Abstract
Covalent modifications of histone N-terminal tails play a critical role in regulating chromatin structure and controlling gene expression. These modifications are controlled by histone-modifying enzymes and read out by histone-binding proteins. Numerous proteins have been identified as histone modification readers. Here we report the family-wide characterization of histone binding abilities of human CW domain-containing proteins. We demonstrate that the CW domains in ZCWPW2 and MORC3/4 selectively recognize histone H3 trimethylated at Lys-4, similar to ZCWPW1 reported previously, while the MORC1/2 and LSD2 lack histone H3 Lys-4 binding ability. Our crystal structures of the CW domains of ZCWPW2 and MORC3 in complex with the histone H3 trimethylated at Lys-4 peptide reveal the molecular basis of this interaction. In each complex, two tryptophan residues in the CW domain form the “floor” and “right wall,” respectively, of the methyllysine recognition cage. Our mutation results based on ZCWPW2 reveal that the right wall tryptophan residue is essential for binding, and the floor tryptophan residue enhances binding affinity. Our structural and mutational analysis highlights the conserved roles of the cage residues of CW domain across the histone methyllysine binders but also suggests why some CW domains lack histone binding ability.
Keywords: chromatin structure, histone methylation, histone modification, post-translational modification (PTM), protein-protein interaction, CW domain, H3K4 methylation, MORC3, ZCWPW2
Introduction
Chromatin structure is dynamically regulated by histone post-translational modifications, such as methylation, acetylation, phosphorylation, ubiquitination, and sumoylation (1). These post-translational modifications constitute the “histone code,” which is written or erased by histone-modifying enzymes and recognized by histone code “reader” proteins (2–4). Histone methylation, such as lysine methylation at the ϵ-amino group at levels from mono- to trimethylation (me1–me3), has received extensive attention (5). A number of domains bind methylated histone tails. Prominent examples include the chromodomain, Tudor domain, MBT domain, PWWP domain, and PHD domain (4, 6, 7). The CW domain has recently been identified as a new member of the lysine methylation reader family (8–11).
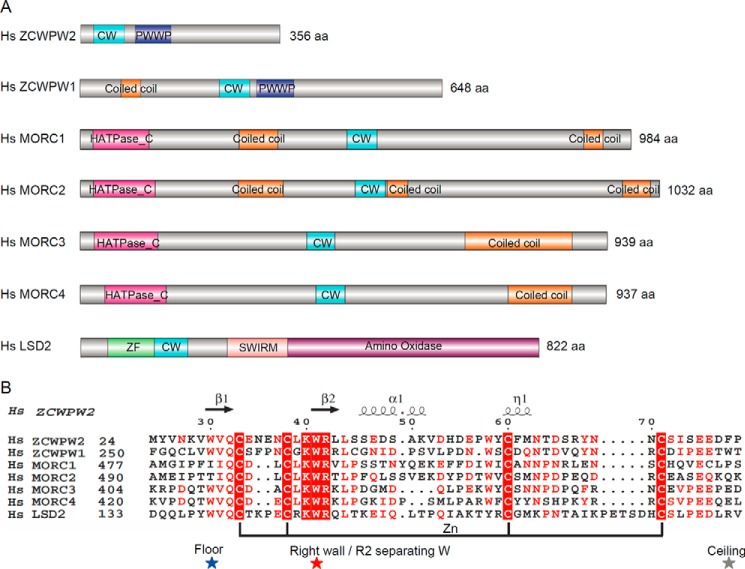
The CW domain is a zinc binding domain, composed of ∼50 amino acid residues with four conserved cysteine (C) and two conserved tryptophan (W) residues, and its name was derived from these conserved residues. CW domains are found in chromatin-associated proteins in animals and plants and grouped into 12 families based on sequence similarity (12). There are seven CW domain-containing proteins in humans, namely ZCWPW1, ZCWPW2, MORC1, MORC2, MORC3, MORC4, and LSD2 (Fig. 1A). Prior studies have shown that the CW domains of ZCWPW1 (8), MORC3 (10), and MORC4 (9) are readers of H3K43 methylated histones with differing preferences for histone H3K4 methylation states (i.e. ZCWPW1 and MORC3 preferentially recognize histone H3K4me3 (8, 10), whereas the CW domain of human MORC4 prefers dimethylated H3K4 (9)). The LSD2 CW domain is required for the demethylation function of LSD2 but does not bind to any H3K4 peptides (13). However, the underlining mechanism for these differences in the CW domain's histone binding abilities is unclear.
FIGURE 1.
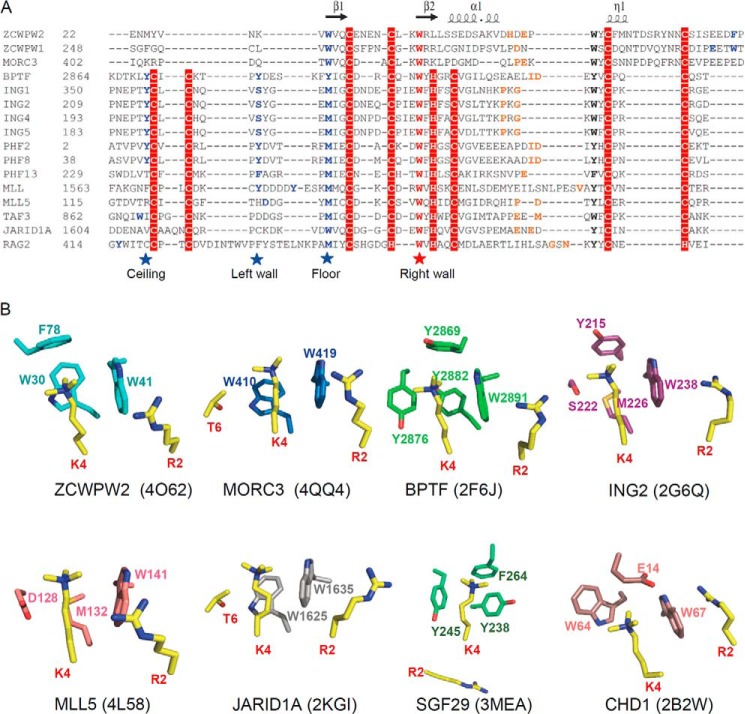
Comparison of human zinc finger CW domain-containing proteins. A, schematic representation of the domain structure of these proteins. B, structure-based sequence alignment of these CW domains. Secondary structure elements and residue numbers of the ZCWPW2 CW domain are indicated above the sequence alignment. The highly conserved “right wall” residues are marked by a red star; the partly conserved “floor” residues are marked by a blue star; the non-conserved “ceiling” residues are marked by a gray star. The alignments were constructed with ClustalW (45) and refined with ESPript (46).
Among the seven known human CW domains, two had been structurally characterized in detail before (i.e. ZCWPW1 and LSD2). In this study, we quantitatively characterized the histone binding selectivity of the CW domains of the other five human proteins, ZCWPW2, MORC1, MORC2, MORC3, and MORC4. From our quantitative fluorescence polarization (FP) and isothermal titration calorimetry (ITC) binding assays, we found that the CW domains of ZCWPW2, MORC3, and MORC4 preferentially bound to H3K4me3, whereas MORC1 and MORC2 did not bind to any histone peptides, regardless of the methylation status of H3K4. We further determined the crystal structures of human ZCWPW2 and MORC3 CW domains in complex with H3K4me3 peptide and provide a structural explanation for the differences in the CW domain's histone binding abilities. Generally, an aromatic cage is a signature feature of methylation reader proteins that recognize methylated lysine, arginine, and even N6-methyladenosine (14), and it is normally composed of 2–4 aromatic residues. However, our case study of ZCWPW2 revealed that only the invariant tryptophan, a cage-forming residue of the CW domain, was essential for recognition of the methyllysine residue, whereas the other cage-forming residues play roles in enhancing binding or filtering methylation states.
