Abstract
Cytochrome c oxidase assembly requires the synthesis of the mitochondria-encoded core subunits, Cox1, Cox2, and Cox3. In yeast, Pet54 protein is required to activate translation of the COX3 mRNA and to process the aI5β intron on the COX1 transcript. Here we report a third, novel function of Pet54 on Cox1 synthesis. We observed that Pet54 is necessary to achieve an efficient Cox1 synthesis. Translation of the COX1 mRNA is coupled to the assembly of cytochrome c oxidase by a mechanism that involves Mss51. This protein activates translation of the COX1 mRNA by acting on the COX1 5′-UTR, and, in addition, it interacts with the newly synthesized Cox1 protein in high molecular weight complexes that include the factors Coa3 and Cox14. Deletion of Pet54 decreased Cox1 synthesis, and, in contrast to what is commonly observed for other assembly mutants, double deletion of cox14 or coa3 did not recover Cox1 synthesis. Our results show that Pet54 is a positive regulator of Cox1 synthesis that renders Mss51 competent as a translational activator of the COX1 mRNA and that this role is independent of the assembly feedback regulatory loop of Cox1 synthesis. Pet54 may play a role in Mss51 hemylation/conformational change necessary for translational activity. Moreover, Pet54 physically interacts with the COX1 mRNA, and this binding was independent of the presence of Mss51.
Keywords: cytochrome c oxidase (Complex IV), mitochondria, mitochondrial DNA (mtDNA), translation initiation, translation regulation, yeast, Cox1, Mss51
Introduction
Cytochrome c oxidase (CcO)3 is the last electron acceptor of the mitochondrial respiratory chain. In the yeast Saccharomyces cerevisiae, this enzyme contains 12 subunits, three of which (Cox1, Cox2, and Cox3) are encoded by the mitochondrial DNA. Assembly of CcO is a complex process regulated by more than 25 factors and chaperones (for reviews, see Ref. 1). The first steps of CcO biogenesis involve the translational activation of the mitochondria-encoded mRNAs COX1, COX2, and COX3 by mRNA-specific proteins. Translational activation of the COX1 mRNA depends on Pet309 and Mss51 (2, 3), whereas COX2 translation depends on Pet111 (4, 5), and COX3 mRNA translation depends on Pet54, Pet122, and Pet494 (6–9). These proteins act on the target mRNA 5′-UTRs to allow translation by the mitochondrial ribosomes. They interact with each other and with the mitochondrial inner membrane and are thought to tether translation initiation close to the assembly sites of CcO in the membrane (for a review, see Ref. 10) (11). The mitochondria-encoded Cox1, Cox2, and Cox3 subunits are proposed to assemble from three different modules, each containing a specific subset of nucleus-encoded subunits (12–14).
Cox1, the largest subunit of the CcO, carries the heme aa3-CuB center to reduce oxygen. Synthesis of Cox1 inside mitochondria is highly regulated. If CcO assembly is blocked by mutations on either integral subunits or accessory chaperones, Cox1 synthesis is down-regulated (15, 16). It is proposed that by this mechanism, mitochondria avoids accumulation of pro-oxidant Cox1 intermediates (17). In addition to its role as translational activator of COX1 mRNA, Mss51 also physically interacts with Cox1 protein to form the first high molecular weight intermediaries (COA complexes) that include the chaperones Cox14 and Coa3 (15, 18–20). The current model (reviewed in Refs. 1 and 21) states that if CcO assembly is defective, then Mss51 is sequestered on COA intermediaries to reduce the effective concentration of Mss51 as a translational activator of the COX1 mRNA, resulting in a decrease of Cox1 synthesis. In this context, Cox14 and Coa3 are negative regulators, because their deletion restores Cox1 synthesis when assembly of CcO is deficient (15, 18, 19). The C-terminal end of Cox1 is also a negative regulator of Cox1 synthesis. Deletion of the last 11–15 residues of the Cox1 C-terminal end results in normal Cox1 synthesis even if CcO is not assembled (16). In addition to the first intermediates formed by Cox1, Mss51, Cox14, and Coa3, subunit 1 forms subsequent intermediates (12, 22) that include proteins like Coa1 (23, 24), Shy1 (23, 25, 26), subunits Cox5/Cox6 (19), Coa2 (25), and Cox15 (26). Moreover, the Hsp70 chaperone Ssc1 is associated with both Mss51 and Mss51-containing complexes and regulates Cox1 synthesis as well (27). Mss51 contains two heme-regulating motifs. These motifs are important for hemylation of the protein and are involved in Cox1 regulation by Mss51 (28).
During the investigation of the role of the Cox1 C-terminal end in the Cox1 synthesis regulatory loop, we observed that Pet54 showed an unusual pattern. Deletion of PET54 down-regulated Cox1 synthesis; however, in contrast to what was observed for most assembly mutants, deletion of the Cox1 C-terminal end did not recover Cox1 synthesis (16), indicating that Pet54 plays an additional role in this process.
Pet54 is a nuclear encoded, RNA recognition motif (RRM) protein located on the matrix face of the inner mitochondrial membrane as a peripheral protein (29). In addition to its role as translational activator of the COX3 mRNA (7), Pet54 is required for splicing of the aI5β intron on the COX1 pre-mRNA (30). In vitro experiments demonstrated that Pet54 binds with the COX1 pre-mRNA aI5β intron and the COX3 mRNA 5′-UTR, both regions sharing 56% identity (31). In the present work, we demonstrated that Pet54 plays a third, novel role in Cox1 synthesis regulation that is independent of Cox14, Coa3, and the Cox1 C-terminal end. We also provide data showing that Pet54 is necessary for activation of translational competent Mss51, probably by binding to the COX1 mRNA and by modulating the hemylation state and/or conformation of Mss51.
Experimental Procedures
Yeast Strains and Genetic Methods
S. cerevisiae strains used in this study are listed in Table 1. Genetic manipulation methods and media were as described previously (32). Strains were cultured in a complete fermentable medium, YPD or YPGal (1% yeast extract, 2% Bacto-peptone, and 2% glucose or 2% galactose), or synthetic complete medium (0.67% yeast nitrogen base, 2% glucose) lacking the indicated amino acids. Nuclear deletion constructs with URA3, LEU2, or KanMX4 cassettes were made by PCR. In all cases, correct integration of the different constructs into the nuclear genome was confirmed by PCR. For two-hybrid experiments, the plasmids used were as follows: Pet54-AD, pNGB8 (33); Pet54-BD, pNGB67 (29); and Pet122-AD, pNGB11 (33). Mss51-AD and Mss51-BD were cloned onto the ClaI site of pGAD-C1 and pGBDU-C1, respectively (34). Constructs were transformed in the yeast strain Pj69-4a (34), and double-transformed cells were selected on medium lacking leucine and tryptophan or leucine and uracil. Protein interactions were tested by plating on medium lacking histidine supplemented with 3 mm amino-1,2,4-triazole or lacking adenine.
TABLE 1.
Strains used in this study
All of these strains are congenic or isogenic to D273-10B. Mitochondrial genotypes are shown in parenthesis. ΔΣai represents an intronless COX1 gene. cox1Δ::ARG8m-1 has a replacement of the COX1 3′-UTR by the COX2 3′-UTR. cox1Δ::ARG8m-2 has a replacement of the COX1 5′-UTR by the COX2 5′-UTR (40).
| Strain | Nuclear (mitochondrial) genotype | Reference/Source |
|---|---|---|
| XPM10b | Matα, arg8::hisG, leu2-3,112, lys2, ura3-52 (ρ+,cox1 Δ::ARG8m) | Ref. 20 |
| XPM11 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112 (ρ+, COX1(1–512)::ARG8m) | Ref. 20 |
| XPM48 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δmss51::LEU2 (ρ+, COX1(1–512)::ARG8m) | Ref. 20 |
| XPM171 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112 (ρ+, cox1Δ::ARG8m, cox2Δ::COX1, COX2)a | Ref. 20 |
| XPM182 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet111::URA3 (ρ+, cox1Δ::ARG8m, cox2Δ::COX1, COX2)a | Ref. 20 |
| XPM201 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112 (ρ+, ΔΣai) | Ref. 16 |
| XPM209 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112 (ρ+, ΔΣai, COX1ΔC15) | Ref. 16 |
| XPM295 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51–3xHA (ρ+, ΔΣai) | Ref. 16 |
| XPM298 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA (ρ+, ΔΣai, COX1ΔC15) | Ref. 16 |
| XPM315 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX4 (ρ+, COX1(1–512)::ARG8m) | Ref. 16 |
| XPM316 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX4 (ρ+, ΔΣai) | Ref. 16 |
| XPM317 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX4 (ρ+, ΔΣai, COX1ΔC15) | Ref. 16 |
| YC61 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δcox6::KANMX4 (ρ+, COX1(1–512)::ARG8m) | Ref. 16 |
| YC75 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δcox14::URA3 (ρ+, ΔΣai) | Ref. 16 |
| YC76 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δcox14::URA3 (ρ+, ΔΣai, COX1ΔC15) | Ref. 16 |
| YC77 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX4 (ρ+, ΔΣai), Δcox14::URA3 | This work |
| YC78 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX4, cox14Δ::URA3 (ρ+, ΔΣai, COX1ΔC15) | Ref. 16 |
| YC100 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, Δpet54::KANMX (ρ+, ΔΣai) | This work |
| YC102 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet494::KANMX (ρ+, ΔΣai) | This work |
| YC 103 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet494::KANMX (ρ+, ΔΣai, COX1ΔC15) | This work |
| YC104 | Matα, arg8::hisG, leu2-3,112, lys2, ura3-52, Δpet494::KANMX (ρ+, cox1Δ::ARG8m) | This work |
| YC105 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet494::KANMX (ρ+, cox1Δ::ARG8m, cox2Δ::COX1, COX2)a | This work |
| YC106 | Matα, arg8::hisG, leu2-3,112, lys2, ura3-52, Δpet122::KANMX (ρ+, cox1Δ::ARG8m) | This work |
| YC109 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, PET54-3xMyc (ρ+, ΔΣai) | This work |
| YC112 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δcoa3::LEU2 (ρ+, ΔΣai) | This work |
| YC113 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δcoa3::LEU2 (ρ+, ΔΣai, COX1ΔC15) | This work |
| YC114 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX4, Δcoa3::LEU2 (ρ+, ΔΣai, COX1ΔC15) | This work |
| YC118 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX4, Δcoa3::LEU2 (ρ+, ΔΣai) | This work |
| LSR13 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet122::KANMX4 (ρ+, ΔΣai) | Ref. 16 |
| LSR28 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet122::KANMX4 (ρ+, ΔΣai, COX1ΔC15) | Ref. 16 |
| LSR33 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX (ρ+, cox1Δ::ARG8m, cox2Δ::COX1, COX2)a | This work |
| LSR39 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δcox14::URA3 (ρ+, COX1(1–512)::ARG8m) | This work |
| LSR43 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δcox6::KANMM (ρ+, COX1(1–512)::ARG8m) | Ref. 16 |
| LSR44 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet122::KANMX (ρ+, cox1Δ::ARG8m, cox2Δ::COX1, COX2)a | This work |
| SB5 | Matα, ade2, lys2, Δura3, PET309::3xHA (ρ+) | Ref. 60 |
| JPM18 | Matα, arg8::hisG, leu2-3,112, lys2, ura3-52, Δpet54::KANMX (ρ+, cox1Δ::ARG8m) | This work |
| JPM21 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet309::URA3 (ρ+, COX1(1–512)::ARG8m) | This work |
| JPM23 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δcox14::URA3, Δpet54::KANMX (ρ+, COX1(1–512)::ARG8m) | This work |
| JPM30 | Matα, ade2, lys2, Δura3, PET309-3xHA, Δpet54::KANMX (ρ+) | This work |
| JPM40 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, Δcox14::URA3 (ρ+, ΔΣai) | This work |
| JPM42 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, Δpet54::KANMX, Δcox14::URA3 (ρ+, ΔΣai) | This work |
| JPM43 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet111::URA3, Δpet54::KANMX (ρ+, cox1Δ::ARG8m, cox2Δ::COX1, COX2), | This work |
| JPM44 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, PET54-3xMyc, Δmss51::LEU2 (ρ+, ΔΣai) | This work |
| JPM49 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, Δpet122::KANMX (ρ+, ΔΣai) | This work |
| JPM50 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA,Δcox11::KANMX (ρ+, ΔΣai) | This work |
| JPM52 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, PET54-3xMyc, Δpet494::URA3 (ρ+, ΔΣai) | This work |
| JPM53 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, PET54-3xMyc, Δpet309::URA3 (ρ+, ΔΣai) | This work |
| JPM55 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, Δpet122::KANMX (ρ+, ΔΣai) | This work |
| JPM56 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, Δpet54::KANMX (ρ+, ΔΣai, COX1ΔC15) | This work |
| JPM57 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, MSS51-3xHA, Δpet122::KANMX (ρ+, ΔΣai, COX1ΔC15) | This work |
| XPM330 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112 (ρ+, cox1Δ::ARG8m-1, cox2Δ::COX1, COX2)a | This work |
| RGV125 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX (ρ+, cox1Δ::ARG8m-1, cox2Δ::COX1, COX2)a | This work |
| XPM329 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112 (ρ+, cox1Δ::ARG8m-2, cox2Δ::COX1, COX2)a | This work |
| RGV124 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX (ρ+, cox1Δ::ARG8m-2, cox2Δ::COX1, COX2)a | This work |
| AGG81 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, LEU2::YIplac128-mss51F199I (ρ+, ΔΣai) | This work |
| AGG82 | Matα, lys2, arg8::hisG, ura3-52, leu2-3, 112, Δpet54::KANMX4 LEU2::YIplac128-mss51F199I (ρ+, ΔΣai) | This work |
a Ectopic insertion of the chimeric COX1 gene upstream of the COX2 gene.
