Abstract
Perchlorate is an important ion on both Earth and Mars. Perchlorate reductase (PcrAB), a specialized member of the dimethylsulfoxide reductase superfamily, catalyzes the first step of microbial perchlorate respiration, but little is known about the biochemistry, specificity, structure, and mechanism of PcrAB. Here we characterize the biophysics and phylogeny of this enzyme and report the 1.86-Å resolution PcrAB complex crystal structure. Biochemical analysis revealed a relatively high perchlorate affinity (Km = 6 μm) and a characteristic substrate inhibition compared with the highly similar respiratory nitrate reductase NarGHI, which has a relatively much lower affinity for perchlorate (Km = 1.1 mm) and no substrate inhibition. Structural analysis of oxidized and reduced PcrAB with and without the substrate analog SeO32− bound to the active site identified key residues in the positively charged and funnel-shaped substrate access tunnel that gated substrate entrance and product release while trapping transiently produced chlorate. The structures suggest gating was associated with shifts of a Phe residue between open and closed conformations plus an Asp residue carboxylate shift between monodentate and bidentate coordination to the active site molybdenum atom. Taken together, structural and mutational analyses of gate residues suggest key roles of these gate residues for substrate entrance and product release. Our combined results provide the first detailed structural insight into the mechanism of biological perchlorate reduction, a critical component of the chlorine redox cycle on Earth.
Keywords: enzyme mechanism, iron-sulfur protein, molybdenum, structure-function, x-ray crystallography, DMSO reductase superfamily, Mo-bis-MGD, perchlorate reductase, substrate access tunnel, substrate inhibition
Introduction
Perchlorate has recently gained notoriety because of the identified environmental prevalence of this exotic ion (1). Its recently documented widespread distribution in both terrestrial and extraterrestrial planetary and non-planetary samples suggests that perchlorate is prevalent throughout our solar system (1, 2). To date, perchlorate is primarily recognized as a groundwater contaminant resulting from Cold War munition manufacturing (3). However, recent studies have additionally determined a natural atmospheric source (1, 4). Widespread perchlorate abundance has also been identified on Mars (5–7), on our moon, and in meteorites (2). On Mars, the unique perchlorate physical chemistry is responsible for maintaining water in the liquid state regardless of the environmental extremes (8). Furthermore, Martian perchlorate may represent a significant energy and oxygen resource for human exploration (9). Although the exact abiotic mechanisms of natural perchlorate neogenesis are not completely understood, there are no known biogenic mechanisms. In contrast, terrestrial perchlorate diagenesis primarily occurs biologically through microbial respiration (10).
Dissimilatory (per)chlorate-reducing bacteria can use perchlorate or chlorate (collectively (per)chlorate) as a terminal electron acceptor during anaerobic respiration (11). Canonical dissimilatory (per)chlorate-reducing bacteria utilize a highly conserved pathway that is phylogenetically distributed through horizontal gene transfer (3, 12, 13). Perchlorate is sequentially reduced to chlorite by the perchlorate reductase (PcrAB), which is then dismutated into chloride and oxygen by the chlorite dismutase (Cld). The biogenic oxygen is respired by the same microorganism, generally through the use of a high affinity cytochrome cbb3 oxidase (3). Due to oxygen biogenesis, these unique chemotrophs represent a distinctive metabolic juxtaposition between aerobic and anaerobic metabolisms. Although an impressive body of literature focuses on the biochemistry, genetics, and evolution of the Cld, relatively little is known about the PcrAB. We have shown that PcrAB is an oxygen-sensitive heterodimeric periplasmic enzyme that catalyzes the 2e− reduction of perchlorate to chlorate, which is further reduced by 2e− to form chlorite (3, 14). The catalytic subunit PcrA is phylogenetically closely related to the NarG subunit of nitrate reductase (15). Both proteins are in the DMSO4 reductase superfamily and are strongly associated with intermediate electron-shuttling protein subunits (PcrB and NarH, respectively). Given the important and ancient roles of [Fe-S] clusters in biology and electron transfer, it is furthermore important to consider the detailed structural relationships of the five [Fe-S] clusters in PcrAB and NarGH.
Here we perform an unprecedented structure and function characterization of PcrAB. The crystal structures revealed all metal ion cofactors along with a novel gating mechanism at the active site that evidently endow the enzyme with unique biophysical characteristics. These, in addition to comparative phylogenetic and mutational studies, identified the evolutionary basis for this enzyme and revealed the unique and distinguishing features between PcrAB and the closely related NarGH respiratory complex.
Experimental Procedures
Expression and Purification of PcrAB
Cells of wild-type or mutant Azospira suillum PS were initially grown aerobically from single colonies in acetate, lactate, pyruvate growth medium (12) in 5-ml cultures and incubated overnight at 37 °C at 250 rpm. This culture was then used to inoculate 1 liter of the same growth medium and incubated at the previously listed conditions. Anaerobic cultures were prepared by adding 100 ml of the overnight aerobic culture to sterile 1-liter glass bottles, and fresh sterile acetate, lactate, pyruvate growth medium containing 5 mm NaNO3, 10 mm NaClO3, and 10 mm NaClO4 was added until the bottles were completely filled with no headspace. The sealed bottles were then incubated without shaking at 37 °C for 18–24 h. Cultures changed from yellow to pink when all of the electron acceptors were consumed. This typically yielded 5–6 g of wet cell paste/ liter of anaerobic culture. Cells were harvested anaerobically by filling centrifuge tubes with the anaerobic cultures in an anaerobic chamber (Coy Laboratory Products), and the tubes were sealed, centrifuged at 10,000 × g for 10 min, and returned to the anaerobic chamber where the supernatant was decanted. The pink cell pellets were resuspended in anaerobic buffer A (50 mm Tris, pH 8.0, 10% (v/v) glycerol) to a density of 0.4 g of cells/ml, supplemented with 1 mg/ml lysozyme and 0.1 mg/ml DNase I (Sigma), sealed in glass bottles, and frozen at −80 °C until needed.
PcrAB was purified anaerobically using techniques described previously with some modifications (16). Frozen cells were thawed, lysed under a nitrogen stream with an EmulsiFlex-C3 cell homogenizer (Avestin), and centrifuged at 150,000 × g for 45 min. The clarified lysate was kept anaerobic until it was loaded onto three columns connected in tandem and equilibrated with anaerobic buffer A using an ÄKTA Explorer FPLC (GE Healthcare). The tandem series of columns consisted of a 1 × 20-cm S-Sepharose HP column (GE Healthcare), a 2.5 × 40-cm Q-Sepharose FF column (GE Healthcare), and finally a 2.5 × 5-cm CaPure-HA® hydroxyapatite column (Tosoh Bioscience). Once the sample was loaded and the UV/visible traces on the FPLC detector returned to baseline, the first two columns were removed, and PcrAB was eluted from the hydroxyapatite column using a 300-ml linear gradient to buffer B (buffer A with 400 mm NaH2PO4). PcrAB typically eluted at 200–250 mm NaH2PO4. Fractions containing activity as described below were pooled and concentrated in sealed centrifugal concentrators with 100-kDa-molecular mass-cutoff filters (Amicon) to <800 μl. The concentrated sample was loaded onto a HiPrep Sephacryl S-300 HR 16/60 size exclusion column (GE Healthcare) that was equilibrated with anaerobic buffer C (buffer A with 150 mm NaCl) at a flow rate of 0.25 ml/min. PcrAB typically eluted at 50–60 ml. Fractions shown to be pure by SDS-PAGE were pooled and concentrated as described previously to 7 mg/ml, snap frozen in gasketed cryovials with liquid N2, and stored at −80 °C until use. Protein concentrations were determined via the Bradford assay (Sigma). PcrAB containing the PcrA W461E mutation was purified in the same manner as wild-type PcrAB.
Steady-state Kinetics
The cNarGHI from Escherichia coli was a generous gift from the Weiner laboratory (University of Alberta). Activity assays of PcrAB and EccNarGHI were performed in an anaerobic chamber by monitoring the substrate-dependent oxidation of NADH (Sigma) with phenazine methosulfate (Sigma) as an electron shuttle (14). 450 μm NADH was incubated with 100 μm phenazine methosulfate and protein sample in 50 mm MOPS, pH 7.2, and reactions were initiated by the addition of substrate. For reactions containing EccNarGHI, the buffer was supplemented with 0.01% (v/v) Thesit detergent (Sigma) to prevent the enzyme from precipitating. NADH oxidation rates were determined by monitoring the changes in absorbance at 340 nm (A340) using an Infinite F200 Pro plate reader (Tecan) at 25 °C. Reaction rates were determined by fitting the data in Origin 6.0 (MicroCal). Although the EccNarGHI Km value for nitrate established with our NADH/phenazine methosulfate assay was consistent with previous reports, the kcat values were lower (17). This suggests that phenazine methosulfate is not as efficient as an electron donor to EccNarGHI as other electron donors, such as methyl viologen, benzyl viologen, and plumbagin (17, 18), but it does not affect the affinity of the enzyme for its primary substrate.
