Abstract
Ebola virus infection can cause severe hemorrhagic fever with a high mortality in humans. The outbreaks of Ebola viruses in 2014 represented the most serious Ebola epidemics in history and greatly threatened public health worldwide. The development of additional effective anti-Ebola therapeutic agents is therefore quite urgent. In this study, via high throughput screening of Food and Drug Administration-approved drugs, we identified that teicoplanin, a glycopeptide antibiotic, potently prevents the entry of Ebola envelope pseudotyped viruses into the cytoplasm. Furthermore, teicoplanin also has an inhibitory effect on transcription- and replication-competent virus-like particles, with an IC50 as low as 330 nm. Comparative analysis further demonstrated that teicoplanin is able to block the entry of Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS) envelope pseudotyped viruses as well. Teicoplanin derivatives such as dalbavancin, oritavancin, and telavancin can also inhibit the entry of Ebola, MERS, and SARS viruses. Mechanistic studies showed that teicoplanin blocks Ebola virus entry by specifically inhibiting the activity of cathepsin L, opening a novel avenue for the development of additional glycopeptides as potential inhibitors of cathepsin L-dependent viruses. Notably, given that teicoplanin has routinely been used in the clinic with low toxicity, our work provides a promising prospect for the prophylaxis and treatment of Ebola, MERS, and SARS virus infection.
Keywords: antibiotics, Ebola virus, glycoprotein, lysosome, virus entry, MERS-CoV, SARS-CoV, glycopeptide
Introduction
Ebola virus (EBOV)3 is a filamentous-enveloped, single-stranded, and negative-sense RNA virus, which is taxonomically classified to the Filoviridae (1). To date, five species in the Ebolavirus genus have been identified, including Zaire, Sudan, Reston, Tai Forest, and Bundibugyo ebolavirus (2–6). Ebola virus infection leads to severe viral hemorrhagic fever in humans and non-human primates. In March 2014, outbreaks of Ebola viruses began in Guinea and caused over 28,000 cases of infection and over 11,000 deaths, which posed a severe threat to public health worldwide.
The Ebola virus genome contains seven genes that encode the NP, VP35, VP40, glycoprotein (GP), VP30, VP24, and RNA-dependent RNA polymerase (L) virus proteins. To infect host cells, the GPs of Ebola viruses first bind to attachment molecules such as β1 integrins, DC-SIGNs, L-SIGNs, lectins, TIM-1s, Tyro3 family proteins, heparan sulfates, or folate receptor-α (7–13). Ebola viruses are then internalized by macropinocytosis and subsequently transported through the early and late endosomes and the endo/lysosomes (14–16), where the Ebola virus GPs are cleaved by cathepsin L and subsequently cathepsin B to expose the receptor-binding domains (17). After binding the specific receptor NPC1, Ebola viruses release their genomes into the cytoplasm of the host cells (16, 18).
Anti-EBOV vaccines and drugs are under extensive development. Two promising vaccines, rVSVΔG-EBOV-GP and cAd3-EBOV, have been shown to render non-human primates resistant to Ebola virus infections and are currently in clinical trials (19, 20). In addition, the anti-EBOV monoclonal antibody Zmapp, siRNAs, and other compounds that can inhibit Ebola virus infections have been developed (21–24). Furthermore, several clinically approved drugs were also reported to inhibit Ebola virus infections (25, 26). However, because the IC50 values of those drugs were relatively high, more anti-EBOV drugs with potent inhibitory activity are urgently needed. To facilitate their identification, the method of high throughput screening of clinically approved drugs, which could be applied immediately in the clinic, is a reasonable approach. In this study, we identified teicoplanin and several other glycopeptide antibiotics as Ebola virus entry inhibitors with high efficiency and low cytotoxicity, providing a promising means to effect the prophylaxis and treatment of Ebola virus infection.
Experimental Procedures
Cell Culture
HEK293T, A549, HeLa, Huh7.5.1, and Madin-Darby canine kidney cell lines were maintained in Dulbecco's modified Eagle's medium (Gibco) with 10% fetal calf serum (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco) at 37 °C and 5% CO2. THP-1 cell lines were maintained in RPMI1640 medium (Gibco) with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C and 5% CO2. Primary human umbilical vein endothelial cells were maintained in human endothelial-SFM (Gibco) with 30 ng/ml endothelial cell growth supplement (Merck Millipore), 20 ng/ml recombinant human FGF basic (146 amino acids) protein (R&D Systems), 20% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C and 5% CO2.
Plasmids
GP sequence of Zaire EBOV-2014 was chemically synthesized and inserted into pcDNA3.1 plasmid. The pHIV-luciferase and pCMV-VSV-G plasmids were obtained from Addgene, and the pCMV-ΔR8.2 plasmid was kindly provided by Dr. Trono (27). The p4cis plasmid that encodes a Renilla luciferase reporter, VP40, GP and VP24, the pCAGGS-NP, pCAGGS-VP35, pCAGGS-VP30, pCAGGS-L, pCAGGS-T7, and pCAGGS-Tim1 plasmids were produced as described previously (28).
Viruses
Pseudotyped viruses were produced by the co-transfection of pHIV-luciferase, pCMV-ΔR8.2, and different envelope plasmids into HEK293T cells that were 90% confluent in a 10-cm plate with Lipofectamine 2000 by following the manufacturer's instructions (Invitrogen). The amounts of plasmids were listed as follows: HIV-luc/Zaire EBOV-GP(2014) pseudotyped viruses: 4.5 μg of pHIV-luciferase, 4.5 μg of pCMV-ΔR8.2, and 7.65 μg of pcDNA3.1-Zaire EBOV-GP(2014); HIV-luc/VSV-G pseudotyped viruses: 4.5 μg of pHIV-luciferase, 4.5 μg of pCMV-ΔR8.2, and 2.7 μg of pCMV-VSV-G; HIV-luc/SARS-CoV-S pseudotyped viruses: 6.5 μg of pHIV-luciferase, 8 μg of pCMV-ΔR8.2, and 20 μg of pcDNA3.1-SARS-CoV-S; and HIV-luc/MERS-CoV-S pseudotyped viruses: 4.5 μg of pHIV-luciferase, 4.5 μg of pCMV-ΔR8.2, and 10 μg of pcDNA3.1-MERS-CoV-S. After 48 h, the supernatants that contain the pseudotyped viruses were collected and filtered through a 0.45-μm pore-size filter (Pall) and then stored at −80 °C until use. Ebola transcription- and replication-competent virus-like particles (trVLPs) were produced by the co-transfection of 250 ng of p4cis plasmids that encode a Renilla luciferase reporter gene, VP40, GP, and VP24, 250 ng of pCAGGS-T7, 125 ng of pCAGGS-NP, 125 ng of pCAGGS-VP35, 75 ng of pCAGGS-VP30, and 1000 ng of pCAGGS-L plasmids into HEK293T cells that were 50% confluent in a 6-well plate with Lipofectamine 2000 (Invitrogen). After 24 h, the medium was discarded, and the cells were incubated with fresh medium for 48 h. Then the supernatants that contain Ebola transcription- and replication-competent virus-like particles were collected and filtered through a 0.45-μm pore-size filter (Pall) and stored at −80 °C until use.
High Throughput Screening of FDA-approved Drug Library
High throughput screening of FDA-approved drug library (Topscience) was conducted in 96-well plates with 50 μm compounds per well. HEK293T cells were incubated with compounds at 37 °C for 1 h and then infected with 100 μl of p24-normalized (5 ng) HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses containing 10 μg/ml Polybrene. After 12 h, the medium was discarded, and the cells were washed briefly with PBS and incubated with fresh medium for 48 h. The intracellular luciferase activity was examined with GloMax® 96 Microplate luminometer (Promega), and the compounds that have the effects of more than 50% inhibition on the luciferase activity were selected for the secondary screening, which was executed with both HIV-luc/Zaire EBOV-GP (2014) and HIV-luc/VSV-G pseudotyped viruses in a similar procedure.
