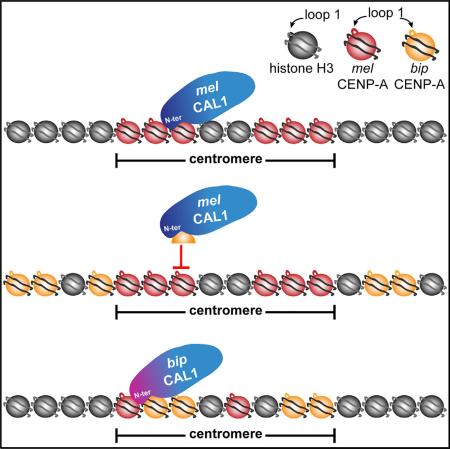
SUMMARY
Centromeres mediate the conserved process of chromosome segregation, yet centromeric DNA and the centromeric histone, CENP-A, are rapidly evolving. The rapid evolution of Drosophila CENP-A loop 1 (L1) is thought to modulate the DNA-binding preferences of CENP-A to counteract centromere drive, the preferential transmission of chromosomes with expanded centromeric satellites. Consistent with this model, CENP-A from Drosophila bipectinata (bip) cannot localize to Drosophila melanogaster (mel) centromeres. We show that this result is due to the inability of the mel CENP-A chaperone, CAL1, to deposit bip CENP-A into chromatin. Co-expression of bip CENP-A and bip CAL1 in mel cells restores centromeric localization, and similar findings apply to other Drosophila species. We identify two co-evolving regions, CENP-A L1 and the CAL1 N terminus, as critical for lineage-specific CENP-A incorporation. Collectively, our data show that the rapid evolution of L1 modulates CAL1-mediated CENP-A assembly, suggesting an alternative mechanism for the suppression of centromere drive.
Graphical Abstract
INTRODUCTION
Centromeres are essential chromosomal structures to which kinetochore proteins and microtubules are recruited during cell division to mediate the accurate distribution of genetic material. While centromere function is highly conserved, centromere size and structure vary greatly between organisms (Fukagawa and Earnshaw, 2014). In complex eukaryotes, the specific DNA sequences found at centromeres are neither necessary nor sufficient for centromere formation (Choo, 2000; Karpen and All-shire, 1997), and centromeres are epigenetically defined by the presence of a centromere-specific histone H3 variant called CENP-A (also called CID in Drosophila) (Earnshaw and Rothfield, 1985; Karpen and Allshire, 1997).
Accurate CENP-A deposition is mediated by specific CENP-A assembly factors (or chaperones). While yeast and humans harbor CENP-A chaperones with common ancestry (called Scm3 and HJURP, respectively) (Bernad et al., 2011; Camahort et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009; Mizuguchi et al., 2007; Pidoux et al., 2009; Sanchez-Pulido et al., 2009), Drosophila employ an evolutionarily distinct CENP-A chaperone called CAL1 (Chen et al., 2014; Erhardt et al., 2008; Phansalkar et al., 2012).
Despite the universally conserved function of centromeres in maintaining genome integrity, both CENP-A (Cooper and Henikoff, 2004; Finseth et al., 2015; Henikoff et al., 2001; Malik and Henikoff, 2001; Malik et al., 2002; Ravi et al., 2010; Schueler et al., 2010; Talbert et al., 2002; Zedek and Bureš, 2012) and centromeric DNA (Melters et al., 2013) are rapidly evolving. This paradox has been explained by the centromere drive hypothesis, which proposes that CENP-A adaptively evolves to maintain meiotic parity by modulating its DNA-binding preferences to counteract the transmission advantage gained by satellite expansion in female meiosis (Henikoff and Malik, 2002; Malik and Henikoff, 2002). In support of this model, adaptive evolution has been observed in both the N-terminal tail and loop 1 (L1) of CENP-A (Cooper and Henikoff, 2004; Finseth et al., 2015; Henikoff et al., 2001; Malik and Henikoff, 2001; Malik et al., 2002; Ravi et al., 2010; Schueler et al., 2010; Talbert et al., 2002; Zedek and Bureš, 2012), both of which are putative DNA-binding regions (Luger et al., 1997; Malik et al., 2002; Vermaak et al., 2002), in plants and animals. The role of CENP-A chaperones in this evolutionary “arms race” has yet to be explored.
Somewhat surprising is the fact that, while Drosophila CENP-A is adaptively evolving (Malik and Henikoff, 2001; Malik et al., 2002), its chaperone CAL1 is highly conserved across both the N-terminal domain, which interacts with CENP-A, and the C-terminal domain, which interacts with CENP-C (Chen et al., 2014; Phansalkar et al., 2012; Schittenhelm et al., 2010). How CAL1 is able to interact with and deposit rapidly evolving CENP-A orthologs, given their different rates of evolution, is unknown.
While several lines of evidence support the rapid evolution of both centromeric DNA and CENP-A in many species (Melters et al., 2013), and also the influence of centromere expansion on meiotic segregation distortion (Chmátal et al., 2014; Daniel, 2002; Fishman and Saunders, 2008; Fishman and Willis, 2005; Pardo-Manuel de Villena and Sapienza, 2001; Wyttenbach et al., 1998), biological data supporting a direct correlation between the evolution of centromeric DNA and CENP-A (the second step in the centromere drive hypothesis (Malik, 2009; Malik and Henikoff, 2002)) are lacking. However, one striking experimental observation supporting centromere drive is that CENP-A from Drosophila bipectinata (bip) expressed in Drosophila melanogaster (mel) tissue culture cells is unable to localize to mel centromeres (Vermaak et al., 2002). This incompatibility is the result of specific amino acid changes in L1 of CENP-A (Vermaak et al., 2002). Because L1 of histone H3 has been shown to interact with DNA (Luger et al., 1997), it was proposed that L1 of CENP-A is adaptively evolving with centromeric DNA satellites to suppress centromere drive (Malik and Henikoff, 2001; Vermaak et al., 2002). However, recent structural studies of human CENP-A octamers and tetramers suggest that L1 of CENP-A does not interact with DNA, and instead is exposed in the nucleosome particle (Sekulic et al., 2010; Tachiwana et al., 2012). Interestingly, in yeast and humans, a domain encompassing L1 known as the CENP-A targeting domain (CATD) is recognized by the assembly factors Scm3 and HJURP, respectively (Bassett et al., 2012; Cho and Harrison, 2011). The CATD is sufficient to confer centromeric localization to histone H3 in both yeast and humans (Black et al., 2004; Shelby et al., 1997). However, the corresponding region of Drosophila CENP-A is not sufficient for the centromeric localization of histone H3 in flies (Moreno-Moreno et al., 2011). How CAL1 recognizes Drosophila CENP-A is unknown.
Here, we use evolutionary cell biology to investigate the relationship between centromere divergence and CENP-A assembly in Drosophila. We reveal that a functional interplay between CAL1 and L1 of CENP-A is both necessary and sufficient for the deposition of orthologous CENP-A proteins at mel native centromeres as well as for de novo CENP-A recruitment to an ectopic locus. Successful CENP-A incorporation requires that L1 and the CAL1 N terminus are compatible, demonstrating that these two domains evolve in concert. These data challenge previous models of centromere drive involving the adaptive evolution of L1 with centromeric DNA in Drosophila and suggest that the evolution of L1 may instead mediate CENP-A centromeric deposition by CAL1.
RESULTS
Inter-species Centromeric Localization of Drosophila CENP-A Orthologs Can Only Partially Be Explained by Phylogenetic Distance
Loop 1 (L1) of CENP-A has long been proposed to be adaptively evolving with centromeric DNA in an “arms race” akin to that occurring between viruses and their hosts (Malik and Henikoff, 2001; Vermaak et al., 2002). A previous study tested the ability of CENP-A orthologs from Drosophila simulans (sim), Drosophila erecta (ere), Drosophila lutescens (lut), Drosophila bipectinata (bip), and Drosophila pseudoobscura (pse) to localize to centromeres in D. melanogaster (mel) cultured Kc cells, to identify CENP-A centromere-targeting motifs (Vermaak et al., 2002). While centromeric localization was observed for CENP-A orthologs from most species, bip CENP-A failed to localize to mel centromeres (12 Ma diverged; Figure 1A), despite the fact that the more divergent pse CENP-A (30 Ma diverged; Figure 1A) was able to localize (Vermaak et al., 2002).