Experimental Procedures
Protein Expression and Purification
The zinc finger CW domains of ZCWPW2 (residues 21–78), MORC3 (residues 400–460), MORC2 (residues 422–485), MORC1 (residues 474–531), and MORC4 (residues 417–472) were subcloned into a modified pET28-MHL or pET28GST-lic vector. Thus encoded, N-terminal His-tagged or GST, His-tagged fusion proteins were overexpressed in Escherichia coli BL21 (DE3) Codon plus RIL (Stratagene) cells at 15 °C and purified by affinity chromatography on nickel-nitrilotriacetic acid resin (Qiagen), followed by tobacco etch virus protease treatment to remove the tag. Proteins were further purified by Superdex75 gel filtration (GE Healthcare). For crystallization experiments, purified proteins were concentrated to 30 mg/ml in a buffer containing 20 mm Tris, pH 7.5, 150 mm NaCl, 50 μm ZnCl2, and 1 mm DTT.
Mutations were introduced with the QuikChange II XL site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing. Mutants were overexpressed and purified as the wild type constructs above. The molecular weight of all protein samples was checked by mass spectrometry.
ITC
Concentrated proteins were diluted with 20 mm Tris, pH 7.5, 150 mm NaCl. The lyophilized H3 peptides (Peptide 2.0 Inc.) were dissolved in the same buffer, and the pH value was adjusted by the addition of a NaOH solution. Peptide concentrations were estimated from the mass of lyophilized material. All measurements except for those of binding between MORC1 or MORC4 and H3K4 peptides were performed at 25 °C, using a VP-ITC microcalorimeter (MicroCal, Inc.). Protein with a concentration of 50 μm was placed in the chamber, and peptide with a concentration of 0.5–1 mm in the syringe was injected in 25 successive injections with a spacing of 180 s and a reference power of 13 μcal/s. Control experiments in the absence of CW protein were performed under identical conditions to determine the heat signals that arise from injection of the peptides into the buffer. Data were fitted using the single-site binding model within the Origin software package (MicroCal, Inc.).
Due to limited protein yield, the MORC1 and MORC4 ITCs were performed using a nano-ITC microcalorimeter (TA, Inc.). Nano-ITC data should be consistent with those from the regular ITC instrument, based on ITC results of ZCWPW2 using both regular and nano-ITC instruments. GST, His-tagged fusion protein with a concentration of 50 μm was placed in the chamber, and H3K4 peptide with a concentration of 0.5 mm was injected in 25 successive injections with a spacing of 120 s at 25 °C. Control experiments in the absence of CW protein were performed under identical conditions to determine the heat signals that arise from injections of the peptide into the buffer or into the GST, His tag only protein. Data were fitted using the independent model within the NanoAnalyze software package (TA, Inc.).
FP Assay
All peptides used for FP were synthesized and purified by Tufts University Core Services (Boston, MA) with fluorescein-labeled C termini. Binding assays were performed in 10 μl at a constant fluorescein-labeled peptide concentration of 40 nm and increasing amounts of protein at concentrations ranging from low to high micromolar in a buffer of 20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm DTT, and 0.01% Triton X-100. All assays were performed in duplicate in 384-well plates, using the Synergy 2 microplate reader (BioTek) with an excitation wavelength of 485 nm and an emission wavelength of 528 nm. Data were corrected by background of the free labeled peptides and fitted to the ligand binding function using GraphPad Prism version 5 software to determine the Kd values.
Protein Crystallization
For cocrystallization, purified protein was mixed with H3K4me3 peptide (residues 1–15) at a molar ratio of 1:3 and crystallized using the sitting drop vapor diffusion method at 20 °C by mixing 0.5 μl of the protein with 0.5 μl of the reservoir solution. The complex crystals were obtained in a buffer containing 2 m (NH4)2SO4, 2% PEG 400, and 0.1 m Na-Hepes, pH 7.5, for ZCWPW2-H3K4me3 and 20% PEG 3350, 0.2 m NH4Cl for MORC3-H3K4me3. Before flash-freezing crystals in liquid nitrogen, crystals were soaked in a cryoprotectant consisting of 85% reservoir solution and 15% glycerol.
Data Collection and Structure Determination
Diffraction data were collected under cooling to 100 K at beam line 19ID of the Advanced Photon Source (Argonne, IL) and reduced with XDS (15) and SCALA/AIMLESS software (16). Additional information on crystallographic experiments and models is listed in Table 1. The ZCWPW2 structure was solved by single wavelength anomalous diffraction (17) with SHELXD (18) and SHELXE (19) programs. The MORC3 structure was solved by molecular replacement with the program PHASER (20) and coordinates of the ZCWPW2 crystallographic model. Arp/Warp was used for phase improvement (21). For both crystal structures, automated model building was performed with Arp/Warp (22). The present models were obtained through iterative manual rebuilding with COOT (23), restrained refinement with REFMAC (24), and model validation with MOLPROBITY (25). CCP4 (26), PHENIX (27), PDB_EXTRACT (28) programs and the IOTBX library (29) were used in preparing model summaries (Table 1) and Protein Data Bank deposition.
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell.
| ZCWPW2-H3K4me3 | MORC3-H3K4me3 | |
|---|---|---|
| PDB code | 4O62 | 4QQ4 |
| Data collection and reduction | ||
| Radiation wavelength (Å) | 1.283 | 0.9792 |
| Space group | H3 | P21212 |
| Cell dimensions a, b, c (Å) | 127.09, 127.09, 63.05 | 62.62, 64.86, 36.02 |