Analysis of Mitochondrial Proteins
Cells were grown in YPGal medium until late log phase. Mitochondria were isolated by disruption of cells with glass beads or by zymolyase 20T treatment as described (35). Proteins were resolved by SDS-PAGE on 12% gels (36) and detected by immunoblotting with antibodies to HA (Roche Applied Science), c-Myc (Roche Applied Science), Cox1, Cox2, Cox3, cytochrome b, Coa3, Tom40, or citrate synthase. Secondary goat IgG anti-mouse or anti-rabbit (Sigma) conjugated to horseradish peroxidase was detected with the ECL (GE Healthcare) or ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore).
Synthesis of Mitochondrial Proteins
In vivo labeling of cells in the presence of [35S]methionine was performed as described previously (20). After 20 min of pulse labeling, cells were chilled on ice and disrupted by vortexing with glass beads to obtain mitochondria by centrifugation. Mitochondrial proteins were resolved on a 16% polyacrylamide gel, transferred to a polyvinylidene fluoride membrane, and analyzed with a Typhoon 8600 PhosphorImager (GE Healthcare).
Blue Native PAGE (BN-PAGE)
BN-PAGE was performed as described previously (37). Samples (100 μg) of mitochondrial protein were washed with 250 mm sorbitol, 50 mm BisTris and lysed with 750 mm aminocaproic acid, 50 mm BisTris, and 1% digitonin for 30 min on ice. Mitochondrial extracts were cleared at 13,200 rpm for 12 min, and the supernatants were mixed with 2.5 μl of 5% Coomassie solution (750 mm aminocaproic acid, 50 mm BisTris). Extracts were loaded on a 5–13% polyacrylamide gel and transferred to a polyvinylidene fluoride membrane. Proteins were detected by immunoblotting with the respective antibodies.
RNA Immunoprecipitation Assay
This technique was performed as described previously (38). Briefly, mitochondria (1 mg) were lysed with 0.7% n-dodecyl-β-d-maltoside, 20 mm Tris-HCl, pH 7.4, 100 mm NaCl, RNaseOUT (Invitrogen), and Minicomplete protease inhibitors (Roche Applied Science). After a clarifying spin, the solubilized fractions were incubated with an anti-Myc antibody coupled to protein A-agarose (Santa Cruz Biotechnology, Inc.). After centrifugation, immunoprecipitates were washed twice with 500 μl of lysis buffer and twice with 1 ml of 20 mm HEPES-KOH, pH 7.4, and then resuspended in 150 μl of the same buffer. One-fourth of the precipitate fractions were saved for Western blotting analysis, and the remainder were used for RNA extraction. RNA from total and immunoprecipitated fractions was extracted by incubation with TRIzol® reagent (Invitrogen). 20 ng of RNA were treated with 1 unit of DNase I (Invitrogen). The first strand of cDNAs were prepared by the addition of primers for COX1, COX3, or VAR1 in the presence of SuperScript III reverse transcriptase (Invitrogen). The resulting cDNAs were used as template for PCRs to amplify COX1, COX3, or VAR1 5′-UTRs. Note that under these conditions, RT-PCRs are not quantitative.
Results
Pet54 Regulates Synthesis of Cox1
We have previously demonstrated that the C-terminal end of Cox1 is involved in the regulation of its own synthesis. In mutants that block CcO assembly, synthesis of Cox1 is down-regulated; however, deletion of the last 15 residues of Cox1 (Cox1ΔC15) recovered normal levels of Cox1 synthesis (16). This pattern was observed in many different mutants on either CcO subunits or chaperones that participate in enzyme assembly. The only exception was a deletion of pet54. In this mutant, synthesis of Cox1 is down-regulated, but deletion of the C-terminal end of Cox1 does not recover synthesis (16).
Pet54 activates translation of the mitochondrially coded COX3 mRNA (7) and also promotes splicing of an intron found in the COX1 transcript of most yeast strains (39). However, Pet54 was not thought to affect expression of COX1 genes lacking this intron (39). In order to investigate whether or not Pet54 played an additional role in Cox1 synthesis, we analyzed mitochondrial translation products by in vivo labeling with [35S]methionine in the presence of cycloheximide to inhibit cytoplasmic translation. We observed that absence of Pet54 reduced by 50% the labeling of wild-type Cox1 encoded by the intronless COX1 gene used here (Fig. 1). This was the only mutant analyzed whose Cox1 labeling was not increased by C-terminal truncation of Cox1, suggesting that this effect might not be due simply to the lack of CcO assembly. Indeed, deletion of Pet122 or Pet494, the other COX3 mRNA translational activators, resulted in a Cox1 labeling pattern similar to that observed for general CcO assembly factor mutants reported previously (16). In CcO assembly mutants, synthesis of Cox1 is down-regulated; however, additional elimination of COX14 or COA3 recovers Cox1 synthesis, even if CcO assembly is impaired (15, 18, 19). To investigate whether Pet54 might be directly promoting Cox1 synthesis, we eliminated the assembly-mediated regulation of Cox1 by removing Cox14 or Coa3. The reduced labeling of Cox1 and Cox1ΔC15 in the double mutants (Δpet54/Δcox14 and Δpet54/Δcoa3) showed a bypass of Cox14 and Coa3 synthesis control (Fig. 2, A and B), in contrast to what has been observed for a Δcox6 mutant (16) or other mutants affecting Cox1 assembly (15, 18, 19). A similar result was observed with strains containing introns in COX1 (data not shown), strongly suggesting that the role of Pet54 in translation of the COX1 mRNA is independent of the role in COX1 intron splicing.
FIGURE 1.
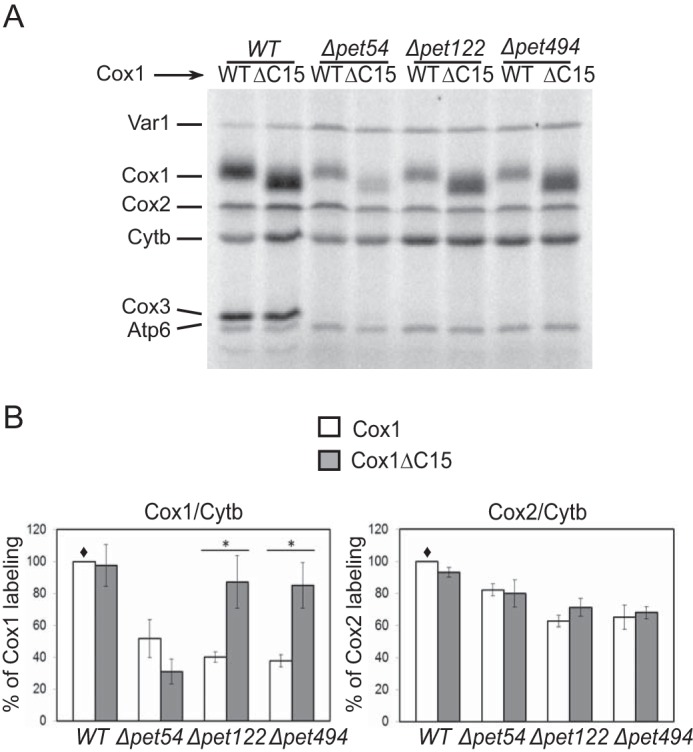
Down-regulation of Cox1 synthesis in Δpet54 mutants is independent of the Cox1 C-terminal end. A, mitochondrial translation products from strains carrying the wild-type Cox1 (WT) or the Cox1 lacking the last 15 residues of the C-terminal end (ΔC15) were pulse-labeled with [35S]methionine in the presence of cycloheximide and analyzed by SDS-PAGE and autoradiography. The Δpet54, Δpet122, and Δpet494 mutants were introduced as indicated. Cytb, cytochrome b; Atp6, subunit 6 of ATPase; Var1, ribosomal protein. B, the intensity of the Cox1 labeling in A was quantified using the ImageJ software and normalized to the cytochrome b signal. It was expressed as a percentage of the wild-type Cox1 signal (♦). Error bars, S.D. values from three independent experiments. We also compared the signals of Cox2 with the cytochrome b signal. In these cases, no significant difference was observed. The relevant significant differences between strains (*) were determined by Student's t test. A p value of <0.01 was considered statistically significant.