Crystallization, Data Collection, and Structure Determination of PcrAB
PcrAB (7 mg/ml) was mixed with an equal volume of the crystallization solution (0.1 m Tris, pH 8.25, 20% (w/v) PEG 6000) using the hanging drop vapor diffusion method in the anaerobic chamber. All crystallization buffers were degassed before use, and crystallization trays were stored in the anaerobic chamber for at least 3 days before use. The brown rod-shaped crystals grew over 6 weeks at ∼20 °C. For W461E PcrA of PcrAB mutant, crystals were grown in 23% (w/v) PEG 3350 and 0.1 m Tris, pH 8.5, with an equal volume of protein and crystallization solution. The hexagonal plate crystals appeared after 1 month at ∼20 °C in the anaerobic chamber. A single crystal of wild-type or mutant PcrAB was transferred into crystallization solution supplemented with cryoprotectant of 20% (v/v) and subsequently 30% (v/v) ethylene glycol for wild-type PcrAB or 25% (v/v) ethylene glycol for mutant PcrAB. The crystal was mounted on a loop on a B3S base (MiTeGen) and inserted in a MicroRT capillary tube (MiTeGen) with 50 μl of reservoir solution in the sealed end of the tube to prevent dehydration. The mounted crystal was then transported outside of the anaerobic chamber and immediately snap frozen in liquid N2.
The reduced PcrAB crystal (Protein Data Bank code 5CH7) was obtained by soaking 4 mm Na2SO3 in the presence of 1.2 mm reduced methyl viologen and 2.8 mm sodium dithionite. This solution was added to a drop containing a single oxidized PcrAB crystal (crystallized in 20% (w/v) PEG 6000, 0.1 m Tris, pH 8.5) in the anaerobic chamber for 18 h at ∼20 °C. Reduced methyl viologen was prepared as a 10 mm stock solution using the reservoir solution and a small amount of zinc powder as a reductant, which was subsequently removed via centrifugation at 20,000 × g for 1 min in the anaerobic chamber. For selenite-bound PcrAB (Protein Data Bank code 5CHC), a PcrAB crystal was soaked with 4 mm Na2SeO3 in the presence of reduced methyl viologen and sodium dithionite for 18 h at ∼20 °C as described above. The reduced PcrAB crystals were then transferred into crystallization solution supplemented with the aforementioned reducing agents and cryoprotectants. The mounting procedure for the reduced crystals was the same as described above.
The x-ray diffraction data of PcrAB crystals were collected at BL12.3.1 SIBYLS beamline (sibyls.als.lbl.gov) at the Advanced Light Source (19). For oxidized PcrAB, two data sets from different spots of the same crystal were collected at 100 K and integrated using the XDS package (20). The data were then merged and scaled using AIMLESS in the CCP4 suite (21). POINTLESS indicates a space group of P212121 for three molecules of PcrAB per asymmetric unit for oxidized and reduced PcrAB.
The oxidized PcrAB structure was determined via molecular replacement using Phaser in the Phenix suite (22) using ethylbenzene dehydrogenase (Protein Data Bank code 2IVF) α- and β-subunits as a search model (23). The molecular replacement solution was then processed with AutoBuild in Phenix (22). The resulting model was manually fit by Coot (24) and refined by Phenix.refine with non-crystallographic symmetry restraints.
The reduced PcrAB structures and W461E PcrA of PcrAB mutant structure were determined by molecular replacement using oxidized PcrAB as a search model. The molecular replacement solutions of reduced PcrAB were processed the same way as oxidized PcrAB except the twin law (−h−l,k,h) was applied for the mutant PcrAB (estimated twin fraction, 0.16) during refinement. The x-ray data collection and refinement statistics are listed in Table 2.
TABLE 2.
X-ray data collection and refinement statistics for the PcrAB crystal structures
CC, cross-correlation; ASU, asymmetric unit; r.m.s.d., root mean square deviation; ALS, Advanced Light Source.
| PcrAB oxidized | PcrAB reduced-Phe164 gate switch | PcrAB reduced-substrate analog bound | PcrAB mutant-W461E PcrA | |
|---|---|---|---|---|
| Data collection | ALS-BL12.3.1 | ALS-BL12.3.1 | ALS-BL12.3.1 | ALS-BL12.3.1 |
| Wavelength (Å) | 1.1158 | 0.9793 | 0.9793 | 1.0062 |
| Space group | P212121 | P212121 | P212121 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 132.82, 175.50, 193.16 | 132.88, 175.67, 193.28 | 133.93, 176.02, 193.69 | 135.59, 253.13, 135.80 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 119.77, 90.00 |
| Resolution (Å)a | 48.35–1.86 (1.89–1.86) | 48.38–2.20 (2.24–2.20) | 48.66–2.38 (2.42–2.38) | 48.32–2.40 (2.44–2.40) |
| Observationsa | 7,305,807 (365,469) | 3,407,133 (169,632) | 2,724,751 (135,221) | 1,141,127 (57,773) |
| Unique observationsa | 372,024 (18,112) | 228,466 (11,210) | 183,135 (8,943) | 306,180 (15,230) |
| Rmergea | 0.178 (1.623) | 0.205 (1.619) | 0.222 (1.607) | 0.145 (0.824) |
| Rpima | 0.059 (0.528) | 0.079 (0.620) | 0.086 (0.618) | 0.136 (0.758) |
| Mean I/σIa | 13.2 (2.1) | 13.6 (2.1) | 13.0 (2.0) | 6.8 (2.0) |
| Completeness (%)a | 99.1 (97.9) | 99.9 (99.7) | 100 (100) | 99.2 (99.8) |
| Multiplicitya | 19.6 (20.2) | 14.9 (15.1) | 14.9 (15.1) | 3.7 (3.8) |
| CC(1/2)a | 0.998 (0.759) | 0.997 (0.683) | 0.996 (0.706) | 0.985 (0.521) |
| Refinement | ||||
| Rwork/Rfree (%)b | 20.0/23.6 | 17.8/23.3 | 17.0/22.7 | 19.5/24.8 |
| mol/ASU | 6 | 6 | 6 | 12 |
| No. of residues/ASU | 3,664 | 3,661 | 3,664 | 7,344 |
| r.m.s.d. bond length (Å) | 0.010 | 0.011 | 0.010 | 0.003 |
| r.m.s.d. bond angle (°) | 1.436 | 1.466 | 1.380 | 0.657 |
| Average B-factor | 29.29 | 35.0 | 36.6 | 38.25 |
| Protein | 29.16 | 35.1 | 36.8 | 38.43 |
| Ligands | 21.90 | 29.8 | 32.0 | 36.03 |
| Solvent | 31.94 | 34.9 | 34.9 | 35.41 |
| Ramachandran (%) | ||||
| Favored | 96.06 | 95.05 | 95.02 | 93.88 |
| Allowed | 3.69 | 4.70 | 4.62 | 5.48 |
| Outliers | 0.25 | 0.25 | 0.36 | 0.64 |
| Protein Data Bank code | 4YDD | 5CH7 | 5CHC | 5E7O |
a Values in parentheses are the statistics for the highest resolution shell of data.
b Rwork = (Σ‖Fobs| − |Fcalc‖)/Σ|Fobs| where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is calculated the same as Rwork, but 5% of the data were excluded from the refinement.
The protein data bank codes are 4YDD (oxidized PcrAB), 5CH7 (reduced PcrAB-Phe164 gate switch), 5CHC (reduced PcrAB-SeO32− bound), and 5E7O (PcrAB mutant-W461E PcrA). All the structure figures are depicted using Chimera (25). The hydrophobicity scale is based on Kyte and Doolittle (26).
PcrAB Site-directed Mutagenesis
The pcrA gene was amplified from A. suillum PS genomic DNA using PcrA_AD-F and PcrA_AD-R primers (supplemental Table S1). This PCR product was purified and combined with the pCR®-Blunt II-TOPO® vector using the Zero Blunt® TOPO PCR cloning kit (Life Technologies) and transformed into XL1 Blue chemically competent E. coli (Stratagene). Positive transformants were grown in LB supplemented with 50 μg/ml kanamycin, and plasmid DNA was isolated using the QIAprep Spin Miniprep kit (Qiagen). This plasmid was used as a template for PCR-based site-directed mutagenesis with primers specific to particular point mutations (supplemental Table S1) (27). These PCRs were digested with DpnI (New England Biolabs) and transformed again into XL1 Blue cells. Positive transformants were grown, plasmid DNA was isolated as done previously, and positive mutations were screened using Sanger sequencing. Plasmids containing the correct mutations were digested with ApaI and BamHI (New England Biolabs) along with the pNPTS138 suicide vector (12), purified, ligated using T4 DNA ligase (New England Biolabs), and transformed into XL1 Blue cells. Transformants were screened via colony PCR, and pNPTS138 plasmids containing correctly mutated pcrA were isolated and confirmed by Sanger sequencing. Allelic replacement of pcrA with the point mutants was performed as described previously (12). Growth curves of all strains were performed anaerobically using a Sunrise 96-well plate reader (Tecan) according to previously published protocols (12). Chlorate accumulation experiments were performed in anaerobic bottles with 10 mm perchlorate and 10 mm acetate and monitored via OD600 and ion chromatography using an ICS1500 (Dionex) outfitted with an AS25 column (Thermo Scientific).