| Resolution limits (Å) | 41.46-1.78 (1.82-1.78) | 45.05-1.75 (1.78-1.75) |
| Unique HKLs | 36,371 (2,051) | 15,427 (838) |
| Completeness (%) | 100.0 (100.0) | 100.0 (99.9) |
| Rsym | 0.070 (1.061) | 0.123 (0.958) |
| I/σ | 20.1 (1.5) | 12.5 (2.3) |
| Redundancy | 5.5 (5.5) | 6.9 (7.1) |
| Model refinement | ||
| Refinement resolution (Å) | 41.46-1.78 | 45.05-1.75 |
| Reflections used/free | 34,782/1,587 | 14,462/928 |
| Number of atoms/average B-factor (Å2) | 1,681/37.0 | 1,045/29.7 |
| Protein | 1,429/35.9 | 796/28.3 |
| Peptide | 51/58.1 | 148/35.0 |
| Water | 171/40.1 | 89/34.3 |
| Others | 30/37.0 | 12/19.3 |
| Rwork/free | 0.165/0.186 | 0.180/0.223 |
| Root mean square deviation | ||
| Bonds (Å) | 0.015 | 0.014 |
| Angles (degrees) | 1.4 | 1.6 |
| Molprobity Ramachandran favored/outliers (%) | 97.09/0.00 | 97.14/0.00 |
Results and Discussion
ZCWPW2, MORC3, and MORC4 Are Histone H3K4me3 Readers
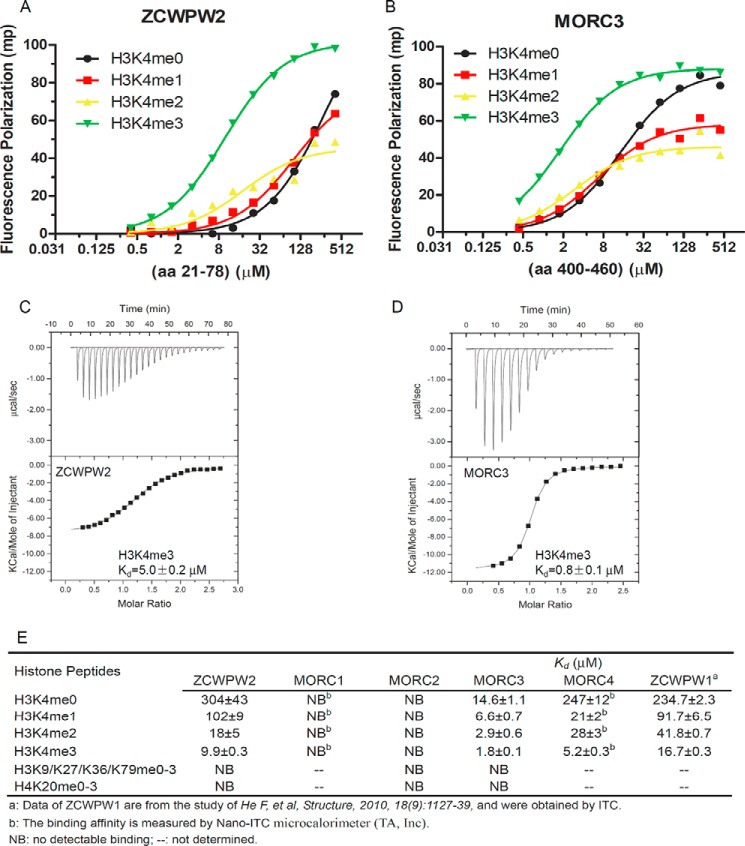
Previous studies have shown that the CW domain is a reader of methylated histone; however, different CW domains behave differently in methylated histone recognition (8–11). In the human genome, there are seven CW domain-containing proteins (Fig. 1A). Although the amino acid sequences of these CW domains are highly conserved overall, only the aromatic cage's “right wall” tryptophan (see below for details) is strictly conserved among these CW domains (Fig. 1B), suggesting divergent histone binding properties. We sought to systematically assess the ligand affinities of these human CW domain proteins and were able to prepare well behaved recombinant samples of the ZCWPW2, MORC2, and MORC3 CW domains. After codon optimization, we were able to express stable GST-tagged MORC1 and MORC4 CW domains, but we were unable to remove the GST tag, and the protein yields were low. Because the CW domain is frequently found in chromatin-associated proteins and the reported CW domains have been shown to bind histones, we screened for ligands of the ZCWPW2, MORC2, and MORC3 CW domains against our histone peptide library of histone H3 Lys-4, Lys-9, Lys-27, Lys-36, and Lys-79 and histone H4 Lys-20 peptides in different methylation states by means of FP (Fig. 2). Our binding results revealed that both ZCWPW2 and MORC3 bound methylated H3K4 peptides and preferred the trimethylated state (me3) (Fig. 2, A and B). We did not observe MORC2 binding any tested peptides, and we also did not observe binding of ZCWPW2 and MORC3 to other tested histone peptides except for methylated H3K4 peptides (Fig. 2E). The interactions of ZCWPW2 and MORC3 with H3K4me3 were confirmed by ITC experiments (Fig. 2, C and D). Due to the limitations of our MORC1 and MORC4 protein preparations, we resorted to use of a nano-ITC microcalorimeter (TA, Inc.) to analyze the preferences of MORC1 and MOCR4 for H3K4 unmodified or methylated peptides. Our binding data showed that MORC1 did not bind any H3K4 peptides, regardless of their methylation states, whereas MORC4 preferred H3K4me3 (Fig. 2E), distinct from a previous pull-down assay, which showed that MORC4 bound dimethylated H3K4 (9). To determine the dependence of affinity values on methods and instrumentation, we analyzed the interaction between ZCWPW2 and H3K4me3 and determined Kd values of 9.9 ± 0.3, 5.0 ± 0.2, and 4.0 ± 0.2 μm by FP, VP-ITC, and nano-ITC assays, respectively, indicating that the binding data are comparable among these different techniques. The GST tag should not affect CW binding ability significantly because the GST tag did not bind any peptides used in the study in control experiments (data not shown). In summary, ZCWPW2 and MORC3 preferred the H3K4me3 peptide with an order of K4me3 > K4me2 > K4me1 > K4me0, similar to that of ZCWPW1 (8), whereas MORC4 exhibited a binding order of K4me3 > K4me2 ≈ K4me1 > K4me0 (Fig. 2E).
FIGURE 2.
ZCWPW2 and MORC3 are H3K4me3 binders. A and B, the binding curves of FP measurements of different H3K4 peptides to CW domains of ZCWPW2 (A) and MORC3 (B). C and D, ITC binding curves of H3K4me3 peptides to CW domains of ZCWPW2 (C) and MORC3 (D). E, binding affinities of selected CW domains to different histone peptides determined by FP or ITC. FP Kd values were calculated from duplicate measurements and fitted to ligand binding function using GraphPad Prism version 5 software. ITC Kd values were calculated from single measurement, and errors were estimated by a fitting curve.
Structural Basis for Selective Binding of H3K4me3 by ZCWPW2 and MORC3
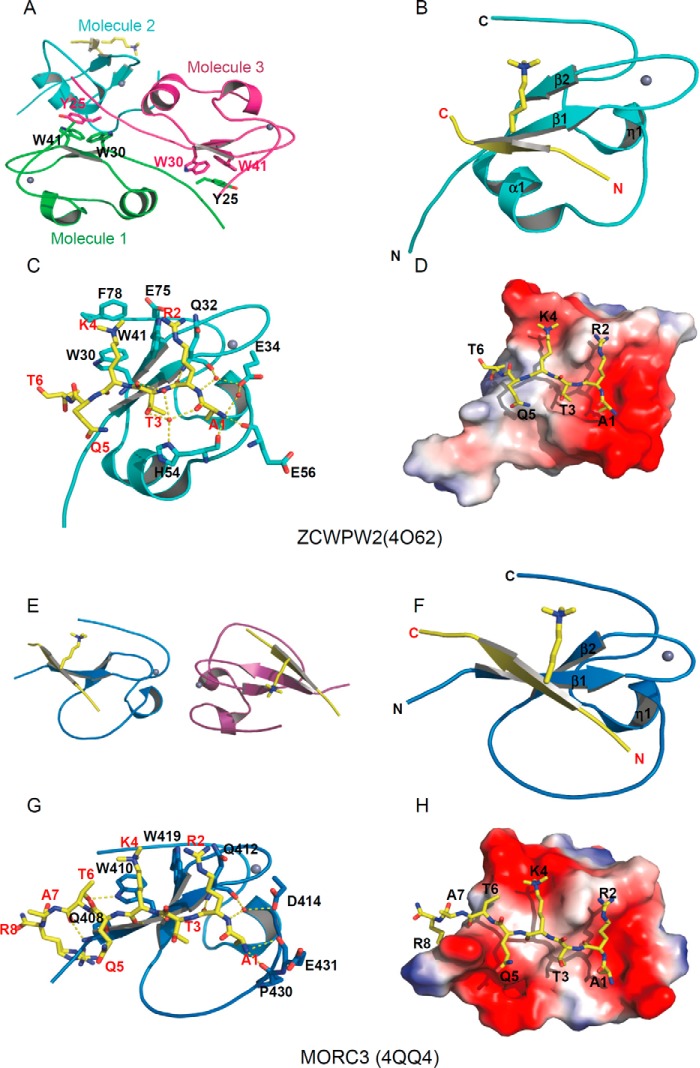
There are only three CW domain structures available in databases: the NMR structure of ZCWPW1-CW in complex with H3K4me3 (8), the NMR structure of AtASHH2-CW in free state (9), and crystal structures of full-length LSD2 including the CW domain (13, 30). To extend our understanding of the molecular mechanism of selective binding of H3K4me3 by the CW family, we solved the crystal structures of ZCWPW2 and MORC3 CW domains in complex with a H3(1–15) K4me3 peptide at 1.78 and 1.75 Å resolution, respectively (Table 1 and Fig. 3). The CW domains of both ZCWPW2 and MORC3 contain a β-hairpin and a zinc finger with the zinc ion coordinated by a quartet of cysteine residues. In addition, the ZCWPW2 CW domain also contains a short α-helix joining the β-hairpin and the zinc finger motifs. The H3 peptide aligns in an anti-parallel orientation with the N-terminal β-strand of the β-hairpin to form a three-stranded β-sheet (Fig. 3, B and F). The overall histone binding mode observed in the ZCWPW2 and MORC3 structures is similar to those of the previously reported ZCWPW1-H3K4me3 (8) and PHD-H3K4me3 complex structures, such as BPTF (31), ING2 (32), JARID1A (33), and MLL5 (34).