Cell Viability Assay
HEK293T cells were seeded in a 96-well plate (2 × 104 per well). After 24 h, the cells were incubated with teicoplanin at different concentrations at 37 °C for 48 h. The cell viability then was determined by the CellTiter® 96 Aqueous One Solution Cell Proliferation Assay (Promega).
Time-of-Addition Assay
HEK293T cells were seeded in a 96-well plate (2 × 104 per well). Twenty four hours later, the cells were infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses and incubated with 50 μm teicoplanin at 0, 2, 4, or 8 h post-infection. After 12 h, the medium was discarded, and the cells were washed briefly with PBS and incubated with fresh medium for 48 h. Then the intracellular luciferase activity was determined. The HIV-luc/VSV-G pseudotyped viruses were used as the controls for specificity.
Virion Entry or Uptake Assay
HEK293T cells were seeded in a 6-well plate (2 × 105 per well). After 24 h, the cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h and then infected with p24-normalized (100 ng) HIV-luc/Zaire EBOV-GP (2014) or HIV-luc/VSV-G pseudotyped viruses per well. For virion entry assay, after 6 h, the cells were washed twice with PBS and incubated with 0.25% trypsin at 37 °C for 2 min to remove the viruses that adhered to the cell surfaces. The cells were then collected and lysed, and the intracellular amount of HIV-1 p24 was then measured by ELISA. For virion uptake assay, after 0.5, 1, or 2 h, the cells were washed twice with PBS and incubated with 0.25% trypsin at 37 °C for 2 min to remove the viruses that adhered to the cell surfaces. The cells were then collected and lysed, and the intracellular amount of HIV-1 p24 was then measured by ELISA.
Viral Entry Inhibition on Various Cell Types
Primary human umbilical vein endothelial cells, A549 cells, and HeLa cells were seeded in a 96-well plate (2 × 104 per well). After 24 h, the cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h. The cells were then infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. After 12 h, the medium was discarded, and the cells were washed briefly with PBS and incubated with fresh medium for 48 h. Then the intracellular luciferase activity was measured. The THP-1 cells were plated in a 48-well format (1.25 × 105 per well) and incubated with teicoplanin at various concentrations at 37 °C for 1 h. The cells were then infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. After 12 h, the supernatants were removed by centrifugation at 300 × g for 10 min, and the cells were suspended and cultured with RPMI 1640 medium at 37 °C for 48 h. The intracellular luciferase activity was determined. HEK293T cells were seeded in a 96-well plate (2 × 104 per well). After 24 h, the cells were incubated with teicoplanin at gradient concentrations at 37 °C for 1 h. The cells were then infected with HIV-luc/SARS-CoV-S or HIV-luc/MERS-CoV-S pseudotyped viruses. After 12 h, the medium was discarded, and the cells were washed briefly with PBS and incubated with fresh medium for 48 h. The intracellular luciferase activity was then measured. For the Ebola trVLPs entry inhibition assay, HEK293T cells were seeded in a 96-well plate (2 × 104 per well). After 24 h, in the pre-transfection experiment, the cells were pre-transfected with 12.5 ng of pCAGGS-NP, 12.5 ng of pCAGGS-VP35, 7.5 ng of pCAGGS-VP30, 100 ng of pCAGGS-L, 25 ng of pCAGGS-T7, and 25 ng of pCAGGS-Tim1 plasmids with Lipofectamine 2000 according to the supplier's protocol (Invitrogen). After 24 h, the medium was discarded, and the cells were incubated with teicoplanin with the gradient concentrations at 37 °C for 1 h. In the non-pre-transfection experiment, HEK293T cells were directly incubated with teicoplanin with the gradient concentrations at 37 °C for 1 h without the pre-transfections of the above plasmids. The cells were then infected with Ebola trVLPs. After 12 h, the medium was discarded, and the cells were washed briefly with PBS and incubated with fresh medium for 48 h. Then the intracellular luciferase activity was measured.
Compound/Virus or Compound/Cell Pre-incubation Assay
p24-normalized (50 ng) HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses were incubated with 50 μm teicoplanin in a 96-well plate at 37 °C for 12 h. Then the compound/virus mixtures were transferred into a Microcon 30-kDa centrifugal filter device (Millipore) and centrifuged (7000 × g) at 4 °C for 15 min. Afterward the fresh medium was twice added onto the filter device to wash the compound/virus mixtures. Then 0.5 ml of DMEM was used to suspend the compound/virus mixtures, and the filter device was centrifuged in reverse (500 × g) at 4 °C for 5 min, and the solution was collected and used to infect HEK293T cells after HIV-1 p24 normalization. After 48 h, the intracellular luciferase activity was measured. For compound/cell pre-treatment assays, HEK293T cells were seeded in a 96-well plate (2 × 104 per well). After 24 h, the cells were incubated with teicoplanin at various concentrations at 37 °C for 12 h. The medium was discarded, and the cells were twice washed with PBS and incubated with fresh medium. Afterward the cells were infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. Twelve hours later, the medium was discarded, and the cells were washed briefly with PBS and incubated with fresh medium for 48 h. The intracellular luciferase activity was then measured.
siRNA Transfection, RNA Isolation, Reverse Transcription, and Quantitative Real Time-PCR
HEK293T cells were seeded in a 96-well plate (1 × 104 per well). After 24 h, the cells were transfected with siRNAs at a final concentration of 200 nm via Lipofectamine® RNAiMAX according to the supplier's protocol (Invitrogen). Forty eight hours later, the cells were lysed by TRIzol® reagent (Invitrogen), and the isolation of total RNAs was conducted according to the supplier's protocol (Invitrogen). The RNAs were reverse-transcripted by PrimeScript RT reagent kit (TaKaRa), and the quantitative RT-PCRs were conducted on a Bio-Rad CFX96 real time-PCR detection system (Bio-Rad) with SYBR Premix Ex Taq (TaKaRa). After the initial denaturation of cDNA was performed at 95 °C for 5 min, 40 cycles of the procedures (10 s of denaturation at 95 °C, 30 s of annealing at 60 °C, and 30 s of extension at 72 °C) were performed with the primers for human GAPDH, ACTB, and a variety of target genes. The smart pools of siRNAs were obtained from RIBOBIO.
Dextran Uptake Assay and Immunofluorescence Assay
The procedures previously described were followed with minor modifications (29). Briefly, HEK293T cells were incubated with 50 μm teicoplanin for 12 h. Then the cells were incubated with 1 mg/ml dextran-Alexa Fluor 568 (Mr 10,000) (Thermo Fisher Scientific) for 2 h in 1% serum-containing DMEM. The cells were washed twice with PBS and incubated with 4% polyformaldehyde at room temperature for 10 min. The cells were washed twice with PBS and incubated with 0.1% Triton X-100 at room temperature for 10 min, followed by incubation with PBS containing 5% bovine serum albumin at room temperature for 1 h. Afterward the cells were washed with PBS and incubated with rabbit anti-LAMP1 antibodies (Proteintech) at room temperature for 1 h. The cells were washed with PBS containing 0.1% Tween 20 three times and incubated with DyLight®488-conjugated goat anti-rabbit IgG (Abcam) at room temperature for 45 min, followed by washing with PBS containing 0.1% Tween 20 three times, and incubated with 5 μg/ml DAPI at room temperature for 5 min. The cells were then washed again with PBS containing 0.1% Tween 20 three times. Images were obtained by Zeiss LSM780 confocal microscopy using Zeiss ZFN software, and the co-localization of dextran and LAMP1 was analyzed from 20 fields (≥5 cells per field) per independent experiment.