Phylogenetic Analysis
Phylogenetic analysis of the catalytic subunit of the perchlorate reductase (PcrA) was performed using the computer cluster at the University of California Berkeley QB3 Computational Genomics Resource Laboratory. The molybdopterin oxidoreductase protein family (Pfam PF00384) formed the starting alignment for a phylogenetic analysis with FastTree2 (version 2.1.7) (28). From the resulting tree, a clade consisting of DmsA and DorA was chosen as an appropriate outgroup to known PcrA proteins. Non-basal sequences were combined with the top 100 blastp hits from both NCBI and Integrated Microbial Genomes (IMG) databases. CD-HIT (29) was used to cluster sequences at a threshold of 0.8. The remaining sequences were aligned using MUSCLE (30) and trimmed with trimAL (31), and these informative positions were used to reconstruct the phylogeny with FastTree2. Salient features of clades, including known function of members, number of sequences containing a predicted twin arginine translocation signal (32), and sequence motifs (33) of proposed active site residues, were visualized on the tree.
Results
Comparative Steady-state Kinetics of PcrAB Reveals Unique Features
PcrAB kinetic analysis revealed nearly identical steady-state parameters for (per)chlorate at substrate concentrations below 200 μm (Table 1A). Although chlorate followed typical Michaelis-Menten kinetics (Fig. 1A), perchlorate showed significant substrate inhibition above 200 μm with an estimated inhibition constant (KI) of ∼7.5 mm (Fig. 1B). Interestingly, a critical inhibitory concentration for perchlorate was not noted over the range analyzed (0–200 mm). PcrAB also mediated turnover of NO3−, BrO3−, and IO3− (Table 1A), but no activity was observed with SO42−, SO32−, SeO42−, SeO32−, AsO43−, AsO33−, NO2−, WO42−, or MoO42−. Neither BrO4− nor IO4− could be investigated due to their reactive nature. The catalytic efficiencies (kcat/Km) for all substrates of PcrAB were within the same order of magnitude (Table 1A), suggesting no evolutionary specificity for any of these oxyanions.
TABLE 1.
Steady-state kinetic parameters and Bond lengths and molecular dipoles of substrates
Steady-state kinetic parameters show relatively high substrate affinity for PcrAB in comparison with EccNarGHI. A, wild-type PcrAB; B, EccNarGHI; C, PcrA W461E. The steady-state kinetics were determined using the NADH/PMS assay. Error represent one standard deviation of triplicate assays. D, Bond lengths and molecular dipoles of substrates (52). N.S., not a substrate. *, Iodate was also previously determined to not be a substrate of EccNarGHI (17). †, Estimated values due to the solubility limitations of NaIO3.
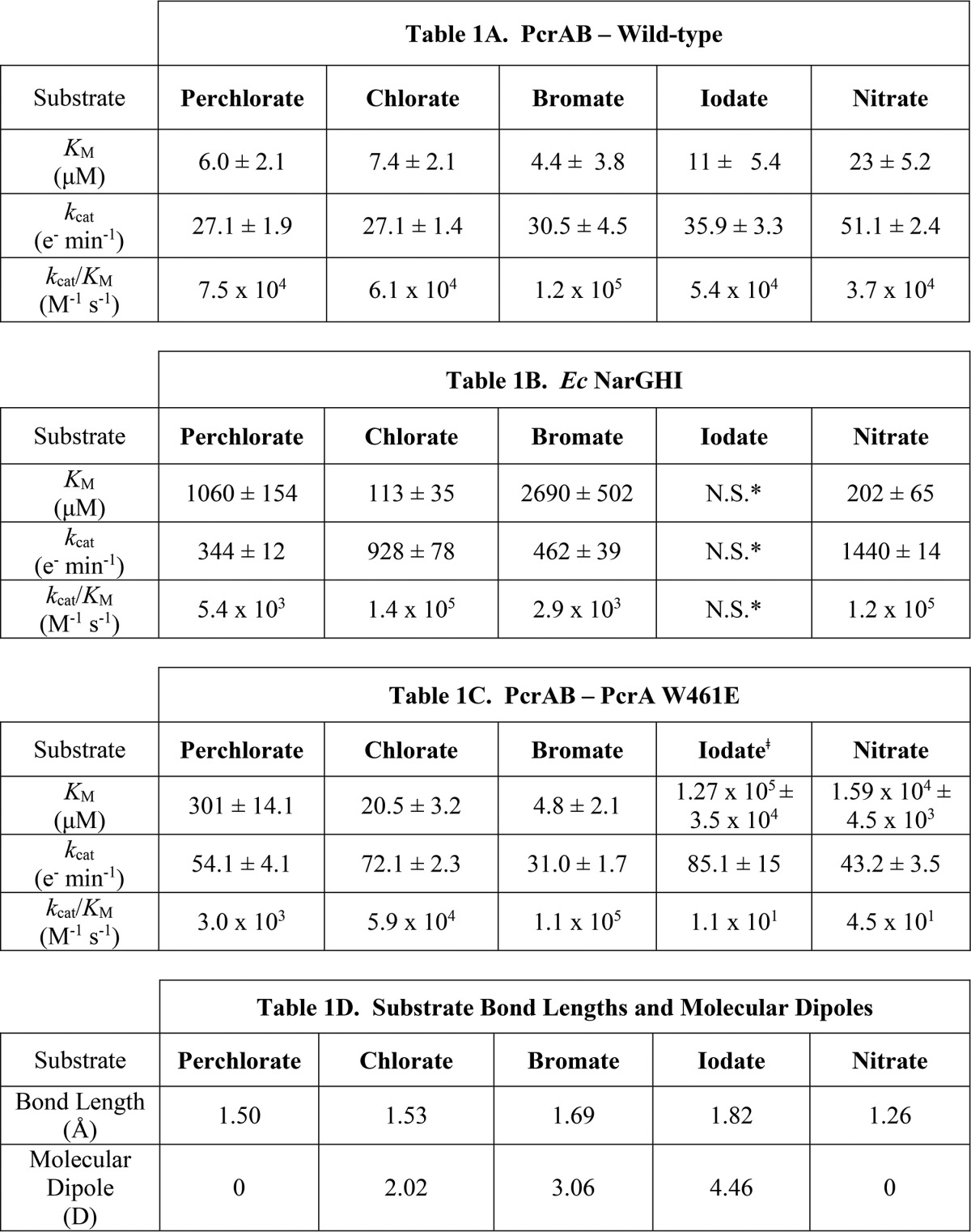
FIGURE 1.

Steady-state kinetic curves for PcrAB and EccNarGHI for chlorate (A), perchlorate (B), and perchlorate (C) at low substrate concentrations (<200 μm). Error bars represent one standard deviation of triplicate assays.
E. coli cytoplasm-facing nitrate reductase (EccNarGHI) also had activity toward (per)chlorate (Table 1B) albeit with very different kinetics. EccNarGHI had a much lower affinity for all substrates analyzed relative to PcrAB and showed ∼200-fold lower affinity toward perchlorate (PcrAB Km = 6 μm; EccNarGHI Km = 1.1 mm). EccNarGHI catalytic efficiencies were inversely proportional to substrate size, being lower for the larger oxyhaloanions (ClO4− and BrO3−) relative to the smaller NO3− and ClO3−. No activity was observed toward IO3− (Table 1B). In contrast to PcrAB, EccNarGHI was uninhibited by increasing concentrations of perchlorate and displayed typical Michaelis-Menten kinetics throughout the range tested (Fig. 1B). Surprisingly, although not considered its natural substrate, EccNarGHI had a much higher maximum perchlorate turnover rate than PcrAB with a kcat nearly 10-fold higher (Table 1, A and B). At ∼125 μm perchlorate, both PcrAB and EccNarGHI had nearly identical turnover rates, whereas below that PcrAB had a turnover rate up to 5-fold greater than EccNarGHI (Fig. 1C), suggesting that it evolved to take advantage of the low concentrations of (per)chlorate naturally found in the environment (4).