FIGURE 3.
Structural basis of selective binding of ZCWPW2 and MORC3 to H3K4me3. A and E, model of the crystal asymmetric unit of the CW domain and H3K4me3 peptide complexes of ZCWPW2 (A) and MORC3 (E). B and F, overall structure of the CW domain and H3K4me3 peptide complexes of ZCWPW2 (B) and MORC3 (F). The CW domains are shown in a schematic representation and colored in cyan and blue, respectively. C and G, detailed interactions of the H3K4me3 peptide with ZCWPW2 CW domain (C) and MORC3 CW domain (G). H3K4me3 peptides are shown in stick mode, and hydrogen bonds are colored in yellow. D and H, electrostatic potential surface representation of CW domain and H3K4me3 peptide complexes of ZCWPW2 (D) and MORC3 (H) (47). Structure figures were generated by using PyMOL. Surface representations were calculated with the built-in protein contact potential function of PyMOL.
The asymmetric unit of our ZCWPW2-H3K4me3 model comprises three CW molecules, with one CW molecule bound to the H3K4me3 peptide and the two other CW molecules forming a homodimer by anti-parallel alignment of their respective β1 strands (Fig. 3A). An aromatic cage, which is normally composed of 2–4 aromatic residues, is a signature feature of methylation reader proteins that recognize methylated lysine, arginine, and even N6-methyladenosine (14). Here, ZCWPW2 used 3 aromatic residues, namely Trp-30, Trp-41, and Phe-78, to read out the trimethyllysine (Fig. 3C). However, in the CW homodimer, the aromatic cage of each monomer is occupied by the Tyr-25 side chain of the respective other monomer (Fig. 3A). The backbones of the ZCWPW2-CW monomers, with or without peptide bound, superimpose well, with root mean square deviations around 0.5 Å (35), indicating that binding of the peptide did not induce significant conformational changes in the protein.
In the ZCWPW2-H3K4me3 complex structure, we were able to trace the N-terminal six residues of the histone peptide. The very N-terminal residue (H3A1) of the peptide is anchored in a small pocket by hydrogen bonds between the amino group of H3A1 and the main chain oxygens of His-54 and Glu-56 and the carboxylate of Glu-34 mediated by solvent (Fig. 3C). Deletion of H3A1 or the addition of another alanyl residue to the peptide's N terminus abrogated H3K4me3 binding, underscoring the importance of this H3A1-binding pocket to histone binding (Figs. 3D and 4 (A and B)). Recognition of the terminal amino group could explain residual affinity of unmethylated H3K4 peptide to many H3K4 binders, such as most PHD domains (6) and SGF29 (36), as well as the CW domains.
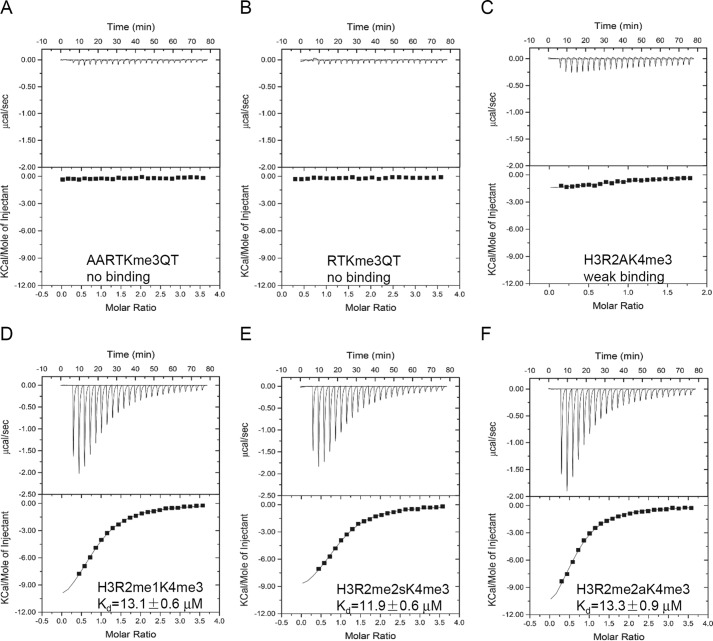
FIGURE 4.
Modification of the H3K4me3 peptide affects binding. A, ITC curve of CW domain of ZCWPW2 binding after N-terminal alanine addition of H3K4me3 peptide. B, ITC curve of CW domain of ZCWPW2 binding after N-terminal alanine deletion from H3K4me3 peptide. C, ITC curve of CW domain of ZCWPW2 binding to histone H3 R2A H3K4me3 peptide. D–F, ITC curves of CW domain of ZCWPW2 binding after H3R2 methylation of H3K4me3 peptide, monomethylation (D), symmetric methylation (E), and asymmetric methylation (F).
We believe that the guanidinium moiety of H3R2, largely unresolved by electron density, is nevertheless close to ZCWPW2-CW residue Gln-32 (Fig. 3C). Although the methylation state of H3R2 did not significantly affect binding, mutation to alanine in this position diminished the binding affinity significantly (Fig. 4, C–F), stressing the importance of the positive charge of the arginine residue in binding.
Trp-30 and Trp-41 provide the floor and the right wall, respectively, of an aromatic cage recognizing trimethylated Lys-4. According to our interpretation of incomplete density, the phenyl ring of Phe-78 may act as the cage's ceiling. Phe-78 appears disordered in the ligand-free protein chains that compose the homodimer. The backbone amino and carbonyl oxygen moieties of trimethylated Lys-4 form hydrogen bonds with the carbonyl oxygen and amino group of Trp-30 in ZCWPW2, respectively. Additional direct or solvent-mediated hydrogen bonds between the peptide and CW domain further stabilize the interaction. Of note, whereas Glu-300 in ZCWPW1 contributes to binding of H3K4me3 (8), the corresponding residue Glu-75 in ZCWPW2 appeared tolerant to mutation to an alanine, suggesting that tiny variations exist in the CW domain family on the spatial level (Fig. 5, A and G).
FIGURE 5.
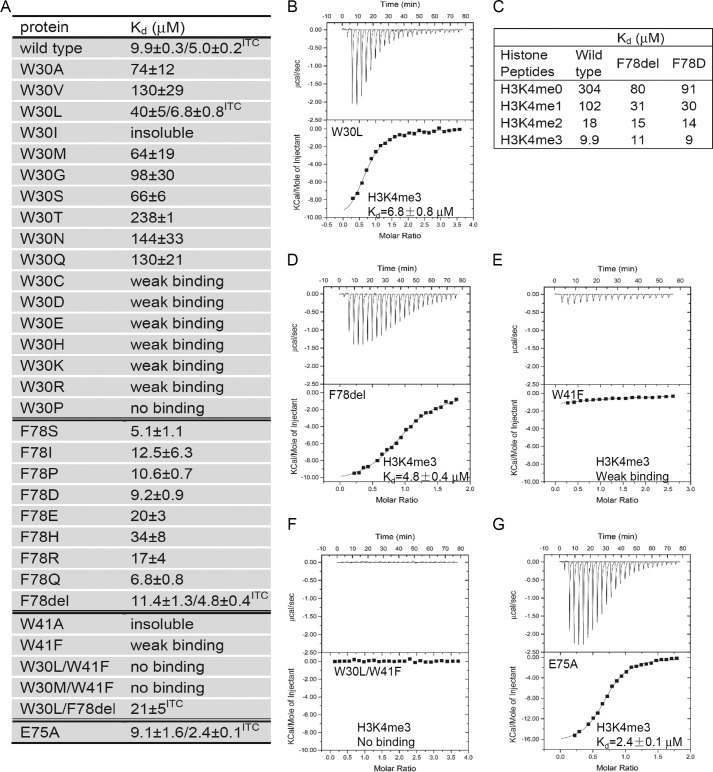
Mutation of the binding cage affects H3K4me3 binding. A, binding affinities of different ZCWPW2 CW mutants to H3K4me3 peptide determined by FP or ITC. F78del, deletion of Phe-78. B and D–G, ITC curves of selected mutants to H3K4me3 peptide. C, binding affinity of selected mutants to H3K4 peptides tested by FP, showing modest methylation selectivity after Phe-78 deletion or mutation. FP Kd values were calculated from duplicate measurements and fitted to a ligand binding curve using GraphPad Prism version 5 software. ITC Kd values were calculated from a single measurement, and errors were estimated by fit to a curve.
