Abstract
We genetically characterized 32 canine rabies viruses isolated in Mali during 2006–2013 and identified 3 subgroups that belonged to the Africa 2 lineage. We also detected subgroup F rabies virus. This information should be useful for development of mass vaccination campaigns for dogs and eventual large-scale control programs in this country.
Keywords: rabies virus, canine rabies virus, viruses, rabies, dogs, molecular analysis, Africa 2 lineage, Mali
Rabies causes an estimated 70,000 human deaths annually worldwide, and >99% occur in developing countries, of which ≈43% occur in Africa, where rabies virus circulates in the dog population (1). A person bitten by a rabid dog, if not given postexposure prophylaxis, has an ≈5% (if bitten on the hand) to 70% (if bitten on the face) probability of showing development of clinical rabies (2). However, postexposure prophylaxis is often unavailable or unaffordable in many developing countries.
Numerous infectious diseases, including tuberculosis, malaria, dengue fever, and rabies, are present in Mali. The domestic dog is the major reservoir and vector of rabies in this country. Although disease surveillance is insufficient throughout Mali, the level of underreporting of rabies cases is unknown. Animal and human cases are recorded mainly in urban and suburban areas. Surveillance data reflect rabies mainly in Bamako (the capital of Mali; population 1.8 million), where rabies diagnostic testing is available.
A standard procedure is in place in Bamako for reporting of an animal bite. The bitten person should immediately contact the Division of Epidemiology, Prevention and Control of Diseases, which is part of the National Directorate of Health. Persons with suspected cases of rabies are referred to a specialized clinic (Lazaret Clinic) in Bamako. Dog owners are requested by the Division of Epidemiology, Prevention and Control of Diseases to bring their dogs to a veterinary clinic for a 15-day quarantine. Rabies diagnosis of suspect animals is made by the Central Veterinary Laboratory (CVL) in Bamako. A diagnosis of rabies in humans is based only on results of a clinical examination because of sociocultural reasons (3). In other cities in Mali, there are reference health centers, hospitals, and veterinary regional services for diagnosis (4).
During 2000–2013, samples from 468 animals showing clinical signs of rabies or to whom humans were exposed were submitted to the CVL for rabies testing by using the fluorescent antibody test (5). Of 468 animals analyzed, 447 (435 dogs, 4 cats, 4 cows, and 4 monkeys) showed positive results for rabies. Twenty-eight human cases of rabies were reported during 2007–2009 in Bamako, which indicated an incidence of 3.3 cases/1,000,000 persons/year despite 141 postexposure prophylaxis vaccinations/1,000,000 persons/year (4). Assuming a dog:human ratio in Bamako of 1:121, the annual incidence of rabies in dogs is ≈2.24 rabid dogs/1,000 dogs during the past 13 years, which is higher than that observed in N’Djaména, Chad (6), which borders Mali.
A total of 306 (45.0%; 95% CI 38%–52%) of 680 dogs were reported as being vaccinated against rabies at least once. However only 59 (19.3%) of the 306 dogs examined had a valid vaccination certificate (4).
In Bamako, an average of 1,470 persons are bitten by animals each year, of whom 1,427 (97.1%) are bitten by dogs (3). A total of 3,544 (60.3%) of 5,870 bitten persons are young adults, including 1,920 (32.73%) children <10 years of age. Men are bitten more often than women.
Four lineages (Africa 1–4) of rabies virus and several subgroups have been detected in Africa. All lineages include classical rabies virus species and vary by geographic area, virus evolution, and reservoir species (7,8). The most comprehensive study of western and central African rabies viruses included some isolates from Mali (7). The purpose of our study was to obtain more detailed information on genetic characteristics of rabies virus circulating in Mali and to clarify the geographic distribution and transboundary spread of this virus in the canine population in Mali.
The Study
During 2002–2013, a total of 468 specimens were submitted from various regions in Mali to the CVL for rabies diagnosis (Figure 1). Samples were tested by using the fluorescent antibody test (5) and stored at −20°C for further analyses. We selected 100 samples (95 with positive results and 5 with negative results) for further testing on the basis of their geographic origin.