FIGURE 2.
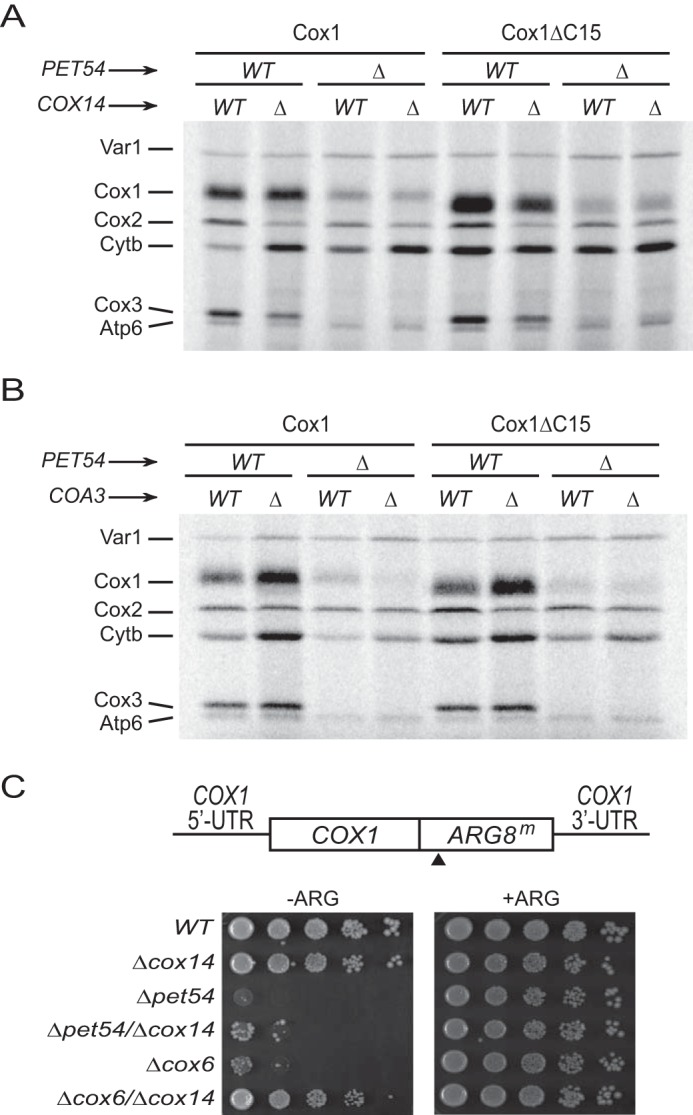
Down-regulation of Cox1 synthesis in Δpet54 mutants is independent of Cox14 and Coa3. Cox1 or Cox1ΔC15 cells with a deletion in PET54, COX14 (A), or COA3 (B), as indicated, were pulse-labeled with [35S]methionine in the presence of cycloheximide. The mitochondrial products were analyzed by SDS-PAGE and autoradiography. C, growth phenotype of strains bearing the COX1(1–512)::ARG8m mitochondrial construct. The ARG8m gene was fused in-frame to the complete COX1 codons, and the black triangle indicates the processing site for the pre-Arg8 matrix-targeting signal. Mutation Δpet54, Δcox14, or Δcox6 was introduced as indicated. Cells were spotted as serial dilutions on medium with arginine (+ARG) or lacking arginine (−ARG) and grown for 2 days at 30 °C.
To confirm that reduction of Cox1 labeling after elimination of assembly-mediated regulation was due to a defect in translation, we used the COX1(1–512)::ARG8m reporter inserted at the COX1 locus. This reporter has been extensively used to differentiate between translation and post-translational defects in Cox1 (16, 20, 25, 28). In this construct, the reporter ARG8m, which codes for an enzyme involved in arginine biosynthesis, is fused in frame to the C-terminal end of Cox1. Because it codes for a mitochondrial targeting and processing sequence, accumulation of mature Arg8 should not be affected by the stability of Cox1. The Cox1 moiety encoded by COX1(1–512)::ARG8m is assembled into active CcO complexes, supporting normal respiratory growth (20). Thus, we combined the Δpet54 and Δcox14 mutations with the COX1(1–512)::ARG8m construct. As observed previously (16), the Δpet54 strain showed reduced growth in medium lacking arginine as compared with wild-type or Δcox14 cells (Fig. 2C). In agreement with the [35S]methionine labeling pattern of Cox1, growth of the double mutant Δpet54/Δcox14 on medium lacking arginine was only slightly improved as compared with the single Δpet54 mutant. In contrast, and as previously observed (16), a Δcox6 mutant showed reduced growth on medium lacking arginine, but the double deletion Δcox6/Δcox14 recovered normal growth levels. These observations show that the CcO assembly defect caused by the loss of Pet54 reduced synthesis of the reporter fused to Cox1 and that this reduction is not recovered on Δcox14 cells.
Taken together, our data indicate that deletion of Pet54 reduced Cox1 synthesis and that this reduction was not dependent on the assembly-regulatory C-terminal end of Cox1 or Cox14/Coa3 regulatory factors, as observed for most CcO assembly mutants. Our results strongly support the idea that Pet54 has a previously undetected direct positive role in promoting Cox1 synthesis.
Pet54 Acts on the 5′-Untranslated Region of COX1 mRNA
Pet54 could regulate synthesis of Cox1 at two different sites. (i) It could have a positive role in COX1 mRNA untranslated regions; this activity could be either directly or indirectly related to Mss51 and Pet309, which are translational activators of COX1 mRNA. (ii) It could act on the COA complexes, where it could contribute to Mss51 release and recycling for translational activation. However, this activity should be independent of both the Cox1 C-terminal end and the Cox14/Coa3-mediated feedback assembly regulatory loop.
To test whether Pet54 regulates Cox1 synthesis by acting through the untranslated regions of the COX1 mRNA, we examined the growth on medium lacking arginine of a strain whose COX1 coding sequence was replaced by the ARG8m gene (cox1Δ::ARG8m). In addition, this strain has the COX1 coding region flanked by both COX2 UTRs inserted at an ectopic site on the mtDNA. In this mtDNA, the levels of Cox1 protein are low as compared with a strain with wild-type mitochondrial DNA (Fig. 3A), and its synthesis depends on the translational activator Pet111 (20). We deleted PET54 as well as the PET122, PET494, and PET111 genes and examined the growth of cells in medium lacking arginine. We observed that Δpet122, Δpet494, and Δpet111 mutants showed a similar growth as compared with a wild-type strain with the cox1Δ::ARG8m construct (Fig. 3B). This is consistent with the idea that in low levels or in the absence of Cox1, Mss51 is not sequestered and therefore is available to support normal COX1 5′-UTR translational activation despite the absence of CcO assembly (15). In contrast, expression of the cox1Δ::ARG8m reporter was reduced by Δpet54 mutation, suggesting that the site of action of Pet54 is in the untranslated region of the mRNA, probably the COX1 5′-UTR. Surprisingly, in the double mutant Δpet54/Δpet111, where Cox1 is not synthesized, expression of the cox1Δ::ARG8m reporter was similar to wild-type levels, suggesting that the effect of Pet54 on the COX1 mRNA untranslated regions depends on the presence of Cox1 protein. To confirm this idea, we first overexpressed PET111, and we observed a decreased growth on medium lacking arginine after pet54 deletion (Fig. 3C). Second, we eliminated PET54 on a cox1Δ::ARG8m strain where the COX1 coding region was absent. In this case, the Δpet54 mutant showed growth levels on medium lacking arginine similar to those of the wild-type, Δpet122, and Δpet494 cells (Fig. 3D).
FIGURE 3.
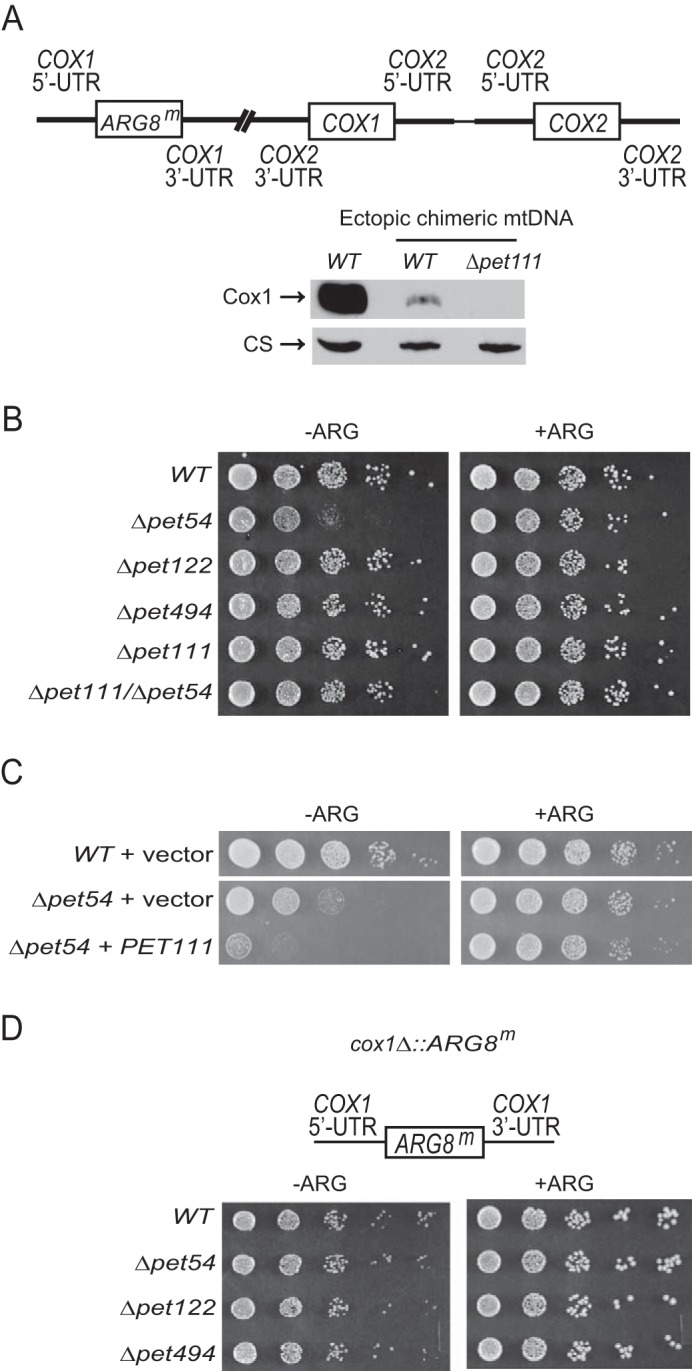
Pet54 plays a role in COX1 mRNA untranslated regions that depends on the presence of the Cox1 protein. A, the cox1Δ::ARG8m construct was inserted at the COX1 locus, where the ARG8m reporter replaced the coding sequence of COX1. In addition, the COX1 coding sequence flanked by the COX2 UTRs was inserted at an ectopic site, 295 bp upstream of COX2 on the mtDNA (20). Mitochondria from cells carrying the wild-type mtDNA or the ectopic chimeric mtDNA were analyzed by SDS-PAGE and Western blotting. The membrane was probed with an antibody for Cox1 and afterward for citrate synthase (CS) as a loading control. The Δpet111 mutation was introduced as indicated. B, serial dilutions of the indicated mutants bearing the ectopic chimeric mtDNA were spotted on medium lacking (−ARG) or containing (+ARG) arginine and were grown for 2 days at 30 °C. C, cells carrying the ectopic chimeric mtDNA and either an empty plasmid or a plasmid expressing a high copy number of PET111 were grown on medium lacking (−ARG) or containing (+ARG) arginine. Serial dilutions were grown for 2 days at 30 °C. D, the COX1 coding region was replaced by the reporter gene ARG8m (cox1Δ::ARG8m); however, in this construct, no chimeric COX1 construct was inserted. The indicated mutants bearing this mtDNA were grown on serial dilutions as in B.