Cathepsin L Enzymatic Inhibition Assay
Two protocols previously described were followed with minor modifications (30, 31). Briefly, HEK293T cells were seeded in a black 96-well plate. After 24 h, the cells were incubated with teicoplanin or Z-Phe-Tyr(t-Bu)-diazomethyl ketones, cathepsin L inhibitor III (Millipore) at various concentrations at 37 °C for 4 h. Then the cells were washed with PBS and incubated with 100 μl of PBS containing 50 μm (Z-Phe-Arg)2-R110 per well at 37 °C for 90 min. Afterward the fluorescence was tested by a fluorometer with excitation at 488 nm and emission at 510 nm. For in vitro enzymatic assay, 20 μl of 2 μm recombinant human cathepsin L (rhCTSL) (Sino Biological Inc) and 60 μl of buffer (400 mm NaOAc, 4 mm EDTA, pH 5.5) containing teicoplanin or Z-Phe-Tyr(t-Bu)-diazomethyl ketones were incubated at 37 °C for 4 h. Then the rhCTSL/compound mixtures were incubated further with 20 μl of 50 μm (Z-Phe-Arg)2-R110. After 4 h, the fluorescence was tested by a fluorometer with excitation at 488 nm and emission at 510 nm.
Results
High Throughput Screening of Ebola Virus Entry Inhibitors
We initiated our screening procedure by generating HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. The viruses were allowed to infect HEK293T cells in the presence of a 1600-member FDA-approved drug library. Compounds that inhibited virus luciferase activity were identified as the initial hits. As shown in Table 1, we identified 133 hits that could inhibit the entry of HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. To exclude the hits that only inhibited early events of the HIV-1 life cycle and to identify EBOV-GP-specific drugs, HIV-luc/VSV-G pseudotyped viruses bearing vesicular stomatitis virus (VSV) glycoproteins were used for secondary screening of the initial hit compounds. Finally, two drugs that act specifically as Ebola virus entry inhibitors were identified, teicoplanin and amiodarone (Fig. 1A).
TABLE 1.
The results of the high throughput screening of the clinically approved drug library
| Parameter | No. |
|---|---|
| Clinically approved drugs screened | 1600 |
| Clinically approved drugs that inhibit the entrances of HIV-luc/Zaire EBOV-GP(2014) pseudotype virusesa | 133 |
| Clinically approved drugs that inhibit the entrances of HIV-luc/VSV-G pseudotype viruses | 131 |
| Clinically approved drugs that specifically inhibit the entrances of HIV-luc/Zaire EBOV-GP(2014) pseudotype viruses | 2 |
a To deal with the threats of the outbreaks of Ebola viral infection in 2014, the pcDNA3.1-Zaire EBOV-GP (2014) plasmids encoding the Zaire Ebola virus glycoproteins were synthesized and transfected into HEK293T cells with pHIV-luciferase plasmids and pCMV-ΔR8.2 plasmids to produce HIV-luc/Zaire EBOV-GP (2014) pseudotype viruses.
FIGURE 1.
Teicoplanin specifically inhibits the entry of Ebola viruses. A, teicoplanin and amiodarone specifically inhibit the entry of Ebola viruses. HEK293T cells were seeded in a 96-well plate, and 24 h later, the cells were incubated with various reagents at 37 °C for 1 h. The cells were then infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. After washing and incubation with fresh medium for 48 h, the intracellular luciferase activity was measured. B, chemical structure of teicoplanin. C, HEK293T cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h. The intracellular luciferase activity was measured at 48 h post-infection. The IC50 was calculated using GraphPad Prism software. D, HEK293T cells were incubated with 500 μm teicoplanin at 37 °C for 48 h. Then the cell viability was determined. E, HEK293T cells were infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses and incubated with 50 μm teicoplanin at 0, 2, 4, and 8 h post-infection. The cells were then incubated for 48 h after which the intracellular luciferase activity was tested. The HIV-luc/VSV-G pseudotyped viruses were used as the controls for specificity. F, HEK293T cells were seeded in a 6-well plate (2 × 105 per well). After 24 h, the cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h and then infected with p24-normalized (100 ng) HIV-luc/Zaire EBOV-GP (2014) or HIV-luc/VSV-G pseudotyped viruses per well. After 6 h, the cells were washed with PBS twice and incubated with 0.25% trypsin at 37 °C for 2 min to remove the viruses that adhered to the cell surfaces. The cells were then collected and lysed, and the intracellular amount of HIV-1 p24 was measured by ELISA. The results are representatives of at least three independent experiments. The bars show the mean values ± S.D. (error bars). The p value was determined by a Student's t test. ***, p < 0.001.
Teicoplanin Specifically Inhibits the Entry of Ebola Viruses
Teicoplanin is a glycopeptide antibiotic that includes five major components based on different side chains (Fig. 1B). It demonstrated an IC50 of 0.34 μm for its inhibitory effect on HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses (Fig. 1C). The cytotoxicity of teicoplanin was also determined using a cell viability assay, and its CC50 was greater than 500 μm (Fig. 1D). To further confirm whether teicoplanin acts as an Ebola virus entry inhibitor, a time-of-addition assay was conducted. The data showed that teicoplanin represses the entry of Ebola viruses at the early stage of Ebola virus infection (Fig. 1E). In addition, a virion entry assay also demonstrated that teicoplanin inhibits the entry of HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses in a dose-dependent manner. However, teicoplanin does not repress the entry of HIV-luc/VSV-G pseudotyped viruses (Fig. 1F).
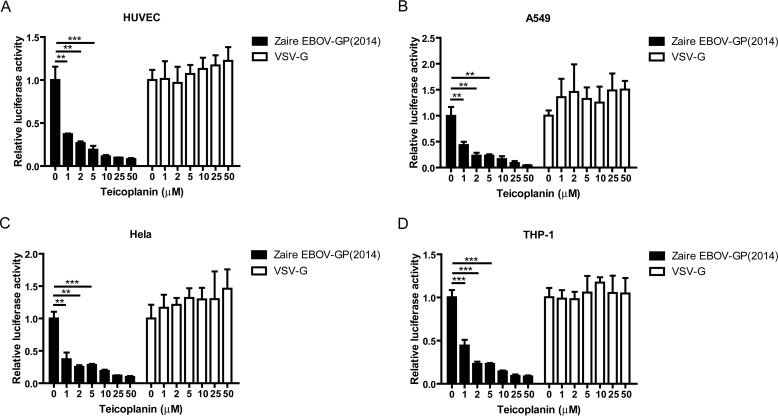
Ebola viruses can infect a wide range of host cells, including vascular endothelial cells, epithelial cells, and monocytes, which account for the pathogenesis observed in Ebola-infected patients (32, 33). Thus, it was important to examine whether teicoplanin could repress the entry of Ebola viruses into different types of cells. The data showed that teicoplanin effectively represses virus entry into primary human umbilical vein endothelial cells (Fig. 2A), human epithelial cell lines such as A549 (Fig. 2B) and HeLa cells (Fig. 2C), and the human acute monocytic leukemia THP-1 cell line (Fig. 2D).
FIGURE 2.
Ebola virus entry into different cell types is repressed by teicoplanin. A, primary human umbilical vein endothelial cells were seeded in a 96-well plate. After 24 h, the cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h. Subsequently, the cells were infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. After washing and incubation with fresh medium for 48 h, the intracellular luciferase activity was measured. B, antiviral entry assay of teicoplanin on the A549 cells was conducted in a similar way. C, antiviral entry assay of teicoplanin on the HeLa cells was conducted in a similar procedure. D, THP-1 cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h. Subsequently, the cells were infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. After 12 h, the supernatants were removed by centrifugation at 300 × g for 10 min, and the cells were suspended and cultured with RPMI 1640 medium at 37 °C for 48 h. The intracellular luciferase activity was then tested. The results are representative of at least three independent experiments. The bars show the mean values ± S.D. (error bars). The p value was determined by a Student's t test. **, p < 0.01; ***, p < 0.001.
Teicoplanin Targets the Host Cells Rather than the Cell-free Viral Particles
To elucidate the molecular mechanisms of the anti-Ebola virus activity of teicoplanin, it is necessary to determine whether the target of teicoplanin is located directly on the virus itself. The compound virus pre-incubation assay demonstrated that when teicoplanin was pre-incubated with Ebola/HIV pseudotyped viruses and then filtered and washed away, the inhibitory effect of the antibiotic on Ebola virus entry did not occur (Fig. 3A). However, the compound cell pre-treatment assay showed that when the host cells were pre-treated with teicoplanin and subsequently washed to remove the drug, its inhibitory effect on Ebola/HIV pseudotyped viruses remained (Fig. 3B). Together, these data indicated that the target of teicoplanin is located on the host cells.