Crystal Structure of Oxidized PcrAB Reveals a Unique Gated Active Site
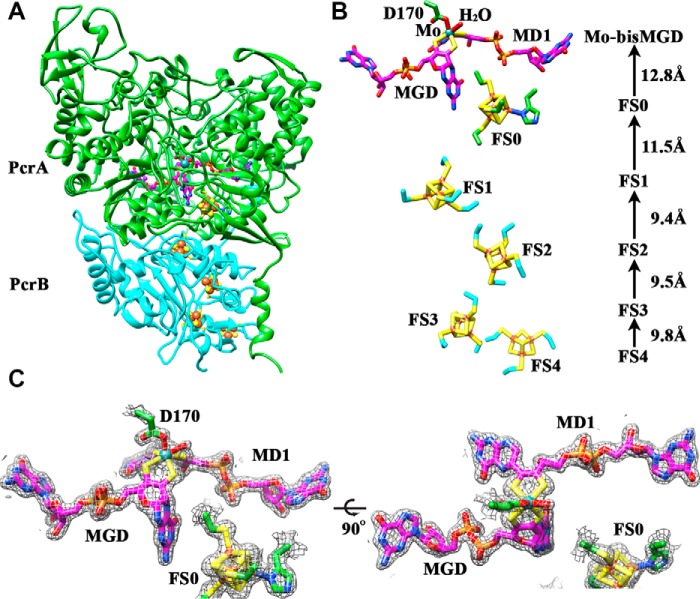
To better understand why PcrAB is more efficient at low perchlorate concentrations, we crystallized the purified enzyme anaerobically and determined the crystal structure of oxidized PcrAB at 1.86-Å resolution (Fig. 2A) along with three reduced or mutant crystal structures (Table 2). PcrAB was crystallized in space group P212121 and formed a trimer of heterodimers. The PISA assembly (34) indicated that PcrAB was stable as a heterodimer with a buried area of 14,000 Å2. The oxidized PcrA structure, where the oxidation state of molybdenum is most likely +5 or +6, resembled other DMSO reductase superfamily enzymes (35–37). PcrAB was composed of one Mo-bis-MGD cofactor and one [4Fe-4S] cluster (FS0) for PcrA and three [4Fe-4S] clusters (FS1–3) and one [3Fe-4S] cluster (FS4) for PcrB with all metal atoms and cofactors having an occupancy of 1 (Fig. 2, A and B). All [Fe-S] clusters were coordinated with Cys residues except the FS0 cluster, which had one iron atom coordinated by the His32 residue. A mutation of this His to Cys in EccNarGHI resulted in loss of activity toward nitrate (38). The molybdenum atom was coordinated in a trigonal prismatic manner with four proximal thiolates of the bis-MGD cofactor (average Mo–S bond distance of 2.4 Å) along with a Asp170 residue and an additional oxygen atom (either an oxo, hydroxyl, or associated water molecule) in the two distal positions (Fig. 2B). The Mo–O bond distances were ∼2.0–2.1 and 2.4–3.1 Å for Asp170 and the additional oxygen atom, respectively, in the three PcrA molecules in the asymmetric unit. The cyclization states of the bis-MGD cofactor were in a configuration with one in a bicyclic dihydropterin and the other in a tricyclic pyranopterin structure (Fig. 2C). Most DMSO reductase superfamily members possess both pterins in the tricyclic state; however, the bicyclic and tricyclic pterins have been noted in NarGHI and ethylbenzene dehydrogenase structures (23, 35), suggesting an interconversion mechanism that controls pterin cyclization states (39). These structural features of PcrAB are similar to EccNarGH (35, 36), which has a root mean square deviation of 0.974 Å between 710 aligned Cα atoms. The positions of cofactors and clusters in PcrAB and EccNarGH were nearly identical with respect to one another, suggesting a strikingly similar pathway of electron transfer to the active site despite 66% sequence disparity.
FIGURE 2.
Crystal structure of oxidized PcrAB heterodimer reveals [Fe-S] clusters are 10–12 Å apart from the adjacent cluster and the Mo-bis-MGD cofactor. A, overall crystal structure of oxidized PcrAB. PcrA and PcrB are shown in green and cyan ribbon, respectively. [Fe-S] clusters are shown in sphere (iron, brown; sulfur, yellow), bis-MGD cofactor is shown in magenta ball-and-stick, and molybdenum is shown in turquoise sphere. B, the relative position of cofactors in PcrAB along with Asp170 and a water molecule that coordinate to the active site molybdenum atom. Edge-to-edge distances between cofactors are shown. Labeling of the [Fe-S] clusters is consistent with previous publications (35, 36). The tricyclic pyranopterin of the molybdopterin cofactor is labeled as MGD, and the bicyclic dihydropterin is labeled as MD1. C, electron density 2|Fo| − |Fc| simulated annealing omit maps (gray) at 1.5 σ contour level of PcrA cofactors and active site for oxidized PcrAB. The tricyclic pyranopterin of the molybdopterin cofactor is labeled as MGD, and the bicyclic dihydropterin is labeled as MD1.
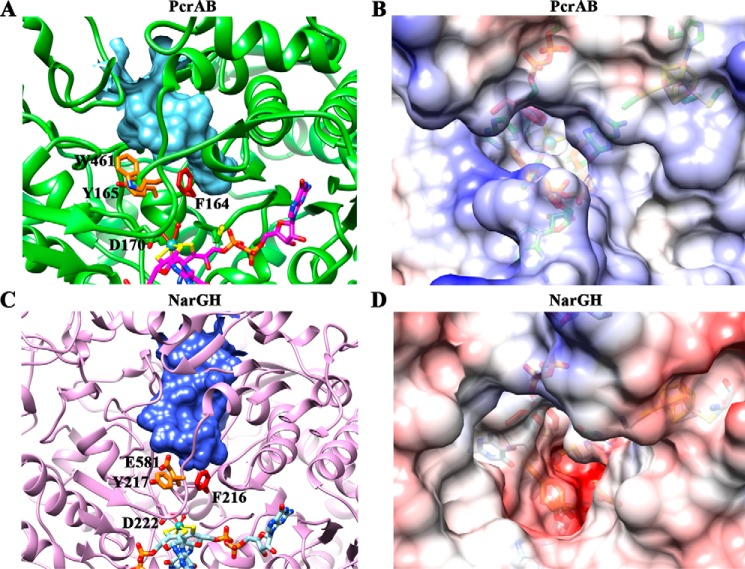
A funnel-shaped substrate access tunnel (volume, ∼1089 Å3) with a wider mouth that narrowed near the active site was detected for PcrAB by the CASTp web server (40) (Fig. 3A). Residue Phe164 in the tunnel near the active site appeared to block substrate access. Two more aromatic residues (Tyr165 and Trp461) were adjacent to Phe164 and together formed a putative hydrophobic gate that controlled active site entry and exit. Calculation of the electrostatic potential of PcrA using Chimera (25) showed the substrate access tunnel was dominated with positive charges (Fig. 3B). This positively charged tunnel creates the relatively high affinity for oxyanions seen in Table 1A. The EccNarG tunnel structure at a similar location formed a much wider pocket toward the active site with a tunnel volume of 1682 Å3 (Fig. 3C). The corresponding gate residues in EccNarG overlay well with the PcrA structure. However, residue Glu581 in EccNarG that is equivalent to Trp461 in PcrA changes the polarity near the gate. The electrostatic potential calculation of EccNarG showed the substrate access tunnel was dominated with negative charges (Fig. 3D), which would generate repulsions for oxyanions, especially for larger anionic molecules, and may explain the relatively low EccNarGHI affinity for these molecules (Table 1B).
FIGURE 3.
Comparisons of substrate accessing tunnels between PcrA and NarG identify the aromatic gate residues and the differences in electrostatic potential. A, structure of the closed tunnel in PcrA from the surface of the enzyme to the active site as seen in the oxidized PcrAB crystal structure (Fig. 2A) forms a funnel shape (cyan). The surface of the closed tunnel was detected by CASTp (40) and depicted using Chimera (25). The gate residues Phe164 (red), Tyr165 (orange), and Trp461 (orange) are shown in stick. B, the electrostatic potential surface of PcrA. The figure is viewed from the mouth of the tunnel down to the active site. The color is shown from negative (red; −10 kcal/mol × e) through white to positive (blue; 10 kcal/mol × e). C, structure of closed tunnel (blue) in EccNarG (Protein Data Bank code 1Q16) (35) forms a wider tunnel (tunnel volume, 1682 Å3) toward the active site than the tunnel in PcrA (tunnel volume, 1089 Å3). The corresponding gate residues in NarG are shown in stick with the same color code. D, the electrostatic potential surface of NarG by viewing from the same angle as B.
Phylogenetic Analysis and Mutational Studies of PcrA Reveal Functional Residues
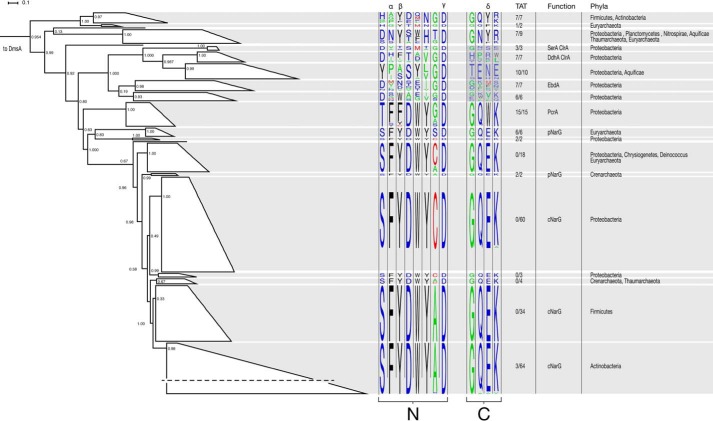
Phylogenetic analysis emphasizing the gate residues (Phe164, Tyr165, and Trp461), an active site residue (Asp170), and adjacent residues was used to compare closely related DMSO reductase superfamily enzymes. This revealed a monophyletic clade for PcrA closely related to the NarG (Fig. 4). NarG-containing enzymes either face the cytoplasm (cNarG) or periplasm (pNarG) and are differentiated by the presence of twin arginine translocation sequences (32) targeting pNarG to the periplasm. Twin arginine translocation sequences are found on all sequenced perchlorate reductases as well as many other type II DMSO reductases (see supplemental Fig. S1 for the complete phylogenetic tree). Sequence alignments of PcrA, pNarG, and cNarG depict several conserved regions. All sequences contain a hallmark Asp residue (41) at the active site that coordinates the molybdenum atom from a bis-MGD (Fig. 2). Additionally, all PcrA, pNarG, and cNarG sequences contain several conserved residues in the N-terminal portion of the molybdopterin-binding domain. The PcrAB crystal structure gate residues (Phe164 and Tyr165) were also conserved in NarG, whereas the PcrA Trp461 gate residue was a Glu in NarG.
