Abstract
Many eukaryotic genes undergo alternative 3′-end poly(A)-site selection producing transcript isoforms with 3′-UTRs of different lengths and post-transcriptional fates. Gene loops are dynamic structures that juxtapose the 3′-ends of genes with their promoters. Several functions have been attributed to looping, including memory of recent transcriptional activity and polarity of transcription initiation. In this study, we investigated the relationship between gene loops and alternative poly(A)-site. Using the KlCYC1 gene of the yeast Kluyveromyces lactis, which includes a single promoter and two poly(A) sites separated by 394 nucleotides, we demonstrate in two yeast species the formation of alternative gene loops (L1 and L2) that juxtapose the KlCYC1 promoter with either proximal or distal 3′-end processing sites, resulting in the synthesis of short and long forms of KlCYC1 mRNA. Furthermore, synthesis of short and long mRNAs and formation of the L1 and L2 loops are growth phase-dependent. Chromatin immunoprecipitation experiments revealed that the Ssu72 RNA polymerase II carboxyl-terminal domain phosphatase, a critical determinant of looping, peaks in early log phase at the proximal poly(A) site, but as growth phase advances, it extends to the distal site. These results define a cause-and-effect relationship between gene loops and alternative poly(A) site selection that responds to different physiological signals manifested by RNA polymerase II carboxyl-terminal domain phosphorylation status.
Keywords: gene transcription, RNA polymerase II, RNA processing, Saccharomyces cerevisiae, yeast
Introduction
Transcription termination is coupled to 3′-end processing, which involves endonucleolytic cleavage of the nascent mRNA and addition of a poly(A) tail (1–3). Many genes exhibit two or more alternative 3′-end processing sites (APAs).2 In human cells, more than half of protein-encoding genes have alternative 3′-end processing sites (4). The cellular choice of 3′-end processing sites is regulated (4–6). Differential 3′-ends often occur within introns, thereby encoding proteins with markedly different functions (7–10). As many of the elements that modulate subsequent steps in mRNA fate are present within 3′-UTRs, mRNA length variation increases regulatory possibilities, including mRNA stability and translational efficiency (9). In humans, APA affects many cellular events, including inflammatory processes (4), and is regulated in a tissue-specific manner (11, 12).
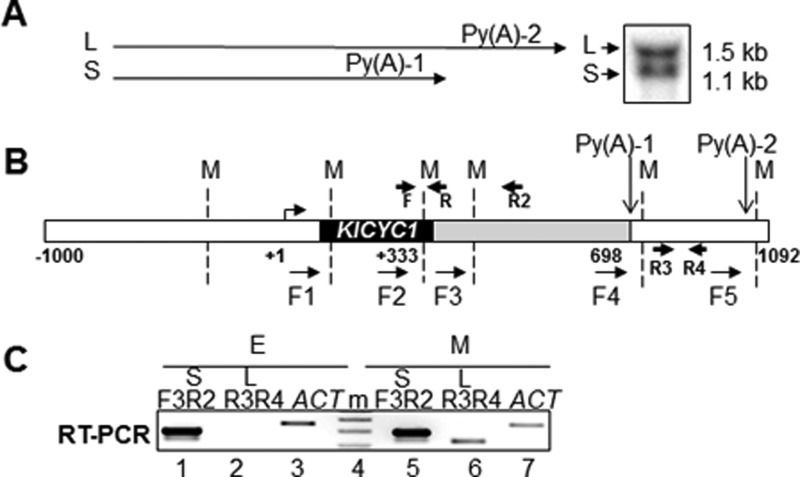
Yeast also exhibits APA sites that have been correlated with distinct regulatory parameters (13–16). As a model gene for APA, we analyzed the CYC1 gene of the yeast Kluyveromyces lactis. KlCYC1 is transcribed as two mRNA isoforms, a consequence of two distinct 3′-end processing sites, generating short (S) and long (L) transcripts. The proximal site yields a transcript of 1.1-kb (S), whereas the distal site is 394 nucleotides downstream, producing a 1.5-kb transcript (L) (Fig. 1A) (15, 17). Alternative KlCYC1 processing is growth phase-dependent and is readily distinguished by changes in the ratio of short/long transcripts (15, 18). Components of Saccharomyces cerevisiae 3′-end processing complexes CF-1A and CPF affect proximal versus distal processing (17, 18).
FIGURE 1.
Alternative polyadenylation of K. lactis CYC1 mRNA (KlCYC1). A, schematic summary of long (L) and short (S) forms of the KlCYC1 mRNA resulting from alternative 3′-end processing at polyadenylation sites Py(A)-1 and Py(A)-2. The Northern blot depicts the L and S transcripts from cells harvested at early log phase growth (A600 = 0.6). B, schematic depiction of the KlCYC1 gene. The numbering system denotes the ATG (+1), termination codon (+333), and endonucleolytic cleavage sites as Py(A)-1 (698) and Py(A)-2 (+1092). Tandem primers used for 3C analysis are F1–F5; these primers are located adjacent to the MspI restriction sites (M) denoted by the dashed vertical lines. The primer pairs F3, R2 and R3, R4 were used to quantify KlCYC1 mRNA by RT-PCR. The F and R primers were used for quantitation of 3C chromatin. C, KlCYC1 mRNA levels as determined by RT-PCR. Lanes 3 and 7 represent mRNA encoded by the ACT1 gene and serve as loading controls. Lane 4 (m) is marker DNA used to estimate DNA length.
CF-IA consists of Rna14, Rna15, Clp1, and Pcf11. CPF consists of an APT (Associated with Pta1) subcomplex that includes Pti1, Swd2, Syc1, and Ref2, two protein phosphatases, Glc7 and Ssu72, and the Pta1 protein, which appears to be a scaffold that bridges the APT subcomplex with core-CPF to form holo-CPF. These complexes and their subunits are conserved among yeast and mammalian cells (2, 19–21). CPF and CF-1A recognize specific RNA sequences in the nascent transcript. Subsequent to assembly of these two complexes, pre-mRNA is endonucleolytically cleaved, followed by addition of a poly(A) tail, catalyzed by poly(A) polymerase (2). 3′-End processing and transcription termination, defined by dissociation of the trimeric DNA-RNA-RNAP II complex, are coupled events, although the mechanism of termination and how it is coupled to 3′-end processing remain to be elucidated.
To address the factors that dictate proximal versus distal 3′-end processing events, we considered the potential effect of gene loops on APA selection. Gene loops juxtapose promoter-terminator regions and have been shown to facilitate the kinetics of transcription reinitiation (22–24) and promoter directionality (25). These effects are likely to be a consequence of RNAP II “hand-off,” resulting in recycling of RNAP II from terminator to promoter.3 Consistent with this model, gene loops are transcription-dependent, requiring components of the transcription initiation and 3′-end processing complexes (22, 23, 26). Accordingly, gene looping implies a second distinct pathway for preinitiation complex (PIC) formation that bypasses the rate-limiting step of de novo recruitment and assembly of RNAP II and the general transcription factors.
