Abstract
ATR (ataxia telangiectasia and Rad-3-related) is a protein kinase that maintains genome stability and halts cell cycle phase transitions in response to DNA lesions that block DNA polymerase movement. These DNA replication-associated features of ATR function have led to the emergence of ATR kinase inhibitors as potential adjuvants for DNA-damaging cancer chemotherapeutics. However, whether ATR affects the genotoxic stress response in non-replicating, non-cycling cells is currently unknown. We therefore used chemical inhibition of ATR kinase activity to examine the role of ATR in quiescent human cells. Although ATR inhibition had no obvious effects on the viability of non-cycling cells, inhibition of ATR partially protected non-replicating cells from the lethal effects of UV and UV mimetics. Analyses of various DNA damage response signaling pathways demonstrated that ATR inhibition reduced the activation of apoptotic signaling by these agents in non-cycling cells. The pro-apoptosis/cell death function of ATR is likely due to transcription stress because the lethal effects of compounds that block RNA polymerase movement were reduced in the presence of an ATR inhibitor. These results therefore suggest that whereas DNA polymerase stalling at DNA lesions activates ATR to protect cell viability and prevent apoptosis, the stalling of RNA polymerases instead activates ATR to induce an apoptotic form of cell death in non-cycling cells. These results have important implications regarding the use of ATR inhibitors in cancer chemotherapy regimens.
Keywords: apoptosis, cell cycle, cell proliferation, cell signaling, DNA damage, DNA damage response, DNA repair, genomic instability, RNA polymerase, transcription
Introduction
Ultraviolet (UV) photoproducts and other bulky DNA lesions block the progression of DNA and RNA polymerases and lead to the activation of various DNA damage responses that are regulated by the action of the ataxia telangiectasia and Rad-3-related (ATR)3 protein kinase. For example, in response to DNA polymerase stalling and uncoupling from DNA helicase activity (1), ATR is thought to be recruited to single-stranded regions of DNA where it promotes a number of DNA metabolic processes that maintain genomic integrity (2–6), including replication fork stabilization, DNA synthesis and lesion bypass, homologous recombination, replication origin firing, and cell cycle delay. The importance of ATR kinase activity in these responses in replicating cells is evidenced by the fate of cells in which the catalytic activity of ATR has been genetically or pharmacologically inhibited following replication stress, which include apoptosis and entry into catastrophic mitoses (7–10). Thus, ATR helps replicating cells cope with DNA damage and replication stress by facilitating cellular processes that maintain genomic stability and avoid cell death.
Given these features of ATR, there has been great interest in the development and use of selective small-molecule inhibitors of ATR kinase activity in cancer chemotherapy regimens in conjunction with compounds that damage DNA and generate replication stress (11–13). For example, several studies have shown that ATR inhibition sensitizes cancer cells to cell killing by ionizing radiation and common cancer chemotherapeutic drugs, including platinum compounds and topoisomerase inhibitors (14–21). Through a related mechanism, ATR inhibition has also been implicated in preventing skin carcinogenesis by promoting the death of cells exposed to environmental carcinogens such as UV light (22–24). Thus, the general consensus view is that ATR protects cells from DNA damage and that ATR inhibition increases the likelihood that DNA damage will result in cell death.
However, the stalling of DNA polymerases by UV photoproducts and other bulky DNA lesions only takes place in cells that are actively replicating their DNA, and thus the function of ATR in response to DNA damage and transcription stress in non-replicating, non-cycling cells is not known. Although previous studies have indicated that ATR can be activated in non-cycling cells in response to DNA repair intermediates (25–32) and in response to the stalling of RNA polymerase (33–35), the functional significance of ATR in these contexts in non-cycling cells has not been examined.
Here, we unexpectedly found that ATR inhibition actually protects non-cycling cells exposed to UV, UV mimetics, and other inducers of transcription stress from undergoing an apoptotic form of cell death. This finding differs significantly from the anti-apoptotic functions of ATR in replicating and cycling cells. Because most cells are in a quiescent or slowly growing state in vivo, we believe that these results have important implications for the use of ATR kinase inhibitors in cancer chemotherapy protocols.
Experimental Procedures
Cell Lines
Human HaCaT keratinocytes, U2OS osteosarcoma cells, mouse embryonic fibroblasts, and hTERT-immortalized normal human fibroblasts (NHF1) were cultured at 37 °C in a 5% CO2 humidified incubator in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 6 mm l-glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Cycling cells were typically plated at 30% confluence and treated the following day. Non-cycling, quiescent cells were plated at 40–60% confluence, grown for 2 days in normal medium until the cells reached confluence, and then maintained for 2–3 days in DMEM containing 0.5% FBS and penicillin/streptomycin before being subjected to experimental treatments as previously described (32, 36). To expose cells to the indicated fluences of UV radiation, media was removed and set aside before placing the cells under a GE germicidal lamp that emits primarily 254-nm UV light (UV-C) that was connected to a digital timer. Treatment with N-acetoxy-2-acetylaminofluorene (AAAF), camptothecin, 5,6-dichloro-1-β-d-ribofuranosyl-1H-benzimidazole (DRB), and triptolide involved adding the compound directly to the culture medium of the cells. Cells were treated with DMSO, the ATR inhibitor VE-821 (10 μm), or other kinase inhibitors for 30–60 min prior to exposure to UV or addition of the indicated drugs.
Chemicals and Reagents
AAAF was purchased from the MRIGlobal Chemical Carcinogen Repository and resuspended in 95% ethanol. Bromodeoxyuridine (BrdU), triptolide, DRB, and inhibitors of the mitogen-activated protein kinases (MAPKs) MEK1/2 (U0126), p38 (SB202190), and Jun kinase (JNK; SP600125), the phosphoinositide 3-kinases (PI3Ks; wortmannin and LY294002), and AMP-dependent protein kinase (dorsomorphin/Compound C) were obtained from Sigma. Inhibitors of the DNA damage kinases ATR (VE-821 and AZD6738), ataxia telangiectasia-mutated (ATM; KU-55933), DNA-dependent protein kinase (DNA-PK; NU7441), and Checkpoint kinase 1 (CHK1; AZD7762) were purchased from Selleckchem. The pan-caspase inhibitor Z-VAD-FMK and inhibitors of the mammalian target of rapamycin kinase (rapamycin) and Tank-binding protein 1 kinase (TBK1; BX795) were obtained from Invivogen.
RNA Interference
Lentiviral DNA particles were generated with HEK293T cells following co-transfection of the packaging plasmid psPAX2 and the envelope plasmid pMD2.G with the appropriate pLKO.1 vector and Lipofectamine 2000. Empty and XPA shRNA-containing pLKO.1 plasmids were from the Open Biosystems TRC1 shRNA library (37). The p53 shRNA plasmids were from Addgene (plasmids 25636 and 25637) (38).
Immunoblotting
Cells were washed with cold PBS, scraped from the plate, and pelleted by gentle centrifugation. Cells were then lysed for 20 min on ice in 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, and 1% Triton X-100 containing a protease inhibitor mixture (Roche Applied Science). Following centrifugation in a microcentrifuge for 10 min at maximum speed, the soluble cell lysates were transferred to new tubes. For experiments examining the phosphorylation of the histone variant H2AX, the insoluble fraction was then resuspended in 1× SDS-PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 5% glycerol, 100 mm DTT, 1% SDS, 0.005% bromphenol blue) and sonicated. Equal amounts of soluble or insoluble cell lysates were separated by SDS-PAGE, transferred to nitrocellulose, and then probed by immunoblotting using standard procedures. Primary antibodies included antibodies against ATR (sc-1887), CHK1 (sc-8408), p53 (sc-6243), proliferating cell nuclear antigen (sc-56), actin (I-19), XPB (sc-293), and XPA (sc-853) from Santa Cruz; RPA70 (A300–421A) and phospho-KAP1 (Ser824; A304–146A) from Bethyl Laboratories; and CHK2 (number 2662), phospho-CHK1 (Ser345; number 2348), phospho-CHK1 (Ser296; number 2349), phospho-CHK2 (Thr68; number 2621), phospho-p53 (Ser15; number 9284), phospho-p53 (Ser20; number 9287), phospho-H2AX (Ser139; number 9718), BCL2 (number 2872), cleaved caspase 3 (number 9661), poly(ADP-ribose) polymerase (PARP) (number 9542), phospho-AKT (Ser473; number 9271), phosphor-c-Jun (Ser63, number 9261), and phospho-ERK1/2 (Thr202/Tyr204; number 4370) from Cell Signaling Technology. All primary antibodies were used at a 1:1000 or 1:2000 dilution in 1× TBST (50 mm Tris-HCl, pH 7.4, 135 mm NaCl, 0.1% Tween 20). Secondary antibodies included horseradish peroxidase-linked anti-rabbit IgG (NA934V) and anti-mouse IgG (NA931V) from GE Amersham Biosciences and anti-goat IgG (number 31402) from Pierce. Chemiluminescence was visualized with Clarity Western ECL Substrate (Bio-Rad) or ECL Prime Western blotting Detection Reagent (GE Amersham Biosciences) using a Molecular Imager Chemi-Doc XRS+ system (Bio-Rad). Chemiluminescent signals within the linear range of detection were quantified using ImageQuant software (GE Healthcare). For each soluble lysate immunoblot, the phosphoprotein signal was quantified and normalized to actin levels in the same protein lysate, and the maximum signal for each blot was set to an arbitrary value of 100. All other phosphoprotein/actin ratios were then normalized to this value for each immunoblot. We note that actin levels did not change during the first few hours following UV and AAAF treatment, and decreased modestly at late time points (8–12 h) when cells began undergoing apoptosis. For analysis of phosphorylated H2AX, the blot was stained with 1% Ponceau red dye (Sigma) in 1% acetic acid (v/v) prior to blocking and immunoblotting. The membrane was then scanned, and the total histone signal quantified for normalization of the phospho-H2AX signals. All experiments analyzing DNA damage response signaling were repeated 2–4 times, and the average (and standard error) of the phosphoprotein/actin and phospho-H2AX/total histone ratios were determined and plotted.