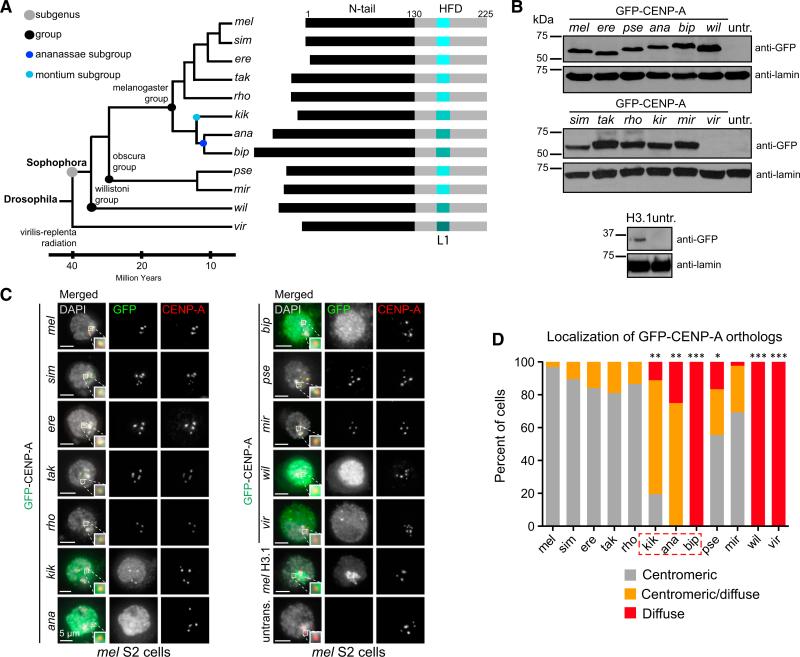
Figure 1. Inter-species Centromeric Localization of Drosophila CENP-A Orthologs Can Only Partially Be Explained by Phylogenetic Distance.
(A) Left: phylogenetic tree of the Drosophila species analyzed in this study (mel, D. melanogaster; sim, D. simulans; ere, D. erecta; tak, D. takahashii; rho, D. rhopalia; kik, D. kikkawai; ana, D. ananassae; bip, D. bipectinata; pse, D. pseudoobscura; mir, D. miranda; wil, D. willistoni; vir, D. virilis). Divergence time and phylogenetic grouping based on Flybase. Right: schematic of CENP-A orthologs from indicated species showing relative differences in protein size. The N terminus is shown in black and the histone fold domain (HFD) is shown in gray. Loop 1 (L1) is shown shades of aqua indicative of divergence in L1. Numbers indicate amino acid positions.
(B) Western blots with anti-GFP (top) and anti-lamin (loading control, bottom) antibodies of total cell extracts showing the expression of GFP-CENP-A orthologs in S2 cells used in (C) and (D). The expression of vir GFP-CENP-A was too low to visualize by western blot.
(C) Immunofluorescence (IF) images of mel S2 interphase cells transiently expressing Drosophila GFP-CENP-A orthologs. Images where endogenous CENP-A is present were chosen to visualize the location of the centromere. DAPI is shown in gray, GFP in green, and mel CENP-A in red. Zoomed insets show representative centromeres with merged colors.
(D) Quantification of (C). Images were manually classified as having either centromeric localization of GFP (gray bars), diffuse localization of GFP (red bars), or centromeric/diffuse (orange bars). Number of transfected cells quantified for each ortholog: 97 for mel, 114 for sim, 94 for ere, 58 for tak, 67 for rho, 36 for kik, 52 for ana, 143 for bip, 90 for pse, 212 for mir, 68 for wil, and 35 for vir. Fisher's two-tailed test p values were ***p < 0.0001 for bip CENP-A, wil CENP-A, and vir CENP-A; **p = 0.0003 for ana; **p = 0.005 for kik; and *p = 0.002 for pse CENP-A compared with mel CENP-A. These data were confirmed by one biological replicate with GFP-tagged constructs, and two additional replicates with HA-tagged constructs.
See also Figures S1–S3.
To better understand the relationship between centromeric localization of CENP-A orthologs in mel cells and their phylogenetic distance from mel, we tested additional CENP-A orthologs from four evolutionarily intermediate species between mel and bip (Drosophila takahashii [tak], Drosophila rhopolia [rho], Drosophila kikkawai [kik], and Drosophila ananassae [ana]), and from three more distant species (Drosophila miranda [mir], Drosophila willistoni [wil], and Drosophila virilis [vir]; Figure 1A), along with mel, sim, ere, bip, and pse as in the original study (Vermaak et al., 2002) for their ability to localize to mel centromeres (Figures 1B–1D and S1). GFP-tagged CENP-A orthologs from these 11 Drosophila species, as well as mel histone H3.1 as a control, were transiently expressed in mel Schneider 2 (S2) cells (Figure 1B). The localization of GFP-CENP-A orthologs was assessed by immunofluorescence (IF) on interphase S2 cells using anti-GFP and anti-mel CENP-A antibodies, which are specific to mel CENP-A and are used as a marker for mel centromeres (Figures 1C and S2).
Localization to mel centromeres was observed for those CENP-A orthologs that are most closely related to mel, namely sim, ere, tak, and rho. In contrast, bip, wil, and vir CENP-A failed to localize, resulting in diffuse GFP signal (p < 0.0001). kik and ana CENP-A partially localized to mel S2 centromeres, displaying both centromeric and diffuse GFP signal (p = 0.005 for kik and p = 0.0003 for ana). Interestingly, centromeric localization was also observed for CENP-A orthologs from the obscura group (pse and mir; 55% [p = 0.002] and 70% centromeric, respectively; Figures 1C, 1D and S1), which is more divergent from mel than either the montium or ananassae subgroups (Figure 1A). The same localization pattern was observed with hem-agglutinin (HA)-tagged CENP-A orthologs (Figures S3A–S3C), indicating that the presence of the GFP tag does not interfere with centromeric localization. Together, these findings confirm and expand upon previous work (Vermaak et al., 2002), and demonstrate that the CENP-A localization pathway is conserved between the melanogaster and obscura groups, but has diverged in the ananassae subgroup. Additionally, the CENP-A localization pathway is not conserved in more divergent lineages (e.g., wil and vir).
Unlike the rapid degradation of mislocalized mel CENP-A after pulse induction (Heun et al., 2006; Olszak et al., 2011), which requires the F-box protein PPA and the CATD (Moreno-Moreno et al., 2011), bip and wil CENP-A proteins appear to persist stably, resulting in higher protein levels compared with those of centromere-localizing CENP-A orthologs (Figure 1C). Perhaps mel PPA cannot recognize bip and wil CENP-A due to their divergent L1 (Vermaak et al., 2002), which is part of the CATD.
We next asked whether the localization of CENP-A orthologs followed a similar pattern at the centromeres of sim, a species closely related to mel (Figure 1A). Transient transfection with sim, mel, ere, bip, and pse GFP-CENP-A constructs in sim M-19 tissue culture cells was followed by IF with anti-GFP and anti-CENP-C antibodies, which recognize sim CENP-C providing a centromere marker (Figures S3D and S3E). Similar to the localization results in mel cells, mel, ere, and pse CENP-A localize to sim centromeres, while bip CENP-A does not (Figures S3D and S3E). These results show that the centromeric localization of CENP-A orthologs to mel and sim centromeres can only partially be explained by phylogenetic distance and that the branch containing the ananassae subgroup is evolving on a separate evolutionary trajectory from that of other close lineages. Furthermore, these experiments demonstrate that the incompatibility between CENP-A and the centromere is not unique to the bip/mel species pair.
D. melanogaster CAL1 Cannot Recruit D. bipectinata CENP-A at an Ectopic Locus
The mislocalization of bip CENP-A in mel cells is due to key amino acid changes between L1 of bip and mel CENP-A (Vermaak et al., 2002). It was originally proposed that this variation in CENP-A L1 is indicative of adaptive evolution with centromeric DNA to suppress drive (Malik and Henikoff, 2001; Vermaak et al., 2002). However, another possibility is that bip CENP-A may be co-evolving (defined here as undergoing coordinated protein evolution) with its loading factor CAL1 (Chen et al., 2014), and that the failure of bip CENP-A to localize to mel centromeres could be due to an incompatibility with mel CAL1.
