Abstract
DNA mismatch repair (MMR) is required for the maintenance of genome stability and protection of humans from several types of cancer. Human MMR occurs in the chromatin environment, but little is known about the interactions between MMR and the chromatin environment. Previous research has suggested that MMR coincides with replication-coupled assembly of the newly synthesized DNA into nucleosomes. The first step in replication-coupled nucleosome assembly is CAF-1-dependent histone (H3-H4)2 tetramer deposition, a process that involves ASF1A-H3-H4 complex. In this work we used reconstituted human systems to investigate interactions between MMR and CAF-1- and ASF1A-H3-H4-dependent histone (H3-H4)2 tetramer deposition. We have found that MutSα inhibits CAF-1- and ASF1A-H3-H4-dependent packaging of a DNA mismatch into a tetrasome. This finding supports the idea that MMR occurs before the DNA mismatch is packaged into the tetrasome. Our experiments have also revealed that CAF-1- and ASF1A-H3-H4-dependent deposition of the histone (H3-H4)2 tetramers does not interfere with MMR reactions. In addition, we have established that unnecessary degradation of the discontinuous strand that takes place in both DNA polymerase δ (Pol δ)- and DNA polymerase ϵ (Pol ϵ)-dependent MMR reactions is suppressed by CAF-1- and ASF1A-H3-H4-dependent deposition of the histone (H3-H4)2 tetramers. These data suggest that CAF-1- and ASF1A-H3-H4-dependent deposition of the histone (H3-H4)2 tetramers is compatible with MMR and protects the discontinuous daughter strand from unnecessary degradation by MMR machinery.
Keywords: cancer, DNA mismatch repair, DNA polymerase, DNA replication, genomic instability, histone, histone chaperone, mutL homolog 1 (MLH1), proliferating cell nuclear antigen (PCNA), replication factor C (RFC)
Introduction
The DNA mismatch repair (MMR)3 system has been conserved from bacteria to humans (1–3). Genetic stabilization provided by the MMR system suppresses both sporadic and inherited cancers (4). The MMR system has multiple functions that are involved in the genome maintenance (1–3, 5–8). Among these functions MMR is the strongest contributor to the suppression of spontaneous mutation rates. Significant progress has been made in understanding MMR in Escherichia coli and eukaryotes (1, 2, 6, 8, 9). In E. coli, MMR is initiated by binding of the mismatch recognition factor MutS to a mismatch (10, 11). After mismatch recognition, MutS recruits MutL in an ATP-dependent manner (12, 13). The MutS-MutL complex activates the MutH endonuclease to nick the newly synthesized DNA strand at a transiently unmethylated GATC site that may be as far as 1 kb away from the mismatch (11). The MutH nick is the loading site for the excision complex that consists of UvrD (helicase II) and one of the four exonucleases (ExoI, ExoVII, ExoX, and RecJ) (14–17). The excision complex unwinds and excises a portion of the newly synthesized strand encompassing the mismatch. The gap is filled in by the DNA polymerase III holoenzyme, and the DNA ligase seals the nick (18).
Eukaryotic MMR is more efficient on the lagging than leading strand (19). One-nucleotide deletion loops, mispaired bases, and one-nucleotide insertion loops are the most common substrates for the eukaryotic MMR reaction (1, 2, 6, 7). In addition to these lesions, the eukaryotic reaction removes oxidized bases, one-nucleotide flaps, and single ribonucleotides that produce mispairs in DNA (20–23). Eukaryotes have two mismatch recognition factors, MutSα (MSH2-MSH6 heterodimer) and MutSβ (MSH2-MSH3 heterodimer); both are homologs of E. coli MutS (24–26). The concentration of MutSα in HeLa cells is ∼10 times higher than that of MutSβ, suggesting that the majority of MMR events in human cells involve MutSα (27). Consistent with this idea, MSH6, but not MSH3, is important for the suppression of carcinogenesis in humans and mice (4, 28). Eukaryotic MMR is initiated by recognition of the mismatch by MutSα or MutSβ. Upon mismatch recognition, the MutS homolog and PCNA (29, 30) loaded by RFC (31) activate the MutL homolog MutLα (MLH1-PMS2 heterodimer; Refs. 32 and 33–36). The activated MutLα incises the discontinuous daughter strand 5′ and 3′ to the mismatch. Incision of the discontinuous strand by MutLα 5′ to the mismatch is required for the excision-dependent and excision-independent MMR pathways (33, 37–44). In the excision-dependent MMR pathway, MutSα-activated Exonuclease 1 (EXO1) removes a DNA segment containing the mismatch in a 5′ → 3′ excision reaction that initiates from a 5′ nick produced by MutLα endonuclease, and the generated gap is repaired by DNA polymerase δ (Pol δ) holoenzyme (21, 33, 37–43, 45, 46). In the excision-independent pathway, a new 3′ end produced by the MutLα incision of the discontinuous daughter strand 5′ to the mismatch is extended by Pol δ holoenzyme in a DNA synthesis reaction that displaces a part of the original strand containing the mismatch (44).
Eukaryotic MMR occurs in the nucleosomal environment. The nucleosome is the major building block of chromatin. Assembly of the nucleosome is a conserved process that can be divided into two major steps. In the first step the tetrasome, a complex of the histone (H3-H4)2 tetramer and ∼146-bp DNA, is formed as a result of deposition of the tetramer onto DNA (47). In the second step the tetrasome is converted into the nucleosome by the addition of two histone H2A-H2B dimers. The daughter strands that emerge from the eukaryotic replication fork are rapidly packaged into nucleosomes by replication-coupled nucleosome assembly (48, 49). The histone chaperone CAF-1 orchestrates replication-coupled nucleosome assembly by depositing the histone (H3-H4)2 tetramers onto the newly synthesized DNA (50–53). The protein-protein interaction between CAF-1 and loaded PCNA ensures that the histone chaperone is able to act behind the replication fork (54–56). The histone chaperones ASF1A and ASF1B (57) assist CAF-1 in replication-coupled deposition of the histone (H3-H4)2 tetramers (58). ASF1A and ASF1B form complexes with newly synthesized histone H3-H4 dimers in cytosol and participate in their transport into the nucleus (59). In the nucleus, the heterotrimeric ASF1A-H3-H4 and ASF1B-H3-H4 complexes are thought to transfer their H3-H4 dimers onto CAF-1 molecules (60). The CAF-1-(H3-H4)2 intermediate next forms a complex with loaded PCNA. After formation of the complex with PCNA, CAF-1 loads the histone (H3-H4)2 tetramer onto the nascent DNA. In addition to their roles in deposition of newly synthesized histone (H3-H4)2 tetramers, ASF1A and ASF1B have also been implicated in CAF-1-dependent loading of parental (H3-H4)2 tetramers (61).
Recent studies have suggested that MMR coincides with CAF-1-dependent assembly of the newly synthesized DNA into nucleosomes (56, 62). In this work we analyzed reconstituted human systems that support MMR and CAF-1- and ASF1A-H3-H4-dependent histone (H3-H4)2 tetramer deposition. We found that there are several interactions between the two processes. The presence of these interactions supports the view that eukaryotic MMR occurs during replication-coupled nucleosome assembly but precedes the incorporation of the mismatch into the tetrasome.
Experimental Procedures
Proteins
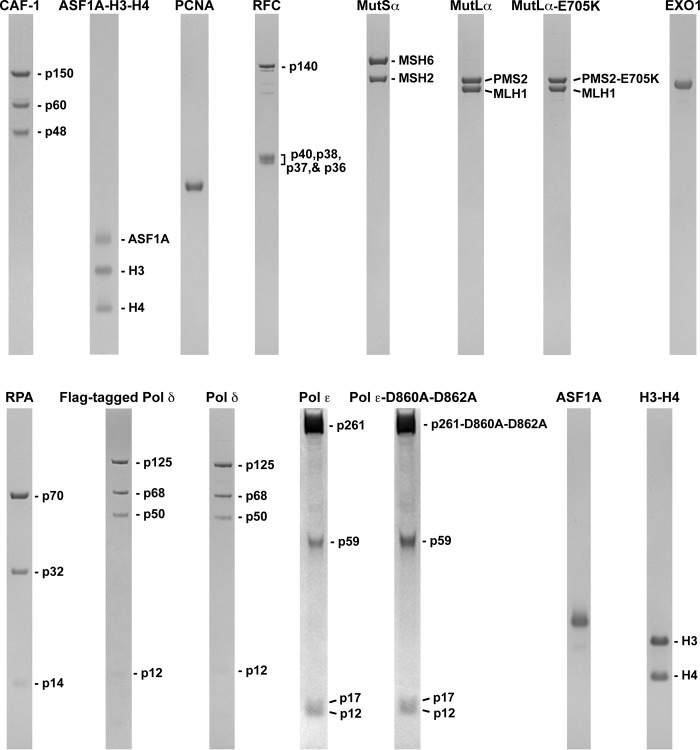
Human ASF1A-H3-H4, CAF-1, histone H3-H4 complex, EXO1, MutLα, MutSα, MutLα-E705K, PCNA, Pol δ, RFC, and RPA were prepared in near homogenous forms as previously described (33, 42, 44, 53, 56). The four-subunit human Pol δ FLAG-tagged at the p125 subunit N terminus and His6-tagged at the p66 subunit N terminus was expressed in baculovirus-infected Sf9 insect cells and purified using anti-FLAG M2 affinity beads (Sigma) and Mono S and Mono Q columns (GE Healthcare). The purity of Pol δ obtained at the final purification step was >95%. Wild-type four-subunit human Pol ϵ and a four-subunit Pol ϵ variant carrying the p261 D860A and D862A substitutions were FLAG-tagged at the N terminus of the p261 subunit, expressed in baculovirus-infected insect Sf9 cells, and purified with anti-FLAG M2 affinity beads (Sigma) and Mono S and Mono Q columns (GE Healthcare). After the final purification step, the wild-type Pol ϵ and the mutant were 90% pure. Human His6-GST-ASF1A was expressed in E. coli (63) and purified on Ni2+ beads (Affymetrix), a Mono Q column (GE Healthcare), and a glutathione-Sepharose column (GE Healthcare). The purified His6-GST-ASF1A was then treated with His6-Tev protease, and ASF1A was separated from the His6-GST tag and protease by chromatography on Ni2+ beads (Affymetrix) and a Mono Q column (GE Healthcare). The purified ASF1A was 85% pure.