FIGURE 3.
Target of teicoplanin is located within the host cells. A, p24-normalized (50 ng) HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses were incubated with 50 μm teicoplanin in a 96-well plate at 37 °C for 12 h. Then the compound virus mixtures were transferred into a Microcon 30-kDa centrifugal filter device (Millipore) and centrifuged (7000 × g) at 4 °C for 15 min. Subsequently, fresh medium was added twice onto the filter device to wash the compound virus mixtures. Then 0.5 ml of DMEM was used to suspend the compound virus mixtures, and the reversed filter device was centrifuged (500 × g) at 4 °C for 5 min. The solution was collected and used to infect HEK293T cells after HIV-1 p24 normalization. After 48 h, the intracellular luciferase activity was measured. B, HEK293T cells were incubated with teicoplanin at various concentrations for 12 h. The cells were then infected with HIV-luc/Zaire EBOV-GP (2014) pseudotyped viruses. After washing and incubation with fresh medium for 48 h, the intracellular luciferase activity was measured. The assay during treatment was the same as the antiviral entry assay. The results are representative of at least three independent experiments. The bars show the mean values ± S.D. (error bars). The p value was determined by a Student's t test. n.s., not significant; **, p < 0.01; ***, p < 0.001.
Teicoplanin Does Not Block Cell Receptors
To clarify the molecular mechanism of teicoplanin action, we tested whether teicoplanin inhibits the entry of other viruses able to utilize similar intracellular transport routes as those employed by Ebola viruses. It has been reported that both Ebola viruses and SARS coronaviruses (SARS-CoVs) need to be transported to the endo/lysosomes to release their genomes (Fig. 4A) (18). Therefore, HIV-luc/SARS-CoV-S pseudotyped viruses that bear the S proteins of SARS-CoVs were generated and used to test whether their entry could be repressed by teicoplanin. The data indicated that teicoplanin also inhibits the entry of SARS-CoVs (Fig. 4B). Given that Niemann-Pick C1 (NPC1) and angiotensin-converting enzyme 2 (ACE2) represent the cell receptors of Ebola viruses and SARS-CoVs, respectively (16, 34, 35), and that teicoplanin inhibits both of these viruses, it is necessary to clarify whether Ebola viruses and SARS-CoVs share cell receptor affinity. Following depletion of the expression of NPC1 and ACE2 in HEK293T cells using siRNAs, the cells were infected with Ebola or SARS-CoV pseudotyped viruses. The results demonstrated that the knockdown of NPC1 affected the infectivity of Ebola but not that of SARS-CoV pseudotyped viruses, whereas the converse was observed following ACE2 knockdown (Fig. 4C). This indicates that these two viruses are each associated with specific receptors and also excludes the possibility that teicoplanin inhibits the interaction between the viruses and their cell receptors or the events following receptor binding. Taken together, these data led us to hypothesize that the target of teicoplanin would be the host factor(s) which is (are) required for both Ebola and SARS-CoV infection. Considering that interferon-inducible transmembrane proteins (IFITMs) represent the first line of anti-viral defense of the cells, we first examined the effect of teicoplanin on the expression of IFITMs. However, the data showed that teicoplanin did not induce the expression of IFITMs (data not shown).
FIGURE 4.
Teicoplanin does not block the cell receptor. A, schematic representation of the entry of Ebola viruses and SARS-CoVs. B, HEK293T cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h. The cells were then infected with HIV-luc/SARS-CoV-S pseudotyped viruses. After washing and incubation with fresh medium for 48 h, the intracellular luciferase activity was measured. C, HEK293T cells were seeded in a 96-well plate. After 24 h, the cells were transfected with 200 nm siRNAs against NPC1 and ACE2, respectively. After 48 h, the cells were infected with HIV-luc/Zaire EBOV-GP (2014) or HIV-luc/SARS-CoV-S pseudotyped viruses. The results are representative of at least three independent experiments. The bars show the mean values ± S.D. (error bars). The p value was determined by a Student's t test. n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Teicoplanin Has No Effect on HOPS Complexes
We further examined the host factors that are required for both Ebola viruses and SARS-CoVs but not VSVs using siRNAs. Through genome-wide haploid genetic screening, several host factors essential for Ebola viral infection have been identified (16). Accordingly, we infected cells with Ebola-, SARS-CoV-, or VSV-pseudotyped HIV-1 viruses after siRNA-mediated knockdown of the expression of 13 individual host factors. We found that cathepsin L (CTSL), VPS11, VPS18, VPS33A, VPS39, and VPS41 are required for both Ebola and SARS-CoV infection (Fig. 5B) rather than VSV infection. These results imply that the host factors might be the targets of teicoplanin. VPS11, VPS18, VPS33A, VPS39, and VPS41 are components of HOPS complexes, which mediate the homotypic fusion between late endosomes and the heterotypic fusion between late endosomes and lysosomes (29, 36, 37). To investigate whether teicoplanin affects the transport of Ebola viruses by inhibiting HOPS complex function and subsequently disturbing endosome maturation and fusion between the late endosomes and the lysosomes, a dextran uptake assay was conducted (Fig. 5C). We found that within 2 h of uptake, the dextran-Alexa Fluor 568 could be transported into lysosome-associated membrane protein 1 (LAMP1)-positive lysosomes. The co-localization coefficient of dextran and LAMP1 was influenced by siRNA knockdown of VPS39 and VPS41, whereas teicoplanin did not exert any effect (Fig. 5D). These data indicated that teicoplanin does not inhibit the transport of dextran-Alexa Fluor 568. Therefore, it is unlikely that teicoplanin inhibits the entry of Ebola viruses by affecting HOPS complexes. In addition, we conducted virion uptake assays to demonstrate that teicoplanin does not inhibit both Ebola- and VSV-pseudotyped virion uptake at 0.5, 1, and 2 h post-infection (Fig. 5, E and F). These data indicated that teicoplanin did not affect the endocytosis of Ebola- and VSV-pseudotyped virions, at least the early phase. In addition, these data also indicated that, except HOPS complexes, teicoplanin has no effect on the other host factors that are involved in the process of virion uptake.
FIGURE 5.
Teicoplanin has no effect on HOPS complexes. A, schematic representation of the entry of Ebola viruses, SARS-CoVs, and VSVs. The host factors thatare essential for the entry of Ebola viruses are illustrated. B, HEK293T cells were transfected with 200 nm siRNAs per well. After 48 h, the cells were infected with HIV-luc/Zaire EBOV-GP (2014), HIV-luc/SARS-CoV-S, or HIV-luc/VSV pseudotyped viruses. After washing and incubation with fresh medium for 48 h, the intracellular luciferase activity was measured. C, HEK293T cells were plated in a glass bottom dish. After 24 h, groups of cells were incubated with 50 μm teicoplanin or control for 12 h. Another group of cells was transfected with siRNAs against VPS39 and VPS41 or the si-control and incubated with these siRNAs at a final concentration of 200 nm for 48 h. Then the cells were incubated with dextran-Alexa Flour 568 (Mr 10,000) for 2 h in 1% serum-containing DMEM. Subsequently, the cells were subjected to immunofluorescence analysis. Images were obtained using Zeiss LSM780 confocal microscopy with Zeiss ZFN software. Scale bar, 10 μm. D, co-localization of dextran and LAMP1 was analyzed from 20 fields (≥5 cells per field) per independent experiment. Pearson's overlap coefficients were used to determine the levels of the co-localization of dextran and LAMP1. E and F, HEK293T cells were incubated with teicoplanin at the various indicated concentrations at 37 °C for 1 h. The cells were then infected with HIV-luc/Zaire EBOV-GP (2014) or HIV-luc/VSV-G pseudotyped viruses, respectively. Subsequently, the cells were incubated with 0.25% trypsin at 37 °C for 2 min to remove the viruses that adhered to the cell surfaces at 0.5, 1, and 2 h post-infection. The intracellular amount of HIV-1 p24 was then measured by ELISA. The results are representative of at least three independent experiments. The bars show the mean values ± S.D. (error bars). The p value was determined by a Student's t test. n.s., not significant; ***, p < 0.001.