Cell Survival Assays
Cell survival assays were performed using 12- or 24-well plates, and survival was typically determined 2 days following treatment unless otherwise indicated. Following aspiration and removal of cell culture media and dead, floating cells and two washes with cold phosphate-buffered saline (PBS), the cells were fixed for 30–60 min in cold 100% methanol at −20 °C. Methanol was then aspirated and replaced with 0.5% crystal violet stain (J.T. Baker) in 25% methanol for 10 min. The plates were then washed extensively in tap water to remove the excess stain. After drying, images of the stained cells were then taken on a Molecular Imager Chemi-Doc XRS+ system (Bio-Rad) for presentation of representative, qualitative results. The crystal violet stain was then solubilized in 1% SDS and the absorbance at 540 nm measured with a SpectraMax M3 spectrophotometer (Molecular Devices). The absorbance of the untreated sample was set to an arbitrary value of 1 for each experiment, and the treatment samples normalized to this value. All cell survival experiments were performed at least 3 times.
Immunoslot Blot Analysis
The level of DNA synthesis in cycling and non-cycling cells was measured with an immunoslot blot assay. BrdU (32.5 μm) was added to the cell culture medium 1 h prior to cell harvesting. Cells were washed in cold PBS, scraped from the plate, centrifuged at 1,600 × g in a microcentrifuge for 5 min at 4 °C, and then frozen on dry ice. Genomic DNA was then purified with a QIAamp DNA Mini kit (Qiagen). Genomic DNA (1 mg) was immobilized on a nitrocellulose membrane with a Bio-Dot SF Cell immunoslot blot apparatus (Bio-Rad) and baked at 80 °C under vacuum for 90 min. Blots were blocked in 5% milk in PBST (phosphate-buffered saline containing 0.1% Tween 20) and probed with an anti-BrdU antibody (Sigma, B2531). Following immunoblotting, the blots were stained with SYBR Gold (Invitrogen) to ensure equal loading of DNA. The experiment was repeated two times, and representative results are presented.
For the analysis of repair of (6-4)pyrimidine-pyrimidone UV photoproducts ((6-4)PPs) (36, 39), cells were harvested at the indicated time points following exposure to 10 J/m2 of UV. The immunoslot blot method was similar to that described above with the exception that BrdU was omitted from the procedure and 250 ng of genomic DNA was immobilized on the nitrocellulose membrane. An anti-(6-4)PP antibody (Cosmo Bio 64 M-2) was used to detect (6-4)PP presence and removal from genomic DNA.
Detection of Excised Oligonucleotide Products of Nucleotide Excision Repair
Nucleotide excision repair activity was visualized as previously described (36, 40–43) with the following modifications. Cells in 10-cm plates were harvested 1 h after irradiation with 20 J/m2 of UV. Cells were lysed in 25 mm HEPES-KOH, 100 mm KCl, 12 mm MgCl2, 0.5 mm EDTA, 12.5% glycerol, and 0.5% Nonidet P-40 for 20 min on ice. Following centrifugation at 16,873 × g for 30 min at 4 °C, the soluble cell lysates were added to a new tube containing 2 μg of anti-XPB antibody (Santa Cruz sc-293) to immunoprecipitate the TFIIH-excised oligonucleotide complexes (40, 43, 44) from the lysates. Following a 1.5-h incubation with the XPB antibody at 4 °C, recombinant protein A/G PLUS-agarose (Santa Cruz) was added and the mixture rotated for 2 h at 4 °C. The immunoprecipitates were then washed three times with lysis buffer. A fraction (25%) of the immunoprecipitated material was saved for immunoblot analysis with an anti-XPB antibody. The excised oligonucleotide products of nucleotide excision repair were purified from the remaining material following incubation at 55 °C for 20 min with elution buffer (50 mm Tris-HCl, pH 8, 250 mm NaCl, 10 mm EDTA, and 0.5% SDS) containing 50 μg of proteinase K (New England Biolabs), phenol-chloroform extraction, and ethanol precipitation. The excised oligonucleotides were resuspended in 10 μl of water, and half of the DNA was 3′-end labeled for 1 h at 37 °C in a 10-μl reaction containing 6 units of terminal deoxynucleotidyl transferase (New England Biolabs), 0.25 mm CoCl2, and 20 μm biotin-11-dUTP (Fermentas) in 1× terminal deoxynucleotidyl transferase buffer (New England Biolabs). Following ethanol precipitation, the biotinylated, TFIIH-associated excised oligonucleotides were separated on a 12% urea-polyacrylamide gel in 1× TBE (300 V, 30–35 min), transferred to a nylon membrane in 0.5× TBE with a Bio-Rad Trans Blot Turbo semi-dry transfer apparatus (25 V, 25 min), and then fixed to the membrane with a UV cross-linker. The membrane was blocked and washed three times for 5 min each with PBS containing 2% SDS before incubation for 1 h with a 1:100,000 dilution of HRP-coupled streptavidin (ThermoFisher Scientific) in PBS containing 0.05% SDS. The membrane was then washed twice for 5 min each with PBS containing 0.05% SDS, TBS containing 0.1% Tween 20, and substrate equilibration buffer (50 mm Tris-HCl, pH 8.8, 150 mm NaCl). Chemiluminescent signals were visualized with Clarity Western ECL Substrate (Bio-Rad) and using a Molecular Imager Chemi-Doc XRS+ system (Bio-Rad). Biotinylated oligonucleotides of known length were resolved on all gels as size markers.
Results
UV-induced DNA Damage Signaling Exhibit Different Profiles in Cycling and Non-cycling Cells
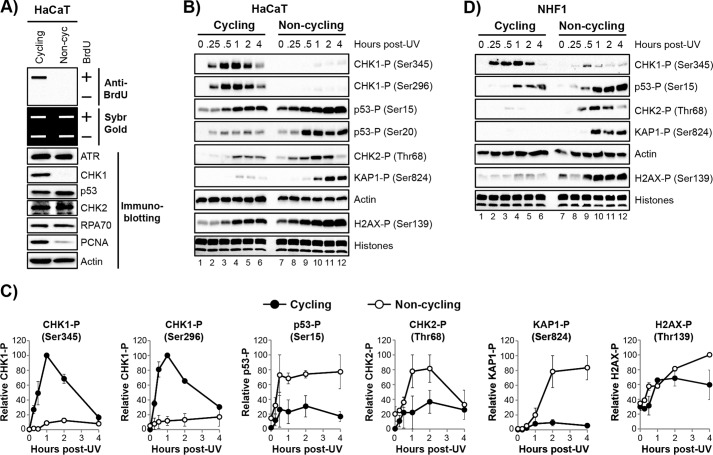
Previous studies comparing UV-induced DNA damage signaling in cycling and non-cycling cells have been rather limited (29, 32). Thus, to examine this issue in greater detail and specifically the contribution of the ATR kinase, we monitored the phosphorylation of several key DNA damage response signaling proteins following UV irradiation of rapidly growing, subconfluent HaCaT keratinocytes (cycling cells) and cells grown to confluence and maintained in low serum (non-cycling cells). To validate that cells grown under these conditions were indeed cycling or non-cycling, we first measured the level of DNA synthesis taking place in the cells. As shown in Fig. 1A (top), only the cycling, sub-confluent HaCaT cells incorporated significant amounts of BrdU into genomic DNA. The examination of protein levels in these two cell populations demonstrated that the DNA polymerase clamp protein proliferating cell nuclear antigen was present at much higher levels in cycling cells than non-cycling cells (Fig. 1A, bottom), which provides additional evidence that the non-cycling cells are not actively replicating their DNA. Importantly, a subset of other DNA damage response proteins, including RPA and ATR, were found to be present at similar levels in cycling and non-cycling cells.
FIGURE 1.
UV-induced DNA damage signaling exhibits different profiles in cycling and non-cycling cells. A, HaCaT cells were plated at low density and maintained in 10% FBS (cycling) or grown to confluence and maintained in 0.5% FBS (non-cycling). DNA synthesis was determined by detecting the level of BrdU incorporation into genomic DNA. Immunoslot blots were stained with SYBR Gold to ensure equal loading of genomic DNA. Results from a representative experiment are shown. Lysates from the cycling and non-cycling cells were examined by immunoblotting with antibodies against the indicated proteins. B, cycling and non-cycling cells HaCaT were exposed to 10 J/m2 of UV. Lysates were prepared from the cells at the indicated time points following irradiation and probed with antibodies against the indicated proteins. C, quantitation of two experiments performed as in B. Phosphoprotein signals were normalized to actin (or total histones, in the case of H2AX phosphorylation), and the highest phosphoprotein/actin ratio for each blot was set to an arbitrary value of 100. All other signals were normalized to this value, and the average and S.E. are shown. D, cycling and non-cycling NHF1 cells were exposed to UV and analyzed as in B.