Figure 1.
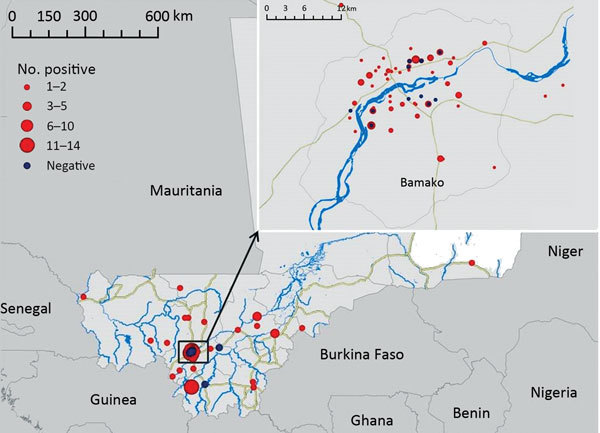
Locations of origin for 100 specimens analyzed in this study (95 with positive results and 5 with negative results) submitted for rabies virus diagnosis, Mali, 2002–2013. Inst shows closer view of the area near the capital city of Bamako.
Supernatants (100 μL) of suspensions (10% wt/vol) of dog brains were deposited on test paper cards, which stabilize nucleic acids. Virus RNA was extracted from stabilized samples by using the Iprep PureLink Virus Kit (Invitrogen, Paris, France) and subjected to partial nucleoprotein gene amplification of a conserved sequence (positions 55–660) (9). Virus RNA was tested by using a hemi-nested reverse transcription PCR (RT-PCR) and a real-time quantitative RT-PCR (10). After amplification, PCR products were sequenced in both directions by Beckman Coulter Genomics (Takeley, UK) and specific primers. A total of 32 stabilized samples showing positive results by hemi-nested RT-PCR and real-time, quantitative RT-PCR were used for phylogenetic analyses (Table 1).
Table 1. Characteristics of 32 rabies virus samples from dogs, Mali, 2006–2013* .
| Virus | GenBank accession no. | Sample ID no. | Region | Quantitative RT-PCR Ct | Subgroup of Africa 2 lineage |
|---|---|---|---|---|---|
| RV01 | KP976113 | 420/2006 | Bamako | 28.51 | G |
| RV04 | KP976114 | 345/2007 | Bamako | 30.19 | H |
| RV05 | KP976130 | 352/2007 | Bamako | 25.35 | G |
| RV06 | KP976125 | 58/2008 | Bamako | 24.09 | G |
| RV09 | KP976126 | 146/2008 | Bamako | 27.51 | G |
| RV10 | KP976122 | 154/2008 | Ségou | 25.75 | G |
| RV11 | KP976124 | 167/2008 | Koulikoro | 23.97 | G |
| RV14 | NA | 259/2008 | Bamako | 31.59 | H |
| RV15 | KP976123 | 261/2008 | Ségou | 26.14 | G |
| RV19 | NA | 530/2008 | Bamako | 27.85 | G |
| RV20 | KP976117 | 003/2009 | Bamako | 32.82 | H |
| RV22 | NA | 69/2009 | Bamako | 26.15 | H |
| RV27 | NA | 118/2009 | Bamako | 32.30 | H |
| RV44 | KP976129 | 587/2009 | Bamako | 26.22 | G |
| RV50 | NA | 19/11/2010 | Bamako | 27.90 | G |
| RV51 | NA | 42/2010 | Bamako | 28.59 | G |
| RV56 | NA | 171/2010 | Koulikoro | 22.20 | G |
| RV57 | KP976121 | 176/2010 | Gao | 21.90 | F |
| RV60 | NA | 221/2010 | Bamako | 24.60 | H |
| RV67 | NA | 603/2010 | Bamako | 21.30 | H |
| RV68 | NA | 137/2011 | Bamako | 20.80 | H |
| RV70 | KP976119 | 149/2011 | Bamako | 21.70 | H |
| RV79 | NA | 339/2011 | Bamako | 24.90 | G |
| RV81 | KP976127 | 357/2011 | Bamako | 34,20 | G |