DNA Substrates and Oligonucleotides
Preparation of 3′-nicked G-T DNA (3′ G-T DNA) and 3′-nicked A-T DNA (3′ A-T DNA), a relaxed covalently closed DNA (ccDNA), and single-stranded DNA primed with 12 oligonucleotides was described previously (33, 42, 53). 3′-Nicked G-T DNA (6.4 kb) carries a strand break that is 141 bp 3′ to a G-T mispair, and 3′-nicked A-T DNA is the same as 3′-nicked G-T DNA except that it contains an A-T pair instead of the G-T mispair (42). DNA sequences of oligonucleotides that were labeled with 32P by T4 polynucleotide kinase (New England BioLabs) and used as hybridization probes are shown in Table 1.
TABLE 1.
DNA sequences of 32P-labeled hybridization probes used in this work
| Oligonucleotide | Oligonucleotide sequence |
|---|---|
| 22-mer v2505 | 5′-CGCTACTGATTACGGTGCTGCT-3′ |
| 22-mer v2531 | 5′-ATGGTTTCATTGGTGACGTTTC-3′ |
| 24-mer v5225 | 5′-GATATTACCAGCAAGGCCGATAGT-3′ |
| 24-mer v5629 | 5′-GCTTTCGAGTCTAGAAATTCGGCT-3′ |
| 24-mer v5690 | 5′-GTTCCGATTTAGTGCTTTACGGCA-3′ |
Histone (H3-H4)2 Tetramer Deposition Assay
Histone tetramer (H3-H4)2 deposition assay was based on a previously described procedure (56). Each histone tetramer (H3-H4)2 deposition reaction was carried out in a 40-μl mixture containing 20 mm HEPES-NaOH (pH 7.4), 110 mm KCl, 5 mm MgCl2, 2 mm ATP, 4 mm DTT, 0.2 mg/ml BSA, 1–3% (v/v) glycerol, and 0.6 nm (0.1 μg) DNA (3′-nicked G-T DNA, 3′-nicked A-T DNA, or relaxed ccDNA). When indicated, human CAF-1 (15 nm), ASF1A-H3-H4 (46 nm), PCNA (21 nm), RFC (3 nm), and MutSα (11 nm, 22 nm, or 44 nm) were present in the reaction mixtures. After a 5-min incubation at 37 °C, 35-μl fraction of each reaction mixture was added to a 5-μl micrococcal nuclease mixture (20 mm HEPES-NaOH (pH 7.4), 20 mm CaCl2, and 2.4 units/μl micrococcal nuclease), and the reaction temperature was changed to 21–23 °C. The DNA cleavage was carried out for 5 min and then terminated by the addition of a 4-μl mixture (0.5% SDS, 70 mm EDTA, 40% glycerol, 2.5 mg/ml proteinase K, and 2.5 μg/ml HindIII-cleaved plasmid DNA pAH1A (a gel loading control)). The proteins were digested at 50 °C for 15–20 min, and DNA products were separated on native 1.8% agarose gels. The separated DNA products were transferred onto nylon membranes and analyzed by Southern hybridizations with the indicated 32P-labeled probes. 32P-Labeled DNA species were visualized with a Typhoon biomolecular imager (GE Healthcare) and quantified using the ImageQuant software (GE Healthcare).
MMR Assay
The MMR assay was carried out according to a published method (44). This assay is based on the observation that nick-directed repair of the G-T mispair on 3′-nicked G-T DNA restores the HindIII site (43, 44, 64). Each MMR reaction was carried out in a 60-μl mixture containing 20 mm HEPES-NaOH (pH 7.4), 110 mm KCl, 5 mm MgCl2, 2 mm ATP, 4 mm DTT, 0.4 mg/ml BSA, 3–5% (v/v) glycerol, 0.2 mm dGTP, 0.2 mm dATP, 0.2 mm dTTP, 0.2 mm dCTP, 0.6 nm (0.15 μg) 3′-nicked G-T DNA, and the indicated purified human proteins. When human ASF1A-H3-H4, CAF-1, EXO1, MutLα, MutSα, PCNA, RFC, and RPA were present in the reaction mixtures, their concentrations were 46, 23, 3, 6, 22, 21, 3, and 52 nm, respectively. The human FLAG-tagged Pol δ concentration in the reaction mixtures was 0, 0.1, 0.2, 1, 5, or 10 nm, and the human Pol ϵ concentration in the reaction mixtures was 0, 1, 2, 5, or 10 nm. MMR reactions were incubated at 37 °C for 20 min, and each reaction was stopped by the addition of a 45-μl solution containing 0.35% SDS, 0.4 m NaCl, 0.3 mg/ml proteinase K, 0.7 mg/ml glycogen, and 13 mm EDTA. After an incubation at 50 °C for 20 min, DNA products were extracted with a phenol/chloroform mixture and precipitated with isopropyl alcohol. The pellets were washed with 75% ethanol and dissolved in the Tris EDTA buffer. To score MMR, a fraction of the recovered DNA was digested with ClaI and HindIII, and the cleavage products separated on a native 1% agarose gel were visualized by ethidium bromide staining. Quantification of the repair products was performed using the ImageJ program. To visualize degradation of the discontinuous strand that occurs in an MMR reaction, a fraction of the recovered DNA was cleaved with ClaI, and the cleaved DNA was resolved on a denaturing agarose gel and hybridized with the indicated 32P-labeled probe. 32P-labeled DNAs were visualized with a Typhoon biomolecular imager (GE Healthcare), and quantification of the discontinuous strand degradation was performed using the ImageQuant program.
DNA Synthesis Assay
Each DNA synthesis reaction was run at 37 °C for 20 min in a 60-μl mixture containing 20 mm HEPES-NaOH (pH 7.4), 110 mm KCl, 5 mm MgCl2, 2 mm ATP, 4 mm DTT, 0.4 mg/ml BSA, 0.2 mm dATP, 0.2 mm dGTP, 0.2 mm dCTP, 0.2 mm dTTP, 3–5% (v/v) glycerol, and 0.6 nm phage MR59 single-stranded DNA primed with 12 oligonucleotides (53). When indicated, the reaction mixtures contained human ASF1A-H3-H4 (46 nm), CAF-1 (23 nm), PCNA (21 nm), RPA (52 nm), RFC (3 nm), Pol ϵ (5 nm), and Pol ϵ-D860A-D862A (5 nm). Each reaction was stopped by the addition of a 15-μl solution containing 0.5% SDS, 1 mg/ml proteinase K, 70 mm EDTA, 40% glycerol, and 48 ng of HindIII-digested pAH1A DNA (used as loading control), and the mixtures were incubated at 50 °C for 20 min. DNAs present in the mixtures were separated on denaturing agarose gels, transferred onto nylon membranes, and hybridized with a 32P-labeled probe. The probe was prepared by 5′-32P labeling of HhaI- and HinfI-digested single-stranded DNA of phage MR59 with T4 polynucleotide kinase. The DNA synthesis products were visualized by phosphorimaging.
Coimmunoprecipitation Assay
α-MSH2 (sc-494; Santa Cruz Biotechnologies), α-ASF1 (sc-53171; Santa Cruz Biotechnologies), and α-histone H3 (sc-8654; Santa Cruz Biotechnologies) antibodies were used in the coimmunoprecipitation assay that was carried out as described below. 5 μl of settled protein A-agarose beads equilibrated with buffer A (20 mm HEPES-NaOH (pH 7.4), 100 mm NaCl, 0.1 mm EDTA, 0.01% Nonidet P-40, 5% (v/v) glycerol, 0.1 mm DTT, and 0.2 mm PMSF) were mixed with 5 μl of buffer A and 25 μl of the indicated antibodies (5 μg), and the mixture was incubated at 4 °C for 16 h with gentle mixing. The beads were then washed with buffer A and incubated at 4 °C for 2 h with an indicated human protein (MutSα (32 pmol), ASF1A-H3-H4 (22 pmol), ASF1A (22 pmol), or histone H3-H4 complex (28 pmol)) in a 30-μl mixture that also contained 20 mm HEPES-NaOH (pH 7.4), 100 mm NaCl (or 20 mm NaCl + 80 mm KCl), 0.1 mm EDTA, 0.01% Nonidet P-40, 5% (v/v) glycerol, 0.1 mm DTT, 0.2 mm PMSF, and 0.2 mg/ml BSA. After the incubation, the beads were washed with buffer A containing 5% nonfat milk. The beads were next incubated at 4 °C for 1.5 h with an indicated human protein (MutSα (17 pmol), ASF1A-H3-H4 (22 pmol), or ASF1A (34 pmol)) in a 30-μl mixture that contained 20 mm HEPES-NaOH (pH 7.4), 100 mm NaCl (or 20 mm NaCl and 80 mm KCl), 0.1 mm EDTA, 0.01% Nonidet P-40, 5% (v/v) glycerol, 0.1 mm DTT, 0.2 mm PMSF, and 5 mg/ml nonfat milk followed by extensive washing of the beads with buffer A. To elute bound proteins, the beads were mixed with 20–40-μl elution buffer (20 mm MOPS-NaOH (pH 7), 10% (v/v) glycerol, 1% SDS, 1% 2-mercaptoethanol, and 5 mm EDTA), and the mixture was incubated at 70 °C for 10 min. The protein input and elution fractions were analyzed by the Western blots using the ECL Plus or ECL Prime kits (GE Healthcare) to detect the signal.