We next exposed cycling and non-cycling cells to UV. As expected, UV induced a rapid and robust but transient phosphorylation of CHK1 in the cycling cells (Fig. 1B, lanes 1–6). CHK1 phosphorylation is the most routinely used biochemical read-out for ATR activation, and its phosphorylation occurs on both an ATR target site (Ser345) as well as a CHK1 autophosphorylation site (Ser296), which requires prior activation of the kinase by ATR. These phosphorylation events are required for CHK1 to suppress replication origin firing and premature entry into mitosis (45, 46).
In contrast, very little CHK1 phosphorylation was observed on either site in non-cycling HaCaT cells (Fig. 1B, lanes 7–12). However, CHK1 protein levels were nearly absent in the non-cycling cells (Fig. 1A), consistent with previous reports in other cell lines (29, 32). Nonetheless, this difference in phosphorylation between cycling and non-cycling cells (Fig. 1C) suggests that the canonical ATR substrate CHK1 is not a physiological target of ATR in non-cycling cells following UV irradiation.
Nonetheless, there are several other potential protein targets for ATR in response to DNA damage, including the tumor suppressor protein p53 (32, 47) and the histone variant H2AX (48). Interestingly, and in striking contrast to the data for CHK1, UV-induced phosphorylation of p53 on Ser15 and H2AX on Ser139 were actually higher in the non-cycling HaCaT cells than in the cycling cells (Fig. 1, B and C). We also observed robust phosphorylation of CHK2 (checkpoint kinase 2) and KAP1 (Krüppel-associated box-associated protein 1) in the non-cycling cells. These proteins are thought to be primarily phosphorylated by the ATM kinase in response to double-strand breaks in DNA (49–52). Although the elevated phosphorylation of p53 may be due in part to moderately higher levels of p53 in non-cycling cells than cycling cells (Fig. 1A), several other proteins, including CHK2, were present at similar levels in both cycling and non-cycling cells (Fig. 1A).
To confirm these distinct DNA damage response substrate phosphorylation profiles in another cell line, we repeated the experiment in cycling and non-cycling telomerase-immortalized normal human fibroblasts (NHF1 cells). As shown in Fig. 1D, higher levels of phosphorylation of p53, H2AX, CHK2, and KAP1 were observed in non-cycling cells than in cycling cells. In addition, much less CHK1 phosphorylation was observed in the non-cycling NHF1 cells relative to the cycling cells (Fig. 1D). However, as was observed in HaCaT cells, CHK1 protein was nearly absent in the non-cycling cells (data not shown). The residual CHK1 phosphorylation that was observed in these experiments is likely due to incomplete growth arrest and to the small percentage of cells that continue to cycle under conditions of confluence and low serum (see Fig. 4B). We conclude that robust DNA damage signaling occurs in non-cycling cells following UV irradiation and that the profile of phosphorylation events is different from in cycling cells.
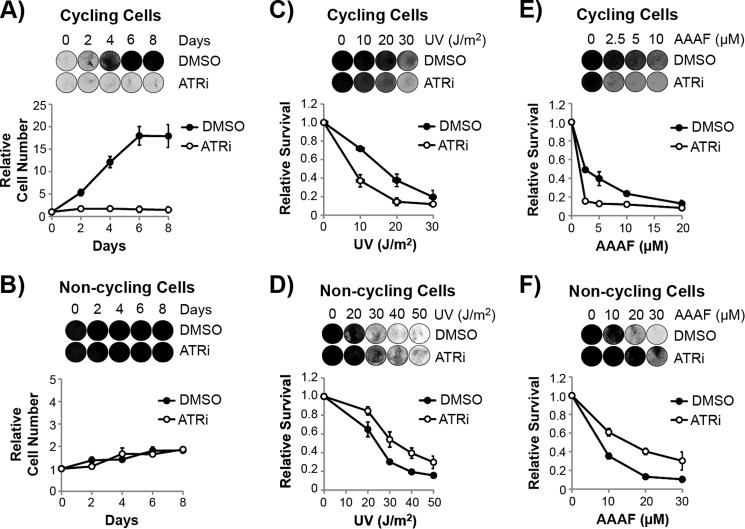
FIGURE 4.
ATR inhibition has opposing effects on viability in cycling and non-cycling cells exposed to UV and UV mimetics. A, HaCaT cells were plated at low density and then treated with either DMSO or ATRi. The culture media was replaced every 2 days with fresh medium containing DMSO or ATRi, and cells were stained with crystal violet at the indicated time points to monitor cell growth. Relative cell number was determined by measuring the absorption of solubilized crystal violet from the stained cells and by normalization to day 0, which was set to an arbitrary value of 1. B, non-cycling HaCaT cells grown to confluence and maintained in low serum were treated and analyzed as in A. The cell number increased less than 2-fold over the course of 8 days, indicating a very low level of cell proliferation under these conditions. C, subconfluent, cycling HaCaT cells were treated with DMSO or ATRi prior to exposure to the indicated fluences of UV. Cells were stained with crystal violet 2 days later, and the incorporated stain was solubilized and quantified with a spectrophotometer. The decrease in cell number is likely due to a combination of cell death and cell cycle arrest in these cells. D, confluent, non-cycling HaCaT cells were treated and analyzed as in C. E, cycling HaCaT cells were treated as in C, except that cells were instead treated with the UV mimetic AAAF. F, non-cycling HaCaT cells were treated as in E. Representative images of the stained cells are shown above the quantitative graphs. ATRi, ATR inhibitor.
ATR Contributes to DNA Damage Response Protein Phosphorylation in Non-cycling Cells Exposed to UV and UV Mimetics
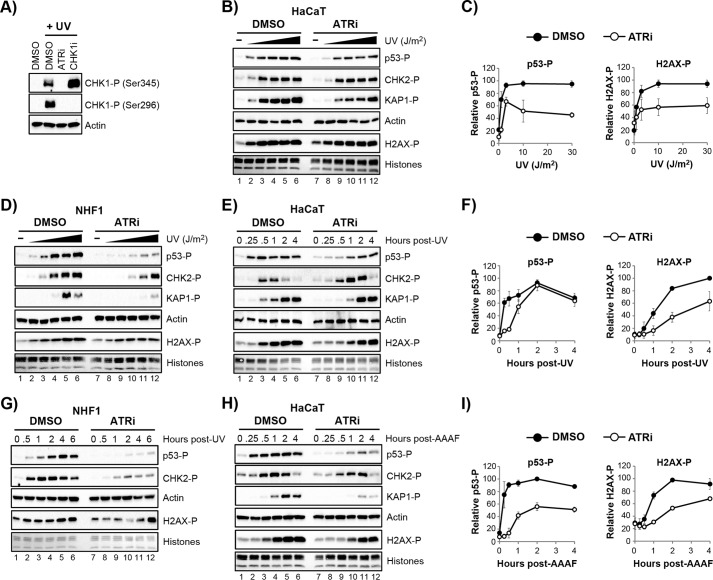
Although ATR activation has been suggested to occur in non-cycling cells (25–33), its signaling function has not been extensively examined and its bona fide substrates are not known. Thus, to examine the role of ATR kinase activity in DNA damage signaling in non-cycling cells, we made use of the highly selective ATR inhibitor VE-821 (21, 53). Consistent with a recent report (21), this compound completely blocked UV-induced phosphorylation of CHK1 in cycling HaCaT cells (Fig. 2A).
FIGURE 2.
ATR contributes to DNA damage signaling in non-cycling cells. A, cycling HaCaT cells were treated with the ATR inhibitor (ATRi) VE-821 (10 μm ATRi) or the CHK1 inhibitor (CHK1i) AZD7762 (300 nm CHK1i) for 30 min prior to irradiation with 10 J/m2 UV. Cell lysates were prepared 30 min post-irradiation and analyzed by immunoblotting. B, non-cycling HaCaT cells were treated with DMSO or the ATRi prior to exposure to increasing fluences of UV (0, 1, 3, 10, 30, and 100 J/m2). Cell lysates were analyzed by immunoblotting. C, quantification of p53 and H2AX phosphorylation from three independent experiments performed as described in B. D, non-cycling NHF1 cells were treated as in B. E, non-cycling HaCaT cells were treated with DMSO or ATRi prior to UV irradiation (10 J/m2). Cell lysates were prepared at the indicated time points and analyzed as in B. F, quantification of results from three independent experiments were performed as shown in E. G, non-cycling NHF1 cells were treated as in E. H, non-cycling HaCaT cells were treated with DMSO or ATRi prior to exposure to the UV mimetic AAAF (10 μm AAAF). Cells lysates were analyzed by immunoblotting. I, quantification of results from three independent experiments performed as in H.
We then took advantage of this inhibitor in non-cycling HaCaT cells exposed to increasing fluences of UV radiation. As shown in Fig. 2, B and C, ATR inhibition partially abrogated the phosphorylation of several DNA damage response substrates, including p53 and H2AX. However, significant levels of phosphorylation remained in the presence of the ATR inhibitor. Importantly, similar results were observed when we treated non-cycling NHF1 cells with the ATR inhibitor and then exposed the cells to increasing amounts of UV (Fig. 2D).