We investigated this possibility by first determining whether bip CENP-A can physically interact with mel CAL1. Immunoprecipitations (IPs) with anti-CAL1 antibodies coupled to beads were performed in two separate chromatin extracts that contained normalized amounts of mel or bip GFP-CENP-A. Quantification of GFP-CENP-A western blot bands indicated that mel CAL1 pulled down approximately 10% of mel GFP-CENP-A and 20% of bip GFP-CENP-A relative to the respective inputs. These experiments indicate that mel CAL1 can form a complex with bip GFP-CENP-A at least as efficiently as with mel GFP-CENP-A and that there is no incompatibility as far as physical interaction between these two proteins is concerned (Figure 2A).
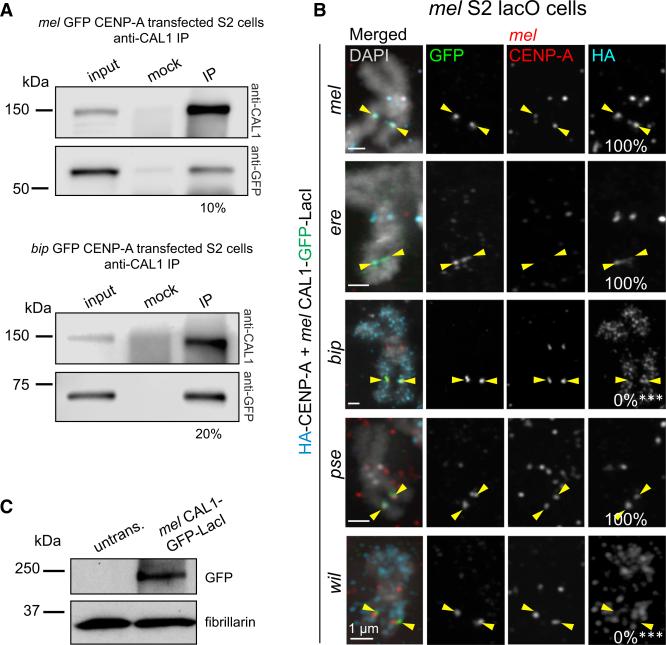
Figure 2. D. melanogaster CAL1 Cannot Recruit D. bipectinata CENP-A to an Ectopic Locus.
(A) Western blots of IPs with anti-CAL1 antibodies from nuclear extracts transiently expressing mel GFP-CENP-A (top) or bip GFP-CENP-A (bottom). IP was confirmed using anti-CAL1 antibody (top blot). Presence of GFP-CENP-A in CAL1 pull-downs was detected with anti-GFP antibody (bottom blot). Shown is the percentage of immunoprecipitated GFP-CENP-A relative to input.
(B) Representative IF images of metaphase chromosome spreads from mel S2 lacO cells transiently co-expressing mel CAL1-GFP-LacI and HA-CENP-A orthologs: mel (top); ere (second); bip (third); pse (fourth); and wil (bottom). Chromosome spreads were quantified for the presence of HA-CENP-A at the lacO site (percentage shown in right column). Endogenous mel CENP-A is shown in red, HA in aqua, GFP in green, and DAPI in gray. Note that mel CENP-A antibodies are specific for this species and that upon expression of CENP-A orthologs that localize to mel centromeres, endogenous CENP-A levels decrease (e.g., ere CENP-A; see also Figure S2). This is not observed for mel HACENP-A, as mel CENP-A antibodies recognize this tagged protein. ***p < 0.0001 (Fisher's two-tailed test) for bip or wil CENP-A compared with mel CENP-A recruitment at the lacO. n = 13 spreads for mel CENP-A recruitment, 8 for ere, 11 for bip, 15 for pse, and 8 for wil. Yellow arrowheads indicate the lacO array. These results were confirmed by one biological replicate with the CAL1-GFP-LacI construct, and two biological replicates with GFP-CENP-A and CAL1-LacI constructs (data not shown).
(C) Western blots with anti-GFP (top) and anti-fibrillarin (loading control, bottom) antibodies of whole-cell extracts showing the expression of induced mel CAL1-GFP-LacI in lacO cells shown in (B).
We next investigated whether the ability of mel CAL1 to interact with bip CENP-A enables its deposition into chromatin. We turned to an ectopic tethering assay, which allows us to interrogate the functional relationship between mel CAL1 and CENP-A from bip and from other representative species without centromeric DNA as a contributing factor. Tethering mel CAL1 via the lac repressor, LacI, at a lacO array stably integrated within a chromosome arm leads to the stable incorporation of mel CENP-A (Chen et al., 2014). We co-expressed HA-tagged mel, ere, pse, bip, or wil CENP-A and an inducible mel CAL1 tagged with GFP and LacI (Figures 2B and 2C). After 24 hr induction of mel CAL1-GFP-LacI, recruitment of HA-CENP-A orthologs to the lacO site was analyzed by IF with anti-HA, anti-GFP (to detect CAL1-GFP-LacI at the lacO site), and anti-mel CENP-A antibodies (to visualize the mel endogenous centromere) on meta-phase chromosomes. This analysis showed that mel CAL1-GFP-LacI successfully recruits sim, ere, and pse CENP-A to the lacO site (Figure 2B). In contrast, bip and wil CENP-A are not recruited to the lacO site and localize all along the chromosome arms in a pattern reminiscent of mel CENP-A overexpression ((Heun et al., 2006); Figure 2B). These experiments suggest that, although mel CAL1 can interact with bip CENP-A (Figure 2A), this interaction is not functional, i.e., mel CAL1 cannot deposit bip CENP-A into chromatin (Figure 2B). Furthermore, they show that the successful ectopic targeting of CENP-A from sim, ere, and pse reflects their competency to localize to mel endogenous centromeres (Figure 1C). These data also demonstrate that the overexpression of mel CAL1-GFP-LacI is not sufficient to promote the centromeric or lacO targeting of bip and wil CENP-A.
Co-expression of CAL1 and CENP-A Ortholog Pairs Rescues Centromeric Localization
While our ectopic targeting assays suggest that mel CAL1 cannot incorporate bip or wil CENP-A into chromatin at the lacO site, they did not allow us to discriminate between defective recruitment by mel CAL1 or an incompatibility between bip or wil CENP-A and DNA sequences present at mel endogenous centromeres. If the failure of bip CENP-A to associate with mel centromeres is solely due to an incompatible assembly factor (mel CAL1), then supplying bip CAL1 should rescue the centromeric localization of bip CENP-A in mel cells (Figure 3A).
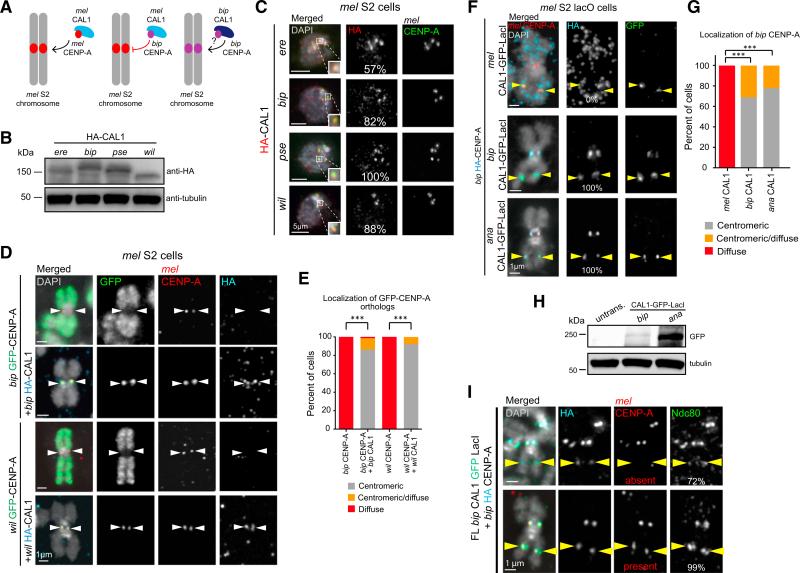
Figure 3. Co-expression of CAL1 and CENP-A Ortholog Pairs Rescues Centromeric Localization.
(A) Schematic of the experiments testing whether co-expression of bip CAL1 and bip CENP-A can result in the localization of bip CENP-A to mel centromeres.