To determine whether the COX1 5′-UTR or 3′-UTR are targets in vivo for Pet54, we created strains with mitochondrial constructs similar to the one indicated in Fig. 3A, except that either the COX1 5′- or 3′-UTR flanking the ARG8m reporter was exchanged with the COX2 5′- or 3′-UTR, respectively. Strains with a wild-type nuclear genome supported growth on medium lacking arginine (Fig. 4), although cells with the substituted COX1 5′-UTR had a weaker growth, probably because in these mitochondria, three copies of the COX2 5′-UTR are present, and therefore Pet111 is limiting for ARG8m expression. Deletion of pet54 did not affect growth in arginine of the strain bearing the COX2 5′-UTR, suggesting that the target of Pet54 was lost in this construct. In contrast, the Δpet54 strain containing the COX2 3′-UTR showed reduced growth in medium lacking arginine. Thus, our data show that the target of Pet54 is localized to the COX1 5′-UTR.
FIGURE 4.
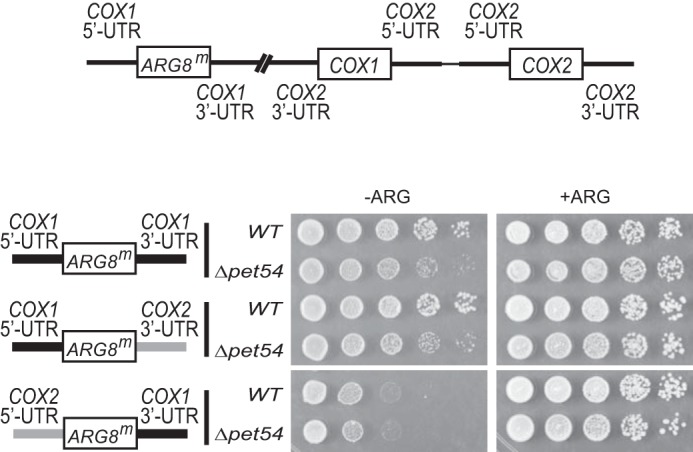
Pet54 acts on the COX1 5′-UTR. Cells containing wild-type Pet54 or the Δpet54 mutation were spotted on serial dilutions on medium lacking (−ARG) or containing (+ARG) arginine and incubated for 2–4 days at 30 °C. The cells contained similar mitochondrial genomes as the one shown in Fig. 3A, except that the cox1Δ::ARG8m gene was flanked by native COX1 5′- and 3′-UTRs (top construct), flanked by the native COX1 5′-UTR and the COX2 3′-UTR (middle), or flanked by the COX2 5′-UTR and the native COX1 3′-UTR (bottom). Black bars, COX1 untranslated regions; gray bars, COX2 untranslated regions. For COX1 3′-UTR replacement, 525 bp of the COX1 downstream sequence were replaced by 118 bp of the COX2 downstream sequence. For COX1 5′-UTR replacement, 505 bp of the COX1 upstream sequence were replaced by 73 bp of the COX2 upstream sequence (40).
Mss51 and Pet309 act on the COX1 mRNA 5′-UTR (2, 40), and both proteins are limiting for translation (40). We investigated whether the steady state levels of these proteins could be affected by deletion of Pet54. Mitochondria from strains with triple hemagglutinin tags fused to the C-terminal end of Pet309 or Mss51 were isolated. These strains did not show any respiratory growth deficiency due to the presence of the tags (data not shown). Western blotting analyses of Mss51–3xHA or Pet309–3xHA revealed that protein levels were not affected by the absence of Pet54 (Fig. 5, A and B). In addition, we asked whether Pet54 might show any genetic interaction with Pet309 or Mss51. As previously observed, the growth in medium lacking arginine of a Δpet54, COX1(1–512)::ARG8m strain was highly reduced (Fig. 2C). We asked whether this phenotype could be alleviated by overexpression of PET309 or MSS51. Each gene was cloned into a 2μ plasmid, and they were used to transform Δpet54, COX1(1–512)::ARG8m cells. Overexpression of PET309 had no compensatory effect, whereas overexpression of MSS51 showed only a very mild effect (Fig. 5C). In contrast, overexpression of MSS51 in a Δcox6 mutant recovered growth on medium lacking arginine, consistent with the observation that the assembly-feedback regulation of Cox1 synthesis can be overcome by expression of additional copies of MSS51 (15).
FIGURE 5.
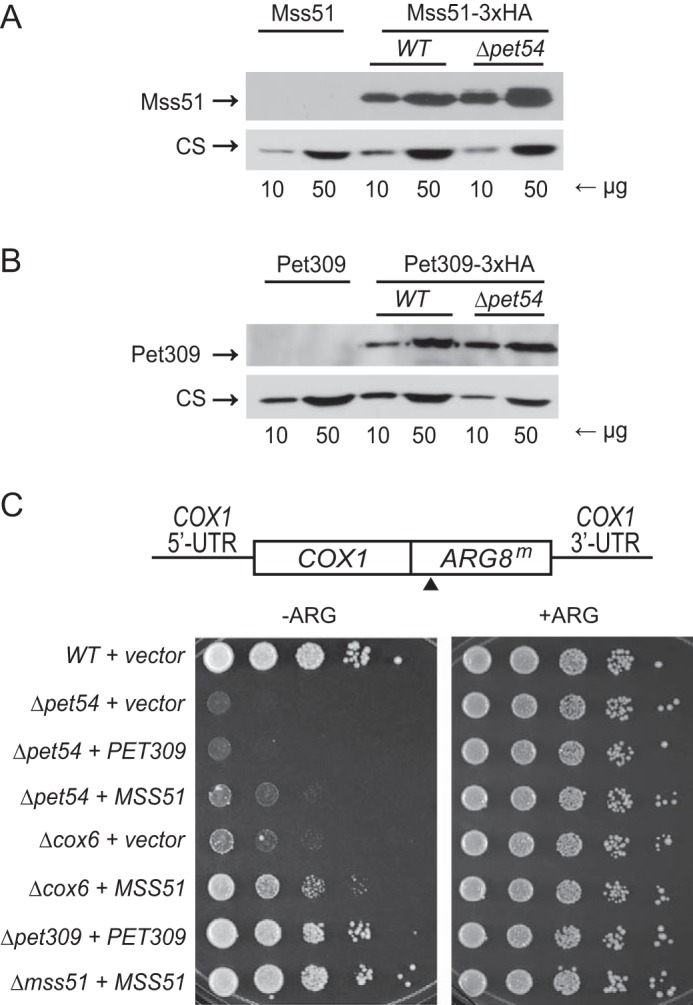
Pet54 does not affect the levels of Pet309 and Mss51 proteins and shows a mild genetic interaction with Mss51. A sample of 10 and 50 μg of mitochondrial protein from strains expressing the Mss51–3xHA (A) or Pet309–3xHA (B) as well as the untagged versions of these genes were separated by SDS-PAGE and analyzed by Western blotting. The membranes were probed with antibodies anti-HA and anti-citrate synthase (CS) as a loading control. C, cells carrying the COX1(1512)::ARG8m construct were transformed with empty plasmid (vector) or with the indicated genes cloned on YEp352 plasmid. The transformants were spotted on synthetic complete medium lacking (−ARG) or containing (+ARG) arginine. 10-Fold serial dilutions were grown for 3 days at 30 °C. The black arrow indicates the processing site for the pre-Arg8 matrix-targeting signal.
Our results suggest that Pet54 acts on the COX1 mRNA 5′-UTR to promote expression. This effect was observed only if Cox1 protein is present, even in small amounts, suggesting that Mss51 could be related to Pet54 function. The absence of Pet54 does not affect steady state levels of the translational activators Pet309 and Mss51, whose activities are limiting for Cox1 synthesis. Moreover, a mild genetic interaction between PET54 and MSS51 was detected because overexpression of the translational activator slightly compensated the growth phenotype of the COX1(1–512)::ARG8m reporter in the absence of Pet54.