To examine the activity of ATR in greater detail, we performed time course experiments with 10 J/m2 of UV in non-cycling HaCaT cells. Fig. 2, E and F, shows that the ATR inhibitor again partially abrogated the phosphorylation of DNA damage response substrates p53 and H2AX. However, we note that each substrate showed unique phosphorylation kinetics, which are impacted in different ways by ATR inhibition. For example, p53 phosphorylation was reduced to a significant extent within the first hour after UV irradiation. In contrast, H2AX phosphorylation was reduced at all time points after UV but continued to become phosphorylated even in the presence of the ATR inhibitor. Importantly, similar results were observed in non-cycling NHF1 cells (Fig. 2G).
To confirm the effects of ATR inhibition on DNA damage signaling with another DNA damaging agent, we treated non-cycling HaCaT cells with the UV mimetic AAAF, which generates bulky, helix distorting lesions on guanine residues in DNA that block RNA polymerase progression (54, 55) and that like UV photoproducts are removed from DNA by the nucleotide excision repair system (42, 56–58). As shown in Fig. 2, H and I, the phosphorylation of p53, KAP1, and H2AX were all reduced following AAAF administration in the presence of the ATR inhibitor. Consistent with the results with UV, however, these proteins continued to be phosphorylated to significant extents when ATR was inhibited. These results suggest that ATR is not the only protein kinase that phosphorylates DNA damage signaling proteins in response to UV and UV mimetics in non-cycling cells.
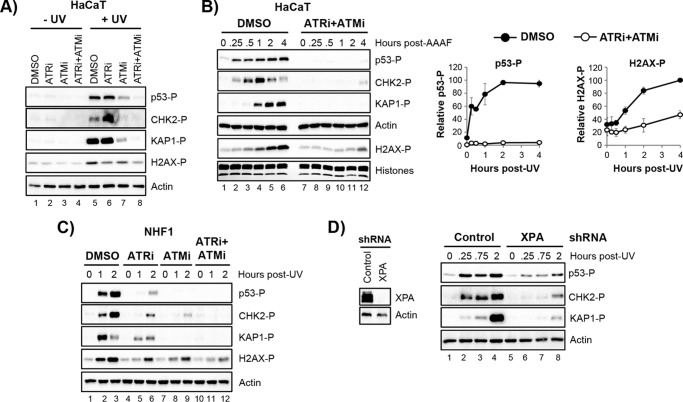
ATM and ATR Both Contribute to Canonical DNA Damage Signaling Events in UV-irradiated Non-cycling Cells
A recent study found that UV irradiation of non-cycling cells leads to formation of double-strand breaks in DNA in a nucleotide excision repair-dependent manner (29). This double-strand break response was associated with the activation of ATM and the phosphorylation of both H2AX and the more specific ATM substrate CHK2. To confirm this previous report and examine additional DNA damage substrates (p53 and KAP1) in this context, we treated cells with an ATR inhibitor, an ATM inhibitor, or a combination of both inhibitors and then examined UV-induced protein phosphorylation. As shown in Fig. 3A, the ATM inhibitor had a more significant effect on protein phosphorylation than the ATR inhibitor 2 h following UV irradiation in non-cycling HaCaT cells. However, near complete loss of p53, CHK2, and KAP1 phosphorylation was only observed when the cells were treated with both inhibitors. Time course experiments further demonstrated a near complete loss of protein phosphorylation in the presence of both ATR and ATM inhibitors (Fig. 3B). There appears to be a small amount of protein phosphorylation that occurs at late time points in these cells, which may be due to DNA-PK activation in response to double-strand break formation (59). Nonetheless, the notion that both ATR and ATM contribute to DNA damage substrate protein phosphorylation at early time points following irradiation was supported by additional experiments in non-cycling NHF1 cells (Fig. 3C).
FIGURE 3.
Both ATR and ATM contribute to canonical DNA damage signaling events in UV-irradiated non-cycling cells. A, non-cycling HaCaT cells were treated with the indicated inhibitors prior to irradiation with UV. Cell lysates were prepared 2 h after irradiation and analyzed by immunoblotting. B, non-cycling cells were treated with DMSO or a combination of an ATR (ATRi) and ATM (ATMi) inhibitors. Protein phosphorylation was analyzed by immunoblotting at the indicated time points following exposure to 10 J/m2 of UV. The graphs on the right show quantifications of the results in B from three independent experiments. C, non-cycling NHF1 cells were treated as in B. D, HaCaT cells were infected with either a control vector or a vector expressing a shRNA against the excision repair factor XPA. Cell lysates from puromycin-selected cells were analyzed by immunoblotting. The cells were then exposed to UV (10 Jm2), and protein phosphorylation was examined by immunoblotting.
The DNA lesions induced by UV and UV mimetics are removed by the process of nucleotide excision repair, which leaves gaps in the DNA that can be enlarged to activate ATR (32, 60, 61) or possibly give rise to double-strand breaks that activate ATM (29). Thus, to determine how nucleotide excision repair affects DNA damage signaling in non-cycling cells, we used lentiviral shRNA to knock down the excision repair factor XPA in HaCaT cells. As shown in Fig. 3D, the phosphorylation of p53, Chk2, and KAP1 were all reduced in non-cycling cells deficient in XPA. However, the residual protein phosphorylation that remains suggests that there are likely excision repair-independent modes of ATR and ATM activation in response to UV in non-cycling cells, which could include direct recognition of the damage by the kinases and associated proteins (62–67) or by stalling the RNA polymerase at UV lesions (33). Nonetheless, the results presented in Figs. 2 and 3 suggest that UV radiation leads to protein phosphorylation events that are performed at least in part by the action of the ATR kinase.
ATR Has Opposing Functions on Cell Viability in Cycling and Non-cycling Cells Exposed to UV Mimetic DNA Damaging Agents
We next wanted to determine the ultimate fate of cells in which ATR kinase activity is inhibited during exposure to DNA damaging agents, which may allow for better definition of possible functions of ATR in non-cycling cells. UV and UV mimetics have the potential to cause a number of cellular responses, including the induction of cell death pathways that lead to a loss in cell viability. Given that ATR kinase has been reported to be essential for cell viability and cell division (68, 69), we therefore next carried out cell survival assays to examine how ATR inhibition affects cell viability in cycling and non-cycling cells exposed to DNA damaging agents.
We first examined the importance of ATR kinase activity on the viability of cells in the absence of any exogenous source of DNA damage. Consistent with the essential nature of ATR (68, 69), HaCaT cells plated at low density and then treated with the ATR inhibitor failed to grow and divide (Fig. 4A). In contrast, when we treated confluent, non-cycling cells with the ATR inhibitor, we did not observe any effect of ATR inhibition on cell viability even after continuous treatment with the ATR inhibitor for more than 1 week (Fig. 4B). These results indicate that ATR kinase activity is dispensable for the viability of non-cycling cells.
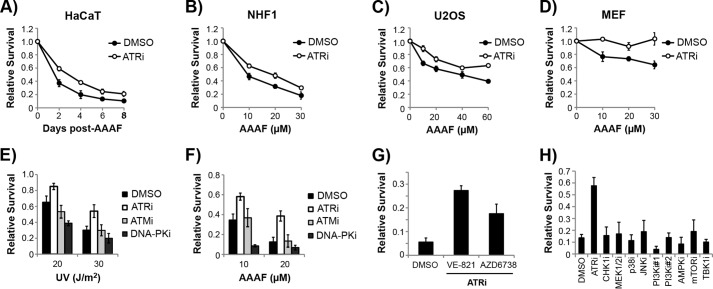
We next investigated the role of the ATR kinase on the viability of cycling and non-cycling cells exposed to DNA damaging agents. As shown in Fig. 4C, inhibition of ATR sensitized cycling HaCaT cells to the killing effects of UV, which is consistent with previous reports (7, 8, 10, 21). In marked contrast, however, the inhibition of ATR in non-cycling HaCaT cells instead protected the cells from the lethal effects of UV (Fig. 4D). Regardless of the UV dose, there were significantly more cells that remained viable and attached to the plate following UV when treated with the ATR inhibitor. This opposite function of ATR on survival in cycling and non-cycling cells was even more evident and striking when we treated cycling and non-cycling cells with the UV mimetic AAAF (Figs. 4, E and F).
To further validate that ATR inhibition protects non-cycling cells from the lethal effects of UV, we performed additional experiments with the UV mimetic AAAF. Time course experiments showed that cell viability was higher in non-cycling cells treated with the ATR inhibitor for several days following AAAF treatment (Fig. 5A). Moreover, the protective effect of the ATR inhibitor was evident in NHF1 cells, U2OS osteosarcoma cells, and mouse embryonic fibroblasts (Fig. 5, B–D). These results show that cell type and genetic background do not explain the phenomenon ATR inhibition protects non-cycling cells from lethality induced by UV and UV mimetics.
FIGURE 5.