(B) Western blots of whole-cell extracts showing expression of HA-CAL1 constructs. Top: anti-HA. Bottom: anti-tubulin (loading control).
(C) Representative IF images of interphase mel S2 cells transiently expressing HA-CAL1 orthologs. DAPI is shown in gray, HA in red, and mel CENP-A in green. The percentage of cells with centromeric HA signal is indicated in the middle column. Zoomed panels show representative centromeres with merged colors.
(D) Representative IF images of metaphase chromosome spreads from S2 cells transiently expressing bip or wil GFP-CENP-A alone (first and third rows, respectively), bip GFP-CENP-A and bip HA-CAL1 (second row), or wil GFP-CENP-A and wil HA-CAL1 (fourth row). DAPI is shown in gray, GFP in green, HA in aqua, and mel CENP-A in red. White arrowheads indicate the position of the centromere.
(E) Quantification of the IF shown in (D). Chromosome spreads were manually classified as having either centromeric (gray bars), diffuse (red), or centromeric/ diffuse (orange) GFP signal. n = 29 spreads for bip CENP-A alone, 73 for bip CENP-A with bip CAL1, 27 for wil CENP-A alone, and 51 for wil CENP-A with wil CAL1. ***p < 0.0001 (Fisher's two-tailed test) for the centromeric localization of bip and wil CENP-A with and without CAL1. These data were confirmed by one biological replicate using HA-tagged CAL1 constructs (data not shown), and two biological replicates using CAL1-GFP-LacI and HA-CENP-A constructs (see F in this figure for bip, and data not shown for wil). See also Figures S4 and S5.
(F) Representative IF images of metaphase chromosome spreads from mel lacO S2 cells transiently co-expressing mel, bip, or ana CAL1-GFP-LacI (first, second, or third row, respectively), and bip HA-CENP-A. GFP is shown in green, HA in aqua, mel CENP-A in red, and DAPI in gray. Yellow arrowheads indicate the position of the lacO site.
(G) Quantification of the images shown in (F). Cells were manually classified as having either exclusively centromeric GFP signal (gray bars), diffuse GFP signal (red bars), or centromeric and diffuse GFP signal (orange bars). n ≥ 30 cells per condition. These data were confirmed by two biological replicates (data not shown). ***p < 0.0001 (Fisher's two-tailed test).
(H) Western blots with anti-GFP (top) and anti-tubulin (loading control, bottom) antibodies of whole-cell extracts showing the expression of induced bip and ana CAL1-GFP-LacI in lacO cells in F.
(I) Representative IF images of metaphase chromosome spreads from mel lacO S2 cells transiently co-expressing bip CAL1-GFP-LacI and bip HA-CENP-A (aqua) showing the lacO recruitment of endogenous mel CENP-A (red) and the outer kinetochore protein Ndc80 (green). GFP fluorescence was quenched with 100% ethanol. DAPI is shown in gray. Percentage of CENP-A positive (bip, or mel and bip) lacO arrays with Ndc80 is indicated in the right column. Yellow arrowheads mark the position of lacO site. These data represent the average of three experiments (two technical replicates and one biological replicate). n = 40 HA-positive cells.
To test this, we first needed to determine whether bip CAL1 can localize to mel centromeres, a necessary prerequisite for the deposition of CENP-A at this location (Chen et al., 2014; Erhardt et al., 2008). HA-tagged bip, ere, pse, or wil CAL1 were transiently expressed in S2 cells (Figure 3B). IF with anti-HA and anti-mel CENP-A antibodies showed that all of the HACAL1 orthologs localize to mel centromeres in at least 50% of cells (Figure 3C). Since CAL1 is recruited to centromeres by CENP-C (Chen et al., 2014), these data suggest that the CENP-C/CAL1 interaction is conserved between mel and bip and, more generally, across the Drosophila phylogeny. The observation that the C terminus of CAL1, which interacts with CENP-C (Chen et al., 2014; Schittenhelm et al., 2010), is under purifying selection (Phansalkar et al., 2012) is consistent with this hypothesis.
Next, we tested whether supplying bip CAL1 enables bip CENP-A to localize to mel centromeres by transiently transfecting mel S2 cells with bip GFP-CENP-A and bip HA-CAL1 constructs (Figure 3D). IF with anti-HA, anti-GFP, and anti-mel CENP-A antibodies on metaphase spreads showed that centromeric targeting of bip CENP-A is completely restored in 86% of cells and partially restored in 12% (Figures 3D and 3E). Furthermore, the observation that the centromeric bip GFP-CENP-A IF signal is resistant to salt extraction demonstrates that it is incorporated into chromatin (Figure S4). The centromeric and lacO targeting of bip CENP-A was also obtained with the reversed tags: bip CAL1-GFP-LacI with bip HA-CENP-A (Figures 3F–3H).
To test whether the functional interaction between bip CAL1 and CENP-A is lineage specific or species specific, we co-expressed bip HA-CENP-A with ana CAL1-GFP-LacI in mel lacO cells and assessed the recruitment of bip HA-CENP-A at the lacO site by IF with anti-HA, anti-GFP, and anti-mel CENP-A antibodies on metaphase spreads. We found that ana CAL1 is competent for bip CENP-A deposition at both mel centromeres (78% fully centromeric and 22% partially centromeric) and the lacO site (100%; Figures 3F–3H). We conclude that the presence of a lineage-specific CAL1 partner can also promote the centromeric targeting of bip CENP-A in mel cells.
To determine whether the centromeric localization of bip CENP-A can also occur in sim cells, we co-expressed bip CENP-A and bip CAL1 in M-19 cells and observed bip CENP-A centromeric targeting in 56% of cells (Figures S5A and S5B). These results are consistent with our findings in mel cells (Figures 3D and 3E) and demonstrate that a similar CENP-A loading defect is present between bip CENP-A and sim CAL1.
Next, we investigated whether a similar mechanism underlies the defective localization of the more divergent wil CENP-A to mel centromeres. We transiently co-expressed wil GFP-CENP-A with wil HA-CAL1, and assessed centromeric localization by IF. As with bip CENP-A, we observed exclusively centromeric localization of wil CENP-A in 92% of cells and partial localization in 8% (Figures 3D and 3E). We conclude that even CENP-A from a species almost 40 Ma diverged from mel can localize to mel centromeres as long as a compatible CAL1 partner is present.
Given the ability of the bip CENP-A/CAL1 complex to localize to mel centromeres, we next asked if this complex can initiate mel kinetochore assembly by assessing the recruitment of the outer kinetochore component Ndc80 (Meraldi et al., 2006). Bip CAL1-GFP-LacI was tethered to the lacO array in mel cells expressing bip HA-CENP-A followed by IF with anti-HA, anti-mel CENP-A, and anti-Ndc80 on metaphase spreads. We noticed that the full-length bip CAL1-GFP-LacI construct recruited mel CENP-A to the lacO site in approximately 50% of chromosome spreads (p < 0.0001 compared with mel CAL1-GFP-LacI recruitment of bip CENP-A), suggesting that there is more functional conservation between mel and bip CAL1 than between mel and bip CENP-A. By scoring bip CENP-A-positive lacO sites for both the presence or absence of mel CENP-A and Ndc80, we found that bip CAL1-GFP-LacI can recruit Ndc80 even when mel CENP-A is absent or nearly undetectable (72% compared with 99% when mel CENP-A is present at the lacO site [p = 0.2]; Figure 3I). We conclude that the bip CENP-A/ CAL1 complex can mediate mel kinetochore formation, bypassing the requirement for mel CENP-A. These results may explain why the co-expression of bip CAL1 and bip CENP-A does not negatively affect chromosome segregation, whereas the expression of ere CENP-A does (Figure S6). In plants, too, centromeric localization of CENP-A orthologs is not a predictor of whether they can form functional kinetochores (Ravi et al., 2010).
CAL1 Recognizes CENP-A via L1
It has previously been shown that replacing L1 of mel CENP-A with the homologous region of bip CENP-A results in a loss of centromeric localization, while substituting L1 of bip CENP-A with mel L1 results in a gain of centromeric localization (Vermaak et al., 2002). Based on these data and our findings so far, we hypothesized that L1 of CENP-A could mediate the functional interaction with CAL1, and that the divergence of bip L1 (Vermaak et al., 2002) results in the failure of mel CAL1 to properly deposit bip CENP-A into chromatin.