FIGURE 4.
Phylogenetic reconstruction of PcrA and closely related DMSO reductase superfamily enzymes using DmsA and DorA as an outgroup. Active site residues identified from the crystal structure of PcrA were visualized for clades with high bootstrap support using sequence motifs. The positions denoted by α, β, and δ in the sequence alignments correspond to the gate residues relative to the A. suillum PS PcrA. The positions denoted by γ in the sequence alignments correspond to the Asp residue that has been shown or is predicted to bind to the molybdenum atom at the active site of the corresponding enzyme. N and C represent the N- and C-terminal portions of the MBD in each enzyme. The fraction of predicted twin arginine translocation (TAT) signal sequences, physiological function of known members, and taxonomic affiliation are superimposed on the tree. For the clades containing SerA, ClrA, and EbdA, the darker highlighting in the C-terminal portion of the aligned sequences denotes lower confidence in the alignment. SerA is selenate reductase, ClrA is chlorate reductase, EbdA is ethylbenzene dehydrogenase, PcrA is perchlorate reductase, pNarG is periplasmic nitrate reductase, and cNarG is cytoplasmic nitrate reductase. A horizontal dash in the lowermost cNar clade denotes the compression of this large group of sequences into a group suitable for visualization.
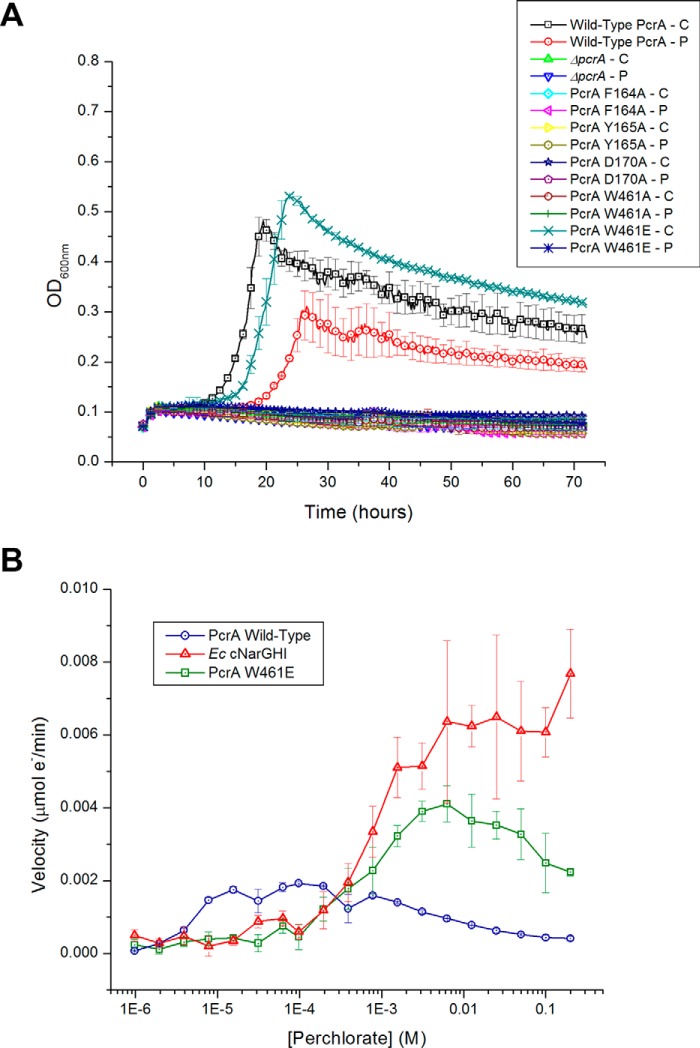
Based on crystal structure and phylogenetic analyses, mutational studies on the gate residues and Asp170 ligand were performed to determine the impact on cell growth and catalytic efficiency. Replacement of the A. suillum wild-type PcrA with PcrA F164A, Y165A, or W461A mutant removed the organism's capacity to grow by (per)chlorate respiration (Fig. 5A), indicating their critical role for PcrAB function. Interestingly, although the PcrA W461A mutant was incapable of growth on (per)chlorate, a W461E mutant retained its capacity to grow with chlorate. Furthermore, the purified W461E mutant PcrAB still displayed activity toward perchlorate albeit with a dramatically increased Km (Table 1C). The iodate Km was also greatly increased, whereas the chlorate and bromate values were only slightly impacted (Table 1C). Overall, the steady-state kinetic parameters for this mutant enzyme resembled a midpoint between wild-type PcrAB and EccNarGHI (Tables 1, A–C). The catalytic efficiency of the PcrA W461E mutant for perchlorate (kcat/Km = 3.0 × 103 m−1 s−1) more resembled that of EccNarGHI (kcat/Km = 5.4 × 103 m−1 s−1) than wild-type PcrAB (kcat/Km = 7.5 × 104 m−1 s−1). However, the catalytic efficiency of the PcrA W461E mutant for nitrate decreased 820- and 2670-fold compared with wild-type PcrAB and EccNarGHI, respectively. Additionally, substrate inhibition for perchlorate was greatly reduced in the W461E mutant, and the kinetics resembled that of EccNarGHI (Fig. 5B). Surprisingly, this Trp to Glu mutation greatly increased the PcrAB nitrate Km almost 700-fold (Table 1C), which was counterintuitive as this mutation resembles the natural NarG sequence.
FIGURE 5.
A, growth curves for wild-type A. suillum PS, ΔpcrA, and PcrA mutant strains under perchlorate or chlorate reducing conditions. C is chlorate, and P is perchlorate. Error bars represent one standard deviation of triplicate growth experiments. B, steady-state kinetics of perchlorate reduction by PcrAB containing the PcrA W461E mutation in comparison with wild-type PcrAB and EcNarGHI. Error bars represent one standard deviation of triplicate assays.
The PcrA W461E crystal structure indicated that Glu461 side chain was highly flexible relative to the corresponding Glu581 residue in NarG (Fig. 6). This flexibility was in part because the modified active site loses the π-stacking interaction with Tyr165 and lack of hydrogen bonding with Ser594 because the location of Ser594 is too far away from Glu461. In EccNarG, Glu581 is stabilized by hydrogen bonding to Asp801 (corresponding to PcrA Ser594; Fig. 6). The mutation of W461E in PcrA not only introduces a negative electrostatic charge in the tunnel but also puts the Glu461 side chain in front of the gate between Tyr165 and Phe164, which affected the open and closed conformation of Phe164. This putatively resulted in the decrease in substrate affinity and catalytic efficiency for perchlorate and nitrate (Table 1C) as well as the substrate inhibition of perchlorate (Fig. 5B).
FIGURE 6.

The comparison of PcrA W461E and EccNarG (Protein Data Bank code 1Q16) in the substrate access tunnel. The overlay of residue W461E PcrA from six chains of PcrA in the asymmetric unit of PcrAB was shown to be highly flexible due to the lack of hydrogen binding with Ser594 and the loss of π-stacking with Tyr165. The corresponding residue Glu581 in EccNarG (magenta) was stabilized by residue Asp801 via hydrogen bonding. The wild-type PcrAB is shown in dark gray color with Trp461 stabilized by π-stacking with Tyr165.
In contrast to perchlorate and nitrate, the W461E mutation had little impact on the chlorate and bromate kinetics (Table 1, A and C). These molecules have significantly longer bond lengths than nitrate and perchlorate and thus a greater molecular size (Table 1D). Consequently, if steric hindrance was the major cause of the increased PcrA W461E Km values, chlorate and bromate would experience a similar shift. This suggests that the mechanism underlying the dramatic kinetic shift (particularly substrate affinities) was not a function of substrate size (Table 1D). The defining feature that differentiates the various substrates is the presence (chlorate and bromate) or absence (perchlorate and nitrate) of a molecular dipole and its interaction with the newly introduced polar Glu461 residue in PcrA. Iodate is the obvious outlier in this proposed trend; however, the significantly larger size of iodate (Table 1D) could represent a ceiling at which advantageous dipole interactions with Glu461 are overcome by unfavorable steric hindrance.