Inhibition of ATR but not other kinases protects mammalian cells from the lethal effects of AAAF. A, non-cycling HaCaT cells were treated with DMSO or ATRi before exposure to AAAF. Cells were stained at the indicated time points following AAAF treatment to monitor cell survival. C and D, cell survival analyses of non-cycling NHF1, U2OS, and MEFs were treated with ATRi and AAAF. E, non-cycling HaCaT cells were treated with the indicated inhibitor and then exposed to UV. Survival was determined with crystal violet staining as previously described and normalized to non-irradiated cells. F, non-cycling cells were treated as in E except that cells were treated with AAAF. G, non-cycling HaCaT cells were treated with DMSO or 10 μm of the ATR inhibitor VE-821 or AZD6738 prior to treatment with 20 μm AAAF. Cell viability was monitored 2 days later by crystal violet staining. H, non-cycling HaCaT cells were treated with DMSO, ATRi (10 μm VE-821), CHK1i (1 μm AZD7762), MEK1/2i (5 μm U0126), p38i (20 μm SB202190), JNKi (50 μm SP600125), PI3Ki#1 (2 μm wortmannin), PI3Ki#2 (10 μm LY294002), AMPKi (5 μm dorsomorphin/Compound C), mammalian target of rapamycin kinase (mTORi (1 μm rapamycin)), or TBK1i (10 μm BX795) for 30 min prior to treatment with 20 μm AAAF. Cells were stained with crystal violet 1 day later. ATRi, ATR inhibitor.
Because ATM is also activated following UV in non-cycling cells, we wanted to determine whether the protective effect of the ATR inhibitor could be seen with other inhibitors of DNA damage response kinases. Thus, we treated cells with inhibitors to ATR, ATM, or DNA-PK before exposure to UV and AAAF. We found that ATM inhibition had little to no effect on viability and that DNA-PK inhibition instead sensitized the non-cycling cells to both UV and AAAF (Fig. 5, E and F). Given that UV induces double-strand breaks in non-cycling cells (29) and that these breaks are expected to be primarily repaired by non-homologous end-joining rather than homologous recombination in non-cycling cells (59), the sensitization of non-cycling cells to DNA-PK inhibition is perhaps expected. Moreover, these results suggest that the ATM-dependent DNA damaging signaling events shown in Fig. 3 are not essential for the short-term viability of UV-irradiated non-cycling cells. However, a previous report suggested that the ATM protein may be necessary for non-cycling cells exposed to UV to proliferate and form colonies once stimulated with serum (29). This result indicates that ATM has an important function in non-cycling cells exposed to UV, potentially related to gene expression and splicing (70). Nonetheless, these results suggest that ATR functions differently than ATM and DNA-PK in the DNA damage response to UV and UV mimetics in non-cycling cells.
To further support a specific role for ATR inhibition in the protection of non-cycling cells from DNA damage, we made use of a second highly specific chemical inhibitor of ATR (AZD6738) (11, 14). As shown in Fig. 5G, the ATR inhibitor AZD6738 partially abrogated the lethal effects of AAAF on non-cycling HaCaT cells, although less efficiently than VE-821.
We next investigated the specificity of ATR inhibition on the survival of non-cycling cells exposed to AAAF. Using inhibitors against a panel of other cell signaling kinases that have the potential to impact cell viability, including CHK1, MAPKs, PI3Ks, AMP-dependent protein kinase, and mammalian target of rapamycin kinase, we found that none of the other inhibitors were able to protect non-cycling HaCaT cells from AAAF treatment (Fig. 5H). We conclude that the effect of ATR inhibition on non-cycling cells exposed to UV and UV mimetics is specific to ATR and unlikely to be mediated by direct or indirect modulation of other cell signaling kinase cascades.
ATR Promotes Apoptotic Signaling in Non-cycling Cells in Response to DNA Damage
To determine the mode of cell death that is affected by ATR kinase function, non-cycling HaCaT cells were treated with a pan-caspase inhibitor during exposure to AAAF. As shown in Fig. 6A, caspase inhibition partially protected the cells from the lethal effects of AAAF, although not as well as the ATR inhibitor. These results indicate that ATR promotes cell death in response to AAAF at least in part through the regulation of apoptosis.
FIGURE 6.
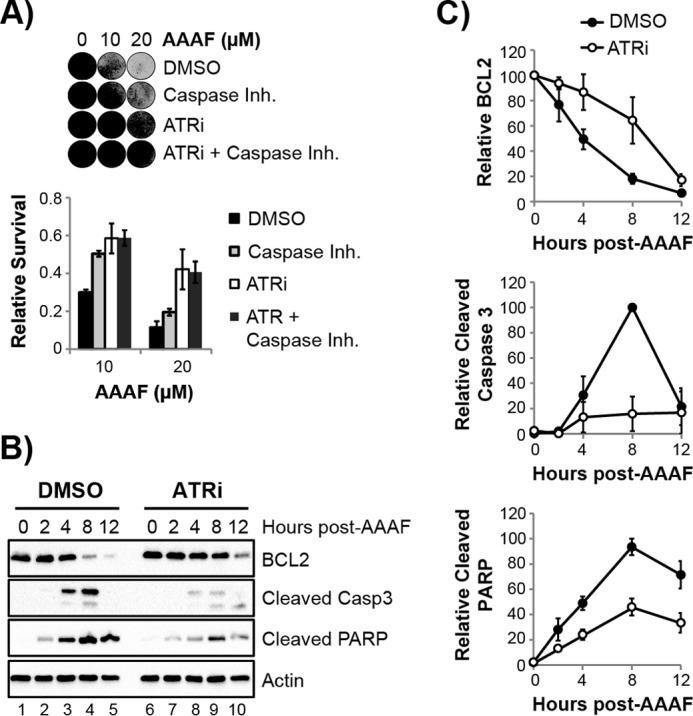
ATR promotes apoptosis in non-cycling cells. A, non-cycling HaCaT cells were treated with the indicated inhibitors prior to exposure to AAAF. Cells were stained with crystal violet 2 days after AAAF treatment, and representative images of the stained cells are shown. The pan-caspase inhibitor Z-VAD-FMK (Caspase Inh.) was added at two time points in these experiments, including immediately prior to AAAF treatment and 24 h later, which was more effective than a single treatment at protecting cells from undergoing cell death. B, non-cycling cells were treated with DMSO or ATRi and then exposed to AAAF. Cell lysates were prepared at the indicated time points and analyzed by immunoblotting with antibodies against the indicated proteins. C, quantitation of immunoblotting data in B from three independent experiments. ATRi, ATR inhibitor.
The induction of apoptosis is associated with the cleavage of various cellular proteins, including the apoptotic effector enzyme Caspase 3 and the DNA repair protein PARP. As shown in Fig. 6, B and C, the ATR inhibitor partially abrogated the cleavage of Caspase 3 and PARP following AAAF treatment. Moreover, the anti-apoptotic protein BCL2, which is destabilized in response to DNA damage and the induction of apoptosis (71), was degraded at a slower rate in response to AAAF when cells were treated with the ATR inhibitor.
These results demonstrate that the ATR kinase promotes (and hence ATR inhibition reduces) the activation of apoptotic signaling in response to AAAF treatment in non-cycling cells. We note that this dependence on ATR kinase activity differentiates our findings from a recent report that a fraction of ATR directly localizes to mitochondria to inhibit apoptosis in a manner that does not require its kinase activity (72). Thus, the pro-apoptotic function of ATR kinase described here is unlikely to be due to a function at the mitochondria.
ATR Promotes Cell Death in Non-cycling Cells Independently of Nucleotide Excision Repair
We next examined the mechanism by which ATR inhibition protects non-cycling cells from undergoing apoptosis in response to UV and AAAF. We first considered that ATR may regulate the removal of bulky DNA adducts that are induced by UV and UV mimetics and that are removed solely by nucleotide excision repair (73, 74). However, using a highly sensitive assay of nucleotide excision repair activity that directly measures the excision of damage-containing DNA oligonucleotides in vivo (40–43), we observed no effect of ATR inhibition on the initial rate of excision repair following UV (Fig. 7A). Furthermore, when we monitored the removal of (6-4)PPs from genomic DNA by immunoslot blot analysis, we observed similar repair rates in the absence and presence of the ATR inhibitor (Fig. 7B). We conclude that the enhanced cell viability provided by ATR inhibition in non-cycling cells is not due to an increase in nucleotide excision repair capacity.
FIGURE 7.
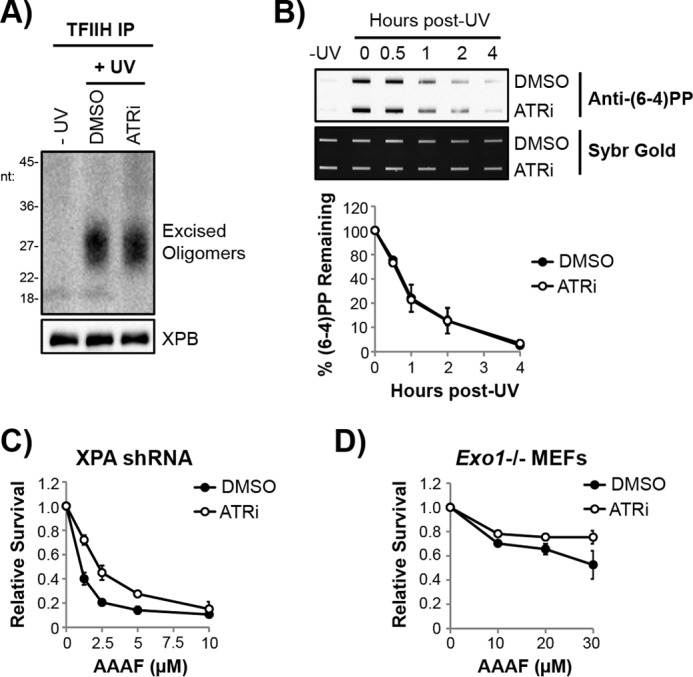
ATR inhibition protects non-cycling cells from UV and UV mimetics in a nucleotide-excision repair-independent manner. A, non-cycling HaCaT cells were treated with DMSO or ATRi and irradiated with 20 J/m2 of UV. Cells lysates were prepared 1 h post-irradiation, and excision repair activity was monitored by detecting the excised oligonucleotide products that associate with the repair factor TFIIH. A fraction of the TFIIH immunoprecipitate was analyzed by immunoblotting for the XPB subunit of TFIIH, and the excised oligonucleotides were extracted from the remaining material and 3′-end labeled with biotin for detection by chemiluminesence. B, immunoslot blotting was used to detect the removal of (6-4)PPs from the genomic DNA of non-cycling cells exposed to UV. The signals from two independent experiments were quantified and plotted in the graph. C, non-cycling HaCaT cells infected with a lentivrus against XPA were treated with DMSO or ATRi and then exposed to increasing concentrations of AAAF. Cell survival was determined by crystal violet staining as previously described. D, MEFs from Exo1−/− knock-out mice were treated as described in C. ATRi, ATR inhibitor; MEF, murine embryonic fibroblasts.