There are two MORC3-CW molecules in the asymmetric unit of the MORC3-H3K4me3 complex structure, and both MORC3-CW molecules bound the H3K4me3 peptide (Fig. 3E). The MORC3-H3K4me3 complex resembles the ZCWPW2-H3K4me3 complex, with some notable differences. First, the aromatic cage of the MORC3-CW domain lacks the ceiling phenylalanine modeled in the ZCWPW2 complex (Fig. 3G). This may explain the reduced MORC3 selectivity between H3K4 methylation states and led us to hypothesize that the ceiling residue was not essential for the histone binding ability of ZCWPW2 either, as discussed below. Second, the short α-helix of the CW domain of ZCWPW2 coincides with a deletion in the MORC3 amino acid sequence (Figs. 1B and 3). Compared with the ZCWPW2 complex structure, electron density for the MORC3 structure allowed us to additionally trace histone H3 Ala-7 and, under increased ambiguity, histone H3 Arg-8. In addition to interactions equivalent to those observed in the ZCWPW2-H3K4me3 complex, in our interpretation of weak electron density in the MORC3-H3K4me3 crystal, histone H3 Arg-8 is surrounded by Pro-406, Asp-407, and Gln-408 (Fig. 3, G and H) near the N terminus of the crystallized MORC3 construct. Overall, the electron density for the H3 ligand was stronger in the MORC3 structure than that in the ZCWPW2 complex structure, consistent with a higher binding affinity for the H3K4 peptides to MORC3.
Cage-forming Residues of the CW Domain Play Distinctive Roles in Recognizing Methylated Lysine
Structural analysis and sequence alignment revealed that the cage residue Trp-41 (right wall) in ZCWPW2 is conserved in all human CW proteins. Trp-30 (floor) is also conserved except in MORC1 and MORC2, but the presumed ceiling Phe-78 residue in ZCWPW2 is divergent among the CW proteins. To test the functional importance of the trimethylated Lys-4 binding residues, we performed mutagenesis in ZCWPW2 and measured the binding affinity for different H3K4 peptides in an FP binding assay (Fig. 5). First, we mutated Trp-30 to non-aromatic amino acids or histidine. We obtained similar protein yields for all of these mutants except for W30I/T/P, which produced much lower yields. In our FP assay, mutating Trp-30 to charged residues or cysteine/proline diminished the binding significantly. The other mutants also exhibited reduced binding affinity but to a lesser extent. Of note, W30L and W30M had the strongest binding affinity among these mutants in the FP assay, which was confirmed by ITC for the W30L mutant (Fig. 5B). The bulkier leucine and methionine residues have also been found to act as cage-forming residues in other histone methyllysine binders, such as MLL5 (34) and L3MBTL1 (37). Taken together, an aromatic residue is preferred at the Trp-30 position but is not essential.
We also mutated Phe-78 to corresponding residue types of other CW members, in addition to outright Phe-78 deletion (F78del) and found that these mutations had little effect on the binding affinity (Fig. 5, A, C, and D). The crystal structure of the F78R mutant-H3K4me3 complex revealed that the histone peptide showed a similar binding mode to that of the wild type protein except that the side chain of the arginine is invisible (data not shown). Tolerance to substitution at this position is also consistent with our binding data of MORC3 and MORC4, which have a glutamic acid residue corresponding to Phe-78 in ZCWPW2 but still bound H3 peptides (Fig. 2E). Interestingly, some selectivity to the methylation state was lost after Phe-78 mutation or deletion (Fig. 5C), resulting in a selectivity pattern similar to the ZCWPW1 W303A mutant (8) or MORC3 but not MORC4 (Fig. 2E). Thus, the ceiling phenylalanine appears dispensable for histone binding by the CW domain but may contribute to methylation state selectivity.
Whereas the W41A mutant was insoluble, the protein yield of W41F was similar to that of the wild type protein. The W41F mutant displayed very weak interaction with H3K4me3 (Fig. 5, A and E), and the double mutants W30L/W41F and W30M/W41F did not detectably bind H3K4me3 (Fig. 5, A and F). The W30L/F78del double mutant with just a single aromatic residue (Trp-41) contributing to the cage still bound H3K4me3 peptide, as determined by ITC (Fig. 5A). These results indicate that the invariant tryptophan cage residue (Trp-41 in ZCWPW2, right wall) is a strong determinant for binding methylated H3K4, similar to the MLL5 PHD domain (Fig. 6D), which uses a cage of a single aromatic residue, Trp, to accommodate H3K4me3 (34). As far as we know, MLL5 is the only identified histone binder that naturally uses a cage with a single aromatic residue to recognize methylated histones.
FIGURE 6.
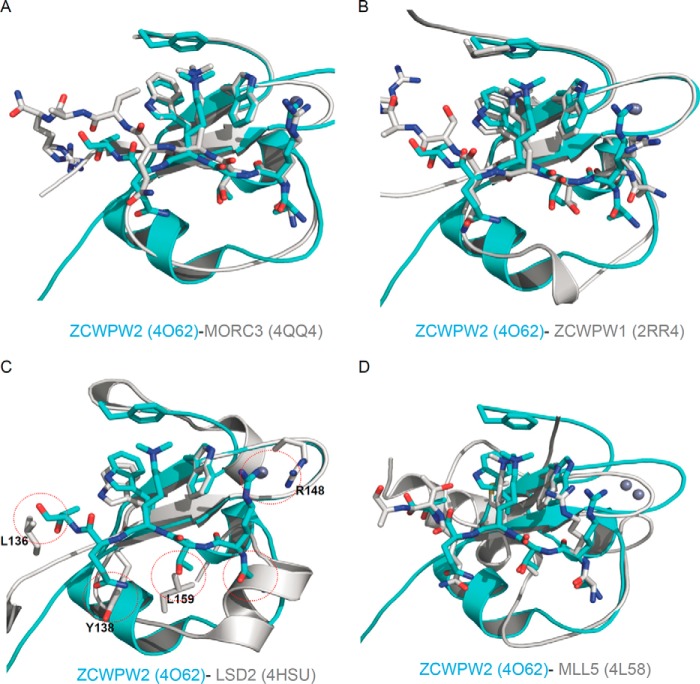
Structural comparison with other members of the CW domain family or the PHD domain of MLL5. A, superposition of the ZCWPW2 CW-H3K4me3 complex (PDB code 4O62; this work) on MORC3 CW-H3K4me3 complex (PDB code 4QQ4; this work). B, superposition of the ZCWPW2 CW-H3K4me3 complex (PDB code 4O62) on ZCWPW1 CW-H3K4me3 complex (PDB code 2RR4). C, superposition of the ZCWPW2 CW-H3K4me3 complex (PDB code 4O62) on LSD2 CW domain (PDB code 4HSU). D, superposition of the ZCWPW2 CW-H3K4me3 complex (PDB code 4O62) on MLL5 PHD-H3K4me3 complex (PDB code 4L58).