Pet54 Could Promote Mss51 Recycling for Translational Activation
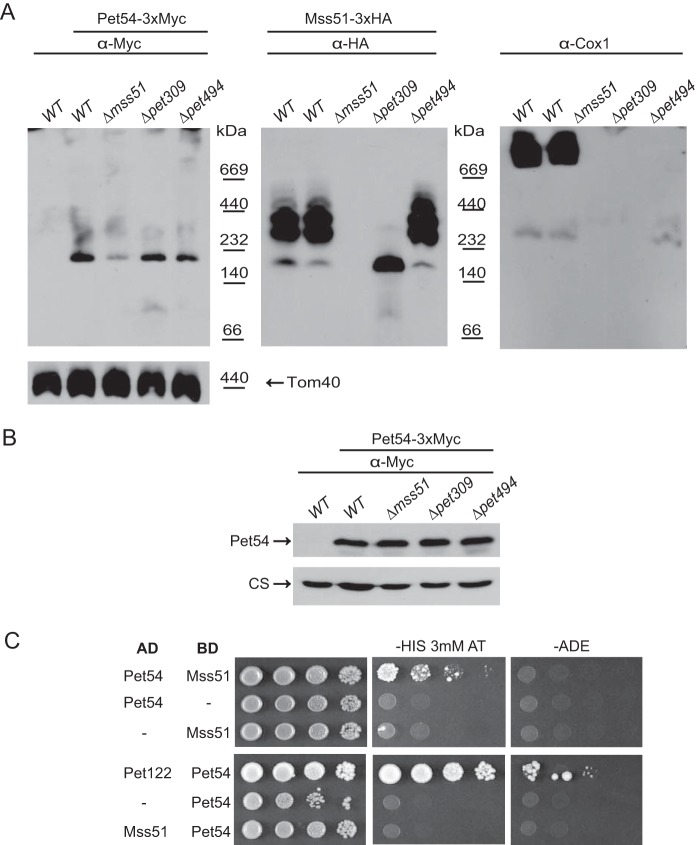
So far, our results have suggested that Pet54 promotes efficient Cox1 synthesis through a mechanism independent of Cox14/Coa3 and the Cox1 C-terminal end. This role involves the COX1 mRNA 5′-UTR. Because the effect of Pet54 on the COX1 5′-UTR was observable only in the presence of the Cox1 protein, there was a possibility that Pet54 could affect either Mss51 or the COA complexes. COA complexes depend on the presence of Cox1 protein, and Mss51 activity in translation activation depends on how much of this activator is released from the COA complexes (1, 15, 19, 21). Therefore, we first analyzed by BN-PAGE whether Mss51 and Pet54 co-migrate. Mss51 has been observed in at least two different complexes, one of ∼120–180 kDa, which has been associated with the translational activator form of Mss51 (translation-effective Mss51, Mss51TE), and two or three complexes from ∼250 to 450 kDa that form the first assembly intermediates of the CcO and represent the COA complexes (19, 24, 27). To detect Pet54, we attached a triple Myc epitope to the C terminus of Pet54. The respiratory competence of the Pet54–3xMyc strain was comparable with wild-type levels, indicating that the tagged protein was functional (data not shown). After BN-PAGE of purified mitochondria, transferring to a PVDF membrane, and Western blotting, Pet54–3xMyc was detected as a ∼190 kDa band that did not co-migrate with any of the complexes observed for Mss51–3xHA (Fig. 6A). Deletion of Mss51 did not affect Pet54 migration; however, we observed a decrease in the band intensity, suggesting that the Pet54 population in the ∼190 kDa band decreased in the absence of Mss51. This decrease was not due to a reduction in the steady state levels of Pet54 on a Δmss51 mutant, as observed in SDS-PAGE and Western blotting (Fig. 6B). Pet54 is associated with the other COX3 translational activators, namely Pet122 and Pet494 (33), and with Pet309 (29). However, the migration of the ∼190 kDa band of Pet54 was not modified after the deletion of Pet494 or Pet309, indicating that this is a different complex (Fig. 6A). As expected, deletion of Pet309 produced only the ∼180-kDa complex of the translational competent Mss51 (19), whereas deletion of Pet494, which blocks CcO assembly, stalled Mss51 on COA complexes. Western blotting experiments of the same membrane using an antibody against Cox1 revealed that supercomplexes were only observed on WT cells, whereas on Δpet494, only assembly intermediaries were detected. We also investigated whether a physical interaction between Mss51 and Pet54 exists. Yeast two-hybrid experiments showed a weak interaction between Pet54-AD and Mss51-BD (Fig. 6C). This interaction was only observable by growth on medium lacking histidine and was undetectable on the more stringent medium lacking adenine. These observations are consistent with a rapid, transient or weak but biologically significant protein-protein interaction, as has been described for different proteins (41, 42).
FIGURE 6.
Pet54 migrates into a high molecular weight complex that is different from that of the Mss51 complexes. A, a sample of 100 μg of mitochondrial proteins from cells carrying the Mss51–3xHA and Pet54–3xMyc or the untagged proteins was solubilized with 1% digitonin and separated on a 5–13% acrylamide blue native gel. Western blotting was performed with the indicated antibodies after consecutive antibody-stripping treatments. An antibody against Tom40 was used as a loading control. B, mitochondria from the strains in A were analyzed by SDS-PAGE and Western blotting using antibodies against the c-Myc epitope (to detect Pet54–3xMyc) and citrate synthase as a loading control (CS). C, yeast two-hybrid plasmids containing the Pet54, Mss51, and Pet122 coding regions fused in frame with the activation domain (AD) or binding domain (BD) of Gal4 were co-transformed into the yeast two-hybrid strain Pj69-4a (34) as indicated. The double transformants were selected on medium lacking leucine and uracil or leucine and tryptophan. Growth was tested on medium lacking histidine (in the presence of 3 mm 3-aminotriazole) and medium lacking adenine. 10-Fold serial dilutions were grown at 30 °C for 7 days. Growth of cells containing Pet122-AD and Pet54-BD was used as a positive control of two proteins previously demonstrated to interact (29, 33).
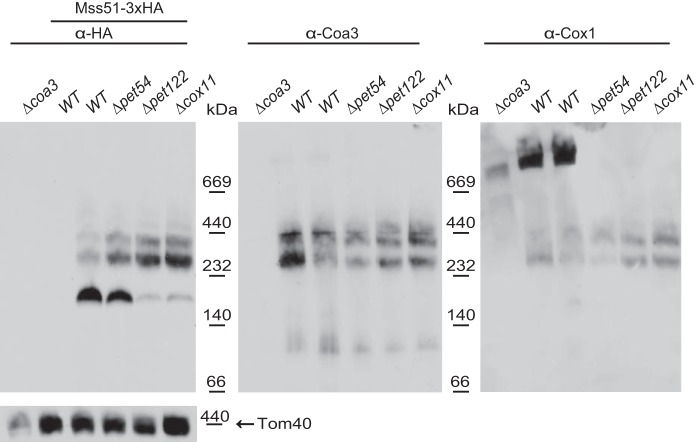
We next asked how Mss51 complexes are affected when Pet54 is deleted. Surprisingly, on a Δpet54 mutant, the ∼180-kDa complex made up by the translational activator form of Mss51 (Mss51TE) was present in similar intensity as compared with the observed levels on wild-type cells (Fig. 7). This is in contrast to the migration of Mss51 on Δpet122 or Δcox11 mutants, where Cox1 synthesis is regulated by the feedback assembly loop (15, 16, 27). In this case, the majority of Mss51 is present on the high molecular mass COA complexes (∼250–450 kDa) and therefore unavailable to promote efficient COX1 mRNA translational activation. This suggests that, in contrast to other mutants blocking CcO assembly, deletion of Pet54 did not trap the most Mss51 on the COA complexes. The same membrane was next decorated with antibodies against Coa3 and Cox1. The migration of Coa3 on COA complexes on BN-PAGE showed a similar pattern in the mutants and in wild-type mitochondria. As reported (19), Cox1 migrated in supercomplexes, together with a population of Coa3 on wild-type cells, whereas in the mutants, only Cox1 assembly intermediaries were observed. In agreement with previous reports, Cox1 migrated with a diffused pattern in the Δcoa3 mutant (22).
FIGURE 7.
Absence of Pet54 has a wild-type migration pattern of Mss51 complexes on blue native PAGE. Mitochondria bearing the untagged Mss51 or the Mss51–3xHA proteins with the indicated mutations Δpet54, Δpet122, or Δcox11 were separated by BN-PAGE. The membrane was probed with antibodies against the HA epitope, Coa3, Cox1, and Tom40 antibodies.
According to the assembly feedback regulatory model, in the absence of Coa3 or Cox14, Mss51 forms the translational active complex. As consequence, the levels of Cox1 synthesis become normal, even if CcO assembly is blocked. An example of this phenotype is observed with the Δpet122 mutant, where synthesis of Cox1 decreased, and after elimination of Cox14, labeling of Cox1 showed normal levels (Fig. 8A). However, as previously demonstrated, an exception was the Δpet54 mutant, where synthesis of Cox1 was not recovered after elimination of Coa3 or Cox14. Thus, we investigated how Mss51 was distributed between the translational active complex and the COA complexes in the double mutant Δpet54/Δcox14. Purified mitochondria from single Δpet54 and Δpet122 mutants, or combined with the Δcox14 deletion were analyzed by BN-PAGE and Western blotting. In the Δcox14 cells as well as in the double Δpet54/Δcox14 and Δpet122/Δcox14 mutants, Mss51 was present as the ∼180-kDa complex representing the translational activator form (Fig. 8B), demonstrating that even when the double mutant Δpet54/Δcox14 renders Mss51 in the ∼180-kDa form, this protein is not efficiently translating the COX1 mRNA.
FIGURE 8.
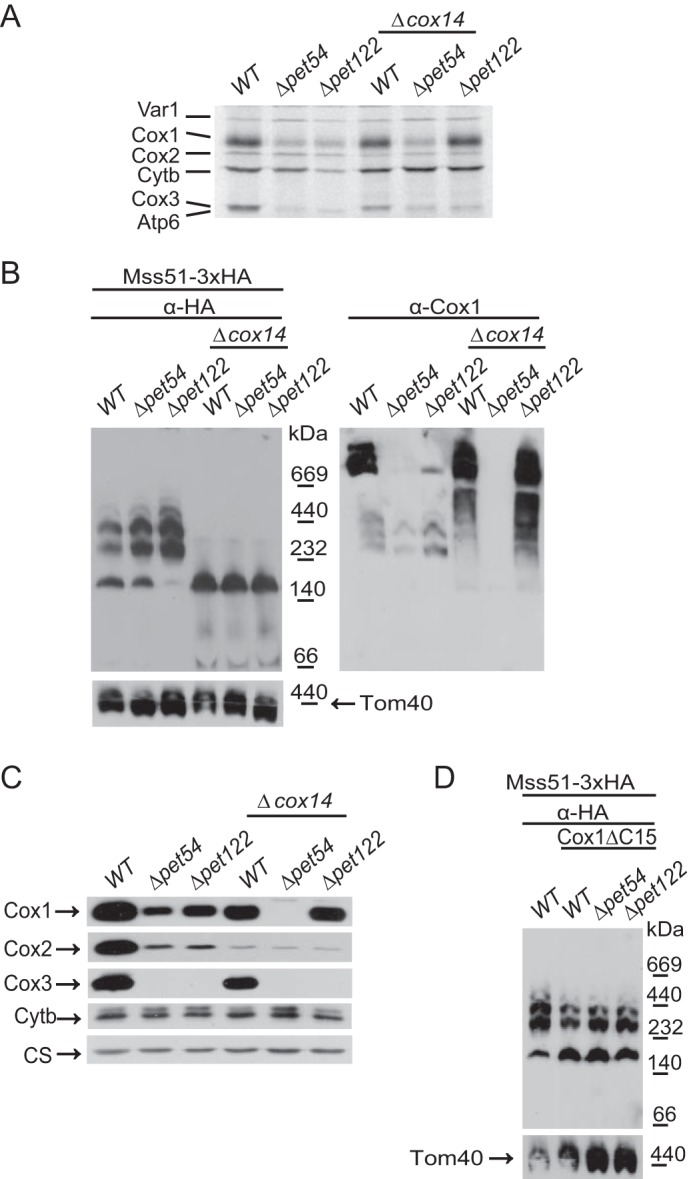
Mss51 is present in the translational active complex but is not competent for efficient synthesis of Cox1 in the absence of Pet54. A, wild type, Δpet54, Δpet122, or the combined mutants with Δcox14 were pulse-labeled with [35S]methionine in the presence of cycloheximide. The mitochondrial products were analyzed by SDS-PAGE and autoradiography. B, mitochondria from the strains in A were separated by BN-PAGE and analyzed by Western blotting using antibodies against the HA epitope (to detect Mss51), Cox1, and Tom40 (as a loading control). C, a sample of 25 μg of mitochondrial proteins was analyzed by SDS-PAGE and Western blotting using antibodies against Cox1, Cox2, Cox3, cytochrome b (Cytb), and citrate synthase as loading control (CS). D, mitochondria from the indicated strains were analyzed by BN-PAGE as in B.