| RV84 | NA | 480/2011 | Bamako | 21.20 | G |
| RV87 | KP976116 | 612/2011 | Bamako | 22.50 | H |
| RV88 | KP976120 | 628/2011 | Koulikoro | 21.70 | H |
| RV89 | NA | 674/2011 | Bamako | 20.20 | H |
| RV90 | KP976118 | 688/2011 | Bamako | 30.80 | H |
| RV93 | KP976115 | 223/2012 | Bamako | 23.20 | H |
| RV95 | NA | 366/2012 | Bamako | 21.00 | G |
| RV96 |
KP976128 |
100/2013 |
Bamako |
29.00 |
G |
| *A fluorescent antibody test was conducted as described by Dean et al. (5). For each tested sample, test paper was impregnated with 100 μL of 10% brain suspension and subjected to molecular biological analysis. Of 100 samples tested, 32 showed positive results by this test. A conventional hemi-nested reverse transcription PCR (RT-PCR) was performed with rabies virus primers JW12–JW6 as described (9). All samples showed positive results by this test. A quantitative RT-PCR was performed with rabies primers JW12–N165-146 (10). This PCR detected >100 RNA copies/µL. The coefficient of determination (R2) was 0.999, the Y intercept was of 36.65, and efficiency was 99%. Samples in bold (n = 15) had duplicate sequences and were not subjected to phylogenetic analysis. ID, identification; Ct, cycle threshold; NA, not available. | |||||
We constructed a maximum-likelihood phylogenetic tree (Figure 2) that excluded 15 duplicate sequences (Table 2) by using MEGA version 6 software (11). We also constructed a maximum-parsimony haplotype network by using TCS version 1.21 software (12).
Figure 2.
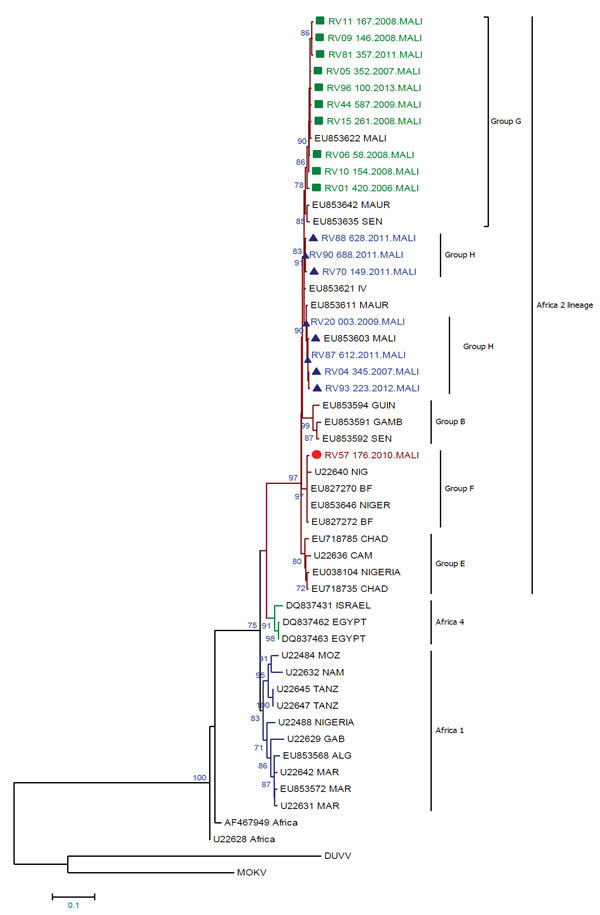
Maximum-likelihood phylogenetic tree based on a 564-nt sequence of nucleoprotein genes of 18 rabies virus sequences from Mali, 2002–2013, and representative sequences from Mali (n = 2), northern Africa (n = 6), South Africa (n = 2), West Africa (n = 32), and central Africa (n = 5). Sequences obtained in this study are identified in green, blue, and red. Green squares indicate genotype G, blue triangles indicate genotype H, and red circles indicate genotype F. The tree is rooted with 2 bat isolates used as outgroups Duvenhage virus (DUVV) (U22848) and Mokola virus (MOKV) (U22843). Bootstrap values (100 replicates) >70% are shown next to nodes. Alg, Algeria; BF, Burkina Faso; Cam, Cameroon; Gab, Gabon; Gamb, Gambia; Guin, Guinea; Maur, Mauritania; Mar, Morocco; Moz, Mozambique; Nig, Niger; Sen, Senegal; Tanz, Tanzania. Scale bar indicates nucleotide substitutions per site.