Results
ASF1A-H3-H4-, CAF-1-, PCNA-, and RFC-dependent Deposition of the Histone (H3-H4)2 Tetramers
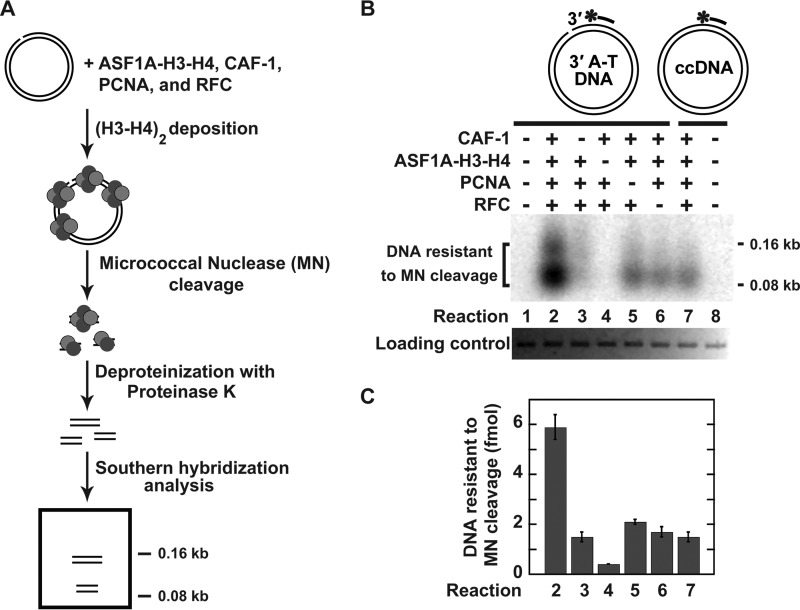
The ASF1A-H3-H4 heterotrimers are a major source of histones H3 and H4 that are deposited onto newly replicated DNA by CAF-1 in S phase (61). We started this work to study whether human MMR interacts with CAF-1- and ASF1A-H3-H4-dependent histone (H3-H4)2 tetramer deposition. In these and other experiments described below we used a set of highly purified human proteins (Fig. 1). We first performed experiments to reconstitute CAF-1-dependent histone (H3-H4)2 tetramer deposition that uses ASF1A-H3-H4 as the only source of histones H3 and H4 (Fig. 2). The experiments were based on previous observations that showed 1) that CAF-1 and PCNA are required for replication-dependent nucleosome assembly (50, 51, 54), 2) that incubation of CAF-1, PCNA, RFC, and the histone H3-H4 complex with a nicked circular DNA leads to deposition of (H3-H4)2 tetramers (56), 3) that a deposited (H3-H4)2 tetramer binds ∼146-bp DNA (47), and 4) that cleavage of DNA containing histone (H3-H4)2 tetrasomes with micrococcal nuclease produces ∼70–150-bp fragments (47, 56). Our experiments revealed that the micrococcal nuclease cleavage of the nicked DNA that had been incubated in the reaction mixture containing near homogeneous human CAF-1, ASF1A-H3-H4, PCNA, and RFC produced the ∼70-bp and 150-bp fragments (Fig. 2, B and C, Reaction 2). The omission experiments showed that the yield of the ∼70- and 150-bp fragments decreased 3–4-fold when micrococcal nuclease cleaved the nicked DNA that had been incubated in the reaction mixture lacking CAF-1, PCNA, or RFC (Fig. 2, B and C, Reactions 3, 5, and 6). The replacement of the nicked DNA with the relaxed ccDNA decreased the yield of the ∼70- and 150-bp fragments by 4-fold (Fig. 2, B and C, Reaction 7). No 70- and 150-bp fragments were formed when micrococcal nuclease cleaved the nicked DNA that had been incubated in the reaction mixture lacking ASF1A-H3-H4 (Fig. 2B, Reaction 4). Taken together, the results of these experiments demonstrated that CAF-1, ASF1A-H3-H4, PCNA, and RFC form a 4-protein system that deposits the histone (H3-H4)2 tetramers onto DNA in a nick-dependent manner.
FIGURE 1.
Human proteins used in this work. The proteins were purified as described under “Experimental Procedures.” The purified proteins were separated in SDS gels, and the protein bands were visualized with Coomassie Blue R-250 staining.
FIGURE 2.
CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent deposition of the histone (H3-H4)2 tetramers in a defined system. The data were obtained using the histone (H3-H4)2 tetramer deposition assay (“Experimental Procedures”). A, outline of the histone (H3-H4)2 tetramer deposition assay. B, representative image showing DNAs protected from micrococcal nuclease cleavage by the histone (H3-H4)2 tetramer deposition. The reaction mixtures contained the indicated proteins and DNA substrates (0.6 nm). When ASF1A-H3-H4, CAF-1, PCNA, and RFC were present in the reaction mixtures, their concentrations were 46, 15, 21, and 3 nm, respectively. The reaction products were analyzed by Southern hybridization with a 32P-labeled 24-mer oligonucleotide v5690 used as a probe. The diagrams outline the DNA substrates and also show the relative position of the hybridization probe (a bar with an asterisk), which is complementary to the top strand. The top strand in 3′-nicked A-T DNA contains a strand break, which is absent in the relaxed ccDNA. C, graphical representation of the results shown in B. The data are the averages ± 1 S.D. (n = 4).
MutSα Inhibits CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent Formation of Mismatch-containing Tetrasomes
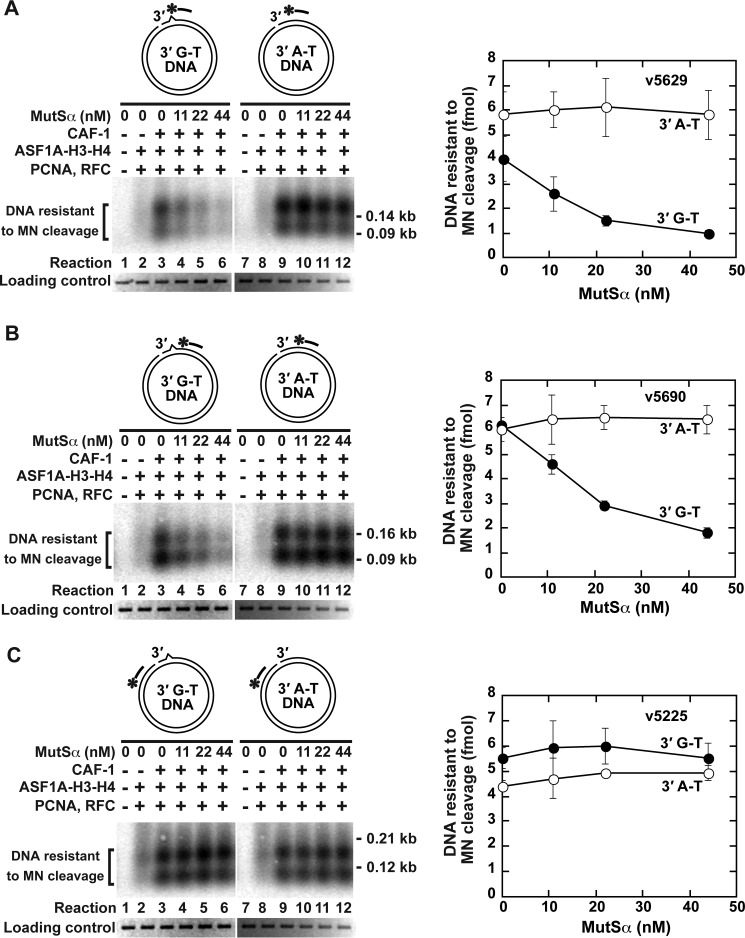
Having reconstituted CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent deposition of histone (H3-H4)2 tetramers onto a nicked DNA, we carried out a series of experiments to investigate whether this process is affected by the MMR system (Fig. 3). In these experiments we utilized two different DNA substrates; 3′-nicked A-T DNA that lacked a DNA mismatch and 3′-nicked G-T DNA that contained a G-T mismatch. Analysis of the reactions that occurred on 3′-nicked G-T DNA revealed that MutSα, a key component of the MMR system, inhibited CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition both on a DNA site containing the mispair (Fig. 3A) and on a nearby site ∼40 bp to the right of the mispair (Fig. 3B). However, MutSα did not affect the histone (H3-H4)2 tetramer deposition on a site that was ∼450 bp to the left from the mispair (Fig. 3C). Furthermore, the results of the control reactions indicated that MutSα did not impact the formation of tetrasomes at any of the three sites on 3′-nicked A-T DNA (Fig. 3, A–C). These data implied that the MMR system inhibits CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent formation of mismatch-containing tetrasomes.
FIGURE 3.
Mismatch-dependent inhibition of CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent deposition of the histone (H3-H4)2 tetramers by the mismatch recognition factor MutSα. The data were obtained using the histone (H3-H4)2 tetramer deposition assay. The reaction mixtures contained the indicated proteins and DNA substrates (0.6 nm). When ASF1A-H3-H4, CAF-1, PCNA, and RFC were present in the reaction mixtures, their concentrations were 46, 15, 21, and 3 nm, respectively. A–C, analysis of the histone deposition reactions by Southern hybridizations with 32P-labeled probes v5629 (A), v5690 (B), and v5225 (C). The diagrams outline the DNA substrates and also show the relative positions of the hybridization probes (bars with asterisks), which are complementary to the discontinuous (top) strand. The data shown in the graphs are the averages ± 1 S.D. (n = 3) and were obtained by quantification of images including those in the figure.
The Effects of CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent Formation of Tetrasomes on Excision-dependent and Excision-independent MMR Reactions
Current evidence suggests that mismatch-containing DNA that is packaged into nucleosomes is resistant to the action of MMR (56, 65). Based on this evidence and the observations that MutSα suppresses two subpathways of the histone (H3-H4)2 tetramer deposition (Ref. 56 and Fig. 3), we hypothesized that a significant fraction of the MMR events occurs during replication-coupled nucleosome assembly but before the nascent mismatch-containing DNA is packaged into nucleosomes. Because the assembly of the tetrasome is the first step in the assembly of the nucleosome, this hypothesis predicted that reconstituted MMR reactions would be able to take place during CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition (Fig. 3). In experiments that are described below we tested and confirmed this prediction. In addition, we analyzed the effects of histone (H3-H4)2 tetramer deposition on reconstituted MMR reactions.