The removal of damaged nucleotides by the nucleotide excision repair system leaves 30-nt long gaps in DNA that can be enlarged by Exo1 (exonuclease 1). Indeed, in vitro studies have shown that this processing of excision gaps leads to robust activation of ATR (32). To determine whether this mode of ATR activation is responsible for the pro-apoptotic function of ATR in non-cycling cells exposed to UV and UV mimetics, we examined how ATR inhibition affected the viability of cells deficient in either the excision repair factor XPA or the gap enlargement nuclease Exo1. As shown in Fig. 7, C and D, ATR inhibition partially rescued the viability of both HaCaT cells depleted of XPA by RNA interference and fibroblasts from Exo1−/− knock-out mice following treatment with AAAF. We conclude that the pro-apoptotic signaling function of ATR occurs independently of either excision gap formation or enlargement in non-cycling cells and that ATR inhibition does not promote repair of bulky DNA lesions.
ATR Promotes Cell Death in Non-cycling Cells Exposed to Transcription Inhibitors
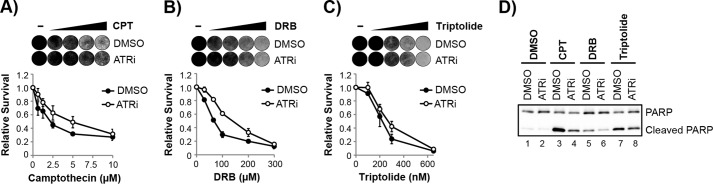
RNA polymerase stalling at DNA lesions has been suggested to be a strong trigger for both apoptosis and the activation of ATR (33–35). We therefore next considered whether transcription arrest by UV and UV mimetics may be the signal for the pro-apoptotic function of ATR. To test this hypothesis, we used several compounds that interfere with transcription and the progression of RNA polymerases and then examined how ATR inhibition affected cell viability in response to such treatment.
We first considered the activity of topoisomerase I, which is required to relieve torsional stress that is generated in DNA during transcription. As shown in Fig. 8A, ATR inhibition partially protected non-cycling cells from the lethal effects of the topoisomerase I inhibitor and cancer chemotherapeutic drug camptothecin. Interestingly, and consistent with the opposing function of ATR on cycling and non-cycling cell viability in response to UV and AAAF (Fig. 4), ATR inhibition was reported to instead potentiate the cell killing effect of camptothecin in cycling cells (19).
FIGURE 8.
ATR inhibition protects non-cycling cells from the lethal effects of transcription stress. A, non-cycling HaCaT cells were treated with DMSO or ATRi before treatment with increasing concentrations of the topoisomerase I inhibitor camptothecin (CPT). Cells were stained with crystal violet 1 day later (representative images are shown), and cell survival was quantified as previously described. B, cells were treated as in A except that cells were treated with the RNA polymerase II inhibitor DRB. C, cells were treated as in A except that cells were treated with the TFIIH inhibitor triptolide. D, cells were treated with DMSO or ATRi and then exposed to 2.5 μm CPT, 100 μm DRB, or 300 nm triptolide for 4 h. Cell lysates were prepared and analyzed by immunoblotting. ATRi, ATR inhibitor; CPT, camptothecin.
The nucleoside analog DRB rapidly inhibits transcription and stalls RNA polymerase II by affecting its phosphorylation status, which prevents transition of RNA polymerase II into its productive, elongating form during transcription (75). As shown in Fig. 8B, the cell lethality induced by DRB in non-cycling HaCaT cells was reduced in the presence of the ATR inhibitor.
The natural product triptolide forms a covalent complex with the XPB subunit of TFIIH (75, 76), which is required to unwind DNA at promoters and activate RNA polymerase II for transcription. Thus, although triptolide causes transcription stress, it is thought to primarily prevent transcription initiation and not transcription elongation and therefore acts in a manner distinct from UV, camptothecin, and DRB, which directly stall RNA polymerases on DNA. Consistent with this notion, ATR inhibition had a less dramatic protective effect on cell viability following triptolide treatment (Fig. 8C).
To validate the cell viability studies with another measure of cell death, we examined apoptotic signaling in these cells. As shown in Fig. 8D, a reduction in PARP cleavage was observed in camptothecin- and DRB-treated cells exposed to the ATR inhibitor, similar to the response to AAAF (Fig. 5). We conclude that transcriptional stress caused by diverse mechanisms, including bulky DNA adduct formation, torsional stress, and RNA polymerase pausing, all lead to the activation of a pro-apoptotic form of ATR kinase in non-cycling cells.
ATR Promotes Cell Death in a p53-independent Manner
The tumor suppressor protein p53 has numerous functions in response to DNA damage, including in the regulation of apoptosis (77). Although ATR contributes to p53 phosphorylation following exposure to UV and UV mimetics (Figs. 2 and 3), the fact that ATR inhibition protects both p53-wild-type (NHF1, U2OS) and p53-mutant (HaCaT) cells indicated that the pro-apoptotic function of ATR was likely independent of canonical p53 function.
Nonetheless, even mutant forms of p53 have been reported to promote the survival of cancer cells (78). Thus, it was formally possible that ATR regulation of p53 in HaCaT cells, in which both alleles have mutations within the p53 binding domain (79), contributes to the pro-survival effect of the ATR inhibitor in non-cycling cells exposed to DNA damage and transcription stress. We therefore used lentiviral shRNAs to knockdown p53 expression in HaCaT cells and then exposed the cells to the UV mimetic compound AAAF. However, using two different shRNA constructs, the ATR inhibitor continued to improve the viability of AAAF-treated cells (Fig. 9A). Moreover, similar results were observed when we knocked down p53 in U2OS cells (Fig. 9B). We conclude that p53 is not required for the pro-apoptotic function of ATR in non-cycling cells.
FIGURE 9.
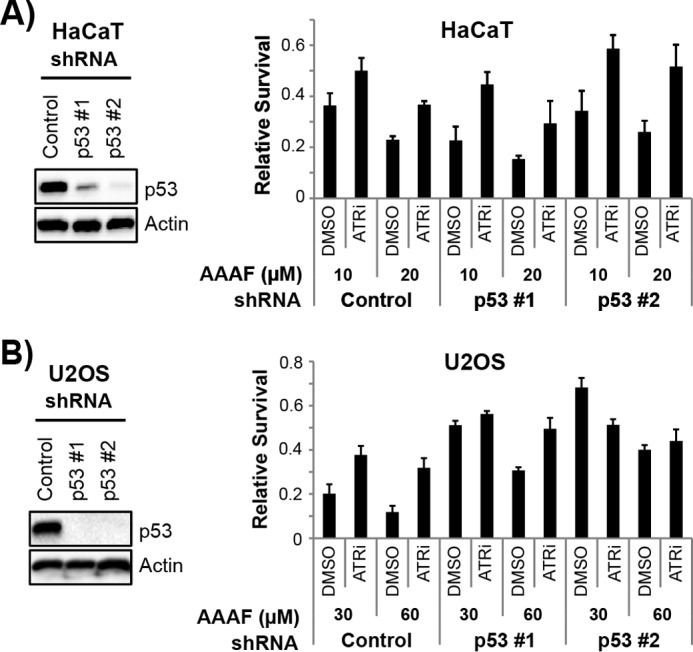
ATR promotes cell death in a p53-independent manner independent of p53. A, HaCaT cells were infected with a control lentivirus or with constructs expressing shRNAs targeting p53 and selected with puromycin. Immunoblotting was used to verify knockdown of p53 in these cells. The cell lines were then treated with DMSO or ATRi and exposed to the indicated concentrations of AAAF. Surviving cells were stained with crystal violet 2 days after treatment to measure cell survival (right). B, U2OS cells were infected with the indicated lentiviruses and processed as described in A.
Discussion
Here we demonstrated that canonical DNA damage response signaling shows striking differences in cycling and non-cycling cells. Because most cells in the body are in a non-cycling or slowly proliferating state, our findings therefore suggest that studies of cellular responses to DNA damage should consider the role of the cell proliferation state in greater detail. We showed that this issue is particularly relevant to the function of the ATR kinase, which is known to have numerous pro-survival roles in the cellular response to replication stress caused by DNA damage (3–5).