The same membrane was next decorated with an antibody against Cox1. As expected, whereas supercomplexes were only present on the wild-type strains, intermediary assembly complexes of Cox1 were observed in the mutants lacking Pet54 and Pet122. In the Δcox14 and the Δpet122/Δcox14 mutants, Cox1 was present as diffuse bands corresponding to aggregation of Cox1 protein with other mitochondrial gene products (22). Interestingly, in the Δpet54/Δcox14 double mutant, no Cox1 signal was detected. This was confirmed by analyzing the steady-state levels of Cox1 by SDS-PAGE and Western blotting experiments. In this case, whereas a reduction of Cox1 levels was observed for the Δpet54, Δpet122, and Δpet122/Δcox14 mutants, no signal of Cox1 was detected in the double deletion of Pet54 and Cox14 (Fig. 8C), even when Cox1 was synthesized at similar levels compared with the single Δpet54 mutant (Fig. 8A).
The assembly feedback regulatory loop of Cox1 synthesis can be interrupted by deletion of Cox14 (15) or the Cox1 C-terminal end (16); however, in the absence of Pet54, synthesis of Cox1 cannot recover. In the Cox1ΔC15 mitochondria, Mss51 was enriched on the ∼180-kDa complex as compared with a strain bearing wild-type Cox1, although some Mss51 is present on the ∼250–450-kDa complexes (43) (Fig. 8D). A Δpet54 mutant carrying Cox1ΔC15 showed a similar enrichment of Mss51 on the ∼180-kDa complex (Fig. 8D), supporting the idea that even if Mss51 is more available for translational activation, Pet54 is necessary to fully activate Mss51.
Mss51 binds heme b, and hemylation is linked to the role of Mss51 in regulation of COX1 mRNA translation. If hemylation of Mss51 is affected by mutation of two heme-binding CPX motifs or by depletion of heme, then synthesis of Cox1 is decreased (28). However, the mss51T167R and mss51F199I variants, originally identified as respiratory suppressors of Δshy1 mutants (44), can bypass the need for Mss51 hemylation. In this case, synthesis of Cox1 has wild-type levels even if hemylation is hampered (28). The binding of hemes to Mss51 can induce conformational changes necessary for function (45). We sought to investigate whether Pet54 could be linked to Mss51 hemylation. We analyzed whether synthesis of Cox1 in the presence of the mss51F199I variant depended on Pet54. in vivo mitochondrial translation assays showed that in cells carrying mss51F199I, synthesis of Cox1 is similar in both wild-type and Δpet54 cells (Fig. 9). These data suggest that the mss51F199I mutation bypassed the need for Pet54 to efficiently translate COX1 mRNA and that Pet54 function might be related to Mss51 hemylation and/or conformational changes.
FIGURE 9.
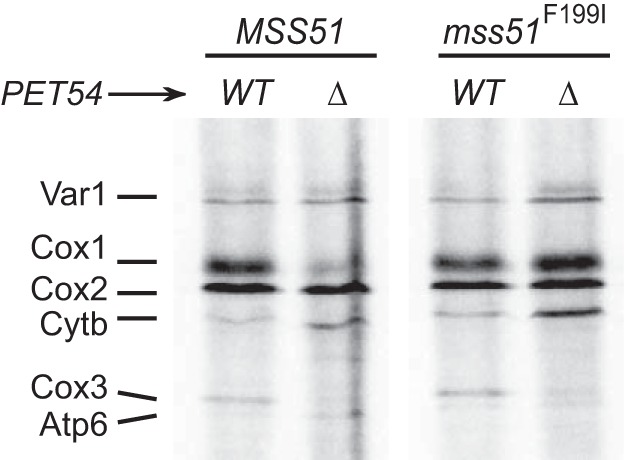
Pet54 might modulate the conformation and/or hemylation of Mss51. Wild-type or Δpet54 cells carrying either the wild-type Mss51 or the mss51F199I variant were pulse-labeled with [35S]methionine in the presence of cycloheximide. The mitochondrial products were resolved by SDS-PAGE and subjected to autoradiography as in Fig. 1.
Pet54 Physically Interacts with the Intronless COX1 mRNA
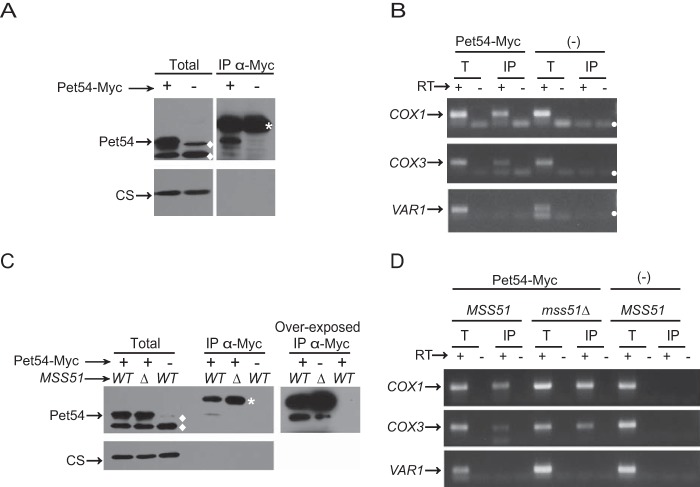
Because Pet54 is an RRM protein, it is possible that the fully translational active form of Mss51 requires binding of Pet54 to the COX1 5′-UTR. To test this idea, we analyzed whether Pet54 could physically interact with the intronless COX1 transcript. Mitochondria carrying the Pet54–3xMyc protein were solubilized with dodecyl maltoside; next, Pet54 was immunoprecipitated with anti-Myc antibodies, and RNA from the immunoprecipitate was isolated and analyzed by reverse transcription and PCR. This technique was used previously to study the Pet309-COX1 mRNA interaction (38). Western blotting analysis showed that Pet54–3xMyc immunoprecipitated. In contrast, the unrelated mitochondrial citrate synthase (negative control) was absent from the immunoprecipitate fraction, indicating that the immunoprecipitation was specific (Fig. 10A). To test whether the COX1 mRNA was associated with Pet54, RNA was purified from the immunoprecipitated Pet54-Myc fraction. Next, it was analyzed by reverse transcription using primers for the COX1 5′-UTR, and the cDNA was amplified by PCR. As a positive control, we analyzed whether COX3 mRNA was also amplified because it was previously reported by in vitro experiments that Pet54 interacts with the COX3 5′-UTR (31). As a negative control, we also amplified VAR1, because translation of this mRNA is independent of Pet54. COX1 mRNA was present in the Pet54–3xMyc immunoprecipitation fraction as well as in the total fraction (Fig. 10B); however, it was absent from the immunoprecipitation fraction from untagged Pet54 mitochondria. As expected, COX3 mRNA was also amplified from the Pet54–3xMyc immunoprecipitate, whereas VAR1 mRNA was absent from the immunoprecipitate. We next asked whether the interaction of Pet54–3xMyc with COX1 mRNA is affected by Mss51. To address this question, Pet54–3xMyc was immunoprecipitated from mitochondria carrying an Δmss51 mutation. RNA analysis from this fraction showed that the COX1 mRNA was still present in the immunoprecipitate, indicating that Pet54 binds to the COX1 mRNA even in the absence of Mss51 (Fig. 10, C and D).
FIGURE 10.
Pet54 interacts with COX1 mRNA, and this interaction is independent of Mss51. A, mitochondria were solubilized with dodecyl maltoside, and Pet54–3xMyc or untagged Pet54 (−) was subjected to immunoprecipitation with antibody anti-Myc. One-fourth of the immunoprecipitate (IP) was resolved by SDS-PAGE and transferred to a PVDF membrane for Western blotting. The membrane was probed with anti-Myc antibody and with anti-citrate synthase antibody (CS) as a negative control for interaction. The total fraction represents 5% of the mitochondrial extract used for immunoprecipitation. *, nonspecific bands from the immunoglobulin heavy chain used for immunoprecipitation. ♦, nonspecific bands when the anti-Myc antibody was used. B, RNA was extracted from the total (T) and immunoprecipitate fractions. Each fraction was divided in two, and cDNA was prepared in the presence (+) or absence (−) of reverse transcriptase (RT) using primers for the COX1 and VAR1 5′-UTRs. The (−) RT lanes represent a negative control for DNA contamination. The PCR products were run on an agarose gel. ●, bands due to primer dimers. C and D, mitochondria from cells carrying Pet54–3xMyc or the untagged Pet54 and either the wild type MSS51 or the Δmss51 deletion were treated and analyzed as in A and B.
Together, our data suggest that in the absence of Pet54, Mss51 is enriched for the translational activator form. However, the protein is not competent to promote efficient translation of COX1 mRNA. Because Pet54 interacts with the COX1 mRNA, this interaction could be important for the activation of Mss51. In addition, the complex formed by the assembly feedback regulator Coa3 was not altered by the Δpet54 mutation. This is not surprising because the phenotype caused by elimination of Pet54 was independent of Coa3/Cox14. Moreover, it was observed that newly made Cox1 is highly unstable after the combined deletion of Pet54 and Cox14.
Discussion
Pet54 was initially described as a translational activator specific for COX3 mRNA (7) and as a factor necessary for COX1 intron aI5β splicing (39). Here we demonstrated that Pet54 plays a third role that is independent of the previous ones. Although this protein is not absolutely required, it promotes efficient Cox1 synthesis.
It is well established that COX1 mRNA translation is reduced in mutants that block CcO assembly (for reviews, see Refs. 18 and 19) and that this regulation can be suppressed by deletion of Cox14 (15) or elimination of the last 15 residues from the C-terminal end of Cox1 (Cox1ΔC15) (16). The third function of Pet54 was discovered because, in contrast to what is observed for most CcO assembly mutants (including the Δpet122 and Δpet494 mutants that, as Δpet54, also impair COX3 mRNA translation), a deletion of Pet54 did not recover Cox1ΔC15 synthesis. This observation suggested that Pet54 could be a positive regulator of Cox1 synthesis, whose function is independent of the Cox1 C-terminal end. Similarly, the function of Pet54 seems to be independent of Cox14 and Coa3 because a double deletion Δpet54,Δcox14 or Δpet54,Δcoa3 did not recover Cox1 synthesis to normal levels.