This model is also supported by the Ssu72 RNAP II CTD phosphatase that genetically and physically interacts with the PIC and is critical for DNA looping (19, 22, 23, 25, 27). Ssu72 catalyzes partial dephosphorylation of the CTD as a prerequisite for phosphorylation of Ser-2 of the CTD and entry into the elongation phase of transcription (28). Ssu72 also catalyzes dephosphorylation of residual Ser(P)-5 at the termination region, presumably enabling recycling of hypophosphorylated RNAP II into the PIC (27–30). According to this model, the Ssu72 phosphatase activity ensures production of initiation-competent, hypo-phosphorylated RNAP II.
Given that gene loops are transcription-dependent dynamic structures defined by juxtaposition of promoter-terminator regions, we asked whether alternative poly(A) site selection correlates with alternative formation of gene loops. KlCYC1 is a model gene for addressing this question because the two poly(A) sites are well separated, and the genetic locus contains multiple MspI restriction sites, which facilitate high resolution mapping of physical interactions along the length of the gene by chromatin conformation capture (3C) (31). Our results clearly demonstrate the formation of two alternative gene loops that correlate with alternative poly(A) site selection and implicate the Ssu72 CTD phosphatase in this process.
Materials and Methods
Strains, Media, and Growth Conditions
The K. lactis yeast strain NRRL-Y1140, which expresses the wild type KlCYC1 gene encoding cytochrome c, was used in this study. The strain ZW13 was used to study KlCYC1 DNA loops in S. cerevisiae (32). The S. cerevisiae isogenic strain pair XH6 (WT) and POC8-23d (pta1-1), described previously (17), was used to analyze the effects of Pta1 on alternative gene loops. Cells were grown in YPD medium (33) to early logarithmic phase (A600 = 0.6) or to mid-logarithmic phase (A600 = 1.5), denoted E or M, respectively, in all experiments.
RT-PCR
Total RNA was isolated using the RNeasy Midi RNA isolation procedure (Qiagen) using cell pellets obtained from 50-ml cultures, grown in parallel with the cultures used for 3C analysis. The F3-R2 primer pairs detected both transcripts (L and S); R3-R4 was specific for the long form. Two actin gene (ACT1) primers were used as an RNA template control. PCR products were analyzed as described for 3C.
3C Assay
Gene loops were assayed as described previously (31), with the following modifications. Transient protein-protein and protein-DNA interactions were fixed by incubation of cells in 2% formaldehyde for 30 min at room temperature with gentle shaking. Cross-links were released by incubation in 0.25 m glycine for 10 min. Cells were washed in TBS buffer containing 1% Triton X-100 and resuspended in FA lysis buffer (50 mm HEPES-KOH, pH 7.9, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate). The chromatin pellet was collected by centrifugation and resuspended thoroughly in 100–120 μl of FA lysis buffer, followed by digestion overnight with the restriction enzyme MspI. The MspI digestion sites within KlCYC1 enable resolution of the promoter, ORF, two fragments of the 3′-UTR, and the two polyadenylation sites, denoted Py(A)-1 and Py(A)-2 (Fig. 1A). MspI digestion fragments were subsequently ligated in dilute solution to maximize intramolecular reaction products (31). Ligation products were assayed by PCR using tandem primers, F1 to F5, that anneal across the length of the KlCYC1 gene (Fig. 1B; Table 1). Primer pair efficiency control PCRs were performed using the KlCYC1 plasmid pCT2 as template DNA (15). pCT2 was digested by MspI and ligated in a 10-μl volume followed by PCR amplification using the same set of primers (Fig. 1B). PCR products were resolved in 1.5% agarose gels containing ethidium bromide. Band intensity was quantified using an AlphaImager 2200 (Alpha Innotech). Quantification was normalized by the ratio of the looping signal (SL) (for L1 or L2) to the control-loading signal obtained from the convergent primers F + R (SL/SF+R).
TABLE 1.
DNA primers used in this study
| Name | Sequence | Positiona |
|---|---|---|
| Primers for DNA-looping experiments | ||
| F1 | 5′ ATCAGTATTAAGGATTGATTCGTC 3′ | P |
| F2 | 5′ CCGGTGGTCCACACAAGG 3′ | ORF |
| F3 | 5′ CGTTACTTACATGTTGAAGGCTTG 3′ | ORF |
| F4 | 5′ GAACGAACCTTTTCATGATAGAAC 3′ | T |
| F5 | 5′ CATCTATATAAACTTCCACAGAATC 3′ | T |
| F | 5′ CTCTTACACTGACGCTAACATC 3′ | ORF |
| R | 5′ CAATCTACCATGCGTGCATG 3′ | T |
| R2 | 5′ AACTTAGAAGAACGTTCATAGACA 3′ | T |
| R3 | 5′ TGTTATCACACATCCCGTCTAT 3′ | T |
| R4 | 5′ TGTAAATCACACGACGCATAC 3′ | T |
| ACTF | 5′ AACCGAAGCTCCAATGAA 3′ | ORF |
| ACTR | 5′ TCAACAGCGGATGATTGA 3′ | ORF |
| Primers for ChIP experiments | ||
| C | 5′ ATGAATCTTCTCCGCAGCAT 3′ | Co |
| C | 5′ CACGTGACTTTTGGTGTGCT 3′ | Co |
| P1F | 5′ ACACCAAAAGTCACGTGTGC 3′ | P |
| P1R | 5′ ACTGTGAACGGAAGCGATCT 3′ | P |
| P2F | 5′ ACATCAGCCAACCAATCAGA 3′ | P |
| P2R | 5′ TCTATCATTTCGGGAACATCG 3′ | P |
| P3F | 5′ ATGCGCACTATGGTTTGTTG 3′ | P |
| P3R | 5′ CCTTCTTTTCGGAACCCTTC 3′ | P |
| OR | 5′ GAAGGGTTCCGAAAAGAAGG 3′ | ORF |
| OR | 5′ CCTCTTCAAACCACCGAAA 3′ | ORF |
| OT1 | 5′ CGGTGGTTTGAAGAAGGAAA 3′ | ORF+T |
| OT1 | 5′ GGGAAGGGTGAGATGGTGTA 3′ | ORF+T |
| T1 | 5′ CCGCAAAACAATAAAGAACGA 3′ | T |
| 1 | 5′ GGGGACCGGGTGTTAAGATA 3′ | T |
| T2 | 5′ CATGCTCACTGAGTTTCAAAAGA 3′ | T |
| T2 | 5′ ACTAGTGCTGCTGGTGCTGA 3′ | T |
| I | 5′ CTGGAGTTACCGGAGCTAGAA 3′ | KI |
| I | 5′ GGATGACAATACCCCAGACG 3′ | KI |
Chromatin Immunoprecipitation
Yeast cells were grown under identical conditions as described for 3C. ChIP experiments were performed as a described (28), with the following modifications. Briefly, cells were lysed by vigorous shaking in a minibar beater (MiniBeadBeater-16, Model 607) for four cycles of 1 min each at 4 °C, and chromatin sonication times were increased to 12 pulses at 15 s each with 30-s intervals in ice-cold FA lysis buffer. The following antibodies were used to immunoprecipitate sonicated chromatin: α-Rpb3 (Neoclone), specific for the Rpb3 subunit of RNAPII; 8WG16 (Covance; ChIP grade), specific for hypophosphorylated Ser-2 of the CTD; and polyclonal α-Ssu72, specific for the Ssu72 CTD phosphatase (27, 28). These antibodies are used routinely in ChIP experiments using S. cerevisiae (28, 34, 35). Here, we demonstrate that they are equally effective and specific for ChIP experiments using K. lactis.