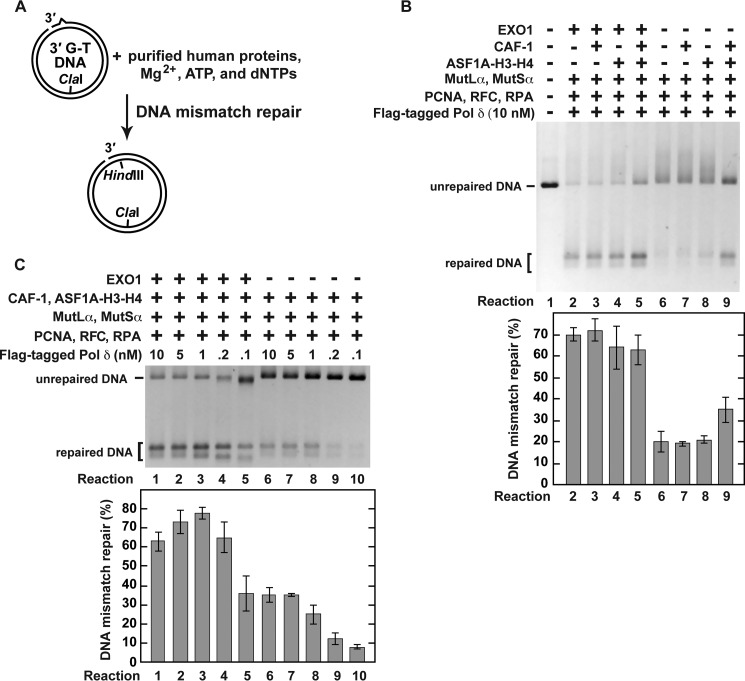
Previous research determined that EXO1, MutLα, MutSα, PCNA, Pol δ, RFC, and RPA form a system that corrects mismatches on nicked DNAs (39, 40, 42–44). A 3′ or 5′ nick-directed MMR reaction carried out by this purified system is dependent on mismatch excision by EXO1 (39, 40, 42–44). Removal of EXO1 from this seven-protein system produces a system that erases mismatches on nicked DNAs via the mismatch excision-independent reaction (44). This and the other (Fig. 2) information allowed us to generate two new reconstituted systems. One of these systems that contained nine human proteins (EXO1, MutLα, MutSα, PCNA, Pol δ, RFC, RPA, CAF-1, and ASF1A-H3-H4) supported the excision-dependent MMR and the four protein-dependent histone (H3-H4)2 tetramer deposition (i.e. a histone (H3-H4)2 tetramer deposition dependent on CAF-1, ASF1A-H3-H4, PCNA, and RFC) and the other that contained eight human proteins (MutLα, MutSα, PCNA, Pol δ, RFC, RPA, CAF-1, and ASF1A-H3-H4) supported the excision-independent MMR and the four protein-dependent histone (H3-H4)2 tetramer deposition (Figs. 2 and 4). In these two systems, Pol δ was the only source of DNA polymerase activity. We utilized these two systems to investigate whether the excision-dependent and excision-independent MMR were affected by the four protein-dependent histone (H3-H4)2 tetramer deposition (Figs. 4 and 5). We first studied how the four protein-dependent histone (H3-H4)2 tetramer deposition influenced the efficiencies of the excision-dependent and excision-independent MMR reactions (Fig. 4). The results revealed that the four protein-dependent histone (H3-H4)2 tetramer deposition did not change the efficiency of the mismatch excision-dependent MMR but modestly increased the efficiency of the excision-independent MMR (Fig. 4B, Reactions 2, 5, 6, and 9). We also determined the impact of different concentrations of Pol δ on the excision-dependent and excision-independent MMR reactions that took place in the presence of the four protein-dependent histone (H3-H4)2 tetramer deposition. As shown in Fig. 4C, ∼⅓ of 3′-nicked G-T DNA substrate was repaired in the excision-dependent reaction that was supported by 0.1 nm Pol δ (reaction 5), and the same level of the repair in the excision-independent reaction was achieved in the presence of 5 nm Pol δ (reaction 7). This observation indicated that much less Pol δ was required for the excision-dependent MMR reaction than for the excision-independent MMR reaction.
FIGURE 4.
Pol δ-dependent MMR reactions that occur in the presence of CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition. The data were obtained using the MMR assay (“Experimental Procedures”). The reaction mixtures contained the indicated proteins and DNA substrate (0.6 nm). When ASF1A-H3-H4, CAF-1, EXO1, MutLα, MutSα, PCNA, RFC, and RPA were present in the reaction mixtures, their concentrations were 46, 23, 3, 6, 22, 21, 3, and 52 nm, respectively. After incubation at 37 °C for 20 min, the reactions were stopped. DNAs recovered from the reaction mixtures were cleaved with HindIII and ClaI, and the cleavage products separated on a native 1% agarose gel were visualized with ethidium bromide staining. A, outline of the MMR assay. B, MMR products that were formed in the indicated reconstituted reactions. Note that the DNA products that were generated in the reconstituted reactions included those that moved in the gel slower than the unrepaired DNA. These slower moving products are MMR reaction intermediates that were formed as a result of strand-displacement syntheses initiated by Pol δ from both the original 3′ end and MutLα endonuclease-generated 3′ ends. C, effects of the different concentrations of Pol δ on the efficiencies of the excision-dependent and excision-independent MMR reactions. The data shown in the graphs are the averages ± 1 S.D. (n = 3) and were obtained by quantification of images including those in the figure.
FIGURE 5.
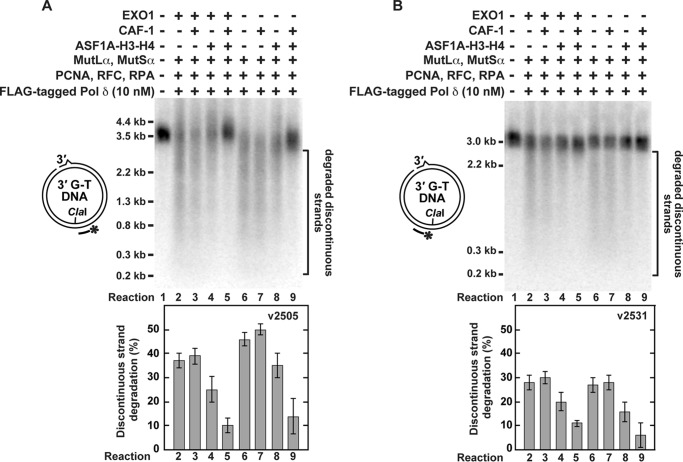
CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition inhibits unnecessary degradation of the discontinuous strand that occurs in the reconstituted Pol δ-dependent MMR reactions. The reaction conditions were the same as in Fig. 4B. DNAs recovered from the indicated reaction mixtures were cleaved with ClaI and then separated on a denaturing agarose gel. After the denaturing gel electrophoresis, the DNAs were analyzed by Southern hybridizations with 32P-labeled probes v2505 (A) and v2531 (B). The data shown in the graphs are the averages ± 1 S.D. (n = 3) and were obtained by quantification of images including those present in this figure.
An earlier study has demonstrated that a strong and unnecessary degradation of the discontinuous strand occurs in the reconstituted excision-dependent MMR reaction (56). We asked whether a similar degradation of the discontinuous strand took place in the reconstituted excision-independent MMR reaction. To this end, DNA products recovered from the reaction mixtures were cleaved with ClaI, separated on denaturing agarose gels, and analyzed by Southern hybridizations with two 32P-labeled oligonucleotide probes. One of the probes, v2505, hybridized to a discontinuous strand sequence located immediately 3′ to the ClaI site (Fig. 5A), and the other, v2531, annealed to a discontinuous strand sequence positioned immediately 5′ to the ClaI site (Fig. 5B). The results of the Southern hybridization analysis showed that the level of degradation of the discontinuous strand in the excision-independent MMR reaction was similar to that in the excision-dependent MMR reaction (Fig. 5, A and B, Reactions 2 and 6). We next studied whether CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition affected the degradation of the discontinuous strands in both the excision-dependent and excision-independent MMR reactions (Fig. 5). Inspection of the data led to the following findings. First, the degradation of the discontinuous strands was barely detectable in both the excision-dependent and excision-independent MMR reactions that took place in the presence of ASF1A-H3-H4 and CAF-1 (Fig. 5, A and B, Reactions 5 and 9). Second, the omission of either ASF1A-H3-H4 or CAF-1 increased the degradation of the discontinuous strands 3–5-fold and 2–3-fold, respectively (Fig. 5, A and B, Reactions 3, 4, 7, and 8). These findings indicated that although ASF1A-H3-H4 partially suppressed the degradation of the discontinuous strands in both the excision-dependent and excision-independent MMR reactions, the most effective suppression of the degradation of the discontinuous strands was observed in the reactions that occurred in the presence of CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent deposition of the histone (H3-H4)2 tetramers.
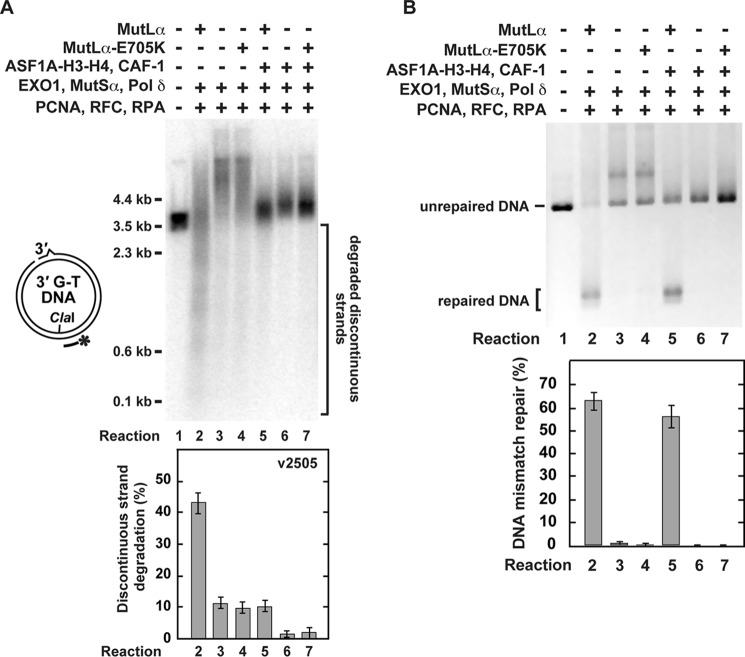
The excision-dependent MMR reaction (43) relies on the action of EXO1, MutLα, MutSα, PCNA, Pol δ, RFC, and RPA and is accompanied by a strong degradation of the discontinuous strand (Ref. 56 and Figs. 4B and 5, Reaction 2). To determine whether the endonuclease activity of MutLα is involved in the degradation of the discontinuous strand, we performed experiments (Fig. 6) that made use of an endonuclease-deficient MutLα-E705K mutant (33). The results of these experiments demonstrated that the endonuclease activity of MutLα produced the majority of discontinuous strand degradation products in the reconstituted excision-dependent MMR reaction (Fig. 6A, Reactions 2 and 4).