Additional mutations were made in pcrA to further elicit the role of the gating residues with respect to trapping transiently produced chlorate generated during perchlorate reduction (Fig. 7). Five pcrA mutations were made in accordance with the phylogenetic analysis such that PcrA mutants were created with one point mutation in the PS PcrA relative to another PcrA in the same clade (Fig. 4). PcrA Y165C and F164S/Y165F show significant growth defects, whereas PcrA G169A and G169S grew identically to wild-type PS (Fig. 7), and PcrA Y165F displayed a minor growth defect. The most dramatic result was that PcrA Y165F accumulated ∼50% more chlorate than wild-type PS, whereas all other strains accumulated the same or lower amounts relative to the wild-type organism. The PcrA Y165F active site mutation is significant as other dissimilatory (per)chlorate-reducing bacteria that have been shown to accumulate chlorate (e.g. Propionivibrio militaris MP (42)) contain active sites with this configuration, suggesting that Tyr165 plays an important role in trapping transiently produced chlorate at the active site of PcrA during perchlorate turnover.
FIGURE 7.
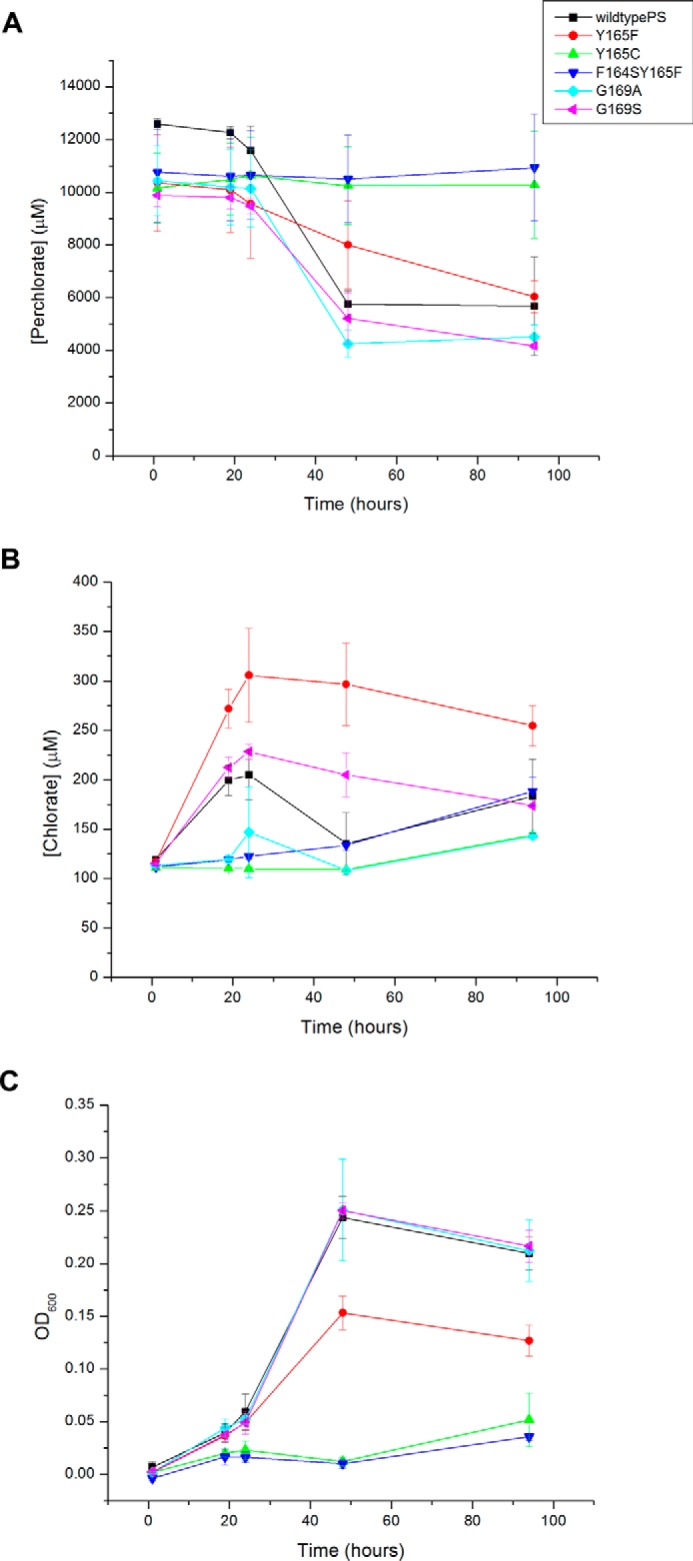
Chlorate accumulation experiments of wild-type A. suillum PS and A. suillum PS cells containing pcrA mutations monitoring chlorate (A), perchlorate (B), and OD600 (C). Error bars represent the standard deviation of three replicate experiments.
Phe164 Gating for Substrate Entry and Product Release
To gain insight into the active site gating mechanism, we obtained and characterized two additional novel crystal structures of reduced PcrAB in comparison with the oxidized structures (Table 2). One of the reduced structures contained the substrate analog SeO32− bound to the active site molybdenum atom (Fig. 8, A and B). The other structure had the gate residue Phe164 in a dual occupancy of both open and closed conformations (Fig. 8, C and D). In the SeO32−-bound structure, one of the selenite oxygen atoms was bound to the molybdenum atom, which was in turn coordinated with Asp170 in a monodentate fashion (Fig. 8A). The bond distances of Mo–O for selenite and Asp170 were 2.0–2.1 and 2.2–2.4 Å, respectively, in the three PcrA molecules in the asymmetric unit. The other oxygen atoms of the selenite molecule were orientated such that they did not coordinate to any active site hydrogen bond acceptors. The cyclization states of the bis-MGD rings in the selenite-bound reduced PcrA structure appeared not to change relative to the oxidized structure (Figs. 2C and 8B). It was proposed in the molybdenum cofactor model complex that the interconversion of bicyclic and tricyclic structures could be solvent-dependent (43), and our results suggest that this interconversion is most likely not redox-dependent.
FIGURE 8.
Reduced PcrAB structures with substrate analog bound and open and closed conformation of PcrA Phe164. A, active site of PcrA (blue) with selenite (SeO32−) coordinated to the molybdenum atom. The occupancy of selenite in the structure is 0.6. The surfaces in A and C are colored in a hydrophobicity scale (26) where red is the most hydrophobic and blue is the most hydrophilic. B, electron density of 2|Fo| − |Fc| simulated annealing omit maps at 1.5 σ contour level of PcrA cofactors and active site for reduced PcrAB with selenite bound. The anomalous map is shown as a green surface at 3 σ contour level. C, overlay of the active sites of oxidized (magenta) and reduced (tan) PcrA along with the tunnel to the active site. The reduced PcrA structure reveals dual conformations of the Phe164 side chain in open and closed states. The tunnel is shown in the open conformation of Phe164. The water molecule in reduced PcrA (tan) is located inside the open tunnel and hydrogen-bonded with residues Asp170 and Asn35. D, electron density of 2|Fo| − |Fc| simulated annealing omit maps at 1.5 σ contour level of PcrA cofactors and active site for reduced PcrAB with the side chain of Phe164 in the open and closed conformations. E, structure of the open tunnel in PcrA with Phe164 in the open tunnel conformation shown as a surface (light purple).
Soaking with and co-crystallization of substrate analogs have previously been tried with NapA, although nitrate and perchlorate were only found near the outside of the tunnel (44). We successfully obtained crystals with the substrate analog bound to the active site by soaking anaerobically under reducing conditions (see “Experimental Procedures”). Intriguingly, Phe164 was in the closed conformation in the selenite-bound structure, suggesting that a molecule is trapped at the active site when it is bound to the molybdenum atom.
We obtained a reduced PcrAB structure with the side chain of Phe164 in dual occupancy of both open (occupancy of 0.45) and closed (occupancy of 0.55) conformations, respectively (Fig. 8, C and D). In this structure, the coordination of Asp170 to the molybdenum atom was shifted from monodentate to bidentate with bond distances of Mo–O1 (2.0–2.2 Å) and Mo–O2 (2.3–2.7 Å) in the three PcrA molecules in the asymmetric unit. This carboxylate shift presumably displaced the water molecule that was originally bound to the molybdenum atom. The water molecule would then interact with one of the carboxylate oxygen atoms in residue Asp170 and the side-chain nitrogen atom in residue Asn35 via hydrogen bonding located in the open tunnel (Fig. 8C). Although monodentate and bidentate coordination of Asp to molybdenum atom have been seen in some independent NarGH structures (35, 36, 45), a carboxylate shift has only previously been proposed to protein metals other than molybdenum, such as iron, manganese, cadmium, and zinc (46). In combination with our results, this suggests that reduced PcrAB enables the Phe164 to switch conformations to open active site access, and this gating may be triggered by the water release from the active site to the tunnel due to the carboxylate shift of the Asp170 residue (Fig. 8E).