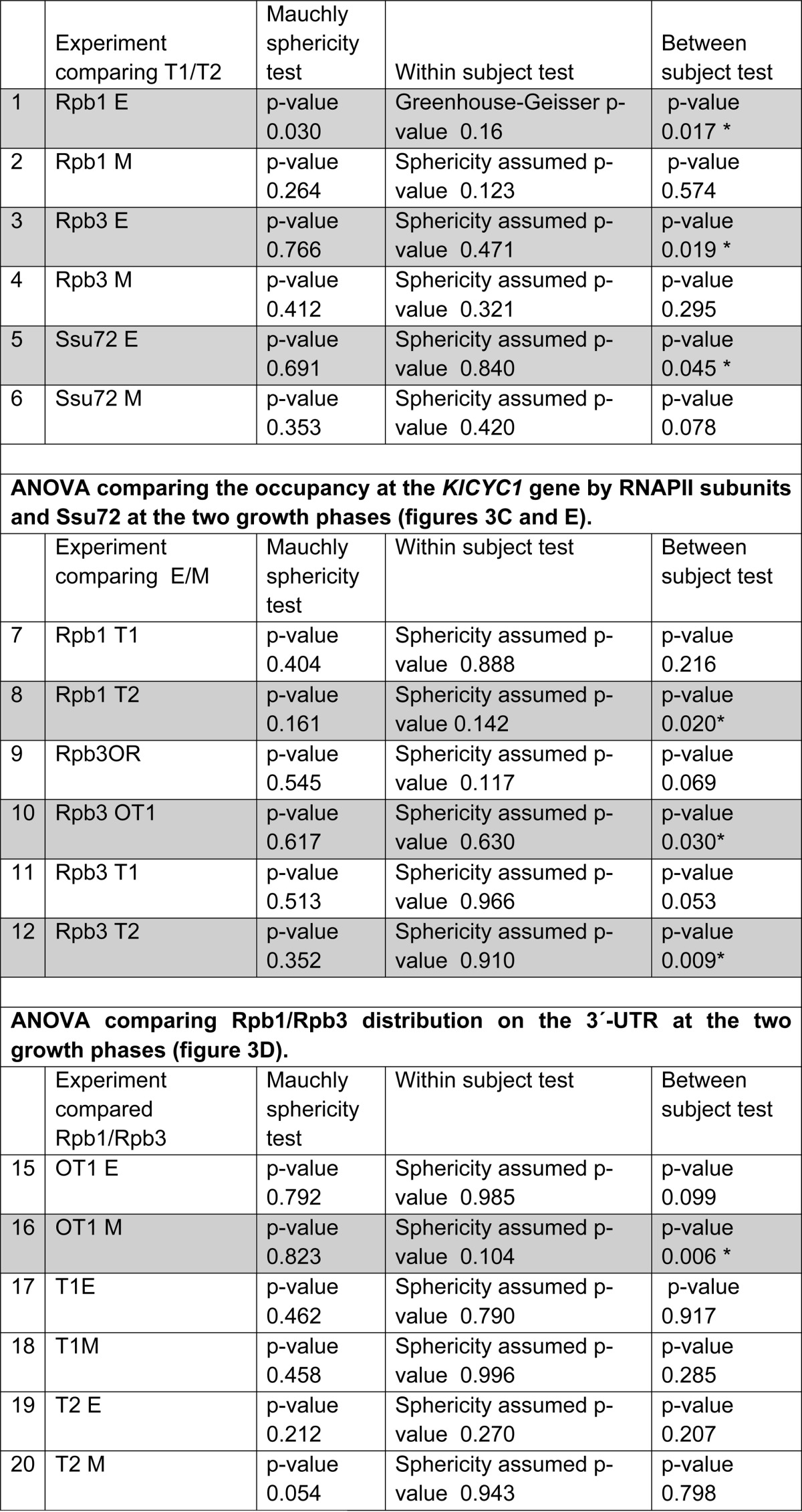
The primer pairs used for PCR amplification are listed in Table 1. PCR products were fractionated and detected as described above for 3C. The data were normalized by determining the IPx/input ratio of each probed region (x) to the same IPI/input ratio of the K. lactis Klla0F intergenic region (I) located 1330 bp from KlCYC1 gene termination codon: ((IPx/inputx)/(IPI/inputI)). Data were quantified in all experiments as the mean of three independent experiments, each done in triplicate, as described (28). Values represent the mean; error bars were calculated as standard deviation of the mean. The statistical significance of means was determined by analysis of variance using SPSS software (IBM) (Table 2).
TABLE 2.
Analysis of variance comparing the usage of the proximal (T1) or distal (T2) processing points at two growth phases
Data corresponds to Fig. 3, C and E.
Results
Experimental Strategy
Transcription of the endogenous KlCYC1 gene of K. lactis yields two distinct transcripts that differ in the lengths of their 3′-UTRs (15). 3′-End processing at the promoter-proximal poly(A) site produces a 1.1-kb transcript (S), whereas processing at the downstream sites yields a clearly discernable 1.5-kb transcript (L) (Fig. 1A). The poly(A) sites producing these transcripts are schematically depicted in Fig. 1B. Synthesis of the S and L forms is differentially regulated in response to cell culture growth phase, i.e. growth phase affects KlCYC1 3′-end formation. The mechanism by which the 3′-end processing machinery differentially selects the poly(A)-1 versus poly(A)-2 site to generate the S versus L forms is related to the interplay between the proximal AU-rich element and the 3′-end processing machinery (17). RT-PCR analyses of the transcript isoforms under different regulatory conditions are shown in Fig. 1C. As expected, during early log phase growth (A600 = 0.6) the S form is predominant, with no detectable synthesis of the L form, and in mid-log phase growth (A600 = 1.5), synthesis of the L form is readily apparent. Note that in M phase, the F3-F2 signal (Fig. 1C, lane 5) corresponds to the two transcripts, whereas R3-R4 (lane 6) is specific to the L form. Whether poly(A) site selection is associated with the establishment of two different DNA loops is unknown. Mechanistically, this is an important issue to understand co-transcriptional RNA processing. These results guided our experimental strategy to investigate the relationship between formation of DNA loops and APA.