FIGURE 6.
The effects of replacement of wild-type MutLα with an endonuclease-deficient MutLα-E705K on the reconstituted excision-dependent MMR reactions. The MMR assay (“Experimental Procedures”) was used to obtain the data. The reaction mixtures contained the indicated proteins and 3′-nicked G-T DNA substrate (0.6 nm). When present in the reaction mixtures, ASF1A-H3-H4, CAF-1, EXO1, MutLα, MutLα-E705K, MutSα, PCNA, Pol δ, RFC, and RPA were at the final concentrations of 46, 23, 3, 6, 6, 22, 21, 10, 3, and 52 nm, respectively. After a 20-min incubation at 37 °C the reactions were stopped, and DNAs present in the reaction mixtures were recovered and analyzed. Analysis of the degradations of the discontinuous strand (A) and MMR (B) is shown in the indicated reactions. To visualize the degradations of the discontinuous strands, the recovered DNAs were cleaved with ClaI, separated on a denaturing agarose gel, and analyzed by a Southern hybridization with the 32P-labeled probe v2505. The data shown in the graphs are the averages ± 1 S.D. (n ≥ 3) and were obtained by quantification of images including those present in A and B.
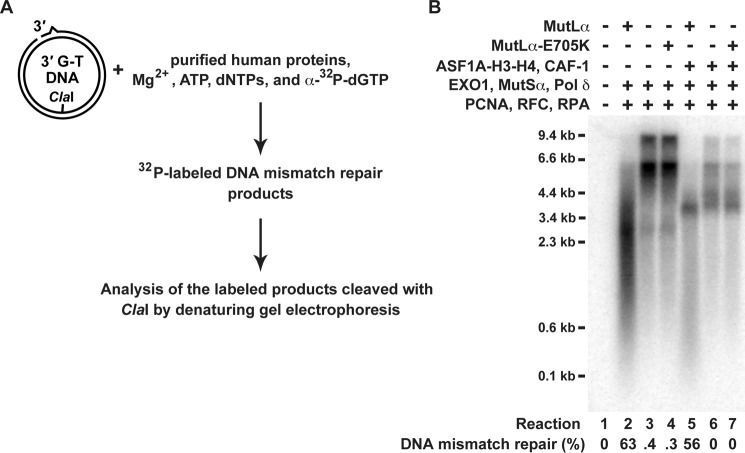
We next investigated whether the sizes of the discontinuous strand products visualized with the indirect labeling (Figs. 5 and 6A) correlated with those visualized with a direct labeling (Fig. 7). A comparison of the results of the indirect (Figs. 5 and 6A) and direct labeling (Fig. 7) experiments indicated that there was a significant correlation between the sizes of the products visualized with the two different methods.
FIGURE 7.
Analysis of DNA molecules labeled with [32P]dGMP during the course of the reconstituted excision-dependent MMR reactions. The reaction mixtures included 33 μCi/ml [α-32P]dGTP (3000 Ci/mmol). The other reaction conditions are described in Fig. 6. A fraction of each recovered DNA was cleaved with ClaI and HindIII to score MMR, and the rest was cleaved with ClaI and separated on a denaturing agarose gel. The gel was dried and exposed to a phosphorimaging screen. The image was generated with a Typhoon biomolecular imager (GE Healthcare). A, outline of the experiment. B, [32P]dGMP-labeled DNA molecules formed in the indicated reactions. The level of MMR in each of the reactions is also shown. The MMR data are the averages ± 1 S.D. (n ≥ 2).
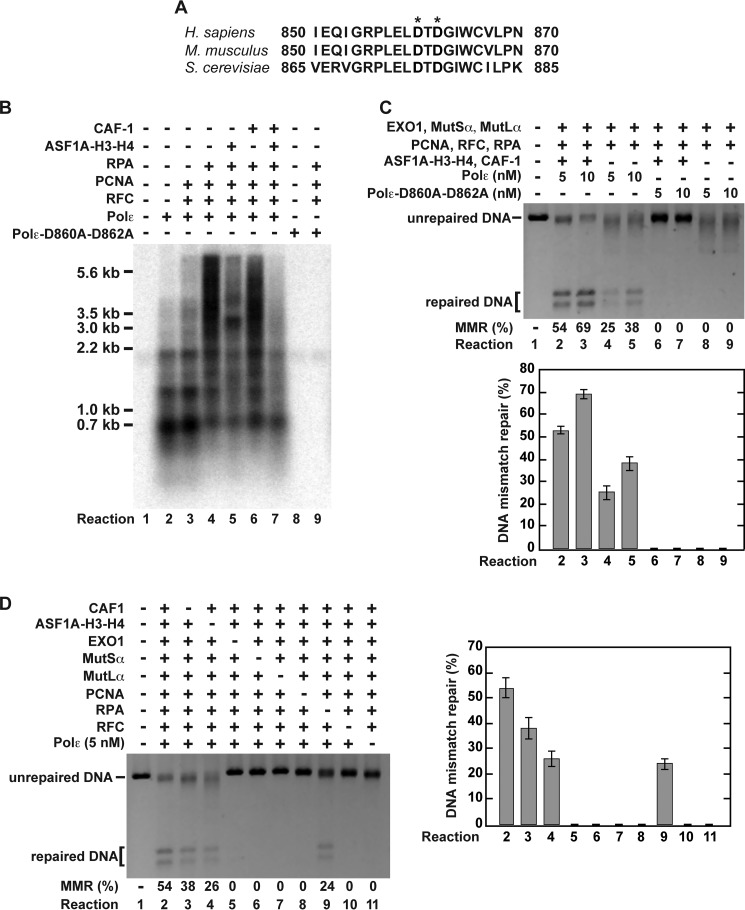
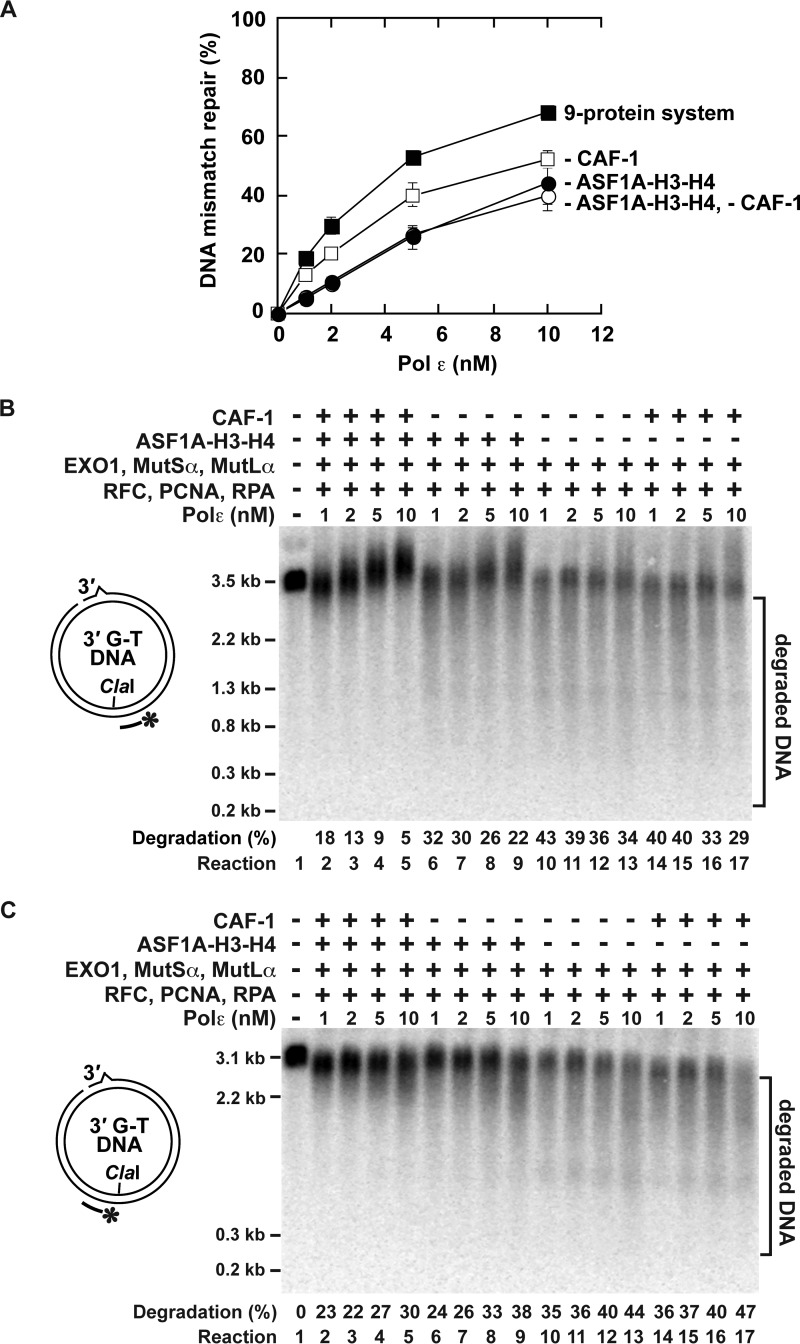
MMR reactions that were analyzed above (Figs. 4–7) depended on Pol δ. We also studied whether the four-protein-dependent histone (H3-H4)2 tetramer deposition affected reconstituted Pol ϵ-dependent MMR reactions (Figs. 8 and 9). We began this series of experiments by purifying recombinant human four-subunit Pol ϵ that was expressed in baculovirus-infected insect Sf9 cells. An electrophoretic analysis (Fig. 1) and a mass spectrometry analysis (data not shown) identified that the purified Pol ϵ contained the p261, p59, p17, and p12 subunits. The purified protein displayed DNA polymerase activity (Fig. 8B, Reaction 2). As expected from previous research (66, 67), PCNA, RFC, and RPA stimulated Pol ϵ to synthesize DNA strands that were significantly longer than those produced in the reaction that occurred in the absence of the accessory proteins (Fig. 8B, Reactions 2 and 4). This stimulation of the Pol ϵ biosynthetic activity required the presence of all three accessory proteins (Fig. 8B, Reactions 2–4, and data not shown). We then performed the reconstitution experiments (Figs. 8, C and D, and 9A). In accord with our knowledge of the eukaryotic MMR, the reconstitution experiments demonstrated that the reactions that occurred in the presence of Pol ϵ, EXO1, MutLα, MutSα, PCNA, RFC, and RPA led to repair of G-T mismatches (Figs. 8C, Reactions 4 and 5, and 9A). Further analysis of the reconstituted Pol ϵ-dependent MMR reaction revealed that it was accompanied by a strong degradation of the discontinuous strand (Fig. 9, B and C, reactions 10–13). Supplementation of the Pol ϵ-containing system with CAF-1 and ASF1A-H3-H4 stimulated the MMR reactions by 2-fold (Figs. 8, C, Reactions 2–3, and 9A) and suppressed the degradation of the discontinuous strands (Fig. 9, B and C, Reactions 2–5). These results suggested that the four protein-dependent histone (H3-H4)2 tetramer deposition stimulated the Pol ϵ-dependent MMR reaction and suppressed the degradation of the discontinuous strand.