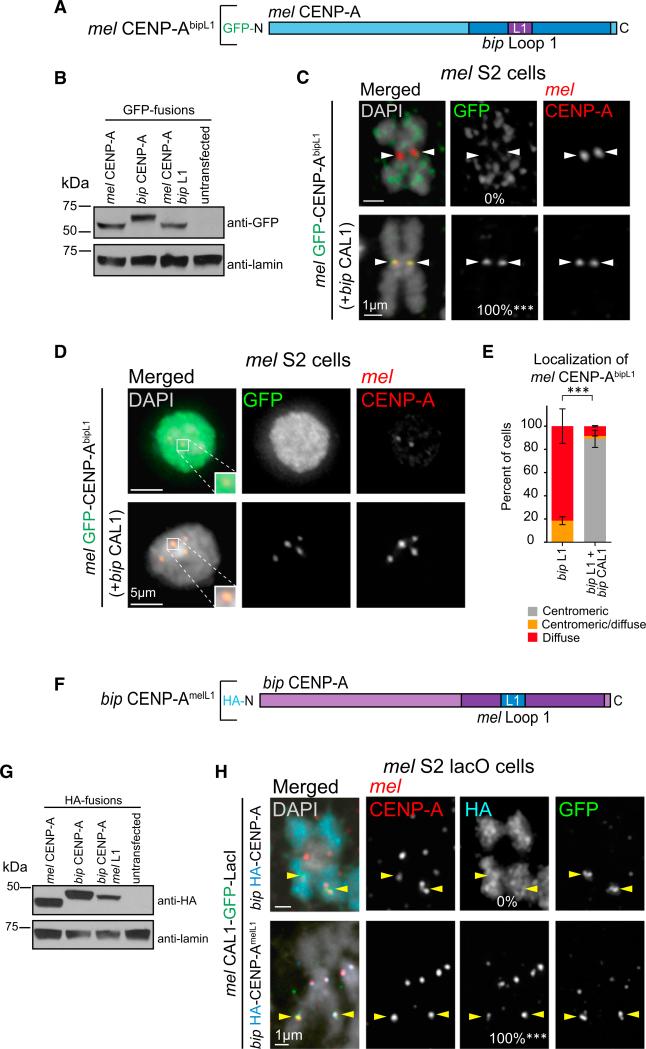
To test this hypothesis, we generated a GFP-tagged mel CENP-A chimera containing L1 from bip CENP-A (mel CENP-AbipL1; Figure 4A) and transiently expressed it in mel S2 cells (Figure 4B) with and without bip CAL1. When expressed alone, mel CENP-AbipL1 is mislocalized in all mitotic chromosome spreads (0% centromeric). However, when mel CENP-AbipL1 is co-expressed with bip CAL1, it becomes centromeric in all spreads (100%; Figure 4C). A similar pattern was observed in interphase cells, where mel CENP-AbipL1 is mislocalized or only partially centromeric (82% and 19% of cells, respectively) when expressed alone, but becomes fully centromeric when co-expressed with bip CAL1 (90%; Figures 4D and 4E). Thus, the mis-localization of the mel CENP-AbipL1 chimera is the result of some sort of dysfunction occurring within the bip CENP-A L1 and the mel CAL1 complex.
Figure 4. CAL1 Recognizes CENP-A via L1.
(A) Schematic of mel CENP-A construct with bip L1 substituted into the mel HFD (mel GFP-CENP-AbipL1). Blue represents the mel CENP-A protein sequence; bip CENP-A amino acids are indicated in purple.
(B) Western blots of whole-cell extracts showing the expression levels and size of GFP-mel CENP-AbipL1 chimera compared with GFP-mel CENP-A and GFP-bip CENP-A. Top: anti-GFP; bottom: anti-lamin (loading control).
(C) IF images of metaphase spreads from S2 cells transiently expressing GFP-mel CENP-AbipL1 chimera alone, or co-expressed with bip CAL1. DAPI is shown in gray, GFP in green, and mel CENP-A in red. White arrowheads indicate position of the centromere. n = 9 for mel CENP-AbipL1 chimera alone and n = 10 for mel CENP-AbipL1 chimera with bip CAL1. The percentage of cells with centromeric GFP signal is as indicated in the middle column. ***p < 0.0001; Fisher's two-tailed test of cells with compared with cells without bip CAL1. These data were confirmed by two biological replicates (data not shown).
(D) IF images of interphase S2 cells transiently expressing GFP-mel CENP-AbipL1 chimera alone or co-expressed with bip CAL1. DAPI is shown in gray, GFP in green, and mel CENP-A in red. Zoomed panels show representative centromeres with merged colors.
(E) Quantification of the IF shown in D. GFP-CENP-AbipL1 chimera localization was classified as centromeric (gray bars), diffuse (red), or centromeric and diffuse (orange). n = 70 cells quantified for mel CENP-AbipL1 chimera and 83 for mel CENP-AbipL1 chimera with bip CAL1. Error bars denote the SD of three biological replicates. ***p < 0.0001; Fisher's two-tailed test comparing cells with and without bip CAL1.
(F) Schematic of bip CENP-A construct with mel L1 substituted into the HFD (bip CENP-AmelL1; HA-tagged). mel CENP-A residues are represented in blue and bip CENP-A amino acids in purple. HFDs here and in (A) are shown in darker shades of the respective colors.
(G) Western blots of whole-cell extracts showing the expression levels and of the bip HA-CENP-AmelL1 chimera compared with mel HA-CENP-A and bip HA-CENP-A. Top: anti-HA; bottom: anti-lamin (loading control).
(H) IF images of metaphase chromosome spreads from mel lacO S2 cells transiently expressing mel CAL1-GFP-LacI (green) and HA-tagged bip CENP-A or bip CENP-AmelL1 chimera (aqua). Endogenous mel CENP-A is shown in red, DAPI in gray. Yellow arrowheads indicate the position lacO array. n = 20 for HA-tagged bip CENP-A and 16 for HA bip CENP-AmelL1 chimera. The recruitment efficiency to the lacO site for each HA-tagged construct is indicated at the bottom of the HA panel. ***p < 0.0001; Fisher's two-tailed test. These results were confirmed by two biological replicates (data not shown).
If L1 is critical for the function of CENP-A and CAL1 complexes, the recruitment of bip CENP-A to the lacO site is expected to be restored if L1 from bip CENP-A is replaced with L1 from mel CENP-A (bip CENP-AmelL1 chimera; Figure 4F) (Vermaak et al., 2002). To test this prediction, we transiently transfected mel CAL1-GFP-LacI and HA-tagged bip CENP-AmelL1 in S2 lacO cells (Figure 4G), and assessed recruitment to the lacO site by IF on metaphase spreads. In agreement with previous data (Vermaak et al., 2002), bip CENP-AmelL1 chimera localizes to mel centromeres. Furthermore, mel CAL1-GFP-LacI recruits bip CENP-AmelL1 to the lacO array with the same efficiency as mel CENP-A (100%; Figure 4H) (Chen et al., 2014). These data demonstrate that the centromeric localization gained by the addition of mel L1 to bip CENP-A is a result of its restored ability to be incorporated into chromatin by mel CAL1.