Gene Loops Correlate with Formation of the S and L Forms of the KlCYC1 Transcript
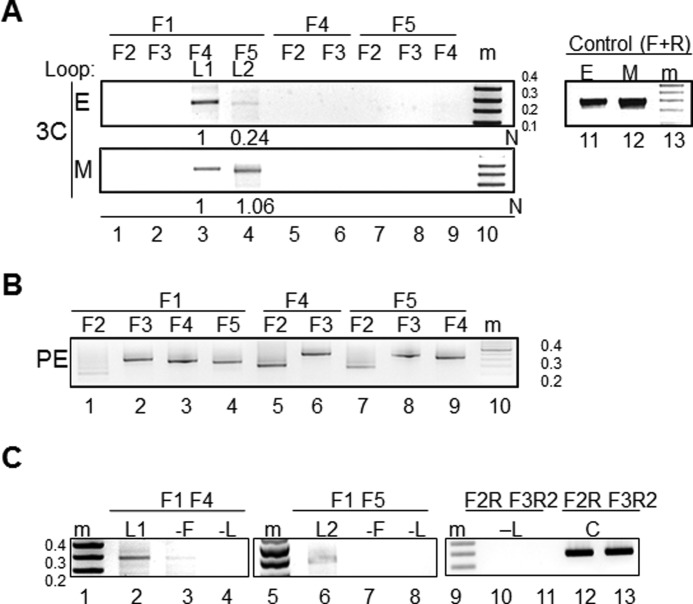
Juxtaposition of promoter-terminator regions to form gene loops raises the question of whether alternative poly(A) site selection affects differential loop formation. To address this issue, we first assayed gene loops at the KlCYC1 locus in K. lactis. Results are presented in Fig. 2A. Cells harvested in early log phase growth (Fig. 2A, panel E) displayed the most intense 3C signal using primer pair F1 and F4, which anneals to the KlCYC1 promoter and Py(A)-1 sites to form the gene loop labeled L1 (Fig. 2A, lane 3). When paired with primer F5, which anneals to the DNA fragment encompassing poly(A)-2, F1 yielded a PCR fragment, corresponding to gene loop L2, but at only ∼24% of the intensity of the F1–F4 fragment (Fig. 2A, lane 4). By contrast, cells harvested in mid-log phase (subpanel M) yielded F1–F4 and F1–F5 PCR bands of approximately equal intensity (loops L1 and L2). To establish that these PCR products were specific to promoter-terminator regions, we demonstrated the absence of PCR products among the other primer pairs that anneal along the length of KlCYC1 (Fig. 2B).
FIGURE 2.
Alternative polyadenylation of KlCYC1 correlates with changes in gene loops. A, PCR products generated by 3C analysis of the KlCYC1 gene are depicted for cells in early log (E) and mid-log (M) phase growth. Primer pairs (F1–F5) are depicted above each lane and shown schematically in Fig. 1B. The F1–F4 and F1–F5 PCR products correspond to looping between the KlCYC1 promoter and the poly(A)-1 (L1) and the poly(A)-2 sites (L2), respectively. Results are shown for a single experiment. The mean value of three independent experiments was determined (see under “Materials and Methods”). B, primer pair efficiencies. PCR products were generated with the indicated primer pairs. C, 3C PCR products are dependent upon formaldehyde cross-linking. 3C experiments were performed as in A using early log phase cells and the indicated primer pairs. PCR results from cross-linked DNA are shown in lanes 2 and 6 and represent the L1 loop, comparable with A, lanes 3 and 4. Omission of formaldehyde from the same experiment yielded the data shown in lanes 3 and 7 (−F); results from omission of DNA ligase subsequent to MspI digestion of cross-linked chromatin are shown in lanes 4 and 8 (−L). The absence of PCR products from the F2-R and F3-R2 primer pair (lanes 10 and 11, −L) demonstrates the efficiency of MspI digestion. The 3C positive control experiments are depicted in lanes 12 and 13. Molecular markers (m) confirm PCR products of the anticipated length (lanes 1, 5, and 9).
These sets of primer pairs indicate that KlCYC1 looping specifically juxtaposes the promoter and polyadenylation sites and excludes physical interactions among other regions of the gene. These results are not a consequence of different primer pair efficiencies, as a control experiment established that all primer pairs yielded PCR products of comparable intensities using KlCYC1 template DNA that had been digested to completion with MspI and subsequently ligated (see under “Materials and Methods”; results using primer F2 are an exception but do not interfere with interpretation of our results.) As additional controls, we also demonstrated that detection of the L1 and L2 gene loops by 3C are dependent upon both formaldehyde cross-linking and ligation subsequent to MspI digestion (Fig. 2C). Thus, gene loops L1 and L2 correlate specifically with formation of KlCYC1 mRNA termination at the proximal poly(A)-1 site and distal poly(A)-2 site, respectively.
Distribution of RNAP II Along the KlCYC1 Gene during Early and Mid-log Phase Growth
RNAP II occupancy was assayed by immunoprecipitation of the Rpb3 subunit, as described previously; indeed, immunoprecipitation of Rpb3 is universally used to detect occupancy of RNAP II complexes, irrespective of phosphorylation status (36). We studied the presence of Rpb3 at the two growth points of the logarithmic phase and at the two UTR processing regions. Results are shown in Fig. 3, B and C. Gene loops have been proposed to facilitate translocation of RNAP II from the terminator to the promoter for subsequent rounds of transcription (22, 23). Indeed, RNAP II competition experiment indicates that RNAP II is recycled from terminator to promoter in a manner dependent upon looping. Recycling, whether directly from the terminator to the promoter of the same gene or from the nuclear pool of RNAP II, requires dephosphorylation of the RNAP II CTD.
FIGURE 3.
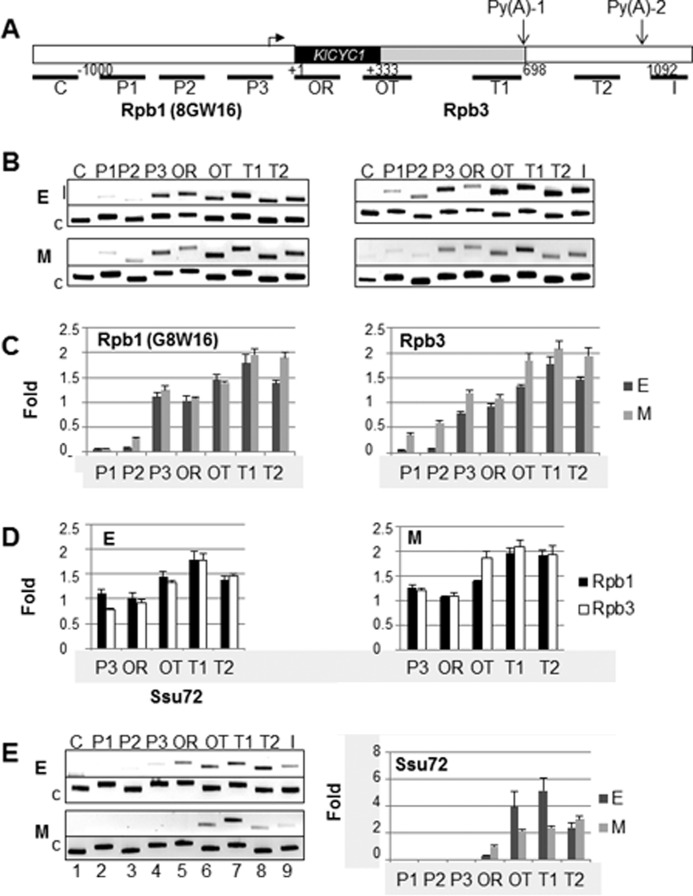
Dependence on growth phase of RNAP II localization along the length of KlCYC1 as determined by ChIP. A, schematic indication of the KlCYC1 locus indicating the regions probed by PCR primers. P1 and P2 are upstream of the promoter; P3 encompasses the promoter; OR and OT1, probe the early ORF and immediate 3′-UTR, respectively; T1 and T2 probe DNA encoding the indicated 3′-end processing sites; I probes the region immediately downstream of the poly(A)-2 site; and C serves as a control, probing a non-transcribed region located upstream of KlCYC1. B, ChIP results for cells harvested in early log (E) and mid-log (L) phase using the 8WG16 or Rpb3 antibodies. C, quantification of Rpb1 and Rpb3 ChIP analysis comparing the growth phase (E and M). D, quantification of ChIP analysis comparing results using the Rpb1 and Rpb3 antibodies. All data are from three independent experiments equivalent to those shown in B. E, ChIP results from cells harvested in early log (E) and mid-log (M). Location of Ssu72 along KlCYC1 and its dependence on the growth phase, using the Ssu72 antibody, are shown. Error bars represent the standard error of the mean of three independent experiments.