We reasoned that Pet54 might act at two different levels of the Cox1 synthesis pathway. (i) Pet54 could affect the formation of the COA complexes by promoting an efficient release of Mss51 from these complexes to be active for COX1 mRNA translation. In the absence of Pet54, Mss51 would be more stably associated with Cox1 and the COA complexes and therefore not accessible as translational activator. However, this possibility is unlikely because we observed in blue native gel electrophoresis that Mss51 is present on the COA complexes similarly in the presence or absence of Pet54. (ii) A possible site of action of Pet54 could involve COX1 5′-UTR, the target for translational activation, which is driven by Pet309 and Mss51 (2, 20, 40). One possibility was that in the absence of Pet54, Mss51 and Pet309 levels decreased, and because these factors are limiting for Cox1 synthesis (40), this would affect the efficiency of COX1 mRNA translation. Alternatively, COX1 mRNA levels could be reduced in the absence of Pet54, decreasing the efficiency of translation. However, none of these possibilities was the case (Fig. 6) (data not shown), indicating that Pet54 did not affect stability of the COX1 translational activators or COX1 transcript. We observed that deletion of Pet54 reduced expression of the reporter ARG8m inserted instead of the COX1 codons. This suggests that Pet54 function is related to the COX1 mRNA UTRs. Indeed, we demonstrated that the function of Pet54 mapped to the COX1 5′-UTR, which is the site of action of Pet309 and Mss51 (2, 40). This effect is observed only in the presence of Cox1 protein, suggesting that Mss51 could be the target for Pet54 function because Mss51 is regulated as a translational activator through the interaction with Cox1 protein and the COA complexes. In addition, we observed a weak genetic interaction between Pet54 and Mss51 rather than with Pet309; overexpression of Mss51 slightly compensated the growth deficiency on medium lacking arginine of Δpet54 mutants with ARG8m located at the COX1 locus.
In the present study, we concluded that Pet54 is necessary for activation of Mss51 as a translational activator and that this function is not related to the role of Mss51 as part of COA complexes. On BN-PAGE and sucrose gradient centrifugation experiments, Mss51 was detected as two different forms: one of high molecular mass (250–450 kDa) that is associated with the COA complexes in early Cox1 assembly intermediates and one of smaller size (around 120–180 kDa) corresponding to a translation-effective Mss51 (Mss51TE), where Mss51 promotes translational activation of the COX1 mRNA (19, 27). In this work, we observed that the sizes and distribution of both Mss51 populations were not affected by the absence of Pet54, indicating that the levels of Mss51TE were similar to wild-type levels. This is in contrast to what is reported for other mutants, like Δpet122 and Δcox11, where the Mss51 population associated with COA complexes is enriched (present work) (27). To confirm our model in which Pet54 positively affects Cox1 synthesis via a mechanism that does not involve assembly intermediates, we observed that when the assembly-feedback regulatory loop is disrupted by a combined deletion of Pet54 and Cox14, Mss51 was exclusively present as the Mss51TE form, yet it is not competent to activate translation, thereby reducing Cox1 synthesis. Moreover, deficiency of Cox1 synthesis in the absence of Pet54 was not compensated by overexpression of PET309 and was slightly compensated by overexpression of MSS51, suggesting that this effect is not only due to a decreased affinity of the translational machinery components Pet309 and Mss51 for the COX1 mRNA. Altogether, our results suggest that Pet54 is necessary to recycle/reactivate Mss51 from the COA state to the “translation-committed” form.
Two examples of similar phenotypes of decreased Cox1 synthesis, even in the presence of Δcox14 or Cox1ΔC15, are reported. The first example is a mutation on the general mitochondrial insertase Oxa1, where Mss51 was observed mainly as the “translation effective” form (15, 27). However, it is unlikely that Pet54 function is related to Oxa1 because in Δpet54 mutants, Mss51 and Cox1 were on COA complexes, which are thought to be inserted in the membrane (27) (present study). The second example includes the point mutations C85S and C96S on Mss51 that alter two CPX motifs. These heme-binding motifs are important for Mss51 function (28). Affinity of heme binding to Mss51 can be modulated by the redox state. Upon oxidation, Mss51 introduces a disulfide bond between Cys-85 and Cys-96 that decreases Mss51 heme affinity (45). In addition, binding of hemes to Mss51 induces conformational changes that may be necessary for function (45).
Pet54 could play a role in the heme addition to Mss51 or could induce a conformational change in Mss51 necessary for translational activation. This idea is supported by the fact that deficiency of COX1 mRNA translation in Δpet54 mutants is bypassed by the presence of the allele mss51F199I. This variant suppressed defects on Cox1 synthesis due to lack of Mss51 hemylation (28).
Pet54 could directly interact with the COX1 mRNA to mediate a more efficient or competent interaction of Mss51 with the translational machinery. In the present work, we have demonstrated that Pet54 physically interacts with the COX1 mRNA. The detected interaction could be either direct or mediated by additional, unknown proteins. However, a direct interaction is more plausible because Pet54 is an RRM protein that directly interacts with both COX3 5′-UTR and the COX1 aI5β intron (31). We observed that the Pet54-COX1 mRNA interaction was not affected by the absence of Mss51, suggesting that the binding of Pet54 to the mRNA is independent of Mss51, probably occurring before Mss51 activates translation.
The role of Pet54 in enhancing Cox1 synthesis does not seem to be related to the generation of mp15, an aberrant peptide derived from the COX1 transcript that is detectable on Δmss51 mutants. This peptide is proposed to be the result of a suboptimal translation initiation by Pet309, maybe by positioning the ribosome on a different translation initiation site on the COX1 transcript (38, 46). In our hands, mp15 was not detectable on Δpet54 cells (not shown), indicating that in the absence of Pet54, translation initiation of COX1 starts on the correct AUG codon.
Our data support the notion that binding of translation-effective Mss51 (Mss51TE) to newly synthesized Cox1 forms the COA complex. Formation of COA complex in turn renders Mss51TE translational inactive (hereby termed Mss51TI) (Fig. 11, step 1). The existence of Mss51TI was proposed previously (19, 21). Mss51 inactivation could be mediated by heme modifications (45) and/or by chaperone Coa1 (19). Assembly of CcO triggers Mss51TI release (Fig. 11, step 2). Because interaction of Pet54 with COX1 mRNA does not require Mss51 (present work), and a physical interaction between Mss51 and COX1 mRNA has not been detected (38), Pet54 might bridge an interaction between Mss51 and COX1 mRNA (Fig. 11, step 3). Binding of Pet54 to the COX1 5′-UTR might be necessary to further activate Mss51TI into its active form (i.e. Mss51TE). Thus, Pet54 might reactivate Mss51TE, probably by mediating hemylation/conformational changes in Mss51 (Fig 11, step 4). As shown previously, many RRM proteins bind RNA along with other proteins for activity (e.g. see Refs. 47 and 48), making it possible that both, Mss51 and the COX1 mRNA are direct targets of Pet54. In the absence of Pet54, Mss51 remains in its inactive form, resulting in a very low COX1 translation. This explains the decreased synthesis of Cox1 observed in the double mutants Δcox14/Δpet54, Δcoa3/Δpet54, and COX1ΔC15/Δpet54, where Mss51 is free but in an inactive state. This model also explains why the presence of the Cox1 protein is necessary to detect a phenotype on Δpet54 mutants, because if Cox1 is absent, then Mss51TE is not inactivated; thus, Pet54 is not required to induce a hemylation-dependent conformational change on Mss51TE.
FIGURE 11.
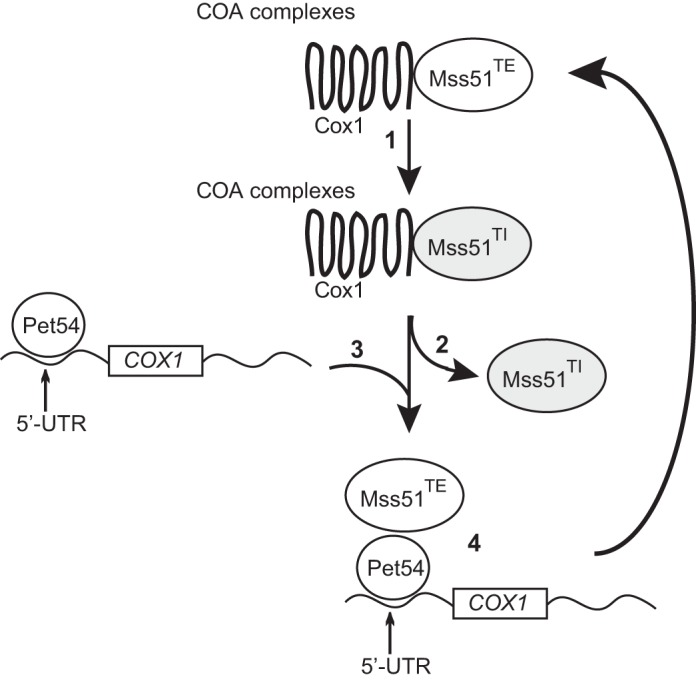
Model of the role of Pet54 on regulation of Cox1 synthesis. Interaction between translational-effective Mss51TE and Cox1 renders Mss51 inactive (Mss51TI, depicted in gray). This might happen in a heme- and/or conformation-dependent manner (step 1). Upon CcO assembly, Mss51TI is released from COA complex (step 2). Pet54 interacts with the COX1 mRNA 5′-UTR (step 3). Via its interaction with Mss51TI, Pet54 may drive Mss51TI reactivation, thus bridging an interaction of Mss51TE with COX1 mRNA (step 4). In the absence of Pet54, Mss51TI is released from the COA complex but is unable to be recycled toward a Mss51TE conformation.
Pet54 is a multifunctional protein with at least three functions. This is not the first case of multifunctional proteins inside mitochondria. Cytochrome c is part of the respiratory chain and also participates in apoptosis (49). Aconitase is involved in mitochondrial DNA maintenance and in the tricarboxylic acid cycle (50, 51). Suv3 is part of the mitochondrial RNA degradosome and is also involved in mitochondrial DNA maintenance and COX1 aI5β intron splicing (52–54). (NAD+)-dependent isocitrate dehydrogenase is known to bind in vitro to mitochondrial mRNAs and probably can modulate translation (55–58). In addition to the three described roles for Pet54, it could be possible that this protein has some function in the assembly of the CcO because it has been detected in supercomplexes (59). As in the case of isocitrate dehydrogenase, we cannot discard the possibility that Pet54 has a broader function and modulates translation of all of the mitochondrial mRNAs.
Author Contributions
X. P.-M. conceived and coordinated the study, analyzed the results, and wrote the manuscript. J. P. M. performed and analyzed the experiments shown in Figs. 1–7 together with M. S.-V. (Fig. 1–3). Y. C.-V. constructed yeast strains, participated in experiments shown in Figs. 1–3, and performed experiments shown in Fig. 10. R. G.-V. created strains and carried out the experiments shown in Fig. 4 and discussed ideas; A. E. G.-G. performed experiments shown in Fig. 9 and discussed ideas; A. Z.-O. performed Northern blotting experiments and discussed ideas; G. H. discussed ideas and wrote parts of the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Antoni Barrientos, Luisa Sandoval-Romero, Gabriel del Río-Guerra, and Teresa Lara-Ortíz for the gift of yeast strains; Miriam Vázquez-Acevedo and Araceli Hernández for technical assistance; and Thomas D. Fox, Antoni Barrientos, and Walter Neupert for the gift of antisera.