In summary, our systematic mutant binding survey revealed that the right wall tryptophan residue (Trp-41 in ZCWPW2) is essential for binding to H3K4me3, Trp-30 maximizes affinity among non-Tyr/non-Phe residues at the cage floor, and the ceiling residue (Phe-78) is not required for histone binding but may be involved in selection between methylation states.
Structural Comparison with Other CW Domains Explains Why Some CW Domains Lack Histone Binding Ability
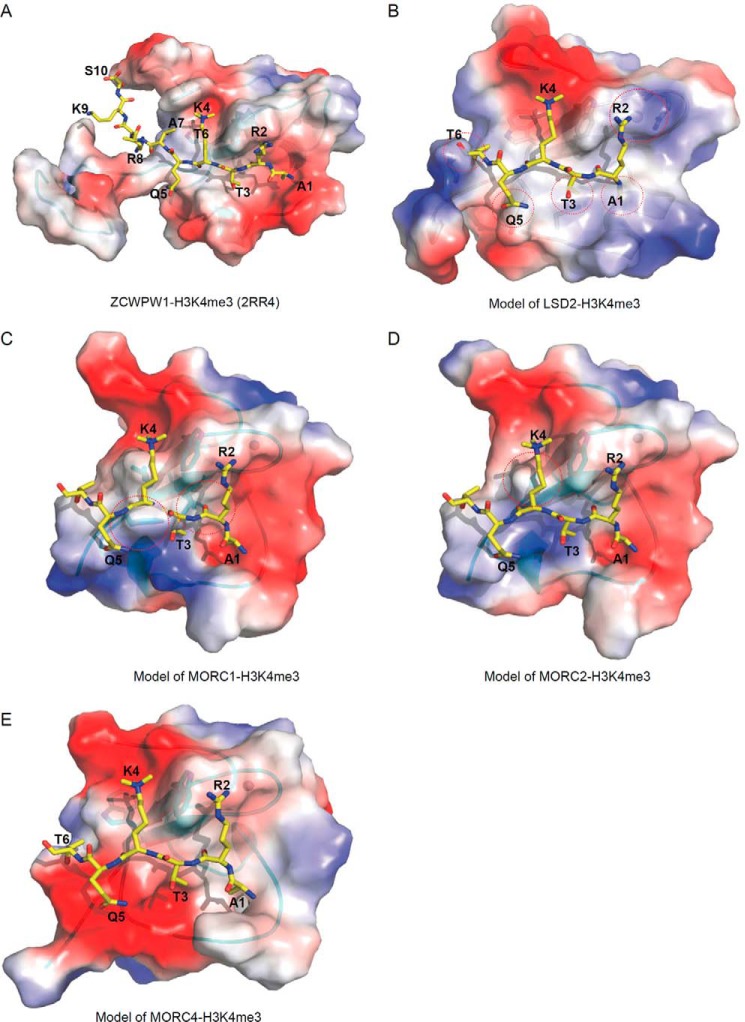
Comparison of our two CW domain structures with the reported human CW domain structures, such as CW domains of ZCWPW1 (PDB code 2RR4) and LSD2 (PDB code 4HSU) confirms common core elements: a β-hairpin and a C4 zinc finger (Fig. 6). The CW domain of ZCWPW2 superimposes readily with the CW domains of MORC3, ZCWPW1, LSD2, and MLL5 PHD domain (Fig. 6), and the electrostatic potential surfaces of ZCWPW1/2 and MORC3 feature a conspicuous and conserved cleft running across the proteins' surfaces, which is either neutral or negatively charged (Figs. 3 (D and H) and 7A). In contrast, comparison with LSD2 structures reveals an α-helix in LSD2, distinct from the α-helix in ZCWPW2, that would obstruct H3A1 binding (Figs. 6C and 7B). Inspection of the sequence alignment and structural superimposition of ZCWPW2 and LSD2 further indicates that the bulky LSD2 side chains of Leu-136, Tyr-138, and Leu-159 could encroach on the H3 binding pocket (Figs. 1B and 6C). Positively charged Arg-148 in LSD2 near the hypothetical H3R2 binding site may repel H3R2 (Fig. 6C). Furthermore, the aromatic cage in the LSD2 CW domain is filled with the side chain of residues Leu-340 and Ile-343 in the adjacent SWIRM domain (30). These observations may explain why LSD2 does not bind the H3 peptide (13).
FIGURE 7.
Electrostatic potential surface representation of other human CW-H3K4me3 complex models. A, ZCWPW1-H3K4me3 complex (PDB code 2RR4). B, model of LSD2-H3K4me3 complex. C, model of MORC1-H3K4me3 complex. D, model of MORC2-H3K4me3 complex. E, model of MORC4-H3K4me3 complex. Theoretical models were built on ZCWPW2 template by SWISS-MODEL (48).
Besides the proteins mentioned above, there are three CW-containing human proteins whose structures have not been determined, namely MORC1, MORC2, and MORC4. To predict the binding properties of these proteins to methylated H3K4 peptides, we built models of these proteins based on our ZCWPW2 structure (Fig. 7, C–E). As shown in the electrostatic potential surfaces, only MORC4 has a binding cleft for the H3K4me3 peptide (Fig. 7E). Sequence alignment, phylogenetic tree of human CW domain-containing proteins (11), and analysis of the homology model's surface suggest that MORC4 closely resembles MORC3 (Figs. 1B and 7E) and should bind Lys-4-methylated H3 peptides, consistent with our ITC data and the published pull-down data (9). MORC1 and MORC2, on the other hand, each only have one of the cage-forming residues (Trp-492 and Trp-505, respectively), corresponding to the invariant residue Trp-41 in ZCWPW2 (Fig. 7, C and D). Ile-483 forms the floor of the MORC1 cage. Whereas we failed to obtain the corresponding ZCWPW2 W30I mutant in a form suitable for analysis, the W30L mutant was compatible with H3 binding (Fig. 5A). Phe-482 (aligned with Tyr-138 in LSD2, which occludes histone binding in LSD2) and Ile-484 in MORC1 probably impede binding of H3K4me3 peptide (Fig. 7C), consistent with our ITC binding data (Fig. 2E). MORC2 residue Thr-496 corresponds to the cage's floor position. The histone binding affinity of the corresponding ZCWPW2 W30T mutant was significantly reduced (Figs. 5A and 7D), which may explain why MORC2 did not bind to H3K4 peptide in our FP assay (Fig. 2E). Taken together, of the seven human CW proteins, ZCWPW1/2 and MORC3/4 are histone H3K4me3 binders, whereas MORC1, MORC2, and LSD2 lack histone H3K4 binding ability.
Structural Comparison with Other H3K4me3 Readers
ZCWPW2 residue Trp-41, separating the side chains of trimethylated H3K4 and H3R2, is invariant in all of the human CW domains (Fig. 1B). Although the sequences and structures of the CW and PHD domains are diverse, they have similar cores (Fig. 8A), and the Lys-4/Arg-2-separating tryptophan is also highly conserved in the PHD domains, where it plays a similarly important role, such as in BPTF (31), ING2 (32), PHF2 (38), and MLL5 (34) (Fig. 8B). Accordingly, the W32F mutation in BPTF reduces affinity >20-fold (31), similar to our data for the ZCWPW2 W41F mutant (Fig. 5E). The W29A mutation in PHF2 (38) and the W141A mutation in MLL5 (34) disrupt the binding. These results indicate that the interaction between the H3K4me3 binder and H3K4me3 depends not only on the aromaticity of this cage residue but also on the type of aromatic ring. In addition to the CW and PHD domains, some proteins of the Royal superfamily also bind to H3K4me3, such as SGF29. However, the aromatic cage of SGF29 does not have a tryptophan residue but rather two tyrosine residues (Fig. 8B), and mutating any aromatic residue to alanine disrupts its histone binding ability (36).