To determine the distribution and phosphorylation status of RNAP II at the KlCYC1 locus, we performed ChIP experiments, using the 8WG16 antibody, which immunoprecipitates RNAP II that is hypophosphorylated at Ser-2 of the CTD, and using antibody to the Rpb3 subunit of RNAPII, which immunoprecipitates RNAP II irrespective of CTD phosphorylation status. Results are presented in Fig. 3. Fig. 3A depicts a schematic of the KlCYC1 locus, indicating the positions of PCR primer pairs used in the ChIP experiments. As expected, RNAP II does not associate with DNA upstream of the promoter but is readily detected at the promoter (P3) and along the length of the gene (OR) in cells that were harvested at early or mid-log phase (Fig. 3B, phases E and M). Downstream of the ORF, however, distinct differences were observed depending on growth phase. Diminished amounts RNAP II were associated with KlCYC1 beyond the proximal polyadenylation site (Py(A)-1) in cells harvested in early log phase (Fig. 3C, T1 versus T2, E phase). Quantitation of ChIP data from three independent experiments indicated ∼23% diminished association of RNAP II with region T2 relative to region T1. Statistical analysis demonstrated that the difference between RNAP II ChIP data between E and M phase cells at T1 and T2 is significant (p value = 0.017, Table 2, row 1). In contrast, the mean association of RNAP II with regions T1 and T2 in M phase cells exhibited no significant difference (p value = 0.57, Table 2, row 2). These results correlate with the mRNA analysis; RNAP II occupancy peaks at the T1 site and trails off at T2 in early log when the S form of KlCYC1 mRNA predominates. Conversely, RNAPII occupancy remains high at T2 in mid-log phase when the S and L forms of mRNA are produced in equal amounts.
A second observation from this experiment is that occupancy of RNAP II associated with KlCYC1 is higher at the termination region than along the length of the gene, an observation that has been seen for RNAP II at other loci (20, 37–39) presumably reflecting stalled RNAP II at poly(A) sites as part of the mechanism of 3′-end processing and transcription termination (Fig. 3, B and C). This ChIP result was substantiated and quantified using anti-Rpb3 antibody (Fig. 3D).
We note the difference at mid-log phase between RNAP II occupancy at T1 and T2; a significant difference (p value = 0.030; Table 2, row 10) at the end of the KlCYC1 ORF was also observed. The Rpb3 antibody signal reveals a significantly higher occupancy of phosphorylated RNAP II at T2 in cells harvested at mid-log phase (Fig. 3B, compare OT in Rpb1 (8WG16) and Rpb3 in E and M). We interpret this difference to elongating RNAP II complexes reaching the distal processing position at mid-log phase. This result is consistent with an increase of 3′-end processing at the distal site (Fig. 1C) and also with formation of the L2 loop at M phase (Fig. 2A).
Alternative Polyadenylation and Ssu72 Occupancy of KlCYC1
Ssu72 is a CTD phosphatase specific for Ser(P)-5 and Ser(P)-7 and an integral component of the CPF 3′-end processing complex (27, 40). Accordingly, Ssu72 is a reliable marker for assembly of CPF at the 3′-ends of genes. We determined the profile of Ssu72 association with KlCYC1 by ChIP. Results are shown in Fig. 3E. In early log phase, Ssu72 is weakly associated with the promoter (Fig. 3E, P3, lane 4), and its occupancy increases substantially at the 3′-end of the gene (Fig. 3E, cf. OT, T1, and T2, lanes 6–8). In mid-log phase, Ssu72 exhibits a 2-fold increase in occupancy of the termination regions OT and T1 (p value = 0.045, Table 2, row 5), coincident with enhanced synthesis of the S form of KlCYC1 mRNA (Fig. 1), and a 4-fold increase in formation of the F1–F4 gene loop relative to the F1–F5 loop (Fig. 2). In contrast, we found no association of Ssu72 with the promoter (P3) in mid-log phase. As RNAP II processed along the gene, Ssu72 occupancy increased, but the relative amounts of Ssu72 are much lower at OT and T1, as expected for processive elongation of RNAP II beyond region T1 into region T2 (Fig. 3E, lanes 6–8). There was no significant difference, however, in Ssu72 association at T2 in early and mid-log phase cells, consistent with enhanced synthesis of the L form of KlCYC1 mRNA and formation of the F1–F5 gene loop at this phase of cell growth (Fig. 2A). Therefore, there is a significant increase of Ssu72 association with the 3′-UTR regions where 3′-end processing and DNA-loop formation are predominant.