Identification of CAL1 Residues Co-evolving with CENP-A L1
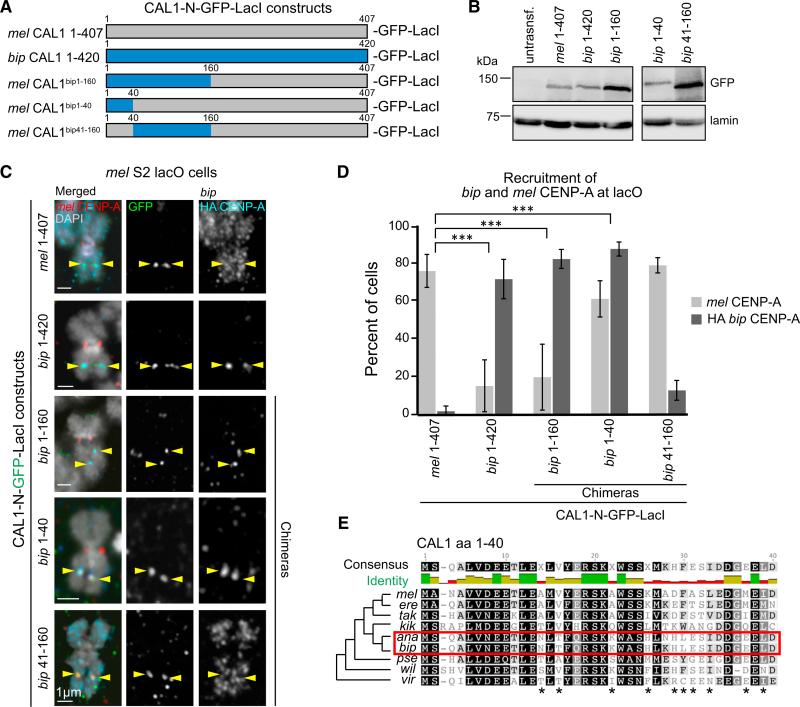
Having determined that the divergence between the L1 of mel and bip CENP-A leads to defective centromeric deposition of bip CENP-A by mel CAL1, we sought to identify the corresponding regions of bip CAL1 that may have adaptively evolved with bip CENP-A. Such a region within bip CAL1 could confer mel CAL1 the ability to deposit bip CENP-A if introduced through amino acid swap experiments. Because the CENP-A interaction domain of CAL1 lies within its N terminus (mel residues 1–407 [Chen et al., 2014; Schittenhelm et al., 2010]; corresponding to 1–420 in bip), we focused on this region to create CAL1 N-terminal bip-mel chimeras (Figures 5A and 5B) and interrogated their competency for bip CENP-A recruitment at the lacO site.
Figure 5. Identification of CAL1 Residues Co-evolving with CENP-A L1.
(A) Schematic of N-terminal CAL1-GFP-LacI constructs. For chimeras, gray indicates mel CAL1 and blue indicates bip CAL1 proteins.
(B) Western blots of whole-cell extracts showing the expression and sizes of the N-CAL1-GFP-LacI constructs used in these experiments (top: anti-GFP; bottom: anti-lamin loading control).
(C) IF images of metaphase chromosome spreads from mel lacO S2 cells transiently expressing the indicated N-CAL1-GFP-LacI constructs from (A), along with bip HA-CENP-A. GFP is shown in green, HA in aqua, mel CENP-A in red, and DAPI in gray. Yellow arrowheads indicate the position of the lacO array. Note that since the N terminus of CAL1 alone cannot localize to centromeres (Chen et al., 2014), these constructs are not expected to deposit bip CENP-A at the endogenous centromere.
(D) Quantification of the IF shown in (C). Error bars represent the SD of three biological replicates. n = 160 spreads for mel CAL1 1–407, 82 for bip CAL1 1–420, 127 for mel CAL1bip1–160, 127 for mel CAL1bip1–40, and 91 for mel CAL1bip41–160. ***p < 0.0001 when comparing mel 1–407 CAL1 recruitment of bip CENP-A with that of bip CAL1 1–420, mel CAL1bip1–160, or mel CAL1bip1–40 (Fisher's two-tailed test).
(E) BLOSUM80 alignment of residues 1–40 of CAL1 from selected species. Shading indicates percent similarity based on the BLOSUM80 score matrix (Henikoff and Henikoff, 1992). Black, 100% similar; dark gray, 80%–100% similar; light gray, 80%–60% similar; white, less than 60% similar. Stars indicate residues that have diverged in the ananassae subgroup (red box) compared with the rest of the melanogaster group and thus are candidates for residues co-evolving with CENP-A L1. Consensus sequence is shown above the alignment. Percent identity is shown as a bar graph below the consensus sequence: green indicates highly conserved, gold indicates somewhat conserved, and red indicates unconserved.
Residues 1–160 of mel CAL1 are sufficient for CENP-A nucleosome assembly in vitro (Chen et al., 2014). Therefore, we created an N-terminal CAL1 (1–407) chimera where the first 160 residues of mel CAL1 were replaced by the homologous region of bip CAL1 (mel CAL1bip1–160; Figure 5A) fused to GFP LacI, and determined whether this construct was able to recruit bip CENP-A to the lacO site. After induction of mel CAL1bip1–160-GFP-LacI in lacO cells co-expressing bip HA-CENP-A, IF on metaphase spreads was performed with anti-HA, anti-GFP, and anti-mel CENP-A antibodies (Figure 5C). We found that mel CAL1bip1–160-GFP-LacI successfully recruits bip CENP-A to the lacO site (82%) while it recruits mel CENP-A inefficiently (20%; Figure 5D). These results indicate that replacing the first 160 residues of mel CAL1 with the corresponding region of bip CAL1 is sufficient to enable the incorporation of bip CENP-A into chromatin and that this region is critical for mel CENP-A recruitment. Furthermore, as we previously observed that full-length bip CAL1 can recruit mel CENP-A to the lacO site in approximately 50% of metaphase spreads (Figure 3I), the lower percentage of recruitment of mel CENP-A by the mel CAL1bip1–160 chimera observed here suggests that the full-length bip CAL1 can engage the endogenous centromere/kinetochore assembly pathway, likely via an interaction between its C terminus and mel CENP-C (Chen et al., 2014; Schittenhelm et al., 2010).
CAL1 contains an “Scm3-like” domain at its N terminus (Figure 5E; residues 1–40 [Phansalkar et al., 2012]), which is essential for ectopic CENP-A deposition (Chen et al., 2014). To further narrow down the region of CAL1 required for CENP-A incorporation, we swapped residues 1–40 and 41–160 of mel CAL1 with the corresponding region of bip CAL1 (mel CAL1bip1–40-GFP LacI and mel CAL1bip41–160-GFP-LacI; Figures 5A and 5B). These chimeras were again transiently expressed in S2 lacO cells along with bip HA-CENP-A, followed by IF on metaphase spreads to assess the presence or absence of bip HA-CENP-A at the lacO (Figure 5C).
We found that mel CAL1bip1–40-GFP-LacI successfully recruits both bip CENP-A and mel CENP-A to the lacO (88% and 61%, respectively). In contrast, mel CAL1bip41–160-GFP-LacI does not efficiently recruit bip CENP-A to the lacO site (13%), but still efficiently recruits mel CENP-A (79%; Figure 5D). These results suggest that residues 1–40 of bip CAL1 are co-evolving with bip CENP-A and that the corresponding mel CAL1 residues are responsible for the incompatibility observed between bip CENP-A and mel centromeres (Figures 1 and 5E) (Vermaak et al., 2002). Furthermore, these findings reveal the conservation of CENP-A recognition mechanisms between the non-homologous CAL1 and Scm3/HJURP chaperones, both of which involve the L1 region of CENP-A (Bassett et al., 2012; Cho and Harrison, 2011).
In summary, L1 of CENP-A is evolving adaptively in Drosophila (Malik and Henikoff, 2001) and has diverged in the branch containing the ananassae subgroup (Vermaak et al., 2002). The Scm3-like region of CAL1 (Phansalkar et al., 2012), which is critical for CENP-A recruitment (Chen et al., 2014), recognizes CENP-A through its L1 and co-evolves with it, thereby maintaining its ability to deposit CENP-A in this branch of the phylogeny. The presence of a competent CAL1 assembly factor (bip CAL1 or a mel CAL1bip1–40 chimera) in mel cells is sufficient to deposit bip CENP-A into chromatin (centromeric or otherwise).
DISCUSSION
Our work sheds light on a puzzling observation in centromere biology: that a CENP-A ortholog is unable localize to the centromeres of a relatively close species (Vermaak et al., 2002). What makes this even more surprising is the report that yeast CENP-A/Cse4 can complement CENP-A knockdown in HeLa cells (Wieland et al., 2004) despite billions of years since these two species last shared a common ancestor. Using coIPs and an ectopic tethering system, we show that mel CAL1 can form a complex with bip CENP-A, but this complex is not competent for bip CENP-A deposition. Centromeric targeting of bip CENP-A can be restored upon co-expression of a functional CAL1 partner in both mel and sim cells. Using CENP-A and CAL1 chimeras we demonstrate that for successful CENP-A deposition into chromatin to occur residues 1–40 of CAL1 and CENP-A L1 must be compatible, suggesting that these regions mediate CAL1/CENP-A function.