Discussion
Our results reveal kinetic and structural features of PcrAB that establish it as a specialized member of the DMSO reductase superfamily of molybdopterin enzymes. We found that, although PcrA and NarG are closely related enzymes, they evolved from a common ancestor and diverged to tune their activities to the environments in which their host organisms lived. In most environments, perchlorate is present in low micromolar concentrations (47). Thus, a perchlorate reductase would experience selective pressure to evolve a relatively high affinity for perchlorate, whereas a nitrate reductase would experience selective pressure to maintain a relatively low affinity for perchlorate due to the toxicity of chlorite in a non-perchlorate-respiring organism lacking a chlorite dismutase. Consistently, below 200 μm perchlorate, PcrA is a more proficient perchlorate reductase than EccNarG. Importantly, the PcrA substrate inhibition by perchlorate does not impact catalytic efficiency at perchlorate concentrations <200 μm.
The unique biophysical parameters (low Km and substrate inhibition by perchlorate) of the PcrAB are putatively associated with differences in key aromatic residues in the substrate access tunnel of each enzyme. Gate residues (Phe164, Tyr165, and Trp461) were identified from crystal structures of PcrAB. The presence of the Phe164 residue in PcrA corresponding to Phe216 in EccNarG suggests this alone cannot explain the low Km and substrate inhibition. However, the additional aromatic residue Trp461 in PcrA, which is a Glu in NarG, might serve as a constrictive factor. A constricted active site with more possible contacts between protein and substrate is associated with higher affinity substrate binding (48). As such, the constrictive gate and tunnel positive electrostatic potential could combine to increase PcrA substrate affinities. Consistently, the PcrA W461E mutation partially shifted the PcrAB steady-state kinetics parameters toward those of EccNarGHI. Interestingly, the W461E mutation not only increased the perchlorate Km by 50-fold but also partially alleviated substrate inhibition. Previously, nitrate inhibition was noted for the periplasmic nitrate reductase NapAB, purportedly due to excess substrate entering the active site tunnel and blocking water release (49). A more constricted active site such as that in PcrA would decrease both the on-rate and off-rate of substrates, products, and intermediates from the active site. Substrate inhibition would therefore be more dramatic when an enzyme must transition through multiple reaction cycles, such as perchlorate reduction, as water must leave the active site after the reduction of perchlorate to chlorate before chlorate can bind to the molybdenum atom for subsequent turnover. The lack of substrate inhibition for chlorate and nitrate turnover in wild-type PcrAB, PcrA W461E, and EccNarGHI is consistent with this hypothesis.
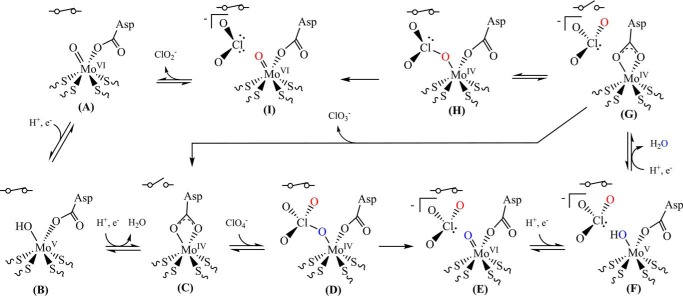
The kinetic observations and unique structural features we identified for PcrAB along with previous publications pertaining to similar molybdenum-containing enzymes (50) support the proposal of a specific mechanism for (per)chlorate reduction by PcrAB (Fig. 9, steps A–I). Initially, the Asp residue (Asp170) is coordinated with an oxygen atom bound to the Mo(VI) atom in a monodentate fashion at the active site with a closed gate (A). Previous work with DMSO reductase from Rhodobacter sphaeroides has shown that this bound oxygen atom is most likely in a Mo(VI)=O configuration (51). Two sequential proton-coupled electron transfer events reduce the active site through a Mo(V)-OH intermediate (B) to Mo(IV) (C). Upon 2e− reduction of the active site, the bound water is released from the molybdenum atom and seems to be trapped at the active site by Asp170 and Asn35 (Fig. 8C). Concomitantly, Asp170 shifts to a bidentate coordination of the Mo(IV) atom as seen in Fig. 7C. Phe164 can now swing to open conformation in the reduced enzyme (Fig. 8C). This bidentate coordination of the Asp170 is a possible trigger that leads to an Phe164 conformational shift and opening of the tunnel to the active site, allowing water exit and substrate entry. At this point, perchlorate can bind to the active site Mo(IV), which causes the Asp residue to shift back to monodentate coordination and the gate to close (D) as seen in Fig. 8A. Next, electrons are transferred from the reduced active site to the bound perchlorate, resulting in an oxygen atom abstraction and generation of chlorate and oxo-Mo(VI) (E). At this point, two additional proton-coupled electron transfer events would occur, forming F and subsequently G. Between F and G, we propose that chlorate and water remain trapped in the active site until Asp170 shifts to a bidentate coordination on the reduced molybdenum and triggers Phe164 to open the gate. Hypothetically, it is this step that is most likely to account for substrate inhibition during perchlorate turnover. Chlorate is a larger product than that formed in the reduction of other oxyanions, and excess perchlorate in the substrate access tunnel may hinder water exit from such a crowded and constricted region. This situation resembles the proposed substrate inhibition mechanism for NapA (49). The chlorate that has been shown to be accumulated during perchlorate turnover by PS is most likely released from G to reform C when the gate reopens as the active site becomes reduced for a second time in the catalytic cycle. When chlorate binds to the reduced active site to form H as seen in Fig. 8A, an additional oxygen atom abstraction occurs, reducing chlorate to chlorite (I), which will diffuse out of the active site to reform A. Notably, this proposed mechanism maintains the molybdenum atom in a six-coordinate geometry throughout the entire catalytic cycle. Although the mechanism is consistent with our results, one main question remains to be answered. Because the open conformation of Phe164 in PcrA was only seen in the reduced structure of PcrAB, is the gating mechanism truly a redox-mediated process? Furthermore, the confirmation of the aforementioned discrete intermediates and the potential roles of other residues or components of the active site during catalysis are all currently being investigated.
FIGURE 9.
Proposed mechanism for (per)chlorate reduction by PcrAB (A–I). The Asp residue corresponds to Asp170 as shown in the oxidized and reduced crystal structures of PcrAB. The symbols for a closed or an open switch represent a closed or open gate, respectively.
Author Contributions
J. D. C. conceived of and coordinated the study. J. D. C. and M. D. Y. co-wrote the paper. M. D. Y. performed all protein purifications and kinetic analysis. C.-L. T. collected and solved all crystallographic data and contributed to writing the paper. I. C. C. performed and analyzed the phylogenetic data shown in Fig. 4. H. K. C., S. A. R., J. A. T., and J. D. C. made significant contributions to experimental design and interpretation. A. P. M. collected and analyzed data shown in Table 1. P. S. G.-P. and A. W. designed, constructed, and tested mutant enzymes. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Joel Weiner's group from University of Alberta for generously providing the cytoplasm-facing nitrate reductase, NarGHI from E. coli (EccNarGHI). We thank SIBYLS beamline 12.3.1 staff scientist Scott Classen for assistance with crystallographic data collection. The crystallography experiments were conducted at the Advanced Light Source, a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program supported by the Department of Energy Office of Biological and Environmental Research.
The work on perchlorate and chlorate respiration in the laboratory of J. D. C. was supported by the Energy Biosciences Institute, University of California, Berkeley, and structural studies were supported by National Institute of Health Project MINOS (Macromolecular INsights Optimized by Scattering) Grant R01GM105404. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. S1 and Table S1.
The atomic coordinates and structure factors (codes 4YDD, 5CH7, 5CHC, and 5E7O) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- DMSO
- dimethylsulfoxide
- [Fe-S]
- iron-sulfur cluster
- [4Fe-4S]
- four-iron/four-sulfur cluster
- [3Fe-4S]
- three-iron/four-sulfur cluster
- MGD
- molybdopterin guanine dinucleotide
- Ec
- E. coli.