Gene Loops Direct Formation of the S and L Forms of the KlCYC1 Transcript Expressed in S. cerevisiae
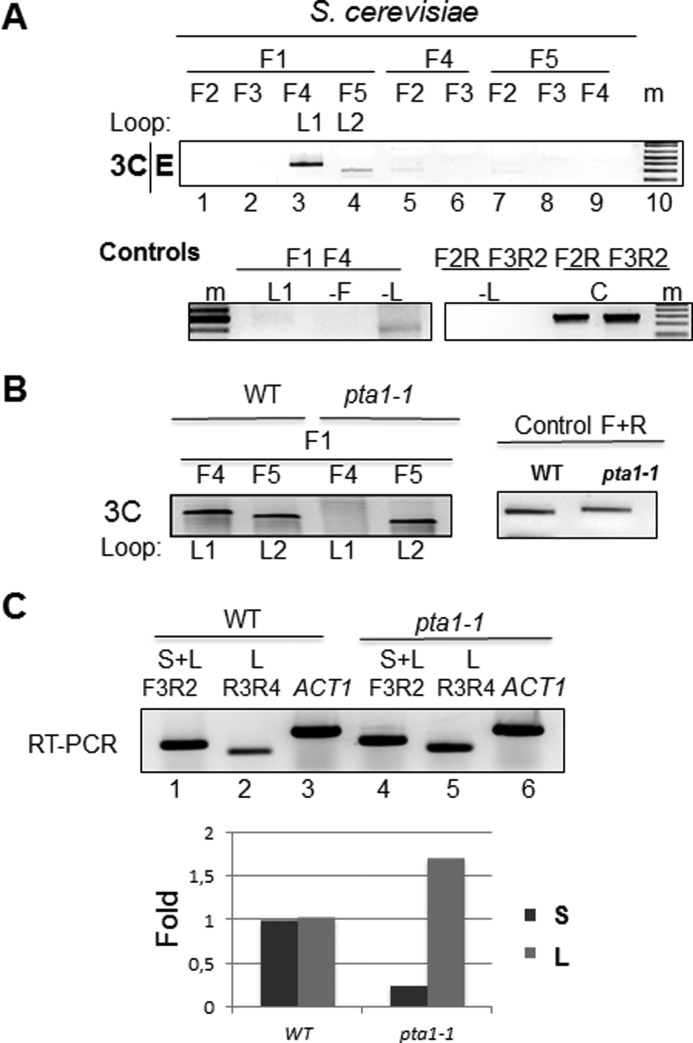
To determine whether the correlation between alternative polyadenylation of KlCYC1 transcripts might be a general effect rather than idiosyncratic to K. lactis, we expressed plasmid-borne KlCYC1 in S. cerevisiae and determined gene loops as described above. We observed the same result; in early log phase (OD = 0.6), looping occurs between the promoter and proximal poly(A) site (F1–F4 primer pairs) and is diminished between the promoter and distal poly(A) site (F1–F5 primer pairs) (Fig. 4A). These data are comparable with that observed for KlCYC1 gene loop formation in K. lactis (Fig. 2A cf. Fig. 5). We did not assay potential changes in looping when cells entered mid-log phase growth (OD = 1.5) as no difference was observed between synthesis of the S and L forms of KlCYC1 expressed in S. cerevisiae during this growth transition.
FIGURE 4.
Alternative gene loops at KlCYC1 expressed in S. cerevisiae. A, PCR products generated by 3C analysis of the KlCYC1 gene are depicted for cells in early log phase growth, E. Primer pairs (F1–F5) are depicted above each lane and shown schematically in Fig. 1B. The F1–F4 and F1–F5 PCR products correspond to looping between the KlCYC1 promoter and the poly(A)-1 (L1) and the poly(A)-2 sites (L2), respectively. B, 3C analysis of the KlCYC1 gene in an isogenic WT and pta1-1 strain pair (C). Control experiments as described in Fig. 2C.
FIGURE 5.
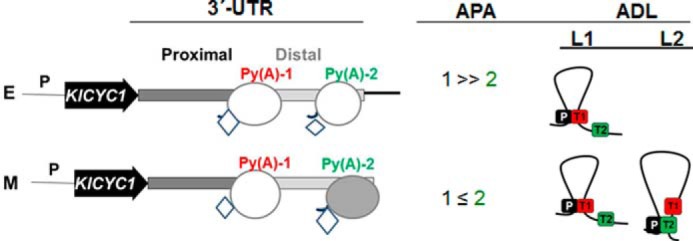
Model for the correlation between APA and alternative gene looping at the KlCYC1 locus. In early (E) log phase cells, 3′-end processing occurs predominantly at the proximal poly(A) site, which is the predominant site for looping between the promoter (P) and poly(A)-1 site (Py(A)-1) (red). Conversely, in mid-log phase (M) cells, RNAP II exhibits enhanced bypass at the proximal poly(A)-1 (Py(A)-1) (red) and processing to the poly(A)-2 site (Py(A)-2) (green), resulting in approximately equal 3′-end processing at the two poly(A) sites, coinciding with comparable looping between these two sites and the promoter. Key: RNAPII (oval) and Ssu72 (diamond) during the different phases of cell growth (E and M). ADL denotes alternative looping being L1 and L2, the two alternatives. RNAPII is in gray color when Rpb3 abundance is higher than hypophosphorylated Rpb1. The increase in diamond size reflects changes of the abundance of this factor.
To address the cause-and-effect relationship between gene looping and alternative polyadenylation, we determined the consequences of a defective form of the CPF 3-end processing complex on poly(A) site selection. This experiment was done using a pta1-1 mutant, shown previously to adversely affect KlCYC1 APA (Seoane et al. (17)). As shown in Fig. 4B, the pta1-1 mutant blocks formation of the L1 loop, resulting in elimination of the S form of KlCYC1 mRNA. Thus, Pta1 is essential for alternative DNA loop formation in S. cerevisiae. Furthermore, the coordinated formation of alternative DNA loops, associated with APA, is conserved between K. lactis and S. cerevisiae. These results define a novel function for gene loops, direction of alternative poly(A) site selection. Given the phylogenetic conservation of eukaryotic 3′-end processing complexes (2), we conclude that gene looping plays a general role in alternative poly(A) site selection.
Discussion
In this study, we demonstrate formation of alternative gene loops (L1 and L2) that juxtapose the KlCYC1 promoter with either the proximal or distal 3′-end processing sites, correlating with formation of KlCYC1 mRNAs that include short (S) and long (L) 3′-UTR regions. To our knowledge, this is the first report establishing a relationship between looping and alternative polyadenylation, suggesting a cause-and-effect relationship between these two events. It is also the first report showing formation of DNA loops in the yeast K. lactis. Moreover, looping and poly(A) site selection are physiologically regulated in response to cellular growth phase, in this case affected by early and mid-log phase growth.
What is the physiological relevance of the S versus L forms of KlCYC1? These isoforms of KlCYC1 mRNA exhibit different fates in response to different physiological conditions, including growth phase, oxygen availability, and the presence or absence of heme (18, 41). As cells progress through log phase growth, the S form is degraded, and the L form increases. The enhanced stability of the L form of KlCYC1 ensures production of cytochrome c in the respiratory phase of growth. This effect is not specific to respiration but also occurs for transcripts encoding proteins of other metabolic processes (17). Accordingly, alternative 3′-UTR processing can be essential for many specific cellular functions.
What is the relationship between synthesis of the L form of KlCYC1 mRNA and the L2 gene loop? De novo recruitment of RNAP II proceeds by assembly of the general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and RNAP II) in a stepwise manner at the DNA promoter to form a transcription PIC. Subsequent to transcription initiation and promoter clearance, some of the general transcription factors dissociate from the promoter, whereas others remain behind, forming a scaffold-like structure (42). A pioneer round of transcription is then required to recruit the 3′-end processing complexes. In yeasts, CPF and CF-1A, which are analogous to the cleavage and polyadenylation-specific factor and cleavage stimulation factor complexes in mammalian cells (2), serve well defined functions in 3′-end processing that link the termination region back to the promoter scaffold.3 Accordingly, transcription reinitiation occurs by a pathway different from de novo PIC assembly. Subsequent to dephosphorylation of the RNAP II CTD, hypo-phosphorylated RNAP II is recycled from the terminator to the promoter. Given that the rate of transcription elongation by RNAP II is estimated at ∼2000 nucleotides/min, yet PIC assembly and recruitment of the 3′-end processing machinery are relatively slow, requiring ∼1 min for functional assembly (43), formation of the initiation and/or termination complexes, rather than transcription itself, is likely to be rate-limiting for gene expression, at least in relatively compact yeast genomes. Accordingly, formation and retention of gene loops that enable recycling of RNAP II would be a novel mechanism to enhance regulatory parameters. This conclusion is consistent with the looping-dependent, rapid kinetics of reactivation of the GAL10 and HXK1 gene following a cycle of activation and repression (24, 39).
