Abstract
Thymine-DNA glycosylase (TDG) plays critical roles in DNA base excision repair and DNA demethylation. It has been proposed, based on structural studies and in vitro biochemistry, that sumoylation is required for efficient TDG enzymatic turnover following base excision. However, whether sumoylation is required for TDG activity in vivo has not previously been tested. We have developed an in vivo assay for TDG activity that takes advantage of its recently discovered role in DNA demethylation and selective recognition and repair of 5-carboxylcytosine. Using this assay, we investigated the role of sumoylation in regulating TDG activity through the use of TDG mutants defective for sumoylation and Small Ubiquitin-like Modifier (SUMO) binding and by altering TDG sumoylation through SUMO and SUMO protease overexpression experiments. Our findings indicate that sumoylation and SUMO binding are not essential for TDG-mediated excision and repair of 5-carboxylcytosine bases. Moreover, in vitro assays revealed that apurinic/apyrimidinic nuclease 1 provides nearly maximum stimulation of TDG processing of G·caC substrates. Thus, under our assay conditions, apurinic/apyrimidinic nuclease 1-mediated stimulation or other mechanisms sufficiently alleviate TDG product inhibition and promote its enzymatic turnover in vivo.
Keywords: base excision repair (BER), enzyme turnover, small ubiquitin-like modifier (SUMO), SUMO-interacting motif (SIM), sumoylation
Introduction
Regulation and coordination of DNA repair mechanisms is essential for maintaining genome integrity and proper cell function. Sumoylation is a post-translational protein modification critical for DNA damage repair, including repair of DNA single- and double-stranded breaks, interstrand cross-links, and nucleotide mismatches (1, 2). Although affecting repair processes through a variety of distinct mechanisms, primary functions of sumoylation at DNA lesions include promotion of protein-protein interactions and stimulation of protein extraction or turnover (1, 3). The exact molecular effects of sumoylation on many modified repair factors, however, remain to be fully understood.
Thymine-DNA glycosylase (TDG)3 is a monofunctional glycosylase involved in DNA repair, DNA demethylation, and transcription activation (4–6). The best studied role of TDG is in base excision repair (BER), where it specifically recognizes G/U and G/T mismatches arising from spontaneous deamination of cytosine or 5-methylcytosine, respectively. BER proceeds through a process involving glycosylase recognition of a specific DNA lesion and removal of the lesion to produce an apurinic/apyrimidinic (AP) site. TDG-mediated BER is of particular interest because of the observed product inhibition caused by high affinity binding of TDG to the AP site following base excision (7, 8). This product inhibition can be relieved by apurinic/apyrimidinic endonuclease 1 (APE1), the enzyme that acts immediately downstream of TDG in the BER pathway (8, 9). Alternatively, in vitro studies and structural analyses have suggested an intriguing model for overcoming product inhibition involving TDG sumoylation (10–14).
TDG is sumoylated at a single lysine residue near its C terminus, Lys330, and also contains a SUMO-interacting motif (SIM) that involves residues ranging from Asp307 to Thr314, including a VQEV motif (residues 308–311) that is similar to that found in SIMs of other proteins (12, 13). When TDG is modified by either SUMO1 or SUMO3 at Lys330, conjugated SUMO interacts noncovalently with the adjacent SIM, thereby promoting formation of a protruded α-helix within the catalytic domain that obstructs DNA binding (10, 11). In addition, sumoylation has been proposed to affect conformational changes within the N-terminal domain of TDG that neutralize nonspecific DNA interactions (14). Consistent with these findings, in vitro binding studies indicate that SUMO-modified TDG has reduced affinity for DNA (12, 14, 15). Based on these observations, it has been proposed that sumoylation serves as a mechanism for promoting enzymatic turnover of TDG by disrupting DNA binding and relieving product inhibition (12, 14).
Despite findings that sumoylation of TDG weakens its binding to abasic DNA in vitro, no studies have explored the question of whether this modification is needed for efficient substrate processing by TDG in vivo. This has been due in part to the redundant substrate specificities of the multiple mammalian DNA glycosylases and the absence of TDG specific activity assays (16). The recently discovered role for TDG in active DNA demethylation, however, provides the basis for developing an assay to selectively interrogate TDG activity. Specifically, DNA demethylation proceeds through an enzymatic pathway that involves the ten-eleven translocation enzymes (TETs) and TDG-initiated BER. The TET enzymes are a family of dioxygenases that iteratively oxidize 5-methylcytosine (5mC) to give 5-hydroxymethylcytosine, 5-formylcytosine, and a final product of 5-carboxylcytosine (5caC) (5). Importantly, TDG is the only glycosylase that recognizes and excises 5-formylcytosine and 5caC from DNA (5, 17–19).
Taking advantage of TDG specificity for 5caC, we developed an in vivo assay to monitor TDG activity and explore requirements for TDG sumoylation and SUMO binding in BER. Our findings reveal that neither sumoylation of TDG nor SUMO binding by TDG are essential for its enzymatic activity under our in vivo assay conditions and thus raise questions about other mechanisms for relieving product inhibition and other possible functions for TDG sumoylation.
Experimental Procedures
Materials
The G·caC DNA for TDG activity assays contained a 40-mer strand, 5′-CAT GTG TCA CCA CTG CTC A−XG TAC AGA GCT GTA GAT GCA C (X = caC), and a 40-mer complement, 5′-GTG CAT CTA CAG CTC TGT ACG TGA GCA GTG GTG ACA CAT G. In this 40-bp duplex, 5caC is paired with G and located in a CpG context (underlined). Oligodeoxynucleotides were obtained from IDT or the Keck Foundation Biotechnology Resource Laboratory of Yale University. Oligodeoxynucleotides were purified by reverse phase HPLC; exchanged into buffer consisting of 0.02 m Tris-HCl, pH 7.5, 0.04 m NaCl; and quantified by absorbance as described (20, 21).
Human APE1 and human TDG were expressed in bacteria (Escherichia coli) and purified essentially as described (9, 21). The enzymes were >99% pure as judged by SDS-PAGE, and the concentrations were determined by absorbance as described (22).
TDG Activity Assays
Enzyme kinetics experiments were performed using either single turnover conditions with a saturating TDG concentration, to obtain the maximal rate of 5caC base excision (kmax), or multiple turnover experiments with a saturating substrate concentration, to obtain the maximal rate of catalytic turnover (kcat), as described (18). The reactions were performed at 37 °C in HEMN.1 buffer (0.02 m HEPES, pH 7.5, 0.1 m NaCl, 0.2 mm EDTA, 2.5 mm MgCl2). Reactions were initiated by adding concentrated TDG to buffered G·caC DNA substrate, and aliquots were removed at the desired time points and quenched with 50% (v/v) 0.3 m NaOH, 0.03 m EDTA. Quenched samples were heated for 15 min at 85 °C to quantitatively cleave the DNA backbone at abasic sites, and the resulting DNA fragments were resolved by anion exchange HPLC (21).
Single turnover experiments were performed with a substrate concentration of 0.5 μm and a saturating concentration of TDG (1.0 μm). Progress curves were fitted by nonlinear regression to Equation 1,
where A is the amplitude, kobs is the rate constant, and t is the reaction time. Given the saturating enzyme conditions employed ([E] ≫ Kd; [E] > [S]), the observed rate constant reflects the maximal rate of product formation (kobs ≈ kmax) and is not influenced by E-S association or by product release or product inhibition. Previous studies show that TDG binds to DNA containing a G/U mispair with a Kd = 0.6 nm (22), and the TDG catalytic domain exhibits 7-fold tighter affinity for G·caC relative to G/U DNA (23).