However, we showed that in contrast to the widely accepted model that ATR inhibition cells to DNA damaging agents that cause replication stress, the opposite cellular outcome is observed upon ATR inhibition in non-cycling cells. Inhibition of ATR instead limits the degree of apoptotic cell death in response to UV, UV mimetics, and other agents that cause transcription stress. The mechanism by which ATR promotes apoptosis in response to transcription stress remains unresolved, although our results and others (33) suggest it to be independent of p53. However, ATR has been reported to have hundreds of potential substrates (80), and so additional work will be needed to identify the pro-apoptotic target(s) of ATR in non-cycling cells. Moreover, it will also be important to determine the molecular mechanism of ATR activation in response to transcription stress to determine how it differs from activation in response to replication stress and DNA excision repair intermediates (3–5).
Finally, our results also have important implications regarding the use of ATR inhibitors in cancer chemotherapy regimens. On the one hand, ATR inhibitors may protect normal, non-replicating cells in various organs and tissues from some of the adverse effects of DNA damaging cancer chemotherapies. However, it is also possible that non-cycling cancer stem cells or other slowly proliferating cells within tumors (81, 82) may be less prone to undergo apoptosis during cancer treatments that utilize an ATR inhibitor. Such treatments could give rise to additional mutations and ultimately to new tumors that may be associated with cancer recurrence. Thus, it may be important to consider these issues as ATR inhibitors progress through clinical trials in human patients.
Author Contributions
M. K. conceived the idea for this project and carried out the experiments. M. K. and A. S. analyzed the results, and M. K. wrote the paper.
Acknowledgments
We thank Laura Lindsey-Boltz and Shobhan Gaddameedhi for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM32833 (to A. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
- ATR
- ataxia telangiectasia-mutated and Rad3-related
- (6-4)PP
- (6-4)pyrimidine-pyrimidone, 6-(1,2)-dihydro-2-oxo-4-pyrimidinyl-5-methyl-2,4-(1H,3H)-pyrimidinediones
- AAAF
- N-acetyoxy-2-acetylaminofluorene
- ATM
- ataxia telangiectasia-mutated
- DNA-PK
- DNA-dependent protein kinase
- CHK1/2
- checkpoint kinase 1/2
- KAP1
- Krüppel-associated box-associated protein 1
- PI3K
- phosphoinositide 3-kinase
- TBK1
- Tank-binding protein kinase
- PARP
- poly(ADP-ribose) polymerase
- DRB
- 5,6-dichloro-1-β-d-ribofuranosyl-1H-benzimidazole
- DMSO
- dimethyl sulfoxide
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone
- Exo1
- exonuclease 1.
References
- 1. Byun T. S., Pacek M., Yee M. C., Walter J. C., and Cimprich K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zou L., and Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 3. Cimprich K. A., and Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nam E. A., and Cortez D. (2011) ATR signalling: more than meeting at the fork. Biochem. J. 436, 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., and Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 6. Choi J. H., Lindsey-Boltz L. A., Kemp M., Mason A. C., Wold M. S., and Sancar A. (2010) Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 13660–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cliby W. A., Roberts C. J., Cimprich K. A., Stringer C. M., Lamb J. R., Schreiber S. L., and Friend S. H. (1998) Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17, 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wright J. A., Keegan K. S., Herendeen D. R., Bentley N. J., Carr A. M., Hoekstra M. F., and Concannon P. (1998) Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc. Natl. Acad. Sci. U.S.A. 95, 7445–7450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nghiem P., Park P. K., Kim Y., Vaziri C., and Schreiber S. L. (2001) ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl. Acad. Sci. U.S.A. 98, 9092–9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heffernan T. P., Kawasumi M., Blasina A., Anderes K., Conney A. H., and Nghiem P. (2009) ATR-Chk1 pathway inhibition promotes apoptosis after UV treatment in primary human keratinocytes: potential basis for the UV protective effects of caffeine. J. Invest. Dermatol. 129, 1805–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karnitz L. M., and Zou L. (2015) Molecular pathways: targeting ATR in cancer therapy. Clin. Cancer Res. 21, 4780–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Llona-Minguez S., Höglund A., Jacques S. A., Koolmeister T., and Helleday T. (2014) Chemical strategies for development of ATR inhibitors. Expert Rev. Mol. Med. 16, e10. [DOI] [PubMed] [Google Scholar]
- 13. Toledo L. I., Murga M., and Fernandez-Capetillo O. (2011) Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Mol. Oncol. 5, 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vendetti F. P., Lau A., Schamus S., Conrads T. P., O'Connor M. J., and Bakkenist C. J. (2015) The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget 6, 44289–44305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pires I. M., Olcina M. M., Anbalagan S., Pollard J. R., Reaper P. M., Charlton P. A., McKenna W. G., and Hammond E. M. (2012) Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. Br. J. Cancer 107, 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prevo R., Fokas E., Reaper P. M., Charlton P. A., Pollard J. R., McKenna W. G., Muschel R. J., and Brunner T. B. (2012) The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol. Ther. 13, 1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fokas E., Prevo R., Pollard J. R., Reaper P. M., Charlton P. A., Cornelissen B., Vallis K. A., Hammond E. M., Olcina M. M., Gillies McKenna W., Muschel R. J., and Brunner T. B. (2012) Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell. Death Dis. 3, e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vávrová J., Zárybnická L., Lukášova E., Řezáčová M., Novotná E., Sinkorová Z., Tichý A., Pejchal J., and Durišová K. (2013) Inhibition of ATR kinase with the selective inhibitor VE-821 results in radiosensitization of cells of promyelocytic leukaemia (HL-60). Radiat. Environ. Biophys. 52, 471–479 [DOI] [PubMed] [Google Scholar]
- 19. Jossé R., Martin S. E., Guha R., Ormanoglu P., Pfister T. D., Reaper P. M., Barnes C. S., Jones J., Charlton P., Pollard J. R., Morris J., Doroshow J. H., and Pommier Y. (2014) ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase i inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 74, 6968–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall A. B., Newsome D., Wang Y., Boucher D. M., Eustace B., Gu Y., Hare B., Johnson M. A., Milton S., Murphy C. E., Takemoto D., Tolman C., Wood M., Charlton P., Charrier J. D., et al. (2014) Potentiation of tumor responses to DNA damaging therapy by the selective ATR inhibitor VX-970. Oncotarget 5, 5674–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reaper P. M., Griffiths M. R., Long J. M., Charrier J. D., Maccormick S., Charlton P. A., Golec J. M., and Pollard J. R. (2011) Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 7, 428–430 [DOI] [PubMed] [Google Scholar]
- 22. Kawasumi M., Lemos B., Bradner J. E., Thibodeau R., Kim Y. S., Schmidt M., Higgins E., Koo S. W., Angle-Zahn A., Chen A., Levine D., Nguyen L., Heffernan T. P., Longo I., Mandinova A., et al. (2011) Protection from UV-induced skin carcinogenesis by genetic inhibition of the ataxia telangiectasia and Rad3-related (ATR) kinase. Proc. Natl. Acad. Sci. U.S.A. 108, 13716–13721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu Y. P., Lou Y. R., Peng Q. Y., Xie J. G., Nghiem P., and Conney A. H. (2008) Effect of caffeine on the ATR/Chk1 pathway in the epidermis of UVB-irradiated mice. Cancer Res. 68, 2523–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu Y. P., Lou Y. R., Peng Q. Y., Nghiem P., and Conney A. H. (2011) Caffeine decreases phospho-Chk1 (Ser317) and increases mitotic cells with cyclin B1 and caspase 3 in tumors from UVB-treated mice. Cancer Prev. Res. (Phila) 4, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marini F., Nardo T., Giannattasio M., Minuzzo M., Stefanini M., Plevani P., and Muzi Falconi M. (2006) DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc. Natl. Acad. Sci. U.S.A. 103, 17325–17330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marti T. M., Hefner E., Feeney L., Natale V., and Cleaver J. E. (2006) H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 103, 9891–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsumoto M., Yaginuma K., Igarashi A., Imura M., Hasegawa M., Iwabuchi K., Date T., Mori T., Ishizaki K., Yamashita K., Inobe M., and Matsunaga T. (2007) Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J. Cell Sci. 120, 1104–1112 [DOI] [PubMed] [Google Scholar]
- 28. Hanasoge S., and Ljungman M. (2007) H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis 28, 2298–2304 [DOI] [PubMed] [Google Scholar]
- 29. Wakasugi M., Sasaki T., Matsumoto M., Nagaoka M., Inoue K., Inobe M., Horibata K., Tanaka K., and Matsunaga T. (2014) Nucleotide excision repair-dependent DNA double-strand break formation and ATM signaling activation in mammalian quiescent cells. J. Biol. Chem. 289, 28730–28737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vrouwe M. G., Pines A., Overmeer R. M., Hanada K., and Mullenders L. H. (2011) UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci. 124, 435–446 [DOI] [PubMed] [Google Scholar]
- 31. Ray A., Milum K., Battu A., Wani G., and Wani A. A. (2013) NER initiation factors, DDB2 and XPC, regulate UV radiation response by recruiting ATR and ATM kinases to DNA damage sites. DNA Repair 12, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindsey-Boltz L. A., Kemp M. G., Reardon J. T., DeRocco V., Iyer R. R., Modrich P., and Sancar A. (2014) Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J. Biol. Chem. 289, 5074–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Derheimer F. A., O'Hagan H. M., Krueger H. M., Hanasoge S., Paulsen M. T., and Ljungman M. (2007) RPA and ATR link transcriptional stress to p53. Proc. Natl. Acad. Sci. U.S.A. 104, 12778–12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindsey-Boltz L. A., and Sancar A. (2007) RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage?. Proc. Natl. Acad. Sci. U.S.A. 104, 13213–13214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ljungman M. (2007) The transcription stress response. Cell. Cycle 6, 2252–2257 [DOI] [PubMed] [Google Scholar]