Differential association of Ssu72 with the 3′-UTR, as well as weaker but substantive association with the promoter (Fig. 3), has been reported previously for Ssu72 in S. cerevisiae (22, 23, 29, 30, 37). As Ssu72 is essential for loop formation (22, 23), this pattern of Ssu72 occupancy has been interpreted as a mechanism to enable loop formation and recycling of RNAP II from the terminator to promoter (44). A novel and significant observation from our studies is that as cells progress through the growth cycle, Ssu72 is displaced, presumably resulting in retention of the RNAP II CTD Ser5-P mark, allowing RNAP II to proceed past the poly(A)-1 site and progress to the poly(A)-2 sites located ∼400 bp downstream (Fig. 3). Accordingly, the Ssu72 CTD phosphatase activity acts as a switch that either enhances or inhibits recognition of the proximal poly(A)-1 site; if inhibited, RNAP II would bypass poly(A)-1, resulting in enhanced processing at the distal poly(A)-2 site, formation of the L2 loop, and release of the L form of KlCYC1 mRNA.
Along the length of the KlCYC1 3′-UTR, there are multiple putative consensus sequences for mRNA processing (15). These discrete 3′-UTR regions were also functional as separate processing units when expressed in S. cerevisiae (18). Indeed, multiple consensus 3′-end processing sites are a common feature of S. cerevisiae genes encoding multiple transcripts (45). Our results shed light on the mechanism of APA selection among consensus processing elements. ChIP analysis of RNAP II and the Ssu72 CTD along the length of KlCYC1 during two different stages of logarithmic cell growth demonstrated that at early log phase, RNAP II accumulates at the poly(A)-1 site and that this form of RNAP II is Ser-5 hypophosphorylated, resulting in processing at poly(A)-1 and formation of the L1 gene loop. By contrast, at mid-log phase, we observed an increase in RNAP II occupancy at the distal poly(A)-2 site that was not coincident with the hypo-phosphorylated form of RNAP II. Accordingly, differential phosphorylation/dephosphorylation of the CTD at Ser-5 and/or Ser-7 is the switch that determines whether RNAP II terminates at the proximal poly(A)-1 site or progresses further downstream to the distal poly(A)-2 site. These observations are not likely to be idiosyncratic to yeast, as recent studies in human cells have identified 3′-end processing proteins that are involved in alternative poly(A) site selection and can be attributed to different physiological conditions (46).
Alternative polyadenylation has been described in all eukaryotes (8). The relative abundance of processing factors affects APA site recognition. Here, we have shown that the AU-rich element at KlCYC1, expressed either endogenously in K. lactis, or ectopically in S. cerevisiae, is the principal determinant of poly(A) site selection. Specifically for KlCYC1 expressed in S. cerevisiae, the AU-rich element at the proximal 3′-UTR determines APA selection, irrespective of growth phase, based on interaction of the RNA-processing factor Pta1. In this case, RNA processing is not dependent upon PTA1 abundance (17). Our ChIP results demonstrate that Ssu72 occupancy at the KlCYC1 locus involves a change in UTR occupancy regulated by the growth phase that correlates with the predominant usage of the distal poly(A) site. Pta1 is very important in APA selection. The formation of the L2 loop also correlates with the predominant processing position. Ssu72 along with Pta1 are principal components of the CPF complex necessary to establish DNA loops (22, 28). Both factors have homologs in the human cleavage and polyadenylation-specific factor complex, which is analogous to yeast CPF (2, 47, 48). In Fig. 5 we present a model that correlates alternative polyadenylation with alternative gene loops, affected by the status of differentially phosphorylated RNAP II CTD phosphorylation. Moreover, changes in these parameters are affected by the physiological status of cell culture, with late log phase correlating with processing at the distal poly(A) site (poly(A)-2) and differentially phosphorylated RNAP II. We suggest that differential CTD phosphorylation is the driving force that distinguishes between poly(A)-1 and the L1 loop versus poly(A)-2 and L2 loop, facilitating an increase in the Py(A)-2 and L2 formation.
These results are likely to be of general significance, not specific to yeast, given the conservation of 3′-end processing components among eukaryotic organisms. As APA has been observed in all eukaryotic organisms, albeit to differing extents depending upon organism complexity, the basic mechanism(s) are likely to be universal. The interplays between APA, alternative gene loop formation, and modulation of RNAP II CTD phosphorylation are critical paradigms in the production of functionally distinct mRNA species encoded by a single gene. Moreover, transcriptional memory, maintained by gene loops, is likely to account for maintenance of functionally distinct mRNA species.
Author Contributions
M. A. F.-P. conceived and coordinated the study and wrote the initial draft of the manuscript. M. L. M. performed all of the experiments. The 3C and ChIP experiments (Fig. 3) were carried out under the guidance of B. N. S. while M. L. M. was working in the M. H. laboratory. M. H. worked with M. A. F.-P. to write the final draft of the manuscript. All four authors read and approved the content of the article.
Acknowledgments
We thank Amalia Jácome (Universidade da Coruña) for advice on statistical analyses of data and Jesús Rosado-Lugo (Rutgers) for technical advice and critical reading of the manuscript.
This work was supported by a José Castillejo fellowship from the Spanish Ministerio de Ciencia y Tecnología (to M. L.-M.) and by National Institutes of Health Grant R01 GM39484 (to M. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
B. N. Singh and M. Hampsey, unpublished data.
- APA
- alternative polyadenylation
- CTD
- carboxyl-terminal domain
- PIC
- preinitiation complex
- RNAP II
- RNA polymerase II
- 3C
- chromatin conformation capture
- E
- early log
- M
- mid-log
- S
- short
- L
- long
- CPF
- cleavage and polyadenylation factor.