Multiple turnover experiments were performed at 37 °C in HEMN.1 buffer (with 0.1 mg/ml BSA), using a limiting concentration of TDG (0.05 μm) and a saturating concentration of G·caC DNA substrate (1.0 μm). The linear portion of the progress curve (product concentration versus time) was used to obtain the initial velocity (v0). Under the saturating substrate conditions used here ([S] ≫ Km), the Michaelis-Menten equation simplifies such that v0 reports on the maximal rate of catalytic turnover (kcat = v0/[E]). Following the same approach, multiple turnover experiments were also performed in the presence of APE1. The reactions were initiated by adding TDG to HEMN.1 buffer (with 5 mm MgCl2) containing G·5caC substrate and APE1, and aliquots were removed and quenched as described above. For experiments collected with APE1, fitting was also limited to the linear region of the progress curve (< 20% product).
Cell Culture and Transfections
HEK293T and U2OS cells were cultured in Dulbecco's modified Eagle's medium-high glucose (DMEM-1×; Life Technologies) containing 4.5 g/liter d-glucose and l-glutamine and supplemented with 10% fetal bovine serum, 1% HEPES, phenol red, and antibiotics and maintained at 37 °C in 5% CO2 incubator. For transient transfections, the cells were plated in 6-well plates with supplemented DMEM-1× culture media absent antibiotics. The Cells were transfected using Lipofectamine 2000 (Life Technologies) for transient overexpression or Lipofectamine RNAiMAX (Life Technologies) for transient knockdowns per manufacturer's instructions. RNAi oligonucleotides were used at final concentrations of 20 nm.
Plasmids and Oligonucleotides
TDG was cloned into the pcDNA3.0 expression vector with an N-terminal FLAG tag. TDG mutant constructs were generated using QuikChange site-directed mutagenesis (Agilent Technologies, Inc.). The TET1catalytic domain (residues 1,418–2,136) was inserted into the pLEXm expression vector and contains an N-terminal histidine tag (generously provided by R. Kohli, University of Pennsylvania). SENP1 was expressed in pEGFP-C1 vectors, as previously described (24). All constructs were confirmed by DNA sequence analysis. siRNA oligonucleotides used were as follows: (SENP1) 5′-UCCUUUACACCUGUCUCGAUGUCUU-3′; (SENP2) 5′-AUAUCUGGAUUCUAUGGGAUU-3′. The commercially available NC1 oligonucleotide (Integrated DNA Technologies) was used as a control.
Cell Lysate Preparation
HEK293T cells were cultured in 6-well plates, washed with 1× PBS, and then lysed with 100 μl of modified radioimmunoprecipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCL, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mm EDTA, 0.1% SDS) supplemented with 1 mm PMSF, 5 mm N-ethylmaleimide, and 1× SigmaFAST protease inhibitor mixture (Sigma-Aldrich). After incubation for 30 min at 4 °C, lysates were sonicated to shear DNA, centrifuged (20,000 × g, 4 °C, 30 min), and stored at −80 °C. Protein concentrations of cell lysates were measured by bicinchoninic acid assay (Pierce, Thermo Scientific).
Immunoblot Analysis
Prepared cell lysates were diluted with 5× SDS loading buffer (300 mm Tris-HCl, pH 6.8, 10% SDS, 50% glycerol, 500 mm 2-mercaptoethanol, and 0.05% bromphenol blue) and incubated for 10 min at 96 °C. For immunoblotting, proteins were resolved by SDS-PAGE and transferred to nitrocellulose membrane. The membranes were blocked with 5% milk in 1× Tris-buffered saline and 0.1% Tween 20 (TBS-T) and then incubated with respective primary antibodies. Antibodies were diluted in TBS-T with the following dilutions: 1:2,000 rabbit polyclonal anti-TDG (Genetex, GTX110473), 1:10,000 mouse monoclonal anti-FLAG (Sigma, F1804), 1:2,000 mouse monoclonal anti-His (GE Life Sciences, 27-4710-01), 1:2,000 mouse monoclonal anti-c-Myc (Santa Cruz, sc-40), 1:5,000 mouse monoclonal anti-GFP (Clontech Living Colors, 632381), 1:5,000 rabbit monoclonal anti-SENP1 (Abcam, ab108981), 1:2,000 mouse monoclonal anti-γH2A.X (Millipore, 05–636), 1:5,000 mouse monoclonal anti-α-tubulin (Sigma, T9026), and 1:1,000 rabbit anti-TET1 (ProSci Inc., 7733). HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were detected by enhanced chemiluminescence (Luminata Western HRP Substrates, Millipore).
Immunofluorescence Microscopy
U2OS cells were cultured on coverslips in 6-well plates. The cells were fixed for 30 min at room temperature with 2% formaldehyde in PBS. The cells were then washed with PBS and permeabilized for 7 min at room temperature with 0.2% Triton X-100 in PBS. The cells were incubated with primary antibody (1:250 rabbit polyclonal anti-FLAG, GenScript, A00170; 1:250 mouse monoclonal anti-SUMO1, 21C7) (25) in TBS-T. After washing with PBS, the cells were incubated with mouse α488 or rabbit α594 Alexa Fluor-conjugated secondary antibodies (Invitrogen). The cells were imaged using a Zeiss Observer Z1 microscope.
Genomic DNA Purification and 5caC Dot Blot Analysis
Genomic DNA (gDNA) was purified from HEK293T cells using GeneJet genomic DNA purification kit per kit instructions for mammalian cell culture (Thermo Scientific). For 5caC analysis, gDNA was denatured by incubation with NaOH (0.4 m final) for 10 min at 95 °C. Immediately following heating, gDNA was neutralized with NH4OAc, pH 7.2 (final concentration, 1 m) and brought to a final concentration of 5 ng/μl. DNA concentrations were measured by NanoDrop 2000 (Thermo Scientific).
Prepared gDNA was subsequently spotted as indicated on nitrocellulose membrane (0.45 μm; Bio-Rad) using a Hybri-dot manifold dot blot apparatus (96-well; Invitrogen, Life Technologies) following standard procedures. The membranes were washed with 2× SSC buffer (300 mm NaCl, 30 mm trisodium citrate, pH 7). The membranes were blocked with 5% milk in TBS-T and subsequently probed with 1:5,000 rabbit polyclonal anti-5caC (Active Motif, 61225). Signals were detected using fluorescent secondary antibodies, and intensities were measured and analyzed using an Odyssey Imaging System (LI-COR Biosciences). To confirm equal loading of gDNA, membranes were briefly incubated with methylene blue (0.02% methylene blue, 300 mm sodium acetate, pH 5.2) at room temperature, destained with double-distilled H2O, and imaged.
Chromatin Fractionation
HEK293T cells were cultured in 6-well plates and washed with PBS and then lysed with 100 μl of Nonidet P-40 lysis buffer (10 mm HEPES, pH 7.4, 10 mm KCl, 0.05% Nonidet P-40) supplemented with 1 mm PMSF, 5 mm N-ethylmaleimide, and 1× SigmaFAST protease inhibitor mixture (Sigma-Aldrich). After 30 min of incubation on ice, lysates were centrifuged (20,000 × g, 4 °C, 10 min), and supernatant was saved as the Nonidet P-40-soluble fraction. The remaining pellet was washed with Nonidet P-40 lysis buffer and then resuspended with low salt buffer (10 mm Tris-HCl, pH 7.4, 0.2 mm MgCl2) and supplemented with 5 mm N-ethylmaleimide and protease inhibitors. After 15 min of incubation on ice, the suspension was centrifuged (20,000 × g, 4 °C, 10 min), and the supernatant was discarded. The remaining pellet was resuspended with 2× sample buffer, briefly sonicated, and saved as the Nonidet P-40-insoluble fraction.