FIGURE 8.
Structural comparison with other H3K4me3 readers. A, sequence alignment of selected CW and PHD domains. Secondary structure elements of the ZCWPW2 CW domain are indicated above the sequence alignment. The highly conserved cage “right wall” residues are marked by a red star; the other cage-forming residues are marked by blue stars or highlighted in blue. The residues recognizing H3A1 are highlighted in orange. The alignments were constructed with ClustalW (45). B, comparison of H3K4me3 binding cage in ZCWPW2 and other H3K4me3 readers' cages.
The H3A1 binding pocket is also conserved in most H3K4me3 readers. This pocket is located on the loop between the second and third zinc-coordinating cysteine residues of the CW domain. The N-terminal amino group of H3A1 forms hydrogen bonds with backbone carbonyl moieties within the CW domain (Fig. 3, C and G). The corresponding loop is also conserved in the PHD domain, where it is located between the fifth and sixth zinc-coordinating cysteine residues (Fig. 8A) (31–34, 38). In these binding pockets, the free N terminus is essential for H3K4me3 recognition. In contrast, the double chromodomains of CHD1, which adopt a shallow and open pocket to interact with the free N-terminal amine of H3K4, tolerate a non-structural protein 1 of influenza virus (NS1) C-terminal mimicking peptide after a conformational change of the peptide (39).
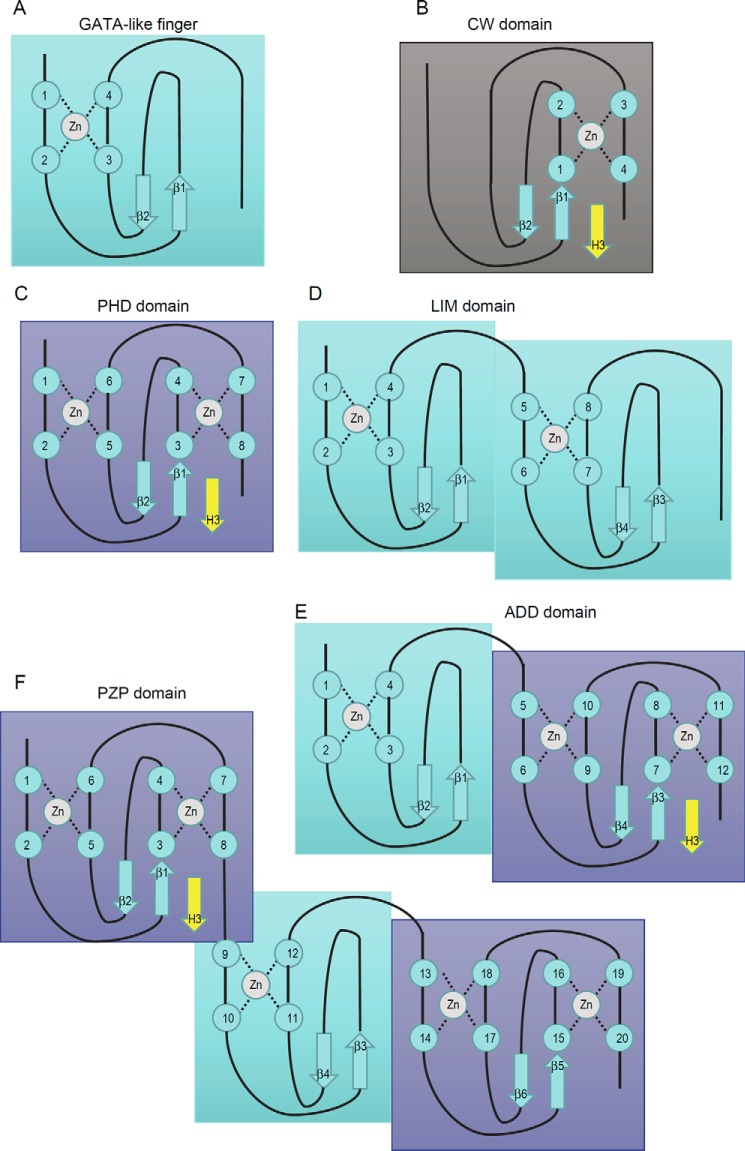
The GATA-like Finger, CW Domain, and PHD Domain Are Basic Structure Units, Which Can Be Further Assembled into the LIM Domain, ADD Domain, and PZP Domain
When the first three-dimensional structure of the CW domain of ZCWPW1 was determined, it was found that the CW domain and PHD domain share a homologous substructure, composed of the β-hairpin and the second zinc knuckle in the PHD domain, and that the CW domain is more closely related to the PHD domain than to other known zinc fingers. More importantly, both the CW domain and PHD domain bind to histone H3K4me3 in a similar manner (8, 31, 32). In addition, we also notice that another type of zinc finger is highly related to the PHD domain (i.e. zinc fingers of the erythroid transcription factor GATA-1, which is composed of the first zinc knuckle in the PHD domain and a β-hairpin and can bind to DNA) (40). To some extent, we can recognize the CW domain and GATA-like finger as two types of variant of the PHD domain (Fig. 9, A–C). From this view, the LIM (Lin11/Isl-1/Mec-3) domain, which shares a similar arrangement of zinc-binding cysteines and histidines with the PHD domain but adopts distinct tertiary structure (41), could be recognized as two tandem GATA-like fingers (Fig. 9D). In addition, the ADD (ATRX-DNMT3-DNMT3L) domain is a fusion of a GATA-like finger and a PHD domain and can bind to unmethylated H3K4 in a similar manner (Fig. 9E) (42). Most recently, two structures of the PZP (PHD-Zn-PHD) domain have been reported, in which two PHD domains are connected by a GATA-like finger (43, 44). For the BRPF1 PZP domain, the first PHD domain is used to bind unmethylated H3K4 (Fig. 9F) (43), whereas the AF10 PZP domain requires all subdomains to bind a region of histone H3 that spans amino acids 15–34 (44), suggesting that intrinsic varieties are present in these domains. Overall, we propose that the GATA-like finger, CW domain, and PHD domain are basic structure units and that they can be further assembled into the LIM domain, ADD domain, and PZP domain. Of all of these structure modules, a CW-type subunit is required (but not sufficient) to bind the N terminus of histone H3 with lysine 4 methylated or unmodified. From structural and functional views, we propose that the CW domain belongs to an extended superfamily.
FIGURE 9.
Schematic representation showing the zinc-binding topology of several CW-related domains. A, GATA-like finger; B, CW domain; C, PHD domain; D, LIM domain; E, ADD domain; F, PZP domain. For the CW domain, PHD domain, ADD domain, and PZP domain, a yellow β-strand is shown to represent the binding N terminus of histone H3.
In conclusion, our combined structural analyses and histone binding experiments for ZCWPW2, MORC1, MORC2, MORC3, MORC4, and systematically designed mutants of ZCWPW2 reveal that, of the seven human CW domains, ZCWPW1/2 and MORC3/4 are capable of binding methylated H3K4 histones, whereas MORC1/2 and LSD2 lack a proper aromatic cage or have structural impediments, preventing methyllysine peptide recognition. Of the three cage-forming residues in the CW domain, only the invariant tryptophan residue of the CW domain is essential for recognition of the methyllysine side chain, whereas the other two cage residues may infer selectivity for specific methylation states or enhance binding, respectively.
Author Contributions
Y. L. purified and crystallized the protein; W. T. collected the data and determined the structures; Q. Z. did partial ITC assays; X. L. did the solubility test; P. L. cloned the constructs; J. M. conceived and designed the study; Y. L., S. Q., and J. M. wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Aiping Dong for reviewing the ZCWPW2 and MORC3 crystallographic models. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357.