Teicoplanin Directly Inhibits the Enzymatic Activity of Cathepsin L
After excluding the possibility that the machinery for vesicle transport is involved in the inhibitory effect of teicoplanin, we assumed that the enzymes in the late endosome/lysosome could be the target of teicoplanin. The proteolysis of glycoprotein by cathepsin L has been reported to be required for the membrane fusion of both Ebola viruses and SARS-CoVs (31). In contract, cathepsin B is required for Ebola virus infection but not SARS-CoV infection (17, 31). As such, we hypothesized that cathepsin L rather than cathepsin B could be the target for teicoplanin. To this end, we examined whether teicoplanin can inhibit the enzymatic activity of cathepsin L (Fig. 6A). Two assays to measure the cathepsin L enzymatic activity were performed (30, 31). We showed that teicoplanin potently inhibits the activity of cathepsin L in a dose-dependent manner (Fig. 6, B and E). Considering that the inhibitory dose of teicoplanin on the activity of cathepsin L is higher than that required for Ebola virus infection inhibition, a cell viability assay was performed to confirm that the inhibitory effect is not due to cytotoxicity (Fig. 6C). In addition, a comparative analysis of the inhibitory dose of teicoplanin and that of previously reported cathepsin L inhibitors, Z-Phe-Tyr(t-Bu)-diazomethyl ketones, on Ebola virus infection was conducted. We demonstrated that both compounds can inhibit Ebola virus infection at low doses. The IC50 dose of teicoplanin required to inhibit the Ebola entry is approximately four times more than that of Z-Phe-Tyr(t-Bu)-diazomethyl ketones (Fig. 6D and Table 2). Similarly, the IC50 dose of teicoplanin required to inhibit the enzymatic activity of cathepsin L was also approximately four times greater (Fig. 6, B and E, and Table 2). These data indicated that the high doses of teicoplanin determined to be required for the inhibition of cathepsin L enzymatic activity could be due to the relatively low sensitivity of the cathepsin L activity assay. These data clearly demonstrate the consistency between the inhibition of virus entry and the inhibition of cathepsin L activity and further support our conclusion that the target molecule of teicoplanin is cathepsin L.
FIGURE 6.
Teicoplanin directly inhibits the activity of cathepsin L. A, schematic representation of the cathepsin L enzymatic inhibition assay. B, HEK293T cells were incubated with teicoplanin or Z-Phe-Tyr(t-Bu)-diazomethyl ketones and then incubated with 50 μm (Z-Phe-Arg)2-R110 at 37 °C for 90 min. The level of fluorescence was determined using a fluorometer with excitation at 488 nm and emission at 510 nm. C, HEK293T cells were incubated with teicoplanin at various concentrations at 37 °C for 48 h. The cell viability was then determined using the CellTiter® 96 Aqueous One Solution Cell Proliferation Assay. D, HEK293T cells were incubated with teicoplanin or Z-Phe-Tyr(t-Bu)-diazomethyl ketones at 37 °C for 1 h. The cells were then infected with HIV-luc/Zaire EBOV-GP (2014) or HIV-luc/VSV-G pseudotyped viruses. After incubation for 48 h, the intracellular luciferase activity was measured. E, recombinant human cathepsin L (rhCTSL), teicoplanin, or Z-Phe-Tyr(t-Bu)-diazomethyl ketones were incubated 37 °C for 4 h. The rhCTSL-compound mixtures were incubated with (Z-Phe-Arg)2-R110 at 37 °C for 4 h. The fluorescence was then tested using a fluorometer with excitation at 488 nm and emission at 510 nm. The results are representative of at least three independent experiments. The bars show the mean values ± S.D. (error bars). The p value was determined by a Student's t test. n.s., not significant, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
TABLE 2.
The IC50 value of glycopeptide antibiotics on Ebola-trVLP entry and cathepsin L activity
| Compound | IC50 on Ebola-trVLP entry (pretreated) | IC50 on CTSL activity (method 1) | IC50 on CTSL activity (method 2) |
|---|---|---|---|
| μm | μm | μm | |
| Teicoplanin | 0.39 ± 0.12 | 208.0 ± 59.8 | 425.3 ± 107.8 |
| Dalbavancin | 1.61 ± 1.26 | 333.7 ± 110.5 | 503.1 ± 102.8 |
| Oritavancin | 1.73 ± 1.24 | 371.8 ± 142.3 | 548.4 ± 126.1 |
| Telavancin | 1.89 ± 1.35 | 377.4 ± 161.3 | 558.8 ± 72.3 |
| Vancomycin | >50 | >2560 | >2560 |
| CTSL inhibitor | 0.10 ± 0.03 | 43.53 ± 18.24 | 127.7 ± 14.2 |
Entry of Ebola trVLPs Is Repressed by Several Glycopeptide Antibiotics with the Exception of Vancomycin
An Ebola trVLP system (28), which can simulate the life cycle of wild-type Ebola viruses to a large extent, was applied to investigate whether teicoplanin and its glycopeptide antibiotic homologs dalbavancin, oritavancin, telavancin, and vancomycin can also inhibit the entry of Ebola trVLPs. Accordingly, the p4cis plasmid encoding Renilla luciferase, VP40, GP, and VP24 was transfected into HEK293T cells along with plasmids expressing T7 RNA polymerase, NP, VP35, VP30, and L viral proteins to produce Ebola trVLPs (Fig. 7A). When the target cells were also pre-transfected with NP, VP35, VP30, L, T7, and Tim1 plasmids, the transcription and replication of the Ebola virus minigenome were actively stimulated to produce infectious Ebola trVLPs. The IC50 value of teicoplanin on Ebola trVLP entry was ∼390 nm under these conditions (Fig. 7B). Additionally, when the target cells were not pre-transfected with the above plasmids, the Ebola viral minigenomes were only weakly transcribed and replicated with minimal production of infectious trVLPs. The IC50 of teicoplanin on the entry of Ebola trVLPs was ∼330 nm under the non-pre-transfection conditions (Fig. 7C). In addition, we also examined whether the homologs of teicoplanin can inhibit Ebola trVLP entry. The data demonstrated that the IC50 values of dalbavancin, oritavancin, and telavancin on Ebola trVLP entry were ∼1.61, 1.73, and 1.89 μm, respectively, under pre-transfection conditions (Fig. 7, D, F, and H) and ∼2.77, 2.13, and 2.30 μm, respectively, under non-pre-transfection conditions (Fig. 7, E, G, and I). However, we found that vancomycin did not inhibit Ebola trVLP entry under either condition (Fig. 7, J and K). The CC50 values of dalbavancin, oritavancin, telavancin, and vancomycin were also determined (data not shown). These data indicated that these homologs might share similar structures that are indispensable for the inhibitory effects on Ebola trVLP entry. However, vancomycin would not be expected to have similar structures (Fig. 9).
FIGURE 7.
Entry of Ebola trVLPs is repressed by glycopeptide antibiotics with the exception of vancomycin. A, schematic representation of the EbolatrVLPs entry assay. B, for the pre-transfection experiment, HEK293T cells were pre-transfected with 12.5 ng of pCAGGS-NP, 12.5 ng of pCAGGS-VP35, 7.5 ng of pCAGGS-VP30, 100 ng of pCAGGS-L, 25 ng of pCAGGS-T7, and 25 ng of pCAGGS-Tim1 plasmids. After 24 h, the medium was discarded, and the cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h. After washing and incubation with fresh medium for 48 h, the intracellular luciferase activity was measured. The IC50 was calculated using GraphPad Prism software. C, for the non-pre-transfection experiment, HEK293T cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h without pre-transfection of the above plasmids. The methods used for conducting Ebola trVLPs infection and measuring the intracellular luciferase activity were the same as those described in B. D and E, IC50 values of dalbavancin on the entry of Ebola trVLPs were determined under pre-transfection and non-pre-transfection conditions, respectively. F and G, IC50 values of oritavancin on the entry of Ebola trVLPs were calculated using pre-transfection or non-pre-transfection conditions, respectively. H and I, IC50 values of telavancin on the entry of Ebola trVLPs were determined under pre-transfection and non-pre-transfection conditions, respectively. J and K, effect of vancomycin on the entry of Ebola trVLPs was determined using pre-transfection or non-pre-transfection conditions, respectively. The results are representative of at least three independent experiments. The bars show the mean values ± S.D. (error bars). The p value was determined by a Student's t test. n.s., not significant.