Comet Assays
HEK293T cells were transfected with indicated expression constructs and analyzed for single and double-stranded DNA breaks using the single cell electrophoresis CometAssay® under alkaline conditions according to the manufacturer (Trevigen, Inc.). As a positive control, untransfected cells were treated with 100 μm hydrogen peroxide for 20 min prior to analysis. Images were collected using a Zeiss Observer Z1 microscope.
Results
Development of an in Vivo TDG Activity Assay
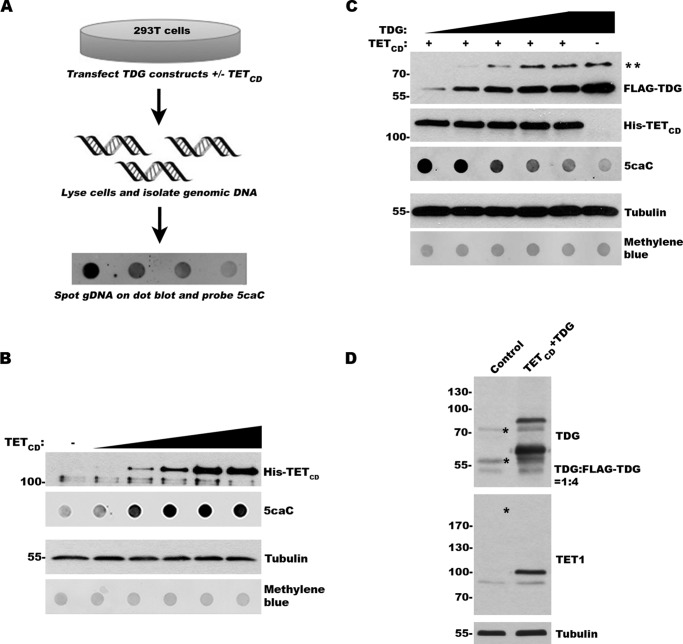
To explore the effects of sumoylation on TDG activity in vivo, we sought to develop a TET-TDG co-expression assay that takes advantage of the unique ability of TDG to excise TET-induced 5caC from DNA (17) (Fig. 1A). First, we transfected 293T cells with a plasmid coding for a His-tagged catalytic domain of human TET1 (TETCD) and analyzed 5caC production by dot blot analysis of purified gDNA using a 5caC-specific antibody. As expected, we observed a dose-dependent increase in 5caC levels with increasing levels of TETCD expression (Fig. 1B). Levels of 5caC detection peaked with intermediate TETCD expression, suggesting saturable levels of cytosine modification by TETCD. Importantly, these results also suggest that endogenous TDG activity is insufficient to repair and effectively reduce the levels of 5caC induced by TETCD expression.
FIGURE 1.
TETCD expression and induction of 5caC can be used to assess TDG activity in vivo. A, schematic of the experimental approach to evaluate TDG activity in vivo. B, expression of TETCD induces formation of 5caC in a dose-dependent manner. The cells were transfected with increasing levels of TETCD expression vector. Protein expression and 5caC levels were monitored by immunoblot analysis. Protein and DNA loading were assessed by tubulin and methylene blue staining, respectively. C, expression of exogenous TDG suppresses 5caC accumulation in a dose-dependent manner. The cells were transfected with TETCD and varying levels of TDG. Protein expression and 5caC levels were monitored as in B. ** indicates sumoylated TDG. D, immunoblot analysis of control transfected cells and cells transfected with TETCD and FLAG-TDG. The blots were probed with TET1- and TDG-specific antibodies for detection of both endogenous and exogenous proteins. Asterisks indicate the positions of endogenous SUMO-TDG, TDG, and the predicted size of endogenous TET1 (not detected). The ratio of TDG to FLAG-TDG was determined using ImageJ.
To investigate the ability of exogenously expressed TDG to excise 5caC in vivo, we next co-transfected 293T cells with plasmids coding for His-tagged TETCD and FLAG-tagged TDG. The cells were transfected so that they expressed a fixed level of TETCD and increasing levels of TDG, as revealed by immunoblot analysis (Fig. 1C). As previously reported, two forms of TDG were detected in transfected cells: an unmodified form migrating at ∼65 kDa and a SUMO-modified form migrating at ∼80 kDa (12, 26, 27). We again used dot blot analysis of gDNA prepared from these cells to detect 5caC levels. This analysis revealed a dose-dependent decrease in 5caC detection, indicative of dose-dependent TDG-mediated excision (Fig. 1C). Under maximum expression conditions, FLAG-TDG levels were ∼4 times above endogenous TDG, whereas relative levels of TETCD expression could not be determined because of the inability to detect endogenous TET1 (Fig. 1D). Our assay provides a quick and robust method for monitoring TDG activity in vivo.
Assessment of Sumoylation and SUMO Binding on TDG Activity in Vivo
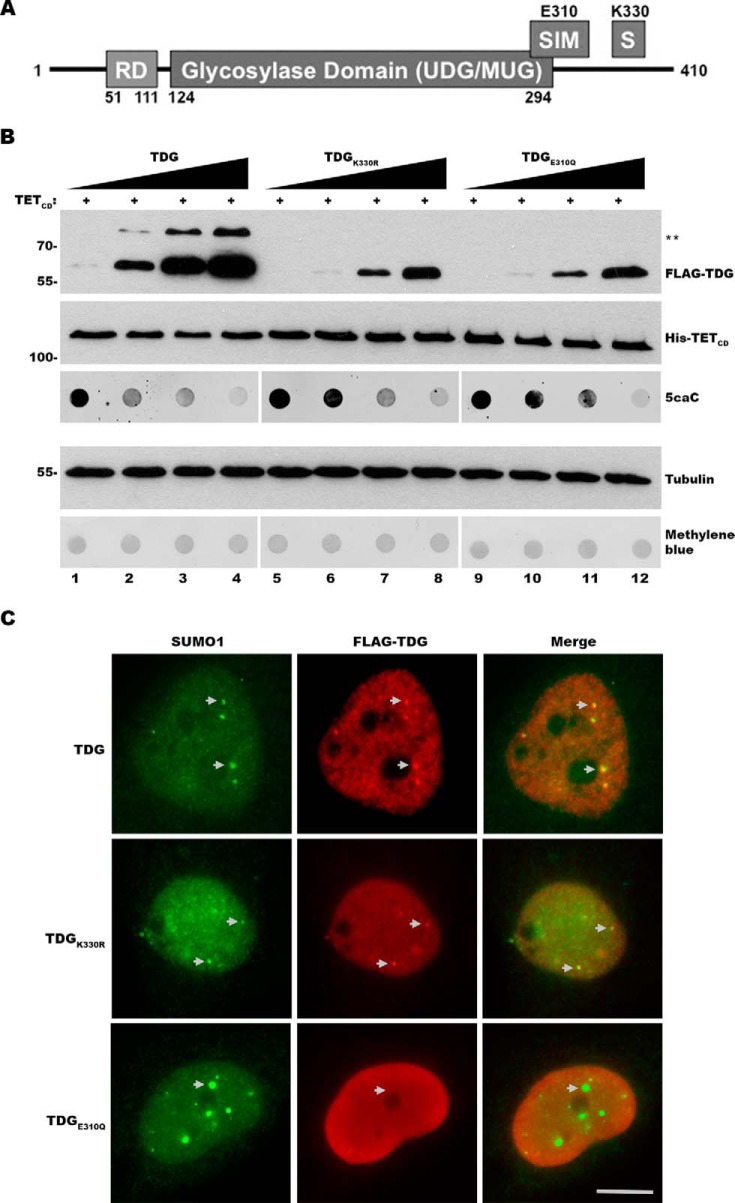
TDG is sumoylated at Lys330 and has a SIM that includes a VQEV motif (residues 308 and 311) (Fig. 2A). Both SUMO conjugation and binding are proposed to affect a conformational change in TDG that promotes enzymatic turnover (10, 12, 14). To evaluate the effects of SUMO modification and SUMO binding on TDG activity in vivo, we generated plasmids coding for FLAG-tagged mutant forms of TDG. One mutant contained a K330R substitution inhibiting covalent sumoylation, and another contained an E310Q substitution previously shown to interrupt noncovalent SUMO binding (10, 26). Plasmids coding for wild type or mutant TDG were transfected into 293T cells with TETCD so that fixed levels of TETCD and increasing levels of TDG expression were achieved, as revealed by immunoblot analysis (Fig. 2B). As anticipated, only unmodified TDG was detected in cells expressing TDGK330R mutant. This was also the case for cells expressing TDGE310Q, consistent with a report that SUMO binding also affects covalent modification of TDG (27).