FIGURE 8.
Pol ϵ-dependent MMR reaction reconstituted with purified human proteins. A, the alignment shows that the catalytic Asp-860 and Asp-862 residues that are marked by the asterisks are present in human Pol ϵ p261 subunit. B, effects of different human proteins on DNA polymerase activities of Pol ϵ and Pol ϵ-D860A-D862A. The data were obtained with the DNA synthesis assay (“Experimental Procedures”). C and D, Pol ϵ-dependent MMR reaction reconstituted with the indicated human proteins. The MMR assay was utilized to acquire the data. The reaction mixtures contained the indicated proteins and 3′-nicked G-T DNA substrate (0.6 nm). When ASF1A-H3-H4, CAF-1, EXO1, MutLα, MutSα, PCNA, RFC, and RPA were present in the reaction mixtures, their concentrations were 46, 23, 3, 6, 22, 21, 3, and 52 nm, respectively. The reactions were incubated at 37 °C for 20 min. The data shown in the graphs are the averages ± 1 S.D. (n ≥ 3) and were obtained by quantification of images including those in C and D.
FIGURE 9.
CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition suppresses unnecessary degradation of the discontinuous strand that occurs in the reconstituted Pol ϵ-dependent MMR reaction. The data were obtained using the MMR assay. The reaction mixtures contained the indicated proteins and 3′-nicked G-T DNA substrate (0.6 nm). When ASF1A-H3-H4, CAF-1, EXO1, MutLα, MutSα, PCNA, RFC, and RPA were included in the reaction mixtures, their concentrations were 46, 23, 3, 6, 22, 21, 3, and 52 nm, respectively. The Pol ϵ concentration was 1, 2, 5, or 10 nm as indicated. The reactions were carried out at 37 °C for 20 min. A, effects of ASF1A-H3-H4, CAF-1, and the different concentrations of Pol ϵ on MMR. The nine-protein MMR system contained ASF1A-H3-H4, CAF-1, EXO1, MutLα, MutSα, PCNA, RFC, RPA, and Pol ϵ. The data are the averages ± 1 S.D. (n ≥ 3). B and C, representative images that show degradations of the discontinuous strands in the reconstituted Pol ϵ-dependent MMR reactions. DNAs recovered from the indicated reaction mixtures were cleaved with ClaI and then separated on a denaturing agarose gel. After the separation, the ClaI-cleaved DNAs were analyzed by Southern hybridizations with 32P-labeled probes v2505 (B) and v2531 (C). The data are averages (n ≥ 3).
To confirm that Pol ϵ was responsible for DNA synthesis in the reconstituted MMR reaction (Fig. 8C), we carried out experiments that took advantage of a Pol ϵ-D860A-D862A mutant protein (Fig. 1) (68). In this mutant protein, the two catalytic aspartate residues are replaced with alanine residues (Fig. 8A). Consistent with a prior work (68), this Pol ϵ mutant protein that was expressed and purified as the wild-type enzyme (Fig. 1) lacked DNA polymerase activity (Fig. 8B, Reactions 8 and 9). Replacement of Pol ϵ with this mutant protein abolished the MMR reactions in both the nine- and seven-protein systems (Fig. 8C, Reactions 6–9). These data indicated that Pol ϵ re-synthesized DNA in the reconstituted MMR reactions (Fig. 8C, Reactions 2–5). We then performed the omission experiments to further characterize the reconstituted Pol ϵ-dependent MMR reaction that occurred in the nine-protein system (Figs. 8D and 9). The omission of MutSα, MutLα, PCNA, RFC, or Pol ϵ completely abolished the Pol ϵ-dependent MMR reaction (Fig. 8D, Reactions 2, 6–8, 10, and 11), but the omission of RPA only decreased the efficiency of the Pol ϵ-dependent MMR reaction (Fig. 8D, Reaction 9). Surprisingly and in contrast to the Pol δ-dependent MMR reaction (44) (Fig. 4), the Pol ϵ-dependent reaction did not occur in the absence of EXO1 (Fig. 8D, Reaction 5). We also found that the efficiency of the Pol ϵ-dependent MMR reaction increased with increasing Pol ϵ concentration and decreased in the absence of ASF1A-H3-H4 or CAF-1 (Fig. 9A). Taken together, the results of the above experiments (Figs. 8 and 9) provided evidence that Pol ϵ can perform DNA synthesis in excision-dependent MMR.
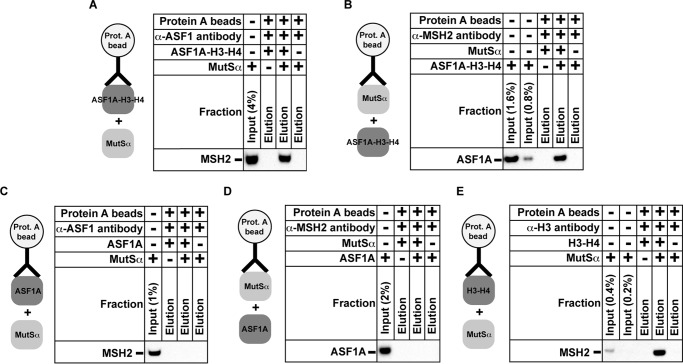
Protein-Protein Interaction between MutSα and ASF1A-H3-H4
Protein-protein interactions coordinate numerous processes that take place on nuclear DNA. Previous research has shown that the mismatch recognition factor MutSα physically interacts with the histone chaperone CAF-1 (62). To determine whether there are additional protein-protein interactions between the components of the MMR system and replication-coupled nucleosome assembly, we carried out the pulldown experiments that involved the purified MutSα and ASF1A-H3-H4 (Fig. 10). We found that the ASF1A-H3-H4 heterotrimers tethered to the protein A beads via the α-ASF1 antibodies pulled down the purified MutSα (Fig. 10A). The possibility of a nonspecific binding of MutSα to the beads was ruled out in an experiment that showed that MutSα molecules were not pulled down by the α-ASF1 antibody-containing beads that lacked ASF1A-H3-H4 (Fig. 10A). Further experiments established that the MutSα-containing beads pulled down the purified ASF1A-H3-H4 (Fig. 10B). Based on these results, we concluded that MutSα physically interacted with ASF1A-H3-H4. To determine whether MutSα forms a complex with ASF1A, the H3-H4 dimer, or both components of the heterotrimer, we conducted pulldown experiments that included these three proteins (Fig. 10, C–E). The results revealed no evidence that MutSα interacted with ASF1A (Fig. 10, C and D) but showed that the histone H3-H4-containing beads pulled down the purified MutSα (Fig. 10E). These findings indicated that MutSα forms a complex with ASF1A-H3-H4, probably via the H3-H4 dimer.
FIGURE 10.
Protein-protein interaction between MutSα and ASF1A-H3-H4. The data were acquired using the coimmunoprecipitation assay (“Experimental Procedures”). Complexes containing protein A beads and the indicated proteins and antibodies were incubated with the purified MutSα (A, C, and E), ASF1A-H3-H4 (B), or ASF1A (D). After the incubation, the beads were extensively washed, and the bound material was eluted. The input and eluted fractions were analyzed by Western blots with α-MSH2 antibodies (A, C, and E) or α-ASF1 antibodies (B and D).
Discussion
The multifunctional MMR system plays a major role in maintaining genome integrity in bacteria and eukaryotes (1–3, 5–8). The genetic stability engendered by the MMR system is required for the suppression of both sporadic and inherited cancers in humans (4). Previous studies have defined the reconstituted human MMR reactions that occur on naked DNA (33, 35, 36, 40, 42–44, 69). However, much less is known about the reactions that repair mismatches in the nucleosomal environment. In this work we have determined that human MMR interacts with CAF-1- and ASF1A-H3-H4-dependent histone (H3-H4)2 tetramer deposition (Figs. 3–10). Our findings are consistent with the idea that a subset of human MMR events coincides with CAF-1- and ASF1A-H3-H4-dependent deposition of histone (H3-H4)2 tetramers onto the newly synthesized DNA (Figs. 3–9). Taken together with the results of previous research (56, 62), our findings support the view that MMR takes place during CAF-1-dependent packaging of the newly replicated DNA into nucleosomes.
Experiments summarized in Fig. 3, A and B, identified that MutSα inhibits CAF-1- and ASF1A-H3-H4-dependent histone (H3-H4)2 tetramer deposition onto DNA at and near a mismatch. A different study showed that MutSα causes a very similar effect on CAF-1-dependent and ASF1A-H3-H4-independent histone (H3-H4)2 tetramer deposition (56). Thus, MutSα interferes with two reconstituted subpathways of CAF-1-dependent histone (H3-H4)2 tetramer deposition that can package DNA mismatches into tetrasomes. Earlier work found that DNA packaged into nucleosomes is refractory to MMR (56) and that nucleosomes inhibit mismatch recognition by MutSα (65). How can these findings be reconciled with the evidence that MMR coincides with CAF-1-dependent assembly of the newly replicated DNA into nucleosomes? We propose that suppression of CAF-1-dependent histone (H3-H4)2 tetramer deposition by MutSα at and around the mismatch maintains the nascent DNA region free of nucleosomes, thereby allowing MMR to occur on the locally naked DNA. Once the mismatch is removed, MutSα no longer blocks CAF-1-dependent histone (H3-H4)2 tetramer deposition, and the repaired DNA is packaged into nucleosomes (Fig. 11).