References
- 1. Jackson W. A., Bohlke J. K., Andraski B. J., Fahlquist L., Bexfield L., Eckardt F. D., Gates J. B., Davila A. F., McKay C. P., Rao B., Sevanthi R., Rajagopalan S., Estrada N., Sturchio N., Hatzinger P. B., Anderson T. A., Orris G., Betancourt J., Stonestrom D., Latorre C., Li Y. H., and Harvey G. J. (2015) Global patterns and environmental controls of perchlorate and nitrate co-occurrence in arid and semi-arid environments. Geochim. Cosmochim. Acta 164, 502–522 [Google Scholar]
- 2. Jackson W. A., Davila A. F., Sears D. W. G., Coates J. D., Mckay C. P., Brundrett M., Estrada N., and Bohlke J. K. (2015) Widespread occurrence of (per)chlorate in the solar system. Earth Planet. Sci. Lett. 430, 470–476 [Google Scholar]
- 3. Coates J. D., and Achenbach L. A. (2004) Microbial perchlorate reduction: rocket-fuelled metabolism. Nat. Rev. Microbiol. 2, 569–580 [DOI] [PubMed] [Google Scholar]
- 4. Rajagopalan S., Anderson T., Cox S., Harvey G., Cheng Q., and Jackson W. A. (2009) Perchlorate in wet deposition across North America. Environ. Sci. Technol. 43, 616–622 [DOI] [PubMed] [Google Scholar]
- 5. Hecht M. H., Kounaves S. P., Quinn R. C., West S. J., Young S. M., Ming D. W., Catling D. C., Clark B. C., Boynton W. V., Hoffman J., Deflores L. P., Gospodinova K., Kapit J., and Smith P. H. (2009) Detection of perchlorate and the soluble chemistry of martian soil at the Phoenix lander site. Science 325, 64–67 [DOI] [PubMed] [Google Scholar]
- 6. Ming D. W., Archer P. D. Jr., Glavin D. P., Eigenbrode J. L., Franz H. B., Sutter B., Brunner A. E., Stern J. C., Freissinet C., McAdam A. C., Mahaffy P. R., Cabane M., Coll P., Campbell J. L., Atreya S. K., et al. (2014) Volatile and organic compositions of sedimentary rocks in Yellowknife Bay, Gale crater, Mars. Science 343, 1245267. [DOI] [PubMed] [Google Scholar]
- 7. Kounaves S. P., Carrier B. L., O'Neil G. D., Stroble S. T., and Claire M. W. (2014) Evidence of martian perchlorate, chlorate, and nitrate in Mars meteorite EETA79001: implications for oxidants and organics. Icarus 229, 206–213 [Google Scholar]
- 8. Ojha L., Wilhelm M. B., Murchie S. L., McEwen A. S., Wray J. J., Hanley J., Massé M., and Chojnacki M. (2015) Spectral evidence for hydrated salts in recurring slope lineae on Mars. Nat. Geosci. 8, 829–832 [Google Scholar]
- 9. Davila A. F., Willson D., Coates J. D., and Mckay C. P. (2013) Perchlorate on Mars: a chemical hazard and a resource for humans. Int. J. Astrobiol.12, 321–325 [Google Scholar]
- 10. Urbansky E. T. (1998) Perchlorate chemistry: implications for analysis and remediation. Bioremed. J. 2, 81–95 [Google Scholar]
- 11. Coates J. D., and Achenback L. A. (2006) The microbiology of perchlorate reduction and its bioremediative application, in Perchlorate: Environmental Occurrence, Interactions and Treatment (Gu B., and Coates J. D., eds) pp. 279–295, Springer, New York [Google Scholar]
- 12. Melnyk R. A., Clark I. C., Liao A., and Coates J. D. (2014) Transposon and deletion mutagenesis of genes involved in perchlorate reduction in Azospira suillum PS. MBio 5, e00769–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melnyk R. A., and Coates J. D. (2015) The perchlorate reduction genomic island: mechanisms and pathways of evolution by horizontal gene transfer. BMC Genomics 16, 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heinnickel M., Smith S. C., Koo J., O'Connor S. M., and Coates J. D. (2011) A bioassay for the detection of perchlorate in the ppb range. Environ. Sci. Technol. 45, 2958–2964 [DOI] [PubMed] [Google Scholar]
- 15. Clark I. C., Melnyk R. A., Engelbrektson A., and Coates J. D. (2013) Structure and evolution of chlorate reduction composite transposons. MBio 4, e00379–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan A., and Kiley P. J. (2009) Techniques to isolate O2-sensitive proteins: [4Fe-4S]-FNR as an example. Methods Enzymol. 463, 787–805 [DOI] [PubMed] [Google Scholar]
- 17. Morpeth F. F., and Boxer D. H. (1985) Kinetic analysis of respiratory nitrate reductase from Escherichia coli K12. Biochemistry 24, 40–46 [DOI] [PubMed] [Google Scholar]
- 18. Rothery R. A., Chatterjee I., Kiema G., McDermott M. T., and Weiner J. H. (1998) Hydroxylated naphthoquinones as substrates for Escherichia coli anaerobic reductases. Biochem. J. 332, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Classen S., Hura G. L., Holton J. M., Rambo R. P., Rodic I., McGuire P. J., Dyer K., Hammel M., Meigs G., Frankel K. A., and Tainer J. A. (2013) Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 46, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kloer D. P., Hagel C., Heider J., and Schulz G. E. (2006) Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure 14, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 26. Kyte J., and Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 27. Novoradovsky A., Zhang V., Ghosh M., Hogrefe H., Sorge J. A., and Gaasterland T. (2005) Computational principles of primer design for site directed mutagenesis, in Technical Proceedings of 2005 NSTI Nanotechnology Conference and Trade Show, Vol. 1, pp. 532–535, Nano Science and Technology Institute, Danville, CA [Google Scholar]
- 28. Price M. N., Dehal P. S., and Arkin A. P. (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu L., Niu B., Zhu Z., Wu S., and Li W. (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Capella-Gutiérrez S., Silla-Martínez J. M., and Gabaldón T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bendtsen J. D., Nielsen H., Widdick D., Palmer T., and Brunak S. (2005) Prediction of twin-arginine signal peptides. BMC Bioinformatics 6, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 35. Bertero M. G., Rothery R. A., Palak M., Hou C., Lim D., Blasco F., Weiner J. H., and Strynadka N. C. (2003) Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 10, 681–687 [DOI] [PubMed] [Google Scholar]
- 36. Jormakka M., Richardson D., Byrne B., and Iwata S. (2004) Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure 12, 95–104 [DOI] [PubMed] [Google Scholar]
- 37. Raaijmakers H., Macieira S., Dias J. M., Teixeira S., Bursakov S., Huber R., Moura J. J., Moura I., and Romão M. J. (2002) Gene sequence and the 1.8 Å crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure 10, 1261–1272 [DOI] [PubMed] [Google Scholar]
- 38. Rothery R. A., Bertero M. G., Spreter T., Bouromand N., Strynadka N. C., and Weiner J. H. (2010) Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation. J. Biol. Chem. 285, 8801–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Enemark J. H., and Garner C. D. (1997) The coordination chemistry and function of the molybdenum centres of the oxomolybdoenzymes. J. Biol. Inorg. Chem. 2, 817–822 [Google Scholar]
- 40. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., and Liang J. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Romão M. J. (2009) Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview. Dalton Trans. 4053–4068 [DOI] [PubMed] [Google Scholar]
- 42. Thrash J. C., Pollock J., Torok T., and Coates J. D. (2010) Description of the novel perchlorate-reducing bacteria Dechlorobacter hydrogenophilus gen. nov., sp. nov. and Propionivibrio militaris, sp. nov. Appl. Microbiol. Biotechnol. 86, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams B. R., Gisewhite D., Kalinsky A., Esmail A., and Burgmayer S. J. (2015) Solvent-dependent pyranopterin cyclization in molybdenum cofactor model complexes. Inorg. Chem. 54, 8214–8222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Najmudin S., González P. J., Trincão J., Coelho C., Mukhopadhyay A., Cerqueira N. M., Romão C. C., Moura I., Moura J. J., Brondino C. D., and Romão M. J. (2008) Periplasmic nitrate reductase revisited: a sulfur atom completes the sixth coordination of the catalytic molybdenum. J. Biol. Inorg. Chem. 13, 737–753 [DOI] [PubMed] [Google Scholar]
- 45. Sparacino-Watkins C., Stolz J. F., and Basu P. (2014) Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 43, 676–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sousa S. F., Cerqueira N. M. F. S. A., Bras N. F., Fernandes P. A., and Ramos M. J. (2014) Enzymatic “tricks”: carboxylate shift and sulfur shift. Int. J. Quantum Chem. 114, 1253–1256 [Google Scholar]
- 47. Gu B., and Coates J. D. (eds) (2006) Perchlorate: Environmental Occurrence, Interactions and Treatment, Springer, New York [Google Scholar]
- 48. Laurie A. T., and Jackson R. M. (2005) Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics 21, 1908–1916 [DOI] [PubMed] [Google Scholar]
- 49. Jacques J. G., Burlat B., Arnoux P., Sabaty M., Guigliarelli B., Léger C., Pignol D., and Fourmond V. (2014) Kinetics of substrate inhibition of periplasmic nitrate reductase. Biochim. Biophys. Acta 1837, 1801–1809 [DOI] [PubMed] [Google Scholar]
- 50. Hille R., Hall J., and Basu P. (2014) The mononuclear molybdenum enzymes. Chem. Rev. 114, 3963–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garton S. D., Hilton J., Oku H., Crouse B. R., Rajagopalan K. V., and Johnson M. K. (1997) Active site structures and catalytic mechanism of Rhodobacter sphaeroides dimethyl sulfoxide reductase as revealed by resonance Raman spectroscopy. J. Am. Chem. Soc. 119, 12906–12916 [Google Scholar]
- 52. Banks A. J., Bollom M. S., Holmes J. L., Jacobsen J. J., Kotz J. C., and Moore J. W. (2015) Models 360, Chemical Education Digital Library, Washington, D. C. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.