This study was supported by National Natural Science Foundation of China Grant 31500613. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from AbbVie, Boehringer Ingelheim, the Canadian Institutes of Health Research, Genome Canada, Ontario Genomics Institute Grant OGI-055, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and Wellcome Trust Grant 092809/Z/10/Z.
The atomic coordinates and structure factors (codes 4O62 and 4QQ4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- H3K4
- histone H3 Lys-4
- H3K4me3
- trimethylated H3K4
- H3A1
- histone H3 Ala-1
- H3R2
- histone H3 Arg-2
- FP
- fluorescence polarization
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank.
References
- 1. Vaquero A., Loyola A., and Reinberg D. (2003) The constantly changing face of chromatin. Sci. Aging Knowledge Environ. 2003, RE4. [DOI] [PubMed] [Google Scholar]
- 2. Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 3. Hake S. B., Xiao A., and Allis C. D. (2004) Linking the epigenetic “language” of covalent histone modifications to cancer. Br. J. Cancer 90, 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams-Cioaba M. A., and Min J. (2009) Structure and function of histone methylation binding proteins. Biochem. Cell Biol. 87, 93–105 [DOI] [PubMed] [Google Scholar]
- 5. Zhang Y., and Reinberg D. (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360 [DOI] [PubMed] [Google Scholar]
- 6. Musselman C. A., Lalonde M.-E., Côté J., and Kutateladze T. G. (2012) Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qin S., and Min J. (2014) Structure and function of the nucleosome-binding PWWP domain. Trends Biochem. Sci. 39, 536–547 [DOI] [PubMed] [Google Scholar]
- 8. He F., Umehara T., Saito K., Harada T., Watanabe S., Yabuki T., Kigawa T., Takahashi M., Kuwasako K., Tsuda K., Matsuda T., Aoki M., Seki E., Kobayashi N., Güntert P., et al. (2010) Structural insight into the zinc finger CW domain as a histone modification reader. Structure 18, 1127–1139 [DOI] [PubMed] [Google Scholar]
- 9. Hoppmann V., Thorstensen T., Kristiansen P. E., Veiseth S. V., Rahman M. A., Finne K., Aalen R. B., and Aasland R. (2011) The CW domain, a new histone recognition module in chromatin proteins. EMBO J. 30, 1939–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li X., Foley E. A., Molloy K. R., Li Y., Chait B. T., and Kapoor T. M. (2012) Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J. Am. Chem. Soc. 134, 1982–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Liu S., Zhang X., Liang X., Zahid K. R., Liu K., Liu J., Deng L., Yang J., and Qi C. (2016) Structure and function of CW domain containing proteins. Curr. Protein Pept. Sci. 10.2174/1389203717666160125115130 [DOI] [PubMed] [Google Scholar]
- 12. Perry J., and Zhao Y. (2003) The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem. Sci. 28, 576–580 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Q., Qi S., Xu M., Yu L., Tao Y., Deng Z., Wu W., Li J., Chen Z., and Wong J. (2013) Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b. Cell Res. 23, 225–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu C., Wang X., Liu K., Roundtree I. A., Tempel W., Li Y., Lu Z., He C., and Min J. (2014) Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 [DOI] [PubMed] [Google Scholar]
- 15. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang B.-C. (1985) Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 115, 90–112 [DOI] [PubMed] [Google Scholar]
- 18. Schneider T. R., and Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 19. Sheldrick G. M. (2002) Macromolecular phasing with SHELXE. Zeitschrift Kristallogr. 217, 644–650 [Google Scholar]
- 20. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phasercrystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perrakis A., Sixma T. K., Wilson K. S., and Lamzin V. S. (1997) wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. D Biol. Crystallogr. 53, 448–455 [DOI] [PubMed] [Google Scholar]
- 22. Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang H., Guranovic V., Dutta S., Feng Z., Berman H. M., and Westbrook J. D. (2004) Automated and accurate deposition of structures solved by x-ray diffraction to the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 60, 1833–1839 [DOI] [PubMed] [Google Scholar]
- 29. Gildea R. J., Bourhis L. J., Dolomanov O. V., Grosse-Kunstleve R. W., Puschmann H., Adams P. D., and Howard J. A. K. (2011) iotbx.cif: a comprehensive CIF toolbox. J. Appl. Crystallogr. 44, 1259–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fang R., Chen F., Dong Z., Hu D., Barbera A. J., Clark E. A., Fang J., Yang Y., Mei P., Rutenberg M., Li Z., Zhang Y., Xu Y., Yang H., Wang P., et al. (2013) LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol. Cell 49, 558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H., Ilin S., Wang W., Duncan E. M., Wysocka J., Allis C. D., and Patel D. J. (2006) Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peña P. V., Davrazou F., Shi X., Walter K. L., Verkhusha V. V., Gozani O., Zhao R., and Kutateladze T. G. (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang G. G., Song J., Wang Z., Dormann H. L., Casadio F., Li H., Luo J.-L., Patel D. J., and Allis C. D. (2009) Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ali M., Rincón-Arano H., Zhao W., Rothbart S. B., Tong Q., Parkhurst S. M., Strahl B. D., Deng L. W., Groudine M., and Kutateladze T. G. (2013) Molecular basis for chromatin binding and regulation of MLL5. Proc. Natl. Acad. Sci. U.S.A. 110, 11296–11301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 36. Bian C., Xu C., Ruan J., Lee K. K., Burke T. L., Tempel W., Barsyte D., Li J., Wu M., Zhou B. O., Fleharty B. E., Paulson A., Allali-Hassani A., Zhou J.-Q., Mer G., Grant P. A., et al. (2011) Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30, 2829–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Min J., Allali-Hassani A., Nady N., Qi C., Ouyang H., Liu Y., MacKenzie F., Vedadi M., and Arrowsmith C. H. (2007) L3MBTL1 recognition of mono- and dimethylated histones. Nat. Struct. Mol. Biol. 14, 1229–1230 [DOI] [PubMed] [Google Scholar]
- 38. Wen H., Li J., Song T., Lu M., Kan P. Y., Lee M. G., Sha B., and Shi X. (2010) Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J. Biol. Chem. 285, 9322–9326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qin S., Liu Y., Tempel W., Eram M. S., Bian C., Liu K., Senisterra G., Crombet L., Vedadi M., and Min J. (2014) Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1. Nat. Commun. 5, 3952. [DOI] [PubMed] [Google Scholar]
- 40. Omichinski J. G., Clore G. M., Schaad O., Felsenfeld G., Trainor C., Appella E., Stahl S. J., and Gronenborn A. M. (1993) NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science 261, 438–446 [DOI] [PubMed] [Google Scholar]
- 41. Capili A. D., Schultz D. C., Rauscher F. J. 3rd, and Borden K. L. (2001) Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20, 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Argentaro A., Yang J.-C., Chapman L., Kowalczyk M. S., Gibbons R. J., Higgs D. R., Neuhaus D., and Rhodes D. (2007) Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc. Natl. Acad. Sci. U.S.A. 104, 11939–11944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klein B. J., Muthurajan U. M., Lalonde M.-E., Gibson M. D., Andrews F. H., Hepler M., Machida S., Yan K., Kurumizaka H., Poirier M. G., Côté J., Luger K., and Kutateladze T. G. (2016) Bivalent interaction of the PZP domain of BRPF1 with the nucleosome impacts chromatin dynamics and acetylation. Nucleic Acids Res. 44, 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen S., Yang Z., Wilkinson A. W., Deshpande A. J., Sidoli S., Krajewski K., Strahl B. D., Garcia B. A., Armstrong S. A., Patel D. J., and Gozani O. (2015) The PZP domain of AF10 senses unmodified H3K27 to regulate DOT1L-mediated methylation of H3K79. Mol. Cell 60, 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thompson J. D., Higgins D. G., and Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bayly C. I., Cieplak P., Cornell W., and Kollman P. A. (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97, 10269–10280 [Google Scholar]
- 48. Schwede T., Kopp J., Guex N., and Peitsch M. C. (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]