References
- 1. Richard P., and Manley J. L. (2009) Transcription termination by nuclear RNA polymerases. Genes Dev. 23, 1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuehner J. N., Pearson E. L., and Moore C. (2011) Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell Biol. 12, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mischo H. E., and Proudfoot N. J. (2013) Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim. Biophys. Acta 1829, 174–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hall-Pogar T., Zhang H., Tian B., and Lutz C. S. (2005) Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 33, 2565–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwartz G. G., Whitlatch L. W., Chen T. C., Lokeshwar B. L., and Holick M. F. (1998) Human prostate cells synthesize 1,25-dihydroxyvitamin D3 from 25-hydroxyvitamin D3. Cancer Epidemiol. Biomarkers Prev. 7, 391–395 [PubMed] [Google Scholar]
- 6. Liu D., Fritz D. T., Rogers M. B., and Shatkin A. J. (2008) Species-specific cis-regulatory elements in the 3′-untranslated region direct alternative polyadenylation of bone morphogenetic protein 2 mRNA. J. Biol. Chem. 283, 28010–28019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Di Giammartino D. C., Li W., Ogami K., Yashinskie J. J., Hoque M., Tian B., and Manley J. L. (2014) RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3′ UTRs. Genes Dev. 28, 2248–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Giammartino D. C., Nishida K., and Manley J. L. (2011) Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gruber A. R., Martin G., Keller W., and Zavolan M. (2014) Means to an end: mechanisms of alternative polyadenylation of messenger RNA precursors. RNA 5, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lutz C. S., and Moreira A. (2011) Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. RNA 2, 22–31 [DOI] [PubMed] [Google Scholar]
- 11. Lianoglou S., Garg V., Yang J. L., Leslie C. S., and Mayr C. (2013) Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miura P., Sanfilippo P., Shenker S., and Lai E. C. (2014) Alternative polyadenylation in the nervous system: to what lengths will 3′ UTR extensions take us? Bioessays 36, 766–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoopes B. C., Bowers G. D., and DiVisconte M. J. (2000) The two Saccharomyces cerevisiae SUA7 (TFIIB) transcripts differ at the 3′-end and respond differently to stress. Nucleic Acids Res. 28, 4435–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim Guisbert K. S., Li H., and Guthrie C. (2007) Alternative 3′ pre-mRNA processing in Saccharomyces cerevisiae is modulated by Nab4/Hrp1 in vivo. PLoS Biol. 5, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Freire-Picos M. A., Lombardía-Ferreira L. J., Ramil E., González-Domínguez M., and Cerdán M. E. (2001) The KlCYC1 gene, a downstream region for two differentially regulated transcripts. Yeast 18, 1347–1355 [DOI] [PubMed] [Google Scholar]
- 16. Yu L., and Volkert M. R. (2013) UV damage regulates alternative polyadenylation of the RPB2 gene in yeast. Nucleic Acids Res. 41, 3104–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seoane S., Lamas-Maceiras M., Rodríguez-Torres A. M., and Freire-Picos M. A. (2009) Involvement of Pta1, Pcf11, and a KlCYC1 AU-rich element in alternative RNA 3′-end processing selection in yeast. FEBS Lett. 583, 2843–2848 [DOI] [PubMed] [Google Scholar]
- 18. Seoane S., Guiard B., Rodríguez-Torres A. M., and Freire-Picos M. A. (2005) Effects of splitting alternative KlCYC1 3′-UTR regions on processing: metabolic consequences and biotechnological applications. J. Biotechnol. 118, 149–156 [DOI] [PubMed] [Google Scholar]
- 19. Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., and Keller W. (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 20. Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C. L., and Greenblatt J. (2003) Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278, 33000–33010 [DOI] [PubMed] [Google Scholar]
- 21. Zhao J., Hyman L., and Moore C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ansari A., and Hampsey M. (2005) A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19, 2969–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh B. N., and Hampsey M. (2007) A transcription-independent role for TFIIB in gene looping. Mol. Cell 27, 806–816 [DOI] [PubMed] [Google Scholar]
- 24. Tan-Wong S. M., Wijayatilake H. D., and Proudfoot N. J. (2009) Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 23, 2610–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tan-Wong S. M., Zaugg J. B., Camblong J., Xu Z., Zhang D. W., Mischo H. E., Ansari A. Z., Luscombe N. M., Steinmetz L. M., and Proudfoot N. J. (2012) Gene loops enhance transcriptional directionality. Science 338, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. El Kaderi B., Medler S., Raghunayakula S., and Ansari A. (2009) Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J. Biol. Chem. 284, 25015–25025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krishnamurthy S., He X., Reyes-Reyes M., Moore C., and Hampsey M. (2004) Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 14, 387–394 [DOI] [PubMed] [Google Scholar]
- 28. Rosado-Lugo J. D., and Hampsey M. (2014) The Ssu72 phosphatase mediates the RNA polymerase II initiation-elongation transition. J. Biol. Chem. 289, 33916–33926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang D. W., Mosley A. L., Ramisetty S. R., Rodríguez-Molina J. B., Washburn M. P., and Ansari A. Z. (2012) Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J. Biol. Chem. 287, 8541–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bataille A. R., Jeronimo C., Jacques P. É., Laramée L., Fortin M. È., Forest A., Bergeron M., Hanes S. D., and Robert F. (2012) A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol. Cell 45, 158–170 [DOI] [PubMed] [Google Scholar]
- 31. Singh B. N., Ansari A., and Hampsey M. (2009) Detection of gene loops by 3C in yeast. Methods 48, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Freire-Picos M. A., González-Siso M. I., Rodríguez-Belmonte E., Rodríguez-Torres A. M., Ramil E., and Cerdán M. E. (1994) Codon usage in Kluyveromyces lactis and in yeast cytochrome c-encoding genes. Gene 139, 43–49 [DOI] [PubMed] [Google Scholar]
- 33. Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 34. Cho E. J., Kobor M. S., Kim M., Greenblatt J., and Buratowski S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15, 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim M., Suh H., Cho E. J., and Buratowski S. (2009) Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284, 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patturajan M., Schulte R. J., Sefton B. M., Berezney R., Vincent M., Bensaude O., Warren S. L., and Corden J. L. (1998) Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273, 4689–4694 [DOI] [PubMed] [Google Scholar]
- 37. O'Sullivan J. M., Tan-Wong S. M., Morillon A., Lee B., Coles J., Mellor J., and Proudfoot N. J. (2004) Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 36, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 38. Kim M., Krogan N. J., Vasiljeva L., Rando O. J., Nedea E., Greenblatt J. F., and Buratowski S. (2004) The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432, 517–522 [DOI] [PubMed] [Google Scholar]
- 39. Lainé J. P., Singh B. N., Krishnamurthy S., and Hampsey M. (2009) A physiological role for gene loops in yeast. Genes Dev. 23, 2604–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiang K., Manley J. L., and Tong L. (2012) The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat. Commun. 3, 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Freire-Picos M. A., Hollenberg C. P., Breunig K. D., and Cerdan M. E. (1995) Regulation of cytochrome c expression in the aerobic respiratory yeast Kluyveromyces lactis. FEBS Lett. 360, 39–42 [DOI] [PubMed] [Google Scholar]
- 42. Yudkovsky N., Ranish J. A., and Hahn S. (2000) A transcription reinitiation intermediate that is stabilized by activator. Nature 408, 225–229 [DOI] [PubMed] [Google Scholar]
- 43. Zenklusen D., Larson D. R., and Singer R. H. (2008) Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hampsey M., Singh B. N., Ansari A., Laine J. P., and Krishnamurthy S. (2011) Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv. Enzymol. Reg. 51, 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sparks K. A., and Dieckmann C. L. (1998) Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 26, 4676–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jimeno S., Tous C., García-Rubio M. L., Ranes M., González-Aguilera C., Marín A., and Aguilera A. (2011) New suppressors of THO mutations identify Thp3 (Ypr045c)-Csn12 as a protein complex involved in transcription elongation. Mol. Cell. Biol. 31, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiang K., Nagaike T., Xiang S., Kilic T., Beh M. M., Manley J. L., and Tong L. (2010) Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467, 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Zhang M., and Zhang Y. (2011) Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem. J. 434, 435–444 [DOI] [PubMed] [Google Scholar]