Given that Drosophila CENP-A L1 is under positive selection (Malik and Henikoff, 2001), one might predict that its binding partner, CAL1, is also adaptively evolving to maintain centro-mere integrity throughout evolution. While we found no evidence of positive selection on CAL1 using standard methods (Phansalkar et al., 2012), the lineage-specific CENP-A/CAL1 compatibility demonstrates that the “Scm3-like” domain of CAL1 is undergoing coordinated protein evolution with CENP-A L1.
Secondary functions of CAL1 may be suppressing its rate of evolution. For example, CAL1 also interacts with the highly conserved FACT complex (Chen et al., 2015) and localizes to the nucleolus (Chen et al., 2012; Lidsky et al., 2013). We hypothesize that the overall CAL1 sequence is under purifying selection (Phansalkar et al., 2012) to preserve its functional interactions with highly conserved partners, while key residues within the N terminus of CAL1 evolve to maintain the functional interaction with CENP-A.
Our experiments focused of the role of L1 in centromere evolution. However, the CENP-A N terminus is also adaptively evolving (Malik et al., 2002). Since our experiments used the full-length bip CENP-A gene, they demonstrate that the divergent N-terminal tail of bip CENP-A does not hinder the ability of bip CENP-A to bind to mel centromeres when bip CAL1 is present, at least in mitosis, challenging the proposal that the N terminus also evolves in conflict with centromeric DNA. However, we cannot exclude the possibility that the adaptive evolution of the CENP-A N terminus may be a contributing factor in modulating the DNA-binding preferences of CENP-A exclusively during meiosis, as the N terminus of CENP-A has been shown to have meiosis-specific functions in Arabidopsis (Lermontova et al., 2006; Ravi and Chan, 2010).
Since we have not directly assayed the CENP-A-associated DNA sequences of any of these Drosophila species, we are unable to completely rule out the divergence of centromeric DNA as a contributing factor in the adaptive evolution of CENP-A L1. Nonetheless, bip CENP-A can localize to both mel and sim centromeres, suggesting that the presence of a functionally compatible CENP-A chaperone is what determines the ability of CENP-A orthologs to be incorporated at the centromeres of both species. Even the more divergent wil CENP-A can localize to mel centromeres in the presence of its CAL1 partner. It is possible that mel, sim, bip, and wil all share the same centromeric sequences. However, such divergent species (spanning 40 million years of evolution), having experienced no changes in centromeric DNA sequences, would go against the fundamental assumption of centromere drive that centromeric satellites are rapidly evolving. Collectively, our data are inconsistent with positive selection of CENP-A L1 affecting its DNA-binding preferences throughout evolution (Malik and Henikoff, 2001; Vermaak et al., 2002).
The question of why CENP-A is rapidly evolving in Drosophila still remains, and experimental evidence that CENP-A evolution is a direct result of conflict with centromeric DNA is lacking. CAL1 is unlikely to drive this rapid evolution, since it is evolving more slowly than CENP-A (Phansalkar et al., 2012). We propose that, in Drosophila, positive selection of CENP-A L1 modulates the efficiency of its centromeric deposition by CAL1 rather than its DNA-binding specificity, as originally proposed (Vermaak et al., 2002). Our analysis of the extreme example of the incompatible bip CENP-A and mel CAL1 suggests that the degree of functional compatibility between these two proteins during intermediate evolutionary times could influence how much CENP-A is incorporated, in turn affecting CENP-C recruitment and kineto-chore assembly (Chen et al., 2014; Erhardt et al., 2008). Thus, the ability to “tune” how much CENP-A is deposited at the centromere via changes in L1 could be a mechanism to curb the increased “kinetochore strength” resulting from centromere satellite expansion during centromere drive (Figure 6), akin to the long-standing model proposed by Henikoff and Malik (Henikoff and Malik, 2002; Malik and Henikoff, 2002). Although our work focuses on the critical role of these co-evolving domains in mitosis, it is important to note that CAL1 is also essential for CENP-A deposition during meiosis (Dunleavy et al., 2012). Therefore it is conceivable that our proposed model would apply to meiosis, the natural battleground of centromere drive.
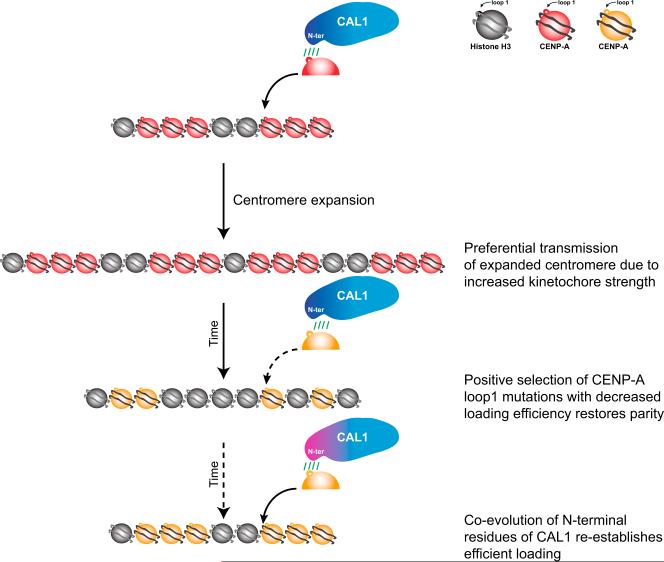
Figure 6. Model for the Co-evolution of CENP-A L1 and CAL1 in the Context of Centromere Drive.
CENP-A centromeric deposition by CAL1 requires compatibility between the CAL1 N terminus and L1 (loop 1) of CENP-A. When centromere expansion occurs as a result of unequal crossover during meiosis, the larger and thus stronger centromere will be preferentially transmitted to the next generation (centromere drive) (Henikoff and Malik, 2002; Malik and Henikoff, 2002). To restore meiotic parity, positive selection of L1 mutations (Malik and Henikoff, 2001) weakens the ability of CAL1 to assemble CENP-A into chromatin, resulting in lower levels of CENP-A being deposited. The N terminus of CAL1 co-evolves, albeit at a slower rate, and re-establishes efficient CENP-A deposition, thereby maintaining centromere identity.
EXPERIMENTAL PROCEDURES
Plasmids
Flies and genomic DNA were obtained from the University of California San Diego Drosophila Species Stock Center or from other laboratories (see Table S1). All non-melanogaster CENP-A and CAL1 orthologs were PCR amplified using Phusion High-Fidelity DNA polymerase (New England Biolabs) from genomic DNA using the primers listed in Table S2. See Supplemental Experimental Procedures for details on cloning.
Cell Culture and Transfections
Drosophila melanogaster Schneider 2 (S2) cells were grown as described previously (Chen et al., 2014; Mellone et al., 2011). S2 cells containing stably integrated LacO arrays (pAFS5 [Straight et al., 1996]) were generated as described previously (Chen et al., 2014; Mendiburo et al., 2011). Drosophila simulans ML82-19a (M-19) cells were purchased from the Drosophila Genomics Resource Center. M-19 cells were grown in Schneider's media with 10% fetal bovine serum at 25°C.
Transient and stable transfections in S2 cells were performed using FuGENE HD Transfection Reagent (Promega) as previously described (Chen et al., 2014). For transient transfection in M-19 cells, 2 × 106 cells were plated in six-well plates and transfected with Cellfectin reagent (Invitrogen) and plasmid DNA. Cells were incubated with the transfection complex in serum-free medium for 3 hr before replacing medium with serum-containing medium. Cells were incubated for 3 days before harvesting for IF.
Metaphase Chromosome Spreads and IF
IF on settled interphase cells and metaphase spreads were performed as previously described (Chen et al., 2014). Primary antibodies: anti-CENP-A (chicken, 1:1,500; Blower and Karpen, 2001) or anti-CID (rabbit, 1:500; Abcam), anti-CENP-C (guinea pig, 1:500; Erhardt et al., 2008), anti-Ndc80 (chicken, 1:200; Cane et al., 2013), anti-GFP Alexa 488-conjugated (rabbit, 1:100; Invitrogen), or anti-GFP (chicken, 1:500; Abcam), and anti-HA (mouse, 1:500; Covance).
For salt extractions, settled cells were incubated with PBS-D (0.1% digitonin) with or without 0.5 M NaCl for 30 min (Perpelescu et al., 2009) before 37% formaldehyde was added to the solution to a final concentration of 3.7% followed by 10 min of incubation before proceeding with IF.