Table 2. Characteristics of representative nucleoprotein gene sequences for rabies virus isolates, Mali, 2006–2013* .
| Isolate | Haplotype | Identical sequences (−546 nt of the N gene) | Phylogroup |
|---|---|---|---|
| RV09 | 2 | RV50, RV56, RV51, RV19, RV79 | G (Africa 2) |
| RV96 | 6 | RV84, RV95 | G (Africa 2) |
| RV90 | 11 | RV67, RV60, RV68, RV88, RV89, RV22 | H (Africa 2) |
| RV87 | 14 | RV14, RV27 | H (Africa 2) |
*All identical sequences have 100% nucleoprotein (N) gene identity on the basis of 546 nt (positions 71–618) compared with the reference isolate. RV, rabies virus.
We analyzed phylogenetic relationships between 18 partial nucleoprotein gene sequences and 31 representative sequences of Africa lineages of rabies virus. This analysis (Technical Appendix Figure 1) showed that all samples that belonged to the Africa 2 lineage were widely distributed in western and central Africa (7), including Mali and neighboring countries (Mauritania, Guinea, Senegal, Niger, Nigeria, Côte d’Ivoire, and Burkina Faso).
We found <2.1% divergence between all isolate sequences. For 17 haplotypes, 10 sequences were identified as belonging to subgroup G; this subgroup also included 3 sequences from Mali, Mauritania, and Senegal. Seven sequences (forming 6 haplotypes; RV88 was identical to RV90) belonged to subgroup H, which contained representative sequences from Côte d’Ivoire, Mauritania and Mali. One sequence from Mali (isolate RV57) belonged to subgroup F, which was similar to sequences from neighboring countries (Niger and Burkina Faso). Our data indicate that subgroup H might contain 2 distinct groups (Technical Appendix Figure 2).
Analysis of the nucleoprotein gene identified canine rabies subgroups G and H in Mali, as reported (7), and subgroup F, which was found throughout Burkina Faso and Niger (8). Subgroup G circulates in Mauritania, Burkina Faso, and Senegal. Subgroup H contains viruses from Mauritania, Mali, Burkina Faso, and Côte d’Ivoire. The RV57 isolate included in subgroup F was isolated from a rabid dog at the border with Niger in 2010. Strong nucleotide identity (99.6%) was shown between RV57 and the strain isolated in Niger in 2010 (Genbank accession no. EU853646).
Conclusions
We identified 3 subgroups of the Africa 2 lineage of rabies virus in Mali. The presence of subgroup F could be explained by the movement of rabid animals across country borders. Previous studies reported rabies virus transmission by human-mediated animal movements (13,14). The information we obtained in this study should be useful for development of mass vaccination campaigns for dogs and eventual large-scale control programs in this country.
Technical Appendix. Additional information on molecular characterization of canine rabies virus, Mali, 2006–2013.
Acknowledgments
This study was supported by the European Union Seventh Framework Program (grant no. 221948, Integrated Control of Neglected Zoonoses) and the French Agency for Food, Environmental, and Occupational Health and Safety.
Biography
Dr. Abdallah Traoré is a research scientist at the Central Veterinary Laboratory, Bamako, Mali. His primary research interest is diagnosis of viral diseases in livestock and poultry.