FIGURE 2.
TDG sumoylation and SUMO binding are not essential for 5caC repair. A, schematic diagram of TDG and the location of the C-terminal SIM and sumoylation site at Lys330. B, cells were co-transfected with expression vectors for TETCD and varying levels of wild type TDG (TDG), or sumoylation (TDGK330R) or SUMO binding (TDGE310Q) mutant TDG. Protein and 5caC levels were monitored by immunoblot analysis. ** indicates sumoylated TDG. Protein and DNA loading were assessed by tubulin and methylene blue staining, respectively. C, analysis of wild type and mutant TDG localization by immunofluorescence microscopy. The cells were transfected with plasmids coding for FLAG-tagged wild type TDG or sumoylation or SUMO binding TDG mutants. TDG localization was assessed using anti-FLAG antibodies. Localization to PML nuclear bodies was assessed by co-localization with SUMO1. Bar, 5 μm.
Because of varying reports (12, 26, 27), we also evaluated the localizations of TDG, TDGK330R, and TDGE310Q by immunofluorescence microscopy to assess possible mutual effects on subcellular targeting (Fig. 2C). Wild type TDG localized predominantly to the nucleoplasm and could also be detected in promyelocytic leukemia (PML) nuclear bodies, as revealed by co-localization with nuclear foci stained with SUMO1. TDGK330R localization was similar to wild type, whereas TDGE310Q localized to the nucleoplasm but was not detected in PML nuclear bodies. The normal heterogeneity in size and number of PML nuclear bodies detected by SUMO1 staining was not affected by expression of wild type or mutant TDGs.
Effects of wild type and mutant TDG expression of 5caC repair were evaluated by dot blot analysis of gDNA. As previously observed, we detected a dose-dependent decrease in 5caC levels with increasing expression of wild type TDG (Fig. 2B). In addition, comparable dose-dependent decreases in 5caC levels were detected in cells expressing TDGK330R and TDGE310Q. Notably, wild type and mutant TDG proteins were equally active under conditions where 5caC levels were in excess (compare lanes 2, 7, and 11, where TDG expression does not fully reduce 5caC detection). Under these conditions, TDGK330R excised 5caC as efficiently as wild type TDG even though it was expressed at slightly lower levels (compare lanes 2 and 7). These findings suggest that product inhibition is not simply overcome by TDG overexpression and, moreover, demonstrate that SUMO modification and SUMO binding are not required for efficient TDG-mediated excision of 5caC under these in vivo conditions.
Effect of APE1 on Catalytic Turnover of TDG
Given our finding that SUMO modification is not needed for efficient TDG excision of 5caC in cells, we sought to determine the effect of APE1 on the catalytic turnover of TDG for processing a G·caC substrate in vitro. APE1 follows TDG in the BER pathway, and it stimulates multiple-turnover processing of G/T and G/U mispairs by TDG (8, 9). To establish a baseline, we examined the activity of TDG in the absence of APE1. Single turnover kinetics experiments collected with a saturating level of TDG give the maximal rate of 5caC excision by TDG; kmax = 0.50 ± 0.03 min−1 (Fig. 3A). Notably, this maximal base excision rate is not altered by the presence of APE1 (1.2 μm), consistent with findings for other substrates that APE1 effects steps of the TDG reaction after chemistry (base excision) (8, 9).
FIGURE 3.
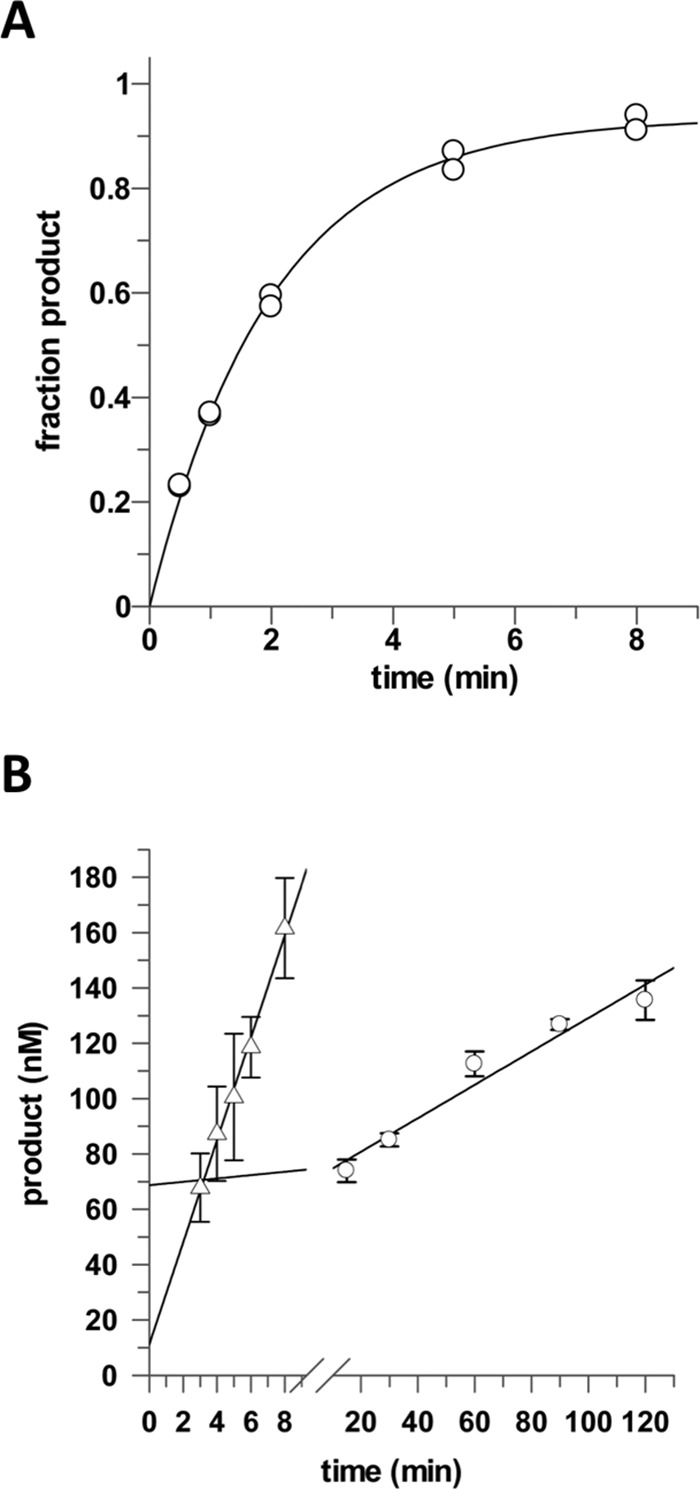
APE1 increases the catalytic turnover of TDG for processing G·caC substrates. A, TDG excision of 5caC from a G·caC DNA substrate exhibits a maximal rate constant of kmax = 0.50 ± 0.02 min−1. Single turnover experiments were performed at 37 °C with 0.5 μm substrate and saturating TDG (1.0 μm). B, catalytic turnover of TDG for a G·5caC substrate and the stimulatory effect of APE1. Multiple turnover activity of TDG is very low in the absence of APE1, kcat = 0.0121 ± 0.0015 min−1 (○), and is dramatically enhanced by APE1, kcat+APE1 = 0.369 ± 0.018 min−1 (▵). Multiple turnover experiments were collected at 37 °C with 1.0 μm DNA substrate, 0.05 μm TDG, and no APE1 or 1.2 μm APE1.
