2006) Involvement of lymphocytes in dextran sulfate sodium-induced experimental colitis. World J. Gastroenterol. 12, 302–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.