Footnotes
Suggested citation for this article: Traoré A, Picard-Meyer E, Mauti S, Biarnais M, Balmer O, Samaké K, et al. Molecular characterization of canine rabies virus, Mali, 2006–2013. Emerg Infect Dis. 2016 May [date cited]. http://dx.doi.org/10.3201/eid2205.150470
References
- 1.World Health Organization. WHO expert consultation on rabies. Second report. World Health Organ Tech Rep Ser. 2013;982:1–139 . [PubMed] [Google Scholar]
- 2.Cleaveland S, Fevre EM, Kaare M, Coleman PG. Estimating human rabies mortality in the United Republic of Tanzania from dog bite injuries. Bull World Health Organ. 2002;80:304–10 . [PMC free article] [PubMed] [Google Scholar]
- 3.Dao S, Abdillahi AM, Bougoudogo F, Toure K, Simbe C. Epidemiological aspects of human and animal rabies in the urban area of Bamako, Mali [in French]. Bull Soc Pathol Exot. 2006;99:183–6 . [PubMed] [Google Scholar]
- 4.Mauti S, Traore A, Hattendorf J, Schelling E, Wasniewski M, Schereffer JL, et al. Factors associated with dog rabies immunisation status in Bamako, Mali. Acta Trop. 2015;12:pii:S0001-706X(15)30139-X. [DOI] [PubMed]
- 5.Dean DJ, Abelseth MK, Atanasiu P. The fluorescent antibody test. In: Meslin FX, Kaplan MM, Koprowski H, editors. Laboratory techniques in rabies. 4th ed. Geneva: World Health Organization; 1996. p. 88–95. [Google Scholar]
- 6.Kayali U, Mindekem R, Yemadji N, Oussiguere A, Naissengar S, Ndoutamia AG, et al. Incidence of canine rabies in N’Djamena, Chad. Prev Vet Med. 2003;61:227–33. 10.1016/j.prevetmed.2003.07.002 [DOI] [PubMed] [Google Scholar]
- 7.Talbi C, Holmes EC, de Benedictis P, Faye O, Nakoune E, Gamatie D, et al. Evolutionary history and dynamics of dog rabies virus in western and central Africa. J Gen Virol. 2009;90:783–91. 10.1099/vir.0.007765-0 [DOI] [PubMed] [Google Scholar]
- 8.De Benedictis P, Sow A, Fusaro A, Veggiato C, Talbi C, Kaboré A, et al. Phylogenetic analysis of rabies viruses from Burkina Faso, 2007. Zoonoses Public Health. 2010;57:e42–6. 10.1111/j.1863-2378.2009.01291.x [DOI] [PubMed] [Google Scholar]
- 9.Picard-Meyer E, Bruyere V, Barrat J, Tissot E, Barrat MJ, Cliquet F. Development of a hemi-nested RT-PCR method for the specific determination of European bat Lyssavirus 1. Comparison with other rabies diagnostic methods. Vaccine. 2004;22:1921–9. 10.1016/j.vaccine.2003.11.015 [DOI] [PubMed] [Google Scholar]
- 10.Picard-Meyer E, Peytavin de Garam C, Schereffer JL, Marchal C, Robardet E, Cliquet F. Cross-platform evaluation of commercial real-time SYBR green RT-PCR kits for sensitive and rapid detection of European bat lyssavirus type 1. Biomed Res Int. 2015;2015:839518. . 10.1016/j.vaccine.2003.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–9. 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- 13.Fèvre EM, Bronsvoort BM, Hamilton KA, Cleaveland S. Animal movements and the spread of infectious diseases. Trends Microbiol. 2006;14:125–31. 10.1016/j.tim.2006.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talbi C, Lemey P, Suchard MA, Abdelatif E, Elharrak M, Jalal N, et al. Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathog. 2010;6:e1001166 . 10.1371/journal.ppat.1001166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Technical Appendix. Additional information on molecular characterization of canine rabies virus, Mali, 2006–2013.