We also collected multiple turnover kinetics experiments, with saturating G·5caC substrate (1.0 μm) and limiting TDG (0.05 μm), finding that TDG turnover is very low, kcat = 0.0121 ± 0.0015 min−1 (Fig. 3B). The observation that kcat is 41-fold lower than kmax indicates that catalytic turnover for G·5caC is severely limited by slow product release and/or strong product inhibition, as observed for TDG processing of other substrates (8, 9, 12, 13).
Using the same multiple turnover experiments, we find that APE1, when present at a 20% molar excess relative to DNA (1.2 μm), greatly enhances the catalytic turnover of TDG, where kcat+APE1 = 0.369 ± 0.018 min−1 (Fig. 3B). Thus, APE1 stimulates the turnover of TDG by 30-fold for G·5caC substrates (kcat+APE1/kcat = 30). Moreover, APE1 provides nearly the maximum possible stimulation of TDG turnover, as indicted by our finding that kcat+APE1 approaches kmax (that is, kcat+APE1/kmax = 0.74). Notably, previous studies have shown that APE1 provides a similarly high enhancement of multiple turnover activity for human alkadenine DNA glycosylase (28).
Assessment of SENP1-mediated Desumoylation on TDG Activity in Vivo
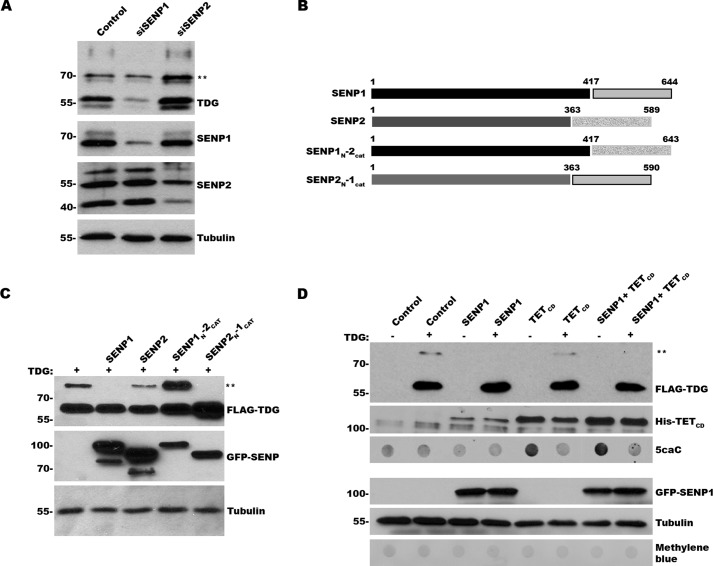
The current paradigm holds that to facilitate enzymatic turnover, TDG is dynamically sumoylated and desumoylated to promote AP-DNA product release and enable substrate binding, respectively. To identify SUMO isopeptidases involved in TDG desumoylation, we used RNAi to knockdown SENP1 and SENP2, the two SUMO isopeptidases that have both SUMO1 and SUMO2 specificity (29). Cell lysates from control and knockdown cells were analyzed by immunoblotting with TDG, tubulin and SENP1 or SENP2 antibodies (Fig. 4A). SENP1 was depleted by ∼90%, whereas each of the three major isoforms of SENP2 recognized by our antibody (30) were depleted by ∼70–80%. Under these knockdown conditions, SENP1 depletion uniquely increased the ratio of SUMO-modified to unmodified TDG. Also of potential significance, SENP1 depletion led to a decrease in overall TDG protein levels.
FIGURE 4.
SENP1 affects TDG sumoylation but not TDG-mediated repair of 5caC. A, cells were transfected with control, SENP1, or SENP2 specific siRNAs, andeffects on protein expression and TDG sumoylation were monitored by immunoblot analysis. ** indicates sumoylated TDG. B, schematic diagram of SENP1, SENP2, and chimeric SENP1–2 proteins. C, cells were transfected with expression vectors for wild type TDG, GFP-SENP1, GFP-SENP2, or chimeric GFP-SENP1–2 proteins, as indicated. Protein expression levels and TDG sumoylation were monitored by immunoblot analysis. ** indicates sumoylated TDG. D, cells were transfected with expression vectors for wild type TDG in the presence or absence of TETCD and GFP-SENP1 expression vectors, as indicated. Protein expression and 5caC levels were monitored by immunoblot analysis. ** indicates sumoylated TDG. Protein and DNA loading were assessed by tubulin and methylene blue staining, respectively.
To compliment RNAi knockdowns, we also overexpressed SENP1 and SENP2 by transient transfection and again evaluated effects on TDG sumoylation by immunoblot analysis. Under these conditions, we found that SENP1 overexpression reduced TDG sumoylation to undetectable levels, whereas SENP2 overexpression reduced but did not eliminate TDG sumoylation (Fig. 4C). Chimeras, in which the catalytic domains of SENP1 and SENP2 were exchanged, revealed that TDG recognition is determined by the SENP1 catalytic domain (Fig. 4, B and C).
Because SENP1 overexpression effectively limits TDG sumoylation, we used this as an alternative approach to explore the functional significance of dynamic sumoylation and desumoylation on TDG-mediated BER activity in vivo. We co-expressed TETCD, TDG, and SENP1 and first evaluated effects on TDG sumoylation by immunoblot analysis. As anticipated, TDG sumoylation was reduced to undetectable levels when co-expressed with SENP1 alone or with SENP1 together with TETCD (Fig. 4D). We next evaluated effects of SENP1 overexpression on TDG-mediated 5caC repair by dot blot analysis of purified gDNA. We observed nearly identical reductions in 5caC levels in cells transfected with TETCD, TDG, and SENP1 compared with cells transfected with only TETCD and TDG. These results further indicate that coordinated sumoylation and desumoylation of TDG is not required for its ability to repair 5caC under these in vivo conditions.
Assessment of Hypersumoylation on TDG Activity in Vivo
To complement studies on SENP1 overexpression and TDG desumoylation, we also sought to identify conditions for promoting and evaluating effects of TDG hypersumoylation. Toward this end, we found that overexpressing SUMO1 in the presence of TDG led to an increase in the ratio of SUMO-modified to unmodified TDG (Fig. 5A). To assess effects of hypersumoylation on TDG BER activity, we evaluated 5caC levels in cells co-expressing TETCD and TDG in the presence and absence of exogenous SUMO1 overexpression. To control for effects of hypersumoylation on proteins other than TDG, analysis was also performed in cells expressing TDGK330R. As observed in SENP1-overexpressing cells, we observed no effect of SUMO1 overexpression and altering the balance of sumoylation and desumoylation on 5caC repair activity (Fig. 5B).
FIGURE 5.