FIGURE 11.

MMR in the presence of CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition. The findings described in this work are consistent with the model in which a subset of human MMR events coincides with CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent deposition of the histone (H3-H4)2 tetramers onto the newly synthesized DNA. The model is in part based on the results of previous studies of human MMR (27, 33, 35, 39, 40, 42–44, 56). The model also depicts that MutSα interacts with ASF1A-H3-H4. As described under “Discussion,” this interaction may be important to facilitate MMR in the presence of CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition. HE, holoenzyme.
Unlike the MMR reaction that occurs in nuclear extracts containing CAF-1, the MMR reaction reconstituted with near homogenous EXO1, MutLα, MutSα, PCNA, Pol δ, RPA, and RFC (43) causes extensive and unnecessary degradation of the discontinuous strands (56). Similar degradation of the discontinuous strands takes place in MMR reactions in a cytosolic extract that is deficient in the replicative histone chaperone CAF-1. Supplementation of the cytosolic extract with purified CAF-1 blocks the unnecessary degradation of the discontinuous strands (56). Analysis of the products formed in the cytosolic extract reactions that occurred in the presence of CAF-1 revealed that the nicked heteroduplex DNA substrate is subjected to both MMR and CAF-1-dependent packaging into nucleosomes. These data support the idea that CAF-1-dependent nucleosome assembly suppresses the unnecessary degradation of the discontinuous strand during MMR. Consistent with this idea, the unnecessary degradation of the discontinuous strand is suppressed in the reconstituted MMR reaction that takes place in the presence of CAF-1-dependent and ASF1A-H3-H4-independent histone (H3-H4)2 tetramer deposition (56). To obtain additional insight into the mechanism of MMR in the chromatin environment, we have studied the impact of the CAF-1- and ASF1A-H3-H4-dependent histone tetramer deposition on MMR in three reconstituted systems. One of the reconstituted systems supports the excision- and Pol δ-dependent MMR and contains EXO1, MutLα, MutSα, PCNA, Pol δ, RFC, RPA, CAF-1, and ASF1A-H3-H4. Another reconstituted system is proficient in the excision-independent and Pol δ-dependent MMR and is composed of MutLα, MutSα, PCNA, Pol δ, RFC, RPA, CAF-1, and ASF1A-H3-H4. The third system supports the excision- and Pol ϵ-dependent MMR and is composed of EXO1, MutLα, MutSα, PCNA, Pol ϵ, RFC, RPA, CAF-1, and ASF1A-H3-H4. In agreement with previous research (56), we have determined that MMR that occurs in the presence of the CAF-1- and ASF1A-H3-H4-dependent histone deposition in the three reconstituted systems causes a very limited degradation of the discontinuous strand, but the omission of CAF-1 and ASF1A-H3-H4 increases the discontinuous strand degradation 2–4-fold (Figs. 5 and 9B). Because the increased degradation of the discontinuous strand does not increase the efficiency of MMR, such DNA degradation is unnecessary (Figs. 4, 5, 6, and 9). We note that although MutSα inhibits CAF-1-dependent histone (H3-H4)2 tetramer deposition in the vicinity of the mismatch, it does not affect the histone tetrasome formation at a site that is located ∼400 bp from the mismatch (56) (Fig. 3C). Our data indicated that histone (H3-H4)2 tetramers deposited at such sites block the unnecessary degradation of the discontinuous strand (Ref. 56 and Figs. 5 and 9B). We have also determined that CAF-1 and ASF1A-H3-H4 stimulate the excision-independent and Pol δ-dependent MMR reaction as well as the excision- and Pol ϵ-dependent MMR reaction (Figs. 4B and 9A). These results suggest that CAF-1-, ASF1A-H3-H4-, PCNA-, and RFC-dependent histone (H3-H4)2 tetramer deposition creates a productive environment for MMR.
Pol δ and Pol ϵ replicate the majority of nuclear DNA (70–72). Fractionation of HeLa nuclear extracts and reconstitution studies determined that Pol δ re-synthesizes DNA in human excision-dependent and excision-independent MMR reactions (43, 44, 46, 69). However, it has remained unknown whether Pol ϵ plays a role in MMR. Our analysis has indicated that Pol ϵ is able to support human MMR (Figs. 8 and 9). The reconstituted Pol ϵ-dependent MMR reaction requires the activities of EXO1, MutLα, MutSα, PCNA, and RFC (Fig. 8D, Reactions 5–8 and 10). This finding indicates that the Pol ϵ-dependent MMR reaction includes the steps of the discontinuous strand incision, mismatch excision, and DNA re-synthesis. The dependence of the Pol ϵ-dependent MMR reaction on EXO1 (Fig. 8D) suggests that unlike human Pol δ (44), human Pol ϵ is not able to drive the excision-independent MMR reaction (Fig. 11). Comparison of the Pol δ- and Pol ϵ-dependent MMR reactions reveals that the specific MMR activity of Pol δ is ∼20 times higher than that of Pol ϵ (Figs. 4C, Reaction 5, and 9A). This supports the view that Pol δ re-synthesizes DNA in the majority of MMR events in human cells. Nevertheless, it remains possible that our biochemical analysis (Figs. 4C and 9A) underestimates the importance of Pol ϵ for human MMR because the reconstituted system (Fig. 8D) lacks one or more factors that are necessary to increase the MMR activity of this DNA polymerase. A recent analysis has shown that the DNA replication factors Ctf4 and GINS stimulate the biosynthetic activity of Pol ϵ (73, 74). Therefore, it may be that Ctf4 and GINS increase the MMR activity of Pol ϵ.
Our pulldown experiments have shown that MutSα forms a complex with ASF1A-H3-H4 (Fig. 10, A and B). Because MutSα is a large protein carrying both negative and positive charges on its surface and the histone H3-H4 dimer is a very basic protein molecule, it may be that the MutSα-ASF1A-H3-H4 complex is simply an in vitro artifact of nonspecific negative-positive charge interaction(s) between MutSα and H3-H4. However, we regard such a conclusion unlikely because in similar experiments we detected no complex formation between ASF1A-H3-H4 and MutLα, a large protein that, like MutSα, contains both negative and positive charges on its surface (data not shown). If the MutSα-ASF1A-H3-H4 interaction happens in vivo, what is the function of this interaction? We hypothesize that this interaction does not allow ASF1A-H3-H4 to participate in packaging of the newly synthesized mismatch-containing DNA into the tetrasomes. Once the mismatch is removed, the local concentration of MutSα sharply decreases, and as a result, ASF1A-H3-H4 is no longer blocked from the involvement in the assembly of the newly replicated DNA into nucleosomes. If the interaction between MutSα and histone H3-H4 (Fig. 10E) occurs in vivo, it may play a role similar to that proposed above for the MutSα-ASF1A-H3-H4 interaction.
In summary, findings described here and previously (56, 62, 65) support the view that the functional interactions between MMR and CAF-1-dependent nucleosome assembly ensure that the mismatch is corrected before it is packaged into the tetrasome and that unnecessary degradation of the discontinuous daughter strand does not occur during MMR.
Author Contributions
F. K. designed the experiments. E. R., L. K., and F. K. performed experiments, contributed reagents, analyzed data, and prepared the figures. F. K. and E. R. wrote the paper.
Acknowledgments
We thank Jerard Hurwitz, Francoise Ochsenbein, and Zachary Pursell for providing the plasmids that were used in this work and Farid F. Kadyrov for critical reading of the manuscript.
This work was supported by National Institutes of Health Grant R01GM095758 (NIGMS). This work was also supported by from the American Cancer Society, Illinois division, Grant 207957. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- MMR
- DNA mismatch repair
- ccDNA
- covalently closed DNA
- Pol δ
- DNA polymerase δ
- Pol ϵ
- DNA polymerase ϵ
- PCNA
- proliferating cell nuclear antigen
- RFC
- replication factor C
- RPA
- replication protein A
- EXO1
- exonuclease 1.