FIGURE 9.
Chemical structures of glycopeptide antibiotics. Teicoplanin, dalbavancin, oritavancin, and telavancin, which inhibit the entry of Ebola trVLPs, MERS-CoVs, and SARS-CoVs, contain key hydrophobic groups. However, vancomycin, which does not exert antiviral activity, does not contain these groups.
MERS-CoV and SARS-CoV Entry Is Also Inhibited by Glycopeptide Antibiotics with the Exception of Vancomycin
Because our data demonstrated that teicoplanin inhibits cathepsin L enzymatic activity and that cathepsin L is also essential for MERS-CoV and SARS-CoV entry (31, 38), we therefore hypothesized that teicoplanin and its homologs could also inhibit the entry of MERS-CoVs and SARS-CoVs. The data illustrated that the IC50 values of teicoplanin, dalbavancin, oritavancin, and telavancin on MERS-CoV entry were ∼0.63, 2.99, 2.12, and 3.24 μm respectively (Fig. 8, A–D). However, vancomycin did not inhibit the MERS-CoV entry (Fig. 8I). Additionally, the IC50 values of teicoplanin, dalbavancin, oritavancin, and telavancin on the entry of SARS-CoVs were ∼3.76, 9.64, 4.96, and 3.45 μm, respectively (Fig. 8, E–H). However, vancomycin did not inhibit the entry of SARS-CoVs (Fig. 8J). These data indicated that glycopeptide antibiotics with the exception of vancomycin exhibit broad antiviral activity because of their inhibitory effects on cathepsin L.
FIGURE 8.
Entry of MERS-CoVs and SARS-CoVs is inhibited by glycopeptide antibiotics with the exception of vancomycin. A and E, HEK293T cells were incubated with teicoplanin at various concentrations at 37 °C for 1 h. The cells were infected with HIV-luc/MERS-CoV-S or HIV-luc/SARS-CoV-S pseudotyped viruses. After washing and incubation with fresh medium for 48 h, the intracellular luciferase activity was measured. The IC50 was calculated using GraphPad Prism software. B and F, IC50 values of dalbavancin on the entry of HIV-luc/MERS-CoV-S and HIV-luc/SARS-CoV-S pseudotyped viruses were determined. C and G, IC50 values of oritavancin on the entry of HIV-luc/MERS-CoV-S and HIV-luc/SARS-CoV-S pseudotyped viruses were calculated. D and H, IC50 values of telavancin on the entry of HIV-luc/MERS-CoV-S and HIV-luc/SARS-CoV-S pseudotyped viruses were determined. I and J, The effect of vancomycin on the entry of HIV-luc/MERS-CoV-S and HIV-luc/SARS-CoV-S pseudotyped viruses was determined. The results are representative of at least three independent experiments. The bars show the mean values ± S.D. (error bars). The p value was determined by a Student's t test. n.s., not significant.
Discussion
In our study, we performed high throughput screening of a clinically approved drug library and identified that teicoplanin not only inhibits the entry of Ebola/HIV-1 pseudotyped viruses but also of transcription- and replication-competent trVLPs. The IC50 value of teicoplanin on the entry of Ebola trVLPs is as low as 330 nm, whereas the CC50 of teicoplanin is over 500 μm. In addition, teicoplanin inhibits the entry of Ebola viruses into different types of host cells, including primary human umbilical vein endothelial cells, A549 cells, HeLa cells, and THP-1 cells. It also inhibits the entry of MERS-CoV/HIV-1 and SARS-CoV/HIV-1 pseudotyped viruses.
Teicoplanin is a glycopeptide antibiotic isolated from Actinoplanes teichomyceticus. Teicoplanin contains five major components (39, 40) that can form complexes with the C-terminal l-Lys–d-Ala–d-Ala subunits of lipid II peptidoglycan precursors. Teicoplanin binding of lipid II inhibits their transglycosylation and transpeptidation, leading to disturbed cell wall synthesis in Gram-positive bacteria (41). Recently, teicoplanin was reported to inhibit Ebola pseudovirus infection in cell culture, which is consistent with our observations. Teicoplanin was also demonstrated to have an effect on a common component used by both enveloped Ebola viruses and human respiratory syncytial viruses but not by non-enveloped viruses (26). However, the mechanism by which teicoplanin inhibits the entry of Ebola remained unresolved. To elucidate the molecular mechanisms underlying this process, we compared the entry of Ebola viruses and SARS-CoVs and the effect of teicoplanin thereon. Teicoplanin represses the entry of both Ebola viruses and SARS-CoVs, which require transport to the endo/lysosomes to deliver their genomes. Because several host factors have been reported to be essential for Ebola and SARS-CoV virus infection, the identification of the common host factors that are required for the entry of both Ebola viruses and SARS-CoVs would likely be helpful to clarify the target of teicoplanin. Our data demonstrated that CTSL, VPS11, VPS18, VPS33A, VPS39, and VPS41 are all indispensable for the entry of both Ebola viruses and SARS-CoVs (Fig. 5B). The elucidation via a dextran uptake assay that teicoplanin does not affect the functions of HOPS complexes suggested that the target of teicoplanin was cathepsin L. In support of this, the results of two cathepsin L enzymatic activity assays indicated that teicoplanin indeed inhibits the activity of cathepsin L, which can explain why teicoplanin inhibited both the entry of Ebola viruses and human respiratory syncytial viruses as shown in a previous study (26) because cathepsin L is required for the infection mechanism of both (17, 42). Considering that cathepsin L is also required for the entry of MERS-CoVs, teicoplanin thus represents a broad virus entry inhibitor. In addition, the inhibitory effects of teicoplanin and its derivatives on HIV-1, HCV, influenza viruses, flaviviruses, FIPV, and SARS-CoVs have also been reported (43–49), further supporting that the antiviral target for glycopeptide antibiotics is a common host factor.
As teicoplanin can bind lipid IIs, we hypothesized that it might interact with the enzymatic domains of cathepsin L and block their functions similar to the reported inhibitory effect of antimicrobial peptide LL-37 on cathepsin L (50). The specific binding sites of teicoplanin on cathepsin L need to be further investigated by molecular docking, amino acid mutation, and surface plasmon resonance to confirm this hypothesis. We also examined the effects of the teicoplanin homologs dalbavancin, oritavancin, telavancin, and vancomycin on the entry of Ebola trVLPs, MERS-CoV/HIV-1, and SARS-CoV/HIV-1 pseudotyped viruses. The data demonstrated that dalbavancin, oritavancin, and telavancin also inhibit the entryof MERS-CoV/HIV-1 and SARS-CoV/HIV-1 pseudotyped viruses. However, vancomycin did not repress their infection. By comparing the structures of these compounds, we found that all the glycopeptide antibiotics that inhibit Ebola trVLP, MERS-CoV/HIV-1, and SARS-CoV/HIV-1 pseudotyped virus entry contain hydrophobic groups at the amidogen domains of their aminosaccharides. These groups might play an important role in the interactions between the glycopeptide antibiotics and cathepsin L. In contrast, vancomycin lacks the hydrophobic groups, which might be the reason why it does not inhibit these viral infections (Fig. 9).