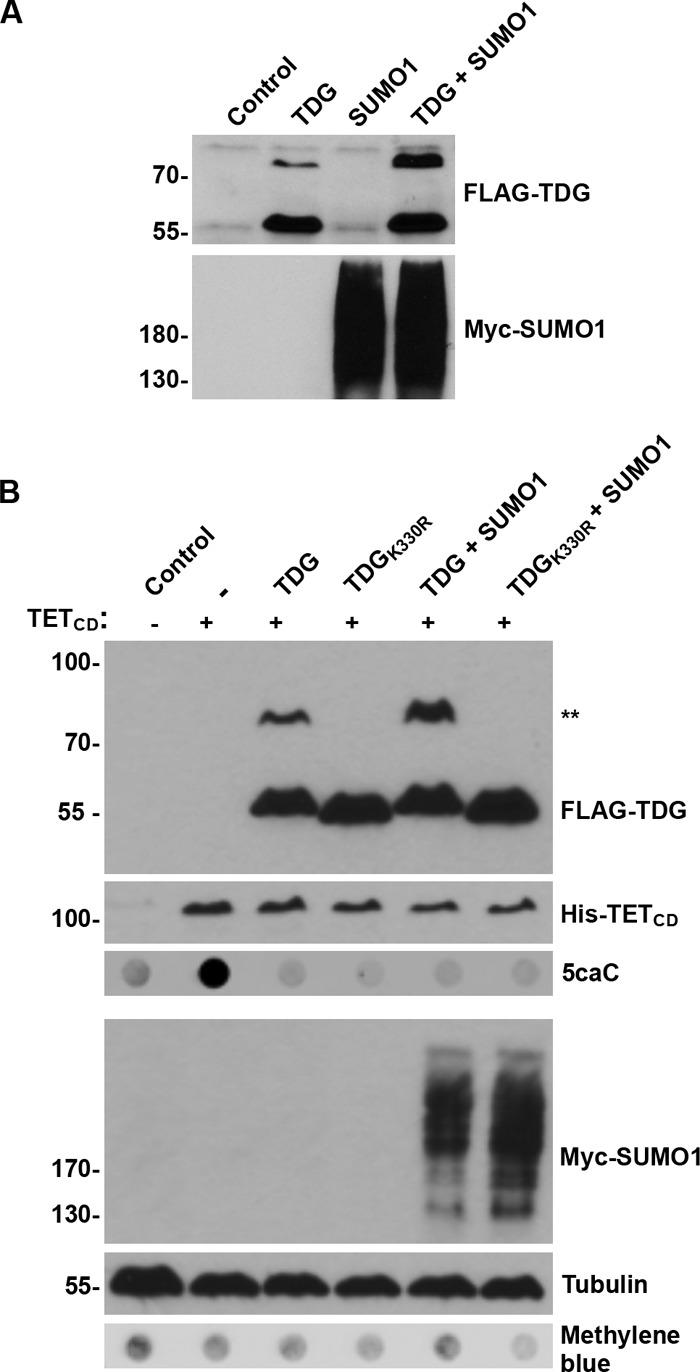
SUMO1 overexpression affects TDG sumoylation but not TDG-mediated repair of 5caC. A, cells were transfected with expression vectors for wild type TDG, Myc-SUMO1, or both vectors, as indicated. Protein expression and TDG sumoylation was monitored by immunoblot analysis. B, cells were transfected with expression vectors for TETCD together with vectors for wild type TDG, SUMO1, or both vectors, as indicated. Protein expression and 5caC levels were monitored by immunoblot analysis. ** indicates sumoylated TDG. Protein and DNA loading were assessed by tubulin and methylene blue staining, respectively.
Assessment of TDG Turnover at AP Sites in Vivo
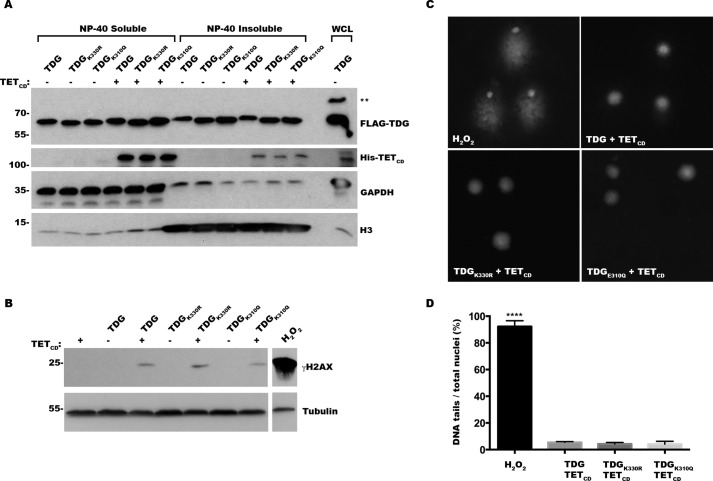
The detection of 5caC levels in purified gDNA provides an indirect measure of the step in BER proposed to be most affected by TDG sumoylation, namely the release of TDG from AP sites following base excision. To investigate more directly whether sumoylation affects the release of TDG from AP sites, we monitored the relative distributions of DNA-bound and soluble TDG in the presence and absence of TETCD expression. The cells were transfected with plasmids coding for TDG, TDGK330R, or TDGE310Q, either alone or together with TETCD, and subsequently fractionated into soluble and chromatin-bound fractions. These fractions were analyzed by immunoblot analysis using antibodies to GAPDH or histone H3, as markers for soluble and chromatin-bound proteins, respectively, and with antibodies to TDG (Fig. 6A). This analysis revealed that wild type and mutant TDGs are equally distributed between soluble and chromatin-bound fractions in the absence of TETCD expression and that the relative distributions do not change significantly in the presence of TETCD. Because mutant TDGs do not accumulate on DNA in the presence of TETCD to a greater extent than wild type TDG, these results support the conclusion that sumoylation and SUMO binding are not required for release of TDG from AP sites or for efficient excision of 5caC under these in vivo conditions.
FIGURE 6.
Sumoylation and SUMO binding mutant TDGs are effectively turned over at AP sites in vivo. A, cells were transfected with constructs for wild type TDGor sumoylation or SUMO binding mutant TDGs in the presence or absence of TETCD, as indicated, and fractionated into chromatin-bound (Nonidet P-40-insoluble) or soluble (Nonidet P-40-soluble) protein fractions. The indicated proteins were detected by immunoblot analysis, with GAPDH used as a marker for soluble cytosolic proteins and histone H3 (H3) as a marker for chromatin-bound proteins. ** indicates sumoylated TDG. Whole cell lysate (WCL) was included as a control. Note that sumoylated TDG was apparently lost during fractionation because of incomplete inhibition of isopeptidases. B, cells were transfected with expression constructs for wild type and mutant TDG in the presence or absence of TETCD, as indicated, and effects on histone H2AX phosphorylation (γH2AX) were monitored by immunoblot analysis. The cells were treated with hydrogen peroxide as a positive control. C, cells were transfected with expression constructs for wild type or mutant TDGs and TETCD, as indicated, or treated with hydrogen peroxide as a positive control. The presence of single- and double-stranded DNA breaks was evaluated using single cell electrophoresis comet assays under alkaline conditions and fluorescence microscopy. D, quantitative analysis of DNA comet assays. The results represent means ± standard deviation from three independent experiments. **** indicates p < 0.0001 for H2O2 control versus transfected cells.