References
- 1. Iyer R. R., Pluciennik A., Burdett V., and Modrich P. L. (2006) DNA mismatch repair: functions and mechanisms. Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 2. Li G. M. (2008) Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 [DOI] [PubMed] [Google Scholar]
- 3. Harfe B. D., and Jinks-Robertson S. (2000) DNA Mismatch Repair and Genetic Instability. Annu. Rev. Genet. 34, 359–399 [DOI] [PubMed] [Google Scholar]
- 4. Lynch H. T., Snyder C. L., Shaw T. G., Heinen C. D., and Hitchins M. P. (2015) Milestones of Lynch syndrome: 1895–2015. Nat. Rev. Cancer 15, 181–194 [DOI] [PubMed] [Google Scholar]
- 5. Surtees J. A., Argueso J. L., and Alani E. (2004) Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res. 107, 146–159 [DOI] [PubMed] [Google Scholar]
- 6. Jiricny J. (2013) Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 5, a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kunkel T. A., and Erie D. A. (2015) Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kadyrova L. Y., and Kadyrov F. A. (2016) Endonuclease activities of MutLα and its homologs in DNA mismatch repair. DNA Repair 38, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Modrich P. (2006) Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 281, 30305–30309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su S.-S., and Modrich P. (1986) Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl. Acad. Sci. U.S.A. 83, 5057–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Au K. G., Welsh K., and Modrich P. (1992) Initiation of methyl-directed mismatch repair. J. Biol. Chem. 267, 12142–12148 [PubMed] [Google Scholar]
- 12. Grilley M., Welsh K. M., Su S.-S., and Modrich P. (1989) Isolation and characterization of the Escherichia coli mutL gene product. J. Biol. Chem. 264, 1000–1004 [PubMed] [Google Scholar]
- 13. Schofield M. J., Nayak S., Scott T. H., Du C., and Hsieh P. (2001) Interaction of MutS and MutL at a DNA mismatch. J. Biol. Chem. 276, 28291–28299 [DOI] [PubMed] [Google Scholar]
- 14. Cooper D. L., Lahue R. S., and Modrich P. (1993) Methyl-directed mismatch repair is bidirectional. J. Biol. Chem. 268, 11823–11829 [PubMed] [Google Scholar]
- 15. Dao V., and Modrich P. (1998) Mismatch, MutS, MutL, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J. Biol. Chem. 273, 9202–9207 [DOI] [PubMed] [Google Scholar]
- 16. Burdett V., Baitinger C., Viswanathan M., Lovett S. T., and Modrich P. (2001) In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 98, 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Viswanathan M., Burdett V., Baitinger C., Modrich P., and Lovett S. T. (2001) Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J. Biol. Chem. 276, 31053–31058 [DOI] [PubMed] [Google Scholar]
- 18. Lahue R. S., Au K. G., and Modrich P. (1989) DNA mismatch correction in a defined system. Science 245, 160–164 [DOI] [PubMed] [Google Scholar]
- 19. Pavlov Y. I., Mian I. M., and Kunkel T. A. (2003) Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr. Biol. 13, 744–748 [DOI] [PubMed] [Google Scholar]
- 20. Earley M. C., and Crouse G. F. (1998) The role of mismatch repair in the prevention of base pair mutations in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 95, 15487–15491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Russo M. T., Blasi M. F., Chiera F., Fortini P., Degan P., Macpherson P., Furuichi M., Nakabeppu Y., Karran P., Aquilina G., and Bignami M. (2004) The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol. Cell. Biol. 24, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen Y., Koh K. D., Weiss B., and Storici F. (2012) Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nat. Struct. Mol. Biol. 19, 98–104 [DOI] [PubMed] [Google Scholar]
- 23. Kadyrova L. Y., Dahal B. K., and Kadyrov F. A. (2015) Evidence that the DNA Mismatch repair system removes 1-nt okazaki fragment flaps. J. Biol. Chem. 290, 24051–24065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drummond J. T., Li G.-M., Longley M. J., and Modrich P. (1995) Isolation of an hMSH2·p160 heterodimer that restores mismatch repair to tumor cells. Science 268, 1909–1912 [DOI] [PubMed] [Google Scholar]
- 25. Palombo F., Gallinari P., Iaccarino I., Lettieri T., Hughes M., D'Arrigo A., Truong O., Hsuan J. J., and Jiricny J. (1995) GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science 268, 1912–1914 [DOI] [PubMed] [Google Scholar]
- 26. Palombo F., Iaccarino I., Nakajima E., Ikejima M., Shimada T., and Jiricny J. (1996) hMutSb, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 6, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 27. Genschel J., Littman S. J., Drummond J. T., and Modrich P. (1998) Isolation of hMutSb from human cells and comparison of the mismatch repair specificities of hMutSb and hMutSa. J. Biol. Chem. 273, 19895–19901 [DOI] [PubMed] [Google Scholar]
- 28. de Wind N., Dekker M., Claij N., Jansen L., van Klink Y., Radman M., Riggins G., van der Valk M., van't Wout K., and te Riele H. (1999) HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat. Genet. 23, 359–362 [DOI] [PubMed] [Google Scholar]
- 29. Umar A., Buermeyer A. B., Simon J. A., Thomas D. C., Clark A. B., Liskay R. M., and Kunkel T. A. (1996) Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87, 65–73 [DOI] [PubMed] [Google Scholar]
- 30. Johnson R. E., Kovvali G. K., Guzder S. N., Amin N. S., Holm C., Habraken Y., Sung P., Prakash L., and Prakash S. (1996) Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem. 271, 27987–27990 [DOI] [PubMed] [Google Scholar]
- 31. Tsurimoto T., and Stillman B. (1989) Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol. Cell. Biol. 9, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li G.-M., and Modrich P. (1995) Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl. Acad. Sci. U.S.A. 92, 1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kadyrov F. A., Dzantiev L., Constantin N., and Modrich P. (2006) Endonucleolytic function of MutLα in human mismatch repair. Cell 126, 297–308 [DOI] [PubMed] [Google Scholar]
- 34. Kadyrov F. A., Holmes S. F., Arana M. E., Lukianova O. A., O'Donnell M., Kunkel T. A., and Modrich P. (2007) Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease. J. Biol. Chem. 282, 37181–37190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pluciennik A., Dzantiev L., Iyer R. R., Constantin N., Kadyrov F. A., and Modrich P. (2010) PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 107, 16066–16071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iyer R. R., Pluciennik A., Genschel J., Tsai M. S., Beese L. S., and Modrich P. (2010) MutLα and proliferating cell nuclear antigen share binding sites on MutSbeta. J. Biol. Chem. 285, 11730–11739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szankasi P., and Smith G. R. (1992) A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J. Biol. Chem. 267, 3014–3023 [PubMed] [Google Scholar]
- 38. Amin N. S., Nguyen M. N., Oh S., and Kolodner R. D. (2001) exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21, 5142–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Genschel J., Bazemore L. R., and Modrich P. (2002) Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 277, 13302–13311 [DOI] [PubMed] [Google Scholar]
- 40. Genschel J., and Modrich P. (2003) Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell 12, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 41. Wei K., Clark A. B., Wong E., Kane M. F., Mazur D. J., Parris T., Kolas N. K., Russell R., Hou H. Jr., Kneitz B., Yang G., Kunkel T. A., Kolodner R. D., Cohen P. E., and Edelmann W. (2003) Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 17, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dzantiev L., Constantin N., Genschel J., Iyer R. R., Burgers P. M., and Modrich P. (2004) A defined human system that supports bidirectional mismatch-provoked excision. Mol. Cell 15, 31–41 [DOI] [PubMed] [Google Scholar]
- 43. Constantin N., Dzantiev L., Kadyrov F. A., and Modrich P. (2005) Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 280, 39752–39761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kadyrov F. A., Genschel J., Fang Y., Penland E., Edelmann W., and Modrich P. (2009) A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 106, 8495–8500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tishkoff D. X., Boerger A. L., Bertrand P., Filosi N., Gaida G. M., Kane M. F., and Kolodner R. D. (1997) Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. U.S.A. 94, 7487–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Longley M. J., Pierce A. J., and Modrich P. (1997) DNA polymerase delta is required for human mismatch repair in vitro. J. Biol. Chem. 272, 10917–10921 [DOI] [PubMed] [Google Scholar]
- 47. Dong F., and van Holde K. E. (1991) Nucleosome positioning is determined by the (H3-H4)2 tetramer. Proc. Natl. Acad. Sci. U .S. A. 88, 10596–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stillman B. (1986) Chromatin assembly during SV40 DNA replication in vitro. Cell 45, 555–565 [DOI] [PubMed] [Google Scholar]
- 49. Lucchini R., Wellinger R. E., and Sogo J. M. (2001) Nucleosome positioning at the replication fork. EMBO J. 20, 7294–7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith S., and Stillman B. (1989) Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25 [DOI] [PubMed] [Google Scholar]
- 51. Smith S., and Stillman B. (1991) Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kaufman P. D., Kobayashi R., and Stillman B. (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11, 345–357 [DOI] [PubMed] [Google Scholar]
- 53. Kadyrova L. Y., Rodriges Blanko E., and Kadyrov F. A. (2013) Human CAF-1-dependent nucleosome assembly in a defined system. Cell Cycle 12, 3286–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shibahara K., and Stillman B. (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 [DOI] [PubMed] [Google Scholar]
- 55. Rolef Ben-Shahar T., Castillo A. G., Osborne M. J., Borden K. L., Kornblatt J., and Verreault A. (2009) Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function. Mol. Cell. Biol. 29, 6353–6365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kadyrova L. Y., Blanko E. R., and Kadyrov F. A. (2011) CAF-I-dependent control of degradation of the discontinuous strands during mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 108, 2753–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Silljé H. H., and Nigg E. A. (2001) Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 58. Groth A., Rocha W., Verreault A., and Almouzni G. (2007) Chromatin challenges during DNA replication and repair. Cell 128, 721–733 [DOI] [PubMed] [Google Scholar]
- 59. Alvarez F., Muñoz F., Schilcher P., Imhof A., Almouzni G., and Loyola A. (2011) Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 286, 17714–17721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu W. H., Roemer S. C., Port A. M., and Churchill M. E. (2012) CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 40, 11229–11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Groth A., Ray-Gallet D., Quivy J. P., Lukas J., Bartek J., and Almouzni G. (2005) Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17, 301–311 [DOI] [PubMed] [Google Scholar]
- 62. Schöpf B., Bregenhorn S., Quivy J. P., Kadyrov F. A., Almouzni G., and Jiricny J. (2012) Interplay between mismatch repair and chromatin assembly. Proc .Natl. Acad. Sci. U.S.A. 109, 1895–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mousson F., Lautrette A., Thuret J. Y., Agez M., Courbeyrette R., Amigues B., Becker E., Neumann J. M., Guerois R., Mann C., and Ochsenbein F. (2005) Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl. Acad. Sci. U.S.A. 102, 5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Holmes J. Jr., Clark S., and Modrich P. (1990) Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc. Natl. Acad. Sci. U.S.A. 87, 5837–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li F., Tian L., Gu L., and Li G. M. (2009) Evidence that nucleosomes inhibit mismatch repair in eukaryotic cells. J. Biol. Chem. 284, 33056–33061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee S.-H., Pan Z.-Q., Kwong A. D., Burgers P. M., and Hurwitz J. (1991) Synthesis of DNA by DNA polymerase e in vitro. J. Biol. Chem. 266, 22707–22717 [PubMed] [Google Scholar]
- 67. Chilkova O., Stenlund P., Isoz I., Stith C. M., Grabowski P., Lundström E. B., Burgers P. M., and Johansson E. (2007) The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 35, 6588–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dua R., Levy D. L., and Campbell J. L. (1999) Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ϵ and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 274, 22283–22288 [DOI] [PubMed] [Google Scholar]
- 69. Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., and Li G. M. (2005) Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 70. Kunkel T. A., and Burgers P. M. (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Burgers P. M. (2009) Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pavlov Y. I., and Shcherbakova P. V. (2010) DNA polymerases at the eukaryotic fork-20 years later. Mutat. Res. 685, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bermudez V. P., Farina A., Tappin I., and Hurwitz J. (2010) Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J. Biol. Chem. 285, 9493–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bermudez V. P., Farina A., Raghavan V., Tappin I., and Hurwitz J. (2011) Studies on human DNA polymerase ϵ and GINS complex and their role in DNA replication. J. Biol. Chem. 286, 28963–28977 [DOI] [PMC free article] [PubMed] [Google Scholar]