For the treatment of Gram-positive bacterial infections in the clinic, teicoplanin can be administered intravenously or intramuscularly once daily following an initial loading dose, which is convenient for outpatient therapy (51). Compared with vancomycin, which was the first discovered glycopeptide antibiotic, teicoplanin is better tolerated with lower nephrotoxicity (52). The Summary of Product Characteristics for teicoplanin in 2014 shows that, after the completion of the loading dose regimens for most Gram-positive bacterial infections, the serum concentrations of teicoplanin are at least 15 mg/liter (8.78 μm), which is ∼27, 14, and 2 times higher than the IC50 values of teicoplanin against the entry of Ebola viruses, MERS-CoV/HIV-1, and SARS-CoV/HIV-1 pseudotyped viruses, respectively. As the toxicity of glycopeptide antibiotics is quite low, we propose that glycopeptide antibiotics might be used in the clinic for Ebola/MERS-CoV/SARS-CoV infection, especially in the case of emergency requirements during outbreaks of these severe viral infections.
Author Contributions
All listed authors contributed to this work and reviewed the manuscript. N. Z. and T. Pan designed the experiments and performed most of these experiments. J. S. Z., X. Z., F. H., T. Peng, L. T., and J. H. Z. performed the different kinds of virus-related experiments. Q. W. L., C. B., and C. L. carried out the high throughput screening of the Food and Drug Administration-approved drug library. N. Z. and H. Z. contributed to the idea generation, experimental design, and manuscript preparation and conceived the project.
Acknowledgments
We thank Dr. Wenlin Huang (Sun Yat-sen University) for providing the pcDNA3.1-SARS-CoV-S plasmid. We thank Dr. Linqi Zhang (Tsinghua University) and Dr. Shibo Jiang (Fudan University) for providing the pcDNA3.1-MERS-CoV-S plasmid. We thank Dr. Jun Li (Sun Yat-sen University) for providing the primary human umbilical vein endothelial cells. We thank Dr. Thomas Hoenen (Federal Research Institute for Animal Health, Germany) for providing several plasmids to generate trVLPs.
This work was supported by National Special Research Program for the Important Infectious Diseases Grant 2013ZX10001004, Guangdong Innovative Research Team Program Grant 2009010058, and National Natural Science Foundation of Key Projects Grant 81590765 (to H. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
- EBOV
- Ebola virus
- trVLPs
- transcription- and replication-competent virus-like particles
- MERS-CoV
- Middle East respiratory syndrome coronavirus
- SARS-CoV
- severe acute respiratory syndrome coronavirus
- VSV
- vesicular stomatitis virus
- GP
- glycoprotein
- rh
- recombinant human
- CTSL
- cathepsin L
- HOPS complex
- homotypic fusion and vacuole protein-sorting complex
- Z
- benzyloxycarbonyl
- t-Bu
- t-butyl
- IFITM
- interferon-inducible transmembrane protein
- FDA
- Food and Drug Administration.
References
- 1. Kiley M. P., Bowen E. T., Eddy G. A., Isaäcson M., Johnson K. M., McCormick J. B., Murphy F. A., Pattyn S. R., Peters D., Prozesky O. W., Regnery R. L., Simpson D. I., Slenczka W., Sureau P., van der Groen G., et al. (1982) Filoviridae: a taxonomic home for Marburg and Ebola viruses? Intervirology 18, 24–32 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization (1978) Ebola haemorrhagic fever in Zaire, 1976. Bull. World Health Organ. 56, 271–293 [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization (1978) Ebola haemorrhagic fever in Sudan, 1976. Report of a WHO/International Study Team. Bull. World Health Organ. 56, 247–270 [PMC free article] [PubMed] [Google Scholar]
- 4. Jahrling P. B., Geisbert T. W., Dalgard D. W., Johnson E. D., Ksiazek T. G., Hall W. C., and Peters C. J. (1990) Preliminary report: isolation of Ebola virus from monkeys imported to U.S.A. Lancet 335, 502–505 [DOI] [PubMed] [Google Scholar]
- 5. Le Guenno B., Formenty P., Formentry P, Wyers M., Gounon P., Walker F., and Boesch C. (1995) Isolation and partial characterisation of a new strain of Ebola virus. Lancet 345, 1271–1274 [DOI] [PubMed] [Google Scholar]
- 6. Towner J. S., Sealy T. K., Khristova M. L., Albariño C. G., Conlan S., Reeder S. A., Quan P. L., Lipkin W. I., Downing R., Tappero J. W., Okware S., Lutwama J., Bakamutumaho B., Kayiwa J., Comer J. A., et al. (2008) Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4, e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alvarez C. P., Lasala F., Carrillo J., Muñiz O., Corbí A. L., and Delgado R. (2002) C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76, 6841–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan S. Y., Empig C. J., Welte F. J., Speck R. F., Schmaljohn A., Kreisberg J. F., and Goldsmith M. A. (2001) Folate receptor-α is a cofactor for cellular entry by Marburg and Ebola viruses. Cell 106, 117–126 [DOI] [PubMed] [Google Scholar]
- 9. Gramberg T., Hofmann H., Möller P., Lalor P. F., Marzi A., Geier M., Krumbiegel M., Winkler T., Kirchhoff F., Adams D. H., Becker S., Münch J., and Pöhlmann S. (2005) LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology 340, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kondratowicz A. S., Lennemann N. J., Sinn P. L., Davey R. A., Hunt C. L., Moller-Tank S., Meyerholz D. K., Rennert P., Mullins R. F., Brindley M., Sandersfeld L. M., Quinn K., Weller M., McCray P. B. Jr., Chiorini J., and Maury W. (2011) T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. U.S.A. 108, 8426–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Hearn A., Wang M., Cheng H., Lear-Rooney C. M., Koning K., Rumschlag-Booms E., Varhegyi E., Olinger G., and Rong L. (2015) Role of EXT1 and glycosaminoglycans in the early stage of filovirus entry. J. Virol. 89, 5441–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimojima M., Takada A., Ebihara H., Neumann G., Fujioka K., Irimura T., Jones S., Feldmann H., and Kawaoka Y. (2006) Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 80, 10109–10116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takada A., Watanabe S., Ito H., Okazaki K., Kida H., and Kawaoka Y. (2000) Downregulation of β1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology 278, 20–26 [DOI] [PubMed] [Google Scholar]
- 14. Saeed M. F., Kolokoltsov A. A., Albrecht T., and Davey R. A. (2010) Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 6, e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nanbo A., Imai M., Watanabe S., Noda T., Takahashi K., Neumann G., Halfmann P., and Kawaoka Y. (2010) Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 6, e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carette J. E., Raaben M., Wong A. C., Herbert A. S., Obernosterer G., Mulherkar N., Kuehne A. I., Kranzusch P. J., Griffin A. M., Ruthel G., Dal Cin P., Dye J. M., Whelan S. P., Chandran K., and Brummelkamp T. R. (2011) Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chandran K., Sullivan N. J., Felbor U., Whelan S. P., and Cunningham J. M. (2005) Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mingo R. M., Simmons J. A., Shoemaker C. J., Nelson E. A., Schornberg K. L., D'Souza R. S., Casanova J. E., and White J. M. (2015) Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J. Virol. 