Unrepaired AP sites induce DNA damage, including DNA strand breaks (31). Thus, we also assayed for accumulation of AP sites in TDG and TETCD expressing cells by measuring levels of DNA breaks using immunoblot analysis for phosphorylated histone H2AX (γH2AX) and DNA comet assays. Modest, but comparable, increases in γH2AX levels were detected in cells co-expressing TETCD together with wild type and mutant TDGs, compared with cells expressing wild type and mutant TDGs alone (Fig. 6B). Similarly, comparable low levels of DNA strand breaks were detected by comet assays using cells transfected with TETCD together with wild type or mutant TDGs (Fig. 6, C and D). Comet assays were performed under alkaline conditions to detect both single and double-stranded breaks (32). The lack of increased DNA damage in cells expressing mutant TDG defective in sumoylation or SUMO binding is consistent with effective turnover of TDG and repair of TDG-generated AP sites. Collectively, these results further support the conclusion that sumoylation of TDG is not required for its enzymatic turnover under our in vivo assay conditions.
Discussion
Relieving TDG AP Site Product Inhibition
We developed an assay that takes advantage of the unique specificity of TDG for excising 5caC to investigate whether sumoylation is required for efficient TDG-mediated BER activity in vivo. Using this assay, we found that TDG mutants defective in covalent SUMO modification and noncovalent SUMO binding have repair activities indistinguishable from wild type TDG. Consistent with the activity of these mutants, we also found that perturbing the dynamics of TDG sumoylation, either by overexpressing SUMO1 or the SENP1 SUMO isopeptidase, had no detectable effects on TDG-mediated repair of 5caC. Thus, we conclude that sumoylation is not essential for TDG-mediated BER repair under our in vivo assay conditions. It should be noted that our assay involves overexpression of both TET and TDG and that further work is needed to validate our findings under conditions of endogenous protein expression. Nonetheless, because or findings also suggest that overexpression of TDG alone is not sufficient to overcome product inhibition (Fig. 2B), they indicate that other mechanisms affecting TDG turnover must be at play.
Previous studies showed that the mammalian AP endonuclease APE1 stimulates multiple turnover activity of TDG in vitro, for G/U and G/T mispairs (8, 9). We show here that APE1 also stimulates TDG catalytic turnover for G·5caC substrates. Indeed, the APE1-stimulated turnover of TDG (kcat+APE1) is ∼74% as fast as the maximal rate of base excision (kmax), indicating that APE1 dramatically enhances the rate of TDG product release and/or relieves product inhibition (by free AP-DNA). Importantly, the result also suggests that efficient processing of G·5caC substrates does not require SUMO modification of product-bound TDG. Because APE1 acts immediately downstream of TDG in the BER pathway, the coupling of TDG release with APE1 binding has the advantage of protecting AP sites from potentially harmful damage, such as strand breaks. In contrast, SUMO-stimulated product release could potentially expose abasic sites to damage, including strand breaks.
Regulation of TDG Sumoylation
Regardless of the effects of sumoylation on TDG function, molecular mechanisms likely regulate its modification both spatially and temporally in the cell. In the case of BER, it was proposed that sumoylation specifically targets TDG that is bound to its reaction product AP sites. In vitro studies, however, have shown that free and DNA-bound TDG are sumoylated at similar rates in the presence of E1 and E2 enzymes (15). Although E3 ligases may more precisely direct TDG sumoylation in vivo, the specific E3 ligases that affect TDG sumoylation, if any, have yet to be identified or investigated.
Isopeptidases also play important roles in controlling the status of protein sumoylation (33). Using both knockdown and overexpression studies, we identified SENP1 as a key regulator of TDG sumoylation levels. Consistent with the observed repair activity of TDG mutants, overexpression of SENP1 had no effect on 5caC repair despite severely limiting TDG sumoylation. We did, however, consistently observe effects of SENP1 depletion of TDG expression levels, suggesting a possible role for sumoylation in affecting TDG stability. TDG stability and expression is controlled in a cell cycle-dependent manner (34–36), but whether sumoylation plays a direct role in this regulation remains to be determined.
The N-terminal domains of the SENP1 and SENP2 isopeptidases determine their subcellular localizations and are predicted to also influence substrate specificities (24). Surprisingly, we found that it is the catalytic domain of SENP1 that mediates TDG recognition rather than the N-terminal domain. Further investigation of the interactions between SENP1 and TDG could therefore provide valuable and unexpected insights into substrate recognition by SUMO isopeptidases.
Other Effects of TDG Sumoylation
Among the most commonly observed effects of sumoylation on target proteins are effects on protein-protein interaction, subcellular localization, and protein stability (1, 2). As reported by others (26, 27), we found that TDG localizes to PML nuclear bodies and that its localization to these bodies is inhibited by a mutation in the SIM (TDGE310Q) that disrupts SUMO binding. This finding has been interpreted as an indication of functional interactions between TDG and PML that are regulated at the level of sumoylation (27). The exact functions of PML nuclear bodies and the possible significance of interactions between TDG and PML, however, remain unknown. Our findings indicate that the TDGE310Q mutant effectively repairs 5caC despite defects in association with PML nuclear bodies. Thus, we conclude that the association of TDG with PML nuclear bodies is not critical for its function in BER.
Many of the factors associated with PML nuclear bodies function in transcription regulation (37), and TDG itself has been implicated as both a positive and negative regulator of transcription (6). Consistent with this function, TDG associates with multiple transcription factors, including RAR and RXR (38), ER (39), CBP/p300 (40), and LEF1/TCF (41). Notably, each of these factors is also regulated through sumoylation (42–45). Thus, it seems very likely that sumoylation and SUMO binding could modulate the interaction of TDG with transcription factors and its functions in regulating gene expression. These interactions and functions could in turn be affected through SUMO-dependent associations of TDG with PML nuclear bodies, as demonstrated for other transcription factors (46). Functional assays evaluating effects of TDG sumoylation and SUMO binding on transcription also represent important future studies.
In conclusion, TDG represents a fascinating and important sumoylation substrate. Past studies of TDG have provided novel conceptual insights into effects of sumoylation on target proteins, and it is anticipated that future studies will continue to reveal important findings both on the effects of sumoylation and the mechanisms underlying its regulation.
Author Contributions
M. J. M. and A. C. D. designed the study and consulted all members of the research team throughout its completion. D. M. developed and performed all assays for detection of 5caC repair in vivo, SUMO isopeptidase analysis, and evaluation of TDG turnover and DNA damage. C. T. C. provided plasmid constructs and expert advice on expression and analysis of TDG and performed all enzymatic assays involving TDG stimulation by APE1. W.-C. Y. provided assistance and expert advice on all transfection studies and related cell culture work. M. J. M. and D .M. wrote the original draft of the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
The plasmid for expressing TET1 catalytic domain was generously provided by R. Kohli (University of Pennsylvania). We are grateful to members of the Matunis and Drohat laboratories for helpful discussions during the course of these studies.
This work was supported in part by National Institutes of Health Grants R01-GM72711 (to A. C. D.) and R01-GM060980 (to M. J. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TDG
- thymine-DNA glycosylase
- SUMO
- small ubiquitin-like modifier
- BER
- base excision repair
- AP
- apurinic/apyrimidinic
- 5caC
- 5-carboxylcytosine
- TET
- ten-eleven translocation
- SENP
- sentrin protease
- SIM
- SUMO-interacting motif
- PML
- promyelocytic leukemia
- APE1
- apurinic/apyrimidinic endonuclease
- gDNA
- genomic DNA.