- 36. Kemp M. G., Gaddameedhi S., Choi J. H., Hu J., and Sancar A. (2014) DNA repair synthesis and ligation affect the processing of excised oligonucleotides generated by human nucleotide excision repair. J. Biol. Chem. 289, 26574–26583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang X., Boehm J. S., Yang X., Salehi-Ashtiani K., Hao T., Shen Y., Lubonja R., Thomas S. R., Alkan O., Bhimdi T., Green T. M., Johannessen C. M., Silver S. J., Nguyen C., Murray R. R., et al. (2011) A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim J. S., Lee C., Bonifant C. L., Ressom H., and Waldman T. (2007) Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol. Cell. Biol. 27, 662–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gaddameedhi S., Kemp M. G., Reardon J. T., Shields J. M., Smith-Roe S. L., Kaufmann W. K., and Sancar A. (2010) Similar nucleotide excision repair capacity in melanocytes and melanoma cells. Cancer Res. 70, 4922–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu J., Choi J. H., Gaddameedhi S., Kemp M. G., Reardon J. T., and Sancar A. (2013) Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying DNA damage in vivo. J. Biol. Chem. 288, 20918–20926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi J. H., Gaddameedhi S., Kim S. Y., Hu J., Kemp M. G., and Sancar A. (2014) Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells. Nucleic Acids Res. 42, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Choi J. H., Kim S. Y., Kim S. K., Kemp M. G., and Sancar A. (2015) An integrated approach for analysis of the DNA damage response in mammalian cells: nucleotide excision repair, DNA damage checkpoint, and apoptosis. J. Biol. Chem. 290, 28812–28821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu J., Adar S., Selby C. P., Lieb J. D., and Sancar A. (2015) Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kemp M. G., Reardon J. T., Lindsey-Boltz L. A., and Sancar A. (2012) Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J. Biol. Chem. 287, 22889–22899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dai Y., and Grant S. (2010) New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin. Cancer Res. 16, 376–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., and Hunter T. (2014) Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 134, 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tibbetts R. S., Brumbaugh K. M., Williams J. M., Sarkaria J. N., Cliby W. A., Shieh S. Y., Taya Y., Prives C., and Abraham R. T. (1999) A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ward I. M., and Chen J. (2001) Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762 [DOI] [PubMed] [Google Scholar]
- 49. Kitagawa R., and Kastan M. B. (2005) The ATM-dependent DNA damage signaling pathway. Cold Spring Harb. Symp. Quant. Biol. 70, 99–109 [DOI] [PubMed] [Google Scholar]
- 50. Bakkenist C. J., and Kastan M. B. (2004) Initiating cellular stress responses. Cell 118, 9–17 [DOI] [PubMed] [Google Scholar]
- 51. Goodarzi A. A., Noon A. T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., and Jeggo P. A. (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 31, 167–177 [DOI] [PubMed] [Google Scholar]
- 52. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D. C., Lukas J., Bekker-Jensen S., Bartek J., and Shiloh Y. (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell. Biol. 8, 870–876 [DOI] [PubMed] [Google Scholar]
- 53. Charrier J. D., Durrant S. J., Golec J. M., Kay D. P., Knegtel R. M., MacCormick S., Mortimore M., O'Donnell M. E., Pinder J. L., Reaper P. M., Rutherford A. P., Wang P. S., Young S. C., and Pollard J. R. (2011) Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J. Med. Chem. 54, 2320–2330 [DOI] [PubMed] [Google Scholar]
- 54. Chen Y. H., Matsumoto Y., Shibutani S., and Bogenhagen D. F. (1991) Acetylaminofluorene and aminofluorene adducts inhibit in vitro transcription of a Xenopus 5S RNA gene only when located on the coding strand. Proc. Natl. Acad. Sci. U.S.A. 88, 9583–9587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Gijssel H. E., Mullenders L. H., van Oosterwijk M. F., and Meerman J. H. (2003) Blockage of transcription as a trigger for p53 accumulation by 2-acetylaminofluorene DNA-adducts. Life Sci. 73, 1759–1771 [DOI] [PubMed] [Google Scholar]
- 56. Gunz D., Hess M. T., and Naegeli H. (1996) Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 271, 25089–25098 [DOI] [PubMed] [Google Scholar]
- 57. Hess M. T., Gunz D., and Naegeli H. (1996) A repair competition assay to assess recognition by human nucleotide excision repair. Nucleic Acids Res. 24, 824–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mu D., Bertrand-Burggraf E., Huang J. C., Fuchs R. P., Sancar A., and Fuchs B. P. (1994) Human and E. coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 22, 4869–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jette N., and Lees-Miller S. P. (2015) The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 117, 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Giannattasio M., Follonier C., Tourrière H., Puddu F., Lazzaro F., Pasero P., Lopes M., Plevani P., and Muzi-Falconi M. (2010) Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol. Cell. 40, 50–62 [DOI] [PubMed] [Google Scholar]
- 61. Sertic S., Pizzi S., Cloney R., Lehmann A. R., Marini F., Plevani P., and Muzi-Falconi M. (2011) Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc. Natl. Acad. Sci. U.S.A. 108, 13647–13652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Unsal-Kaçmaz K., Makhov A. M., Griffith J. D., and Sancar A. (2002) Preferential binding of ATR protein to UV-damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 6673–6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yilmaz S., Sancar A., and Kemp M. G. (2011) Multiple ATR-Chk1 pathway proteins preferentially associate with checkpoint-inducing DNA substrates. PLoS ONE 6, e22986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jiang G., and Sancar A. (2006) Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol. Cell. Biol. 26, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Choi J. H., Lindsey-Boltz L. A., and Sancar A. (2007) Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 104, 13301–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Choi J. H., Lindsey-Boltz L. A., and Sancar A. (2009) Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic Acids Res. 37, 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kemp M. G., Lindsey-Boltz L. A., and Sancar A. (2011) The DNA damage response kinases DNA-dependent protein kinase (DNA-PK) and ataxia telangiectasia mutated (ATM) are stimulated by bulky adduct-containing DNA. J. Biol. Chem. 286, 19237–19246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brown E. J., and Baltimore D. (2000) ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14, 397–402 [PMC free article] [PubMed] [Google Scholar]
- 69. Brown E. J., and Baltimore D. (2003) Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tresini M., Warmerdam D. O., Kolovos P., Snijder L., Vrouwe M. G., Demmers J. A., van IJcken W. F., Grosveld F. G., Medema R. H., Hoeijmakers J. H., Mullenders L. H., Vermeulen W., and Marteijn J. A. (2015) The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dunkern T. R., Fritz G., and Kaina B. (2001) Ultraviolet light-induced DNA damage triggers apoptosis in nucleotide excision repair-deficient cells via Bcl-2 decline and caspase-3/-8 activation. Oncogene 20, 6026–6038 [DOI] [PubMed] [Google Scholar]
- 72. Hilton B. A., Li Z., Musich P. R., Wang H., Cartwright B. M., Serrano M., Zhou X. Z., Lu K. P., and Zou Y. (2015) ATR plays a direct antiapoptotic role at mitochondria, which is regulated by prolyl isomerase Pin1. Mol. Cell. 60, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sancar A., and Reardon J. T. (2004) Nucleotide excision repair in E. coli and man. Adv. Protein Chem. 69, 43–71 [DOI] [PubMed] [Google Scholar]
- 74. Reardon J. T., and Sancar A. (2005) Nucleotide excision repair. Prog. Nucleic Acids Res. Mol. Biol. 79, 183–235 [DOI] [PubMed] [Google Scholar]
- 75. Bensaude O. (2011) Inhibiting eukaryotic transcription: which compound to choose? How to evaluate its activity?. Transcription 2, 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Titov D. V., Gilman B., He Q. L., Bhat S., Low W. K., Dang Y., Smeaton M., Demain A. L., Miller P. S., Kugel J. F., Goodrich J. A., and Liu J. O. (2011) XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7, 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kruiswijk F., Labuschagne C. F., and Vousden K. H. (2015) P53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393–405 [DOI] [PubMed] [Google Scholar]
- 78. Kim M. P., Zhang Y., and Lozano G. (2015) Mutant p53: multiple mechanisms define biologic activity in cancer. Front. Oncol. 5, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lehman T. A., Modali R., Boukamp P., Stanek J., Bennett W. P., Welsh J. A., Metcalf R. A., Stampfer M. R., Fusenig N., and Rogan E. M. (1993) P53 mutations in human immortalized epithelial cell lines. Carcinogenesis 14, 833–839 [DOI] [PubMed] [Google Scholar]
- 80. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R. 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., and Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 81. Moore N., and Lyle S. (2011) Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J. Oncol. 2011, 10.1155/2011/396076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Moore N., Houghton J., and Lyle S. (2012) Slow-cycling therapy-resistant cancer cells. Stem Cells Dev. 21, 1822–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]