89, 2931–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rampling T., Ewer K., Bowyer G., Wright D., Imoukhuede E. B., Payne R., Hartnell F., Gibani M., Bliss C., Minhinnick A., Wilkie M., Venkatraman N., Poulton I., Lella N., Roberts R., et al. (2015) A monovalent chimpanzee adenovirus Ebola vaccine- preliminary report. N. Engl. J. Med. 10.1056/NEJMoa1411627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Regules J. A., Beigel J. H., Paolino K. M., Voell J., Castellano A. R., Munoz P., Moon J. E., Ruck R. C., Bennett J. W., Twomey P. S., Gutierrez R. L., Remich S. A., Hack H. R., Wisniewski M. L., Josleyn M. D., et al. (2015) A recombinant vesicular stomatitis virus Ebola vaccine–preliminary report. N. Engl. J. Med. 10.1056/NEJMoa1414216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J. B., Fausther-Bovendo H., Wei H., Aviles J., Hiatt E., Johnson A., Morton J., Swope K., Bohorov O., Bohorova N., et al. (2014) Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geisbert T. W., Lee A. C., Robbins M., Geisbert J. B., Honko A. N., Sood V., Johnson J. C., de Jong S., Tavakoli I., Judge A., Hensley L. E., and Maclachlan I. (2010) Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet 375, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warren T. K., Wells J., Panchal R. G., Stuthman K. S., Garza N. L., Van Tongeren S. A., Dong L., Retterer C. J., Eaton B. P., Pegoraro G., Honnold S., Bantia S., Kotian P., Chen X., Taubenheim B. R., et al. (2014) Protection against filovirus diseases by a novel broad-spectrum nucleoside analog BCX4430. Nature 508, 402–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolf M. C., Freiberg A. N., Zhang T., Akyol-Ataman Z., Grock A., Hong P. W., Li J., Watson N. F., Fang A. Q., Aguilar H. C., Porotto M., Honko A. N., Damoiseaux R., Miller J. P., Woodson S. E., et al. (2010) A broad-spectrum antiviral targeting entry of enveloped viruses. Proc. Natl. Acad. Sci. U.S.A. 107, 3157–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gehring G., Rohrmann K., Atenchong N., Mittler E., Becker S., Dahlmann F., Pöhlmann S., Vondran F. W., David S., Manns M. P., Ciesek S., and von Hahn T. (2014) The clinically approved drugs amiodarone, dronedarone and verapamil inhibit filovirus cell entry. J. Antimicrob. Chemother. 69, 2123–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y., Cui R., Li G., Gao Q., Yuan S., Altmeyer R., and Zou G. (2016) Teicoplanin inhibits Ebola pseudovirus infection in cell culture. Antiviral Res. 125, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zufferey R., Nagy D., Mandel R. J., Naldini L., and Trono D. (1997) Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15, 871–875 [DOI] [PubMed] [Google Scholar]
- 28. Watt A., Moukambi F., Banadyga L., Groseth A., Callison J., Herwig A., Ebihara H., Feldmann H., and Hoenen T. (2014) A novel life cycle modeling system for Ebola virus shows a genome length-dependent role of VP24 in virus infectivity. J. Virol. 88, 10511–10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pols M. S., ten Brink C., Gosavi P., Oorschot V., and Klumperman J. (2013) The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic 14, 219–232 [DOI] [PubMed] [Google Scholar]
- 30. Ebert D. H., Deussing J., Peters C., and Dermody T. S. (2002) Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277, 24609–24617 [DOI] [PubMed] [Google Scholar]
- 31. Simmons G., Gosalia D. N., Rennekamp A. J., Reeves J. D., Diamond S. L., and Bates P. (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 102, 11876–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ryabchikova E. I., Kolesnikova L. V., and Luchko S. V. (1999) An analysis of features of pathogenesis in two animal models of Ebola virus infection. J. Infect. Dis. 179, S199–S202 [DOI] [PubMed] [Google Scholar]
- 33. Geisbert T. W., Young H. A., Jahrling P. B., Davis K. J., Larsen T., Kagan E., and Hensley L. E. (2003) Pathogenesis of Ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am. J. Pathol. 163, 2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Côté M., Misasi J., Ren T., Bruchez A., Lee K., Filone C. M., Hensley L., Li Q., Ory D., Chandran K., and Cunningham J. (2011) Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477, 344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., and Farzan M. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bröcker C., Kuhlee A., Gatsogiannis C., Balderhaar H. J., Hönscher C., Engelbrecht-Vandré S., Ungermann C., and Raunser S. (2012) Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. U.S.A. 109, 1991–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seals D. F., Eitzen G., Margolis N., Wickner W. T., and Price A. (2000) A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. U.S.A. 97, 9402–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shirato K., Kawase M., and Matsuyama S. (2013) Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 87, 12552–12561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parenti F., Beretta G., Berti M., and Arioli V. (1978) Teichomycins, new antibiotics from Actinoplanes teichomyceticus Nov. Sp. I. Description of the producer strain, fermentation studies and biological properties. J. Antibiot. 31, 276–283 [DOI] [PubMed] [Google Scholar]
- 40. Borghi A., Coronelli C., Faniuolo L., Allievi G., Pallanza R., and Gallo G. G. (1984) Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. IV. Separation and characterization of the components of teichomycin (teicoplanin). J. Antibiot. 37, 615–620 [DOI] [PubMed] [Google Scholar]
- 41. Corti A., Soffientini A., and Cassani G. (1985) Binding of the glycopeptide antibiotic teicoplanin to d-alanyl-d-alanine-agarose: the effect of micellar aggregates. J. Appl. Biochem. 7, 133–137 [PubMed] [Google Scholar]
- 42. Corry J., Johnson S. M., Cornwell J., and Peeples M. E. (2015) Preventing cleavage of the respiratory syncytial virus attachment protein in Vero cells rescues the infectivity of progeny virus for primary human airway cultures. J. Virol. 10.1128/JVI.02351-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Balzarini J., Pannecouque C., De Clercq E., Pavlov A. Y., Printsevskaya S. S., Miroshnikova O. V., Reznikova M. I., and Preobrazhenskaya M. N. (2003) Antiretroviral activity of semisynthetic derivatives of glycopeptide antibiotics. J. Med. Chem. 46, 2755–2764 [DOI] [PubMed] [Google Scholar]
- 44. Preobrazhenskaya M. N., and Olsufyeva E. N. (2006) Polycyclic peptide and glycopeptide antibiotics and their derivatives as inhibitors of HIV entry. Antiviral Res. 71, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bereczki I., Kicsák M., Dobray L., Borbás A., Batta G., Kéki S., Nikodém É. N., Ostorházi E., Rozgonyi F., Vanderlinden E., Naesens L., and Herczegh P. (2014) Semisynthetic teicoplanin derivatives as new influenza virus binding inhibitors: synthesis and antiviral studies. Bioorg. Med. Chem. Lett. 24, 3251–3254 [DOI] [PubMed] [Google Scholar]
- 46. Maieron A., and Kerschner H. (2012) Teicoplanin therapy leading to a significant decrease in viral load in a patient with chronic hepatitis C. J. Antimicrob. Chemother. 67, 2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Obeid S., Printsevskaya S. S., Olsufyeva E. N., Dallmeier K., Durantel D., Zoulim F., Preobrazhenskaya M. N., Neyts J., and Paeshuyse J. (2011) Inhibition of hepatitis C virus replication by semi-synthetic derivatives of glycopeptide antibiotics. J. Antimicrob. Chemother. 66, 1287–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. De Burghgraeve T., Kaptein S. J., Ayala-Nunez N. V., Mondotte J. A., Pastorino B., Printsevskaya S. S., de Lamballerie X., Jacobs M., Preobrazhenskaya M., Gamarnik A. V., Smit J. M., and Neyts J. (2012) An analog of the antibiotic teicoplanin prevents flavivirus entry in vitro. PLoS ONE 7, e37244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Balzarini J., Keyaerts E., Vijgen L., Egberink H., De Clercq E., Van Ranst M., Printsevskaya S. S., Olsufyeva E. N., Solovieva S. E., and Preobrazhenskaya M. N. (2006) Inhibition of feline (FIPV) and human (SARS) coronavirus by semisynthetic derivatives of glycopeptide antibiotics. Antiviral Res. 72, 20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andrault P. M., Samsonov S. A., Weber G., Coquet L., Nazmi K., Bolscher J. G., Lalmanach A. C., Jouenne T., Brömme D., Pisabarro M. T., Lalmanach G., and Lecaille F. (2015) Antimicrobial peptide LL-37 is both a substrate of cathepsins S and K and a selective inhibitor of cathepsin L. Biochemistry 54, 2785–2798 [DOI] [PubMed] [Google Scholar]
- 51. Brogden R. N., and Peters D. H. (1994) Teicoplanin. A reappraisal of its antimicrobial activity, pharmacokinetic properties and therapeutic efficacy. Drugs 47, 823–854 [DOI] [PubMed] [Google Scholar]
- 52. Cavalcanti A. B., Goncalves A. R., Almeida C. S., Bugano D. D., and Silva E. (2010) Teicoplanin versus vancomycin for proven or suspected infection. Cochrane Database Syst. Rev. 16, CD007022. [DOI] [PubMed] [Google Scholar]