References
- 1. Jackson S. P., and Durocher D. (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 [DOI] [PubMed] [Google Scholar]
- 2. Raman N., Nayak A., and Muller S. (2013) The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma 122, 475–485 [DOI] [PubMed] [Google Scholar]
- 3. Jentsch S., and Psakhye I. (2013) Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 47, 167–186 [DOI] [PubMed] [Google Scholar]
- 4. Bellacosa A., and Drohat A. C. (2015) Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites. DNA Repair 32, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohli R. M., and Zhang Y. (2013) TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sjolund A. B., Senejani A. G., and Sweasy J. B. (2013) MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 743–744, 12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hardeland U., Bentele M., Lettieri T., Steinacher R., Jiricny J., and Schär P. (2001) Thymine DNA glycosylase. Prog. Nucleic Acid Res. Mol. Biol. 68, 235–253 [DOI] [PubMed] [Google Scholar]
- 8. Waters T. R., Gallinari P., Jiricny J., and Swann P. F. (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 274, 67–74 [DOI] [PubMed] [Google Scholar]
- 9. Fitzgerald M. E., and Drohat A. C. (2008) Coordinating the initial steps of base excision repair: apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. J. Biol. Chem. 283, 32680–32690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baba D., Maita N., Jee J. G., Uchimura Y., Saitoh H., Sugasawa K., Hanaoka F., Tochio H., Hiroaki H., and Shirakawa M. (2005) Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435, 979–982 [DOI] [PubMed] [Google Scholar]
- 11. Baba D., Maita N., Jee J. G., Uchimura Y., Saitoh H., Sugasawa K., Hanaoka F., Tochio H., Hiroaki H., and Shirakawa M. (2006) Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J. Mol. Biol. 359, 137–147 [DOI] [PubMed] [Google Scholar]
- 12. Hardeland U., Steinacher R., Jiricny J., and Schär P. (2002) Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21, 1456–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smet-Nocca C., Wieruszeski J. M., Léger H., Eilebrecht S., and Benecke A. (2011) SUMO-1 regulates the conformational dynamics of thymine-DNA glycosylase regulatory domain and competes with its DNA binding activity. BMC Biochem. 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steinacher R., and Schär P. (2005) Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 15, 616–623 [DOI] [PubMed] [Google Scholar]
- 15. Coey C. T., Fitzgerald M. E., Maiti A., Reiter K. H., Guzzo C. M., Matunis M. J., and Drohat A. C. (2014) E2-mediated Small Ubiquitin-like Modifier (SUMO) Modification of thymine DNA glycosylase is efficient but not selective for the enzyme-product complex. J. Biol. Chem. 289, 15810–15819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krokan H. E., and Bjørås M. (2013) Base excision repair. Cold Spring Harb. Perspect. Biol. 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He Y. F., Li B. Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C. X., Zhang K., He C., and Xu G. L. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maiti A., and Drohat A. C. (2011) Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen L., Wu H., Diep D., Yamaguchi S., D'Alessio A. C., Fung H. L., Zhang K., and Zhang Y. (2013) Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malik S. S., Coey C. T., Varney K. M., Pozharski E., and Drohat A. C. (2015) Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA. Nucleic Acids Res. 43, 9541–9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morgan M. T., Bennett M. T., and Drohat A. C. (2007) Excision of 5-halogenated uracils by human thymine DNA glycosylase. Robust activity for DNA contexts other than CpG. J. Biol. Chem. 282, 27578–27586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morgan M. T., Maiti A., Fitzgerald M. E., and Drohat A. C. (2011) Stoichiometry and affinity for thymine DNA glycosylase binding to specific and nonspecific DNA. Nucleic Acids Res. 39, 2319–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang L., Lu X., Lu J., Liang H., Dai Q., Xu G. L., Luo C., Jiang H., and He C. (2012) Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 8, 328–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cubeñas-Potts C., Goeres J. D., and Matunis M. J. (2013) SENP1 and SENP2 affect spatial and temporal control of sumoylation in mitosis. Mol. Biol. Cell 24, 3483–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matunis M. J., Coutavas E., and Blobel G. (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohan R. D., Rao A., Gagliardi J., and Tini M. (2007) SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment. Mol. Cell. Biol. 27, 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takahashi H., Hatakeyama S., Saitoh H., and Nakayama K. I. (2005) Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J. Biol. Chem. 280, 5611–5621 [DOI] [PubMed] [Google Scholar]
- 28. Baldwin M. R., and O'Brien P. J. (2010) Nonspecific DNA binding and coordination of the first two steps of base excision repair. Biochemistry 49, 7879–7891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mikolajczyk J., Drag M., Békés M., Cao J. T., Ronai Z., and Salvesen G. S. (2007) Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J. Biol. Chem. 282, 26217–26224 [DOI] [PubMed] [Google Scholar]
- 30. Goeres J., Chan P. K., Mukhopadhyay D., Zhang H., Raught B., and Matunis M. J. (2011) The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107–160 nucleoporin subcomplex. Mol. Biol. Cell 22, 4868–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boiteux S., and Guillet M. (2004) Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 3, 1–12 [DOI] [PubMed] [Google Scholar]
- 32. Calini V., Urani C., and Camatini M. (2002) Comet assay evaluation of DNA single- and double-strand breaks induction and repair in C3H10T1/2 cells. Cell Biol. Toxicol. 18, 369–379 [DOI] [PubMed] [Google Scholar]
- 33. Nayak A., and Müller S. (2014) SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 15, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hardeland U., Kunz C., Focke F., Szadkowski M., and Schär P. (2007) Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 35, 3859–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shibata E., Dar A., and Dutta A. (2014) CRL4Cdt2 E3 ubiquitin ligase and proliferating cell nuclear antigen (PCNA) cooperate to degrade thymine DNA glycosylase in S phase. J. Biol. Chem. 289, 23056–23064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Slenn T. J., Morris B., Havens C. G., Freeman R. M. Jr., Takahashi T. S., and Walter J. C. (2014) Thymine DNA glycosylase is a CRL4Cdt2 substrate. J. Biol. Chem. 289, 23043–23055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lallemand-Breitenbach V., and de Thé H. (2010) PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Um S., Harbers M., Benecke A., Pierrat B., Losson R., and Chambon P. (1998) Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 273, 20728–20736 [DOI] [PubMed] [Google Scholar]
- 39. Chen D., Lucey M. J., Phoenix F., Lopez-Garcia J., Hart S. M., Losson R., Buluwela L., Coombes R. C., Chambon P., Schär P., and Ali S. (2003) T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor α. J. Biol. Chem. 278, 38586–38592 [DOI] [PubMed] [Google Scholar]
- 40. Tini M., Benecke A., Um S. J., Torchia J., Evans R. M., and Chambon P. (2002) Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell 9, 265–277 [DOI] [PubMed] [Google Scholar]
- 41. Jia Y., Nie F., Du A., Chen Z., Qin Y., Huang T., Song X., and Li L. (2014) Thymine DNA glycosylase promotes transactivation of β-catenin/TCFs by cooperating with CBP. J. Mol. Cell Biol. 6, 231–239 [DOI] [PubMed] [Google Scholar]
- 42. Burrage P. S., Schmucker A. C., Ren Y., Sporn M. B., and Brinckerhoff C. E. (2008) Retinoid X receptor and peroxisome proliferator-activated receptor-γ agonists cooperate to inhibit matrix metalloproteinase gene expression. Arthritis Res. Ther. 10, R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Girdwood D., Bumpass D., Vaughan O. A., Thain A., Anderson L. A., Snowden A. W., Garcia-Wilson E., Perkins N. D., and Hay R. T. (2003) P300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11, 1043–1054 [DOI] [PubMed] [Google Scholar]
- 44. Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., and Grosschedl R. (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15, 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sentis S., Le Romancer M., Bianchin C., Rostan M. C., and Corbo L. (2005) Sumoylation of the estrogen receptor α hinge region regulates its transcriptional activity. Mol. Endocrinol. 19, 2671–2684 [DOI] [PubMed] [Google Scholar]
- 46. Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., and Shih H. M. (2006) Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]