This work was supported by Consejo Nacional de Ciencia y Tecnología Grant 47514 (to X. P.-M.) and Fellowships 238399 (to J. P. M.), 298954 (to A. Z.-O.), 255917 (to A. E. G.-G.), and 250726 (to R. G.-V.); the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica; Universidad Nacional Autónoma de México Grants IN208711 and IN204414 (to X. P.-M.); the Fundación Miguel Alemán, A.C. (to X. P.-M.); and the National Institute of Cancer (to G. H.). This manuscript is part of the Ph.D. thesis of J. P. M. from the Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México. The authors declare that they have no conflicts of interest with the contents of this article.
- CcO
- cytochrome c oxidase
- ARG8m
- mitochondria-encoded acetylornithine aminotransferase
- RRM
- RNA recognition motif
- Mss51TE
- translation-effective Mss51
- Mss51TI
- translational inactive Mss51
- BN-PAGE
- blue native PAGE
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- COA
- cytochrome oxidase assembly.
References
- 1. Soto I. C., Fontanesi F., Liu J., and Barrientos A. (2012) Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta 1817, 883–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Manthey G. M., and McEwen J. E. (1995) The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14, 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siep M., van Oosterum K., Neufeglise H., van der Spek H., and Grivell L. A. (2000) Mss51p, a putative translational activator of cytochrome c oxidase subunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr. Genet. 37, 213–220 [DOI] [PubMed] [Google Scholar]
- 4. Poutre C. G., and Fox T. D. (1987) PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics 115, 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mulero J. J., and Fox T. D. (1993) PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics 133, 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cabral F., and Schatz G. (1978) Identification of cytochrome c oxidase subunits in nuclear yeast mutants lacking the functional enzyme. J. Biol. Chem. 253, 4396–4401 [PubMed] [Google Scholar]
- 7. Costanzo M. C., Seaver E. C., and Fox T. D. (1986) At least two nuclear gene products are specifically required for translation of a single yeast mitochondrial mRNA. EMBO J. 5, 3637–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costanzo M. C., and Fox T. D. (1986) Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol. Cell. Biol. 6, 3694–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kloeckener-Gruissem B., McEwen J. E., and Poyton R. O. (1988) Identification of a third nuclear protein-coding gene required specifically for posttranscriptional expression of the mitochondrial COX3 gene is Saccharomyces cerevisiae. J. Bacteriol. 170, 1399–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herrmann J. M., Woellhaf M. W., and Bonnefoy N. (2013) Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim. Biophys. Acta 1833, 286–294 [DOI] [PubMed] [Google Scholar]
- 11. Ott M., Amunts A., and Brown A. (2016) Organization and regulation of mitochondrial protein synthesis. Annu. Rev. Biochem. 10.1146/annurev-biochem-060815-014334 [DOI] [PubMed] [Google Scholar]
- 12. McStay G. P., Su C. H., Thomas S. M., Xu J. T., and Tzagoloff A. (2013) Characterization of Cox1p assembly intermediates in Saccharomyces cerevisiae. J. Biol. Chem. 288, 26546–26556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McStay G. P., Su C. H., and Tzagoloff A. (2013) Modular assembly of yeast cytochrome oxidase. Mol. Biol. Cell 24, 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Su C. H., McStay G. P., and Tzagoloff A. (2014) The Cox3p assembly module of yeast cytochrome oxidase. Mol. Biol. Cell 25, 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barrientos A., Zambrano A., and Tzagoloff A. (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23, 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shingú-Vázquez M., Camacho-Villasana Y., Sandoval-Romero L., Butler C. A., Fox T. D., and Pérez-Martínez X. (2010) The carboxyl-terminal end of Cox1 is required for feedback assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J. Biol. Chem. 285, 34382–34389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khalimonchuk O., Bird A., and Winge D. R. (2007) Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282, 17442–17449 [DOI] [PubMed] [Google Scholar]
- 18. Fontanesi F., Clemente P., and Barrientos A. (2011) Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 286, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mick D. U., Vukotic M., Piechura H., Meyer H. E., Warscheid B., Deckers M., and Rehling P. (2010) Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 191, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perez-Martinez X., Broadley S. A., and Fox T. D. (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mick D. U., Fox T. D., and Rehling P. (2011) Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McStay G. P., Su C. H., and Tzagoloff A. (2013) Stabilization of Cox1p intermediates by the Cox14p-Coa3p complex. FEBS Lett. 587, 943–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mick D. U., Wagner K., van der Laan M., Frazier A. E., Perschil I., Pawlas M., Meyer H. E., Warscheid B., and Rehling P. (2007) Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26, 4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., and Winge D. R. (2007) Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26, 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pierrel F., Khalimonchuk O., Cobine P. A., Bestwick M., and Winge D. R. (2008) Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis facilitating the maturation of Cox1. Mol. Cell. Biol. 28, 4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bareth B., Dennerlein S., Mick D. U., Nikolov M., Urlaub H., and Rehling P. (2013) The heme a synthase Cox15 associates with cytochrome c oxidase assembly intermediates during Cox1 maturation. Mol. Cell. Biol. 33, 4128–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fontanesi F., Soto I. C., Horn D., and Barrientos A. (2010) Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell. Biol. 30, 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soto I. C., Fontanesi F., Myers R. S., Hamel P., and Barrientos A. (2012) A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab. 16, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naithani S., Saracco S. A., Butler C. A., and Fox T. D. (2003) Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell 14, 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valencik M. L., Kloeckener-Gruissem B., Poyton R. O., and McEwen J. E. (1989) Disruption of the yeast nuclear PET54 gene blocks excision of mitochondrial intron aI5 β from pre-mRNA for cytochrome c oxidase subunit I. EMBO J. 8, 3899–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaspar B. J., Bifano A. L., and Caprara M. G. (2008) A shared RNA-binding site in the Pet54 protein is required for translational activation and group I intron splicing in yeast mitochondria. Nucleic Acids Res. 36, 2958–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burke D., Dawson D., and Stearns T. (2000) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Brown N. G., Costanzo M. C., and Fox T. D. (1994) Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. James P., Halladay J., and Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diekert K., de Kroon A. I., Kispal G., and Lill R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65, 37–51 [DOI] [PubMed] [Google Scholar]
- 36. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 37. Wittig I., Braun H. P., and Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 38. Zamudio-Ochoa A., Camacho-Villasana Y., Garcïa-Guerrero A. E., and Përez-Martïnez X. (2014) The Pet309 pentatricopeptide repeat motifs mediate efficient binding to the mitochondrial COX1 transcript in yeast. RNA Biol. 11, 953–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valencik M. L., and McEwen J. E. (1991) Genetic evidence that different functional domains of the PET54 gene product facilitate expression of the mitochondrial genes COX1 and COX3 in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 2399–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perez-Martinez X., Butler C. A., Shingu-Vazquez M., and Fox T. D. (2009) Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol. Biol. Cell 20, 4371–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leung G. P., Lee L., Schmidt T. I., Shirahige K., and Kobor M. S. (2011) Rtt107 is required for recruitment of the SMC5/6 complex to DNA double strand breaks. J. Biol. Chem. 286, 26250–26257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao X., Muller E. G., and Rothstein R. (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2, 329–340 [DOI] [PubMed] [Google Scholar]
- 43. Khalimonchuk O., Kim H., Watts T., Perez-Martinez X., and Winge D. R. (2012) Oligomerization of heme o synthase in cytochrome oxidase biogenesis is mediated by cytochrome oxidase assembly factor Coa2. J. Biol. Chem. 287, 26715–26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barrientos A., Korr D., and Tzagoloff A. (2002) Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 21, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soto I. C., and Barrientos A. (2016) Mitochondrial cytochrome c oxidase biogenesis is regulated by the redox state of a heme-binding translational activator. Antioxid. Redox Signal. 24, 281–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zambrano A., Fontanesi F., Solans A., de Oliveira R. L., Fox T. D., Tzagoloff A., and Barrientos A. (2007) Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 18, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chuang T. W., Chang W. L., Lee K. M., and Tarn W. Y. (2013) The RNA-binding protein Y14 inhibits mRNA decapping and modulates processing body formation. Mol. Biol. Cell 24, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lau C. K., Diem M. D., Dreyfuss G., and Van Duyne G. D. (2003) Structure of the Y14-Magoh core of the exon junction complex. Curr. Biol. 13, 933–941 [DOI] [PubMed] [Google Scholar]
- 49. Liu X., Kim C. N., Yang J., Jemmerson R., and Wang X. (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 50. Chen X. J., Wang X., and Butow R. A. (2007) Yeast aconitase binds and provides metabolically coupled protection to mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 104, 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen X. J., Wang X., Kaufman B. A., and Butow R. A. (2005) Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307, 714–717 [DOI] [PubMed] [Google Scholar]
- 52. Golik P., Szczepanek T., Bartnik E., Stepien P. P., and Lazowska J. (1995) The S. cerevisiae nuclear gene SUV3 encoding a putative RNA helicase is necessary for the stability of mitochondrial transcripts containing multiple introns. Curr. Genet. 28, 217–224 [DOI] [PubMed] [Google Scholar]
- 53. Turk E. M., and Caprara M. G. (2010) Splicing of yeast aI5β group I intron requires SUV3 to recycle MRS1 via mitochondrial degradosome-promoted decay of excised intron ribonucleoprotein (RNP). J. Biol. Chem. 285, 8585–8594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guo X. E., Chen C. F., Wang D. D., Modrek A. S., Phan V. H., Lee W. H., and Chen P. L. (2011) Uncoupling the roles of the SUV3 helicase in maintenance of mitochondrial genome stability and RNA degradation. J. Biol. Chem. 286, 38783–38794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dekker P. J., Stuurman J., van Oosterum K., and Grivell L. A. (1992) Determinants for binding of a 40 kDa protein to the leaders of yeast mitochondrial mRNAs. Nucleic Acids Res. 20, 2647–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elzinga S. D., Bednarz A. L., van Oosterum K., Dekker P. J., and Grivell L. A. (1993) Yeast mitochondrial NAD+-dependent isocitrate dehydrogenase is an RNA-binding protein. Nucleic Acids Res. 21, 5328–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Papadopoulou B., Dekker P., Blom J., and Grivell L. A. (1990) A 40 kd protein binds specifically to the 5′-untranslated regions of yeast mitochondrial mRNAs. EMBO J. 9, 4135–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. de Jong L., Elzinga S. D., McCammon M. T., Grivell L. A., and van der Spek H. (2000) Increased synthesis and decreased stability of mitochondrial translation products in yeast as a result of loss of mitochondrial (NAD+)-dependent isocitrate dehydrogenase. FEBS Lett. 483, 62–66 [DOI] [PubMed] [Google Scholar]
- 59. Vukotic M., Oeljeklaus S., Wiese S., Vögtle F. N., Meisinger C., Meyer H. E., Zieseniss A., Katschinski D. M., Jans D. C., Jakobs S., Warscheid B., Rehling P., and Deckers M. (2012) Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab 15, 336–347 [DOI] [PubMed] [Google Scholar]
- 60. Tavares-Carreón F., Camacho-Villasana Y., Zamudio-Ochoa A., Shingú-Vázquez M., Torres-Larios A., and Pérez-Martínez X. (2008) The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 283, 1472–1479 [DOI] [PubMed] [Google Scholar]