Imaging
Images were acquired on a wide-field fluorescence microscope (PersonalDV; GE Healthcare) equipped with a 60Å~/1.42 NA or a 100 Å~/1.40 NA oil-immersion objective (Olympus) and a CoolSnap HQ2 camera (Photometrics), keeping exposure conditions constant between all samples. Images were acquired and processed in softWoRx (Applied Precision), maintaining the scaling constant between samples, and saved as PSD files. Figures were assembled in Adobe Illustrator. For quantification, see Supplemental Experimental Procedures.
Western Blots and IPs
Whole-cell lysates and western blots were prepared as previously described (Chen et al., 2014). Membranes were incubated with either anti-GFP (Goat, 1:150; Rockland), anti-CAL1 (rabbit, 1:000; gift from Aaron Straight), anti-HA (mouse, 1:500; Covance), anti-tubulin (mouse, 1:500; Sigma-Aldrich), anti-fibrillarin (mouse, 1:1,000; Cytoskeleton), or anti-lamin (mouse, 1:1,000; Hybridoma Bank, University of Iowa) primary antibodies. Blots were imaged on an Odyssey Fc (LI-COR Biosciences) using chemiluminescent substrate for detection of horseradish peroxidase-labeled secondary antibodies, or were developed on X-ray films.
IPs were performed from nuclear extracts as previously described (Chen et al., 2012), using 5 μg of anti-CAL1 antibody or 5 μg of anti-immunoglobulin G antibody. For normalization, whole-cell lysates were prepared from 1 × 106 cells expressing either mel or bip GFP-CENP-A, and total GFP protein levels were quantified by western blotting using Image Studio software (LI-COR) and normalized compared with a loading control (lamin). Nuclear extracts were performed from the same cells and were diluted in resuspension buffer (0.29 M sucrose, 0.5 mM Tris-HCl [pH 7.4], 1.5 mM NaCl, 5 mM MgCl2, 1 mM EGTA, 0.04% Triton X-100, 1× EDTA-free protease inhibitors, and 1 mM DTT) so that the levels of GFP-CENP-A in all samples were equal. 150 μl of diluted bip or undiluted mel nuclear extract were loaded onto antibody-conjugated beads for IP. See Supplemental Experimental Procedures for quantification of IPs.
Supplementary Material
Highlights.
Rapidly evolving CENP-A loop 1 displays species-specific centromere incompatibility
The incompatibility reflects a mismatch between CENP-A and its assembly factor CAL1
The N terminus of CAL1 mediates CENP-A assembly in a lineage-specific manner
Compatible CENP-A loop 1 and CAL1 N terminus are critical for CENP-A deposition
ACKNOWLEDGMENTS
We thank Gary Karpen, Aaron Straight, Patrick Heun, and Tom Maresca for re-agents; Rachel O'Neill and Eric Joyce for critically reading the manuscript; Jason Palladino and Chin-Chi Chen for discussions and suggestions; Harmit Malik for fly genomic DNA; Doris Bachtrog, Rich Meisel, and Andy Clark for flies; Patrick Lenehan, Chin-Chi Chen, and Ankita Chavan for technical help; and Fly-base, the UCSD Stock Center, and the DGRC for fly stocks and other Drosophila resources. L.R. was supported by NSF award MCB1330667; B.G.M. was supported by NSF award MCB1330667 and NIH award GM108829.
Footnotes
AUTHOR CONTRIBUTIONS
B.G.M. and L.R. conceived the project; L.R. conducted experiments; B.G.M. and L.R. wrote the manuscript.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2016.03.021.
REFERENCES
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centro-mere assembly. Dev. Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R, Sánchez P, Rivera T, Rodríguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, et al. Xenopus HJURP and condensin II are required for CENP-A assembly. J. Cell Biol. 2011;192:569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kineto-chore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Cane S, Ye AA, Luks-Morgan SJ, Maresca TJ. Elevated polar ejection forces stabilize kinetochore-microtubule attachments. J. Cell Biol. 2013;200:203–218. doi: 10.1083/jcb.201211119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Greene E, Bowers SR, Mellone BG. A role for the CAL1-partner Modulo in centromere integrity and accurate chromosome segregation in Drosophila. PLoS One. 2012;7:e45094. doi: 10.1371/journal.pone.0045094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG. CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 2014;204:313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Bowers S, Lipinszki Z, Palladino J, Trusiak S, Bettini E, Rosin L, Przewloka MR, Glover DM, O'Neill RJ, et al. Establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev. Cell. 2015;34:73–84. doi: 10.1016/j.devcel.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U-S, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl. Acad. Sci. USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH. Centromerization. Trends Cell Biol. 2000;10:182–188. doi: 10.1016/s0962-8924(00)01739-6. [DOI] [PubMed] [Google Scholar]
- Cooper JL, Henikoff S. Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 2004;21:1712–1718. doi: 10.1093/molbev/msh179. [DOI] [PubMed] [Google Scholar]
- Daniel A. Distortion of female meiotic segregation and reduced male fertility in human Robertsonian translocations: consistent with the centromere model of co-evolving centromere DNA/centromeric histone (CENP-A). Am. J. Med. Genet. 2002;111:450–452. doi: 10.1002/ajmg.10618. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Beier NL, Gorgescu W, Tang J, Costes SV, Karpen GH. The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol. 2012;10:e1001460. doi: 10.1371/journal.pbio.1001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finseth FR, Dong Y, Saunders A, Fishman L. Duplication and adaptive evolution of a key centromeric protein in mimulus, a genus with female meiotic drive. Mol. Biol. Evol. 2015;32:2694–2706. doi: 10.1093/molbev/msv145. [DOI] [PubMed] [Google Scholar]
- Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- Fishman L, Willis JH. A novel meiotic drive locus almost completely distorts segregation in mimulus (monkeyflower) hybrids. Genetics. 2005;169:347–353. doi: 10.1534/genetics.104.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Malik HS. Centromeres: selfish drivers. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell. 2006;18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidsky PV, Sprenger F, Lehner CF. Distinct modes of centro-mere protein dynamics during cell cycle progression in Drosophila S2R+ cells. J. Cell Sci. 2013;126:4782–4793. doi: 10.1242/jcs.134122. [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Malik HS. The centromere-drive hypothesis: a simple basis for centro-mere complexity. Prog. Mol. Subcell. Biol. 2009;48:33–52. doi: 10.1007/978-3-642-00182-6_2. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Adaptive evolution of Cid, a centromerespecific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Conflict begets complexity: the evolution of centromeres. Curr. Opin. Genet. Dev. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- Malik HS, Vermaak D, Henikoff S. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA. 2002;99:1449–1454. doi: 10.1073/pnas.032664299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, Sebra R, Peluso P, Eid J, Rank D, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centro-mere evolution. Genome Biol. 2013;14:R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centro-mere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O, Medina-Giró S, Torras-Llort M, Azorín F. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3CID. Curr. Biol. 2011;21:1488–1493. doi: 10.1016/j.cub.2011.07.041. [DOI] [PubMed] [Google Scholar]
- Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat. Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Female meiosis drives karyotypic evolution in mammals. Genetics. 2001;159:1179–1189. doi: 10.1093/genetics/159.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phansalkar R, Lapierre P, Mellone BG. Evolutionary insights into the role of the essential centromere protein CAL1 in Drosophila. Chromosome Res. 2012;20:493–504. doi: 10.1007/s10577-012-9299-7. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JKR, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M, Chan SWL. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–618. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- Ravi M, Kwong PN, Menorca RMG, Valencia JT, Ramahi JS, Stewart JL, Tran RK, Sundaresan V, Comai L, Chan SW-L. The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics. 2010;186:461–471. doi: 10.1534/genetics.110.120337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm RB, Althoff F, Heidmann S, Lehner CF. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J. Cell Sci. 2010;123:3768–3779. doi: 10.1242/jcs.067934. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Comparative Sequencing Program NISC, Green ED. Adaptive evolution of foundation kinetochore proteins in primates. Mol. Biol. Evol. 2010;27:1585–1597. doi: 10.1093/molbev/msq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr. Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Kurumizaka H. Comparison between the CENP-A and histone H3 structures in nucleosomes. Nucl. Austin Tex. 2012;3:6–11. doi: 10.4161/nucl.18372. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of CID. Mol. Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol. Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach A, Borodin P, Hausser J. Meiotic drive favors Robertsonian metacentric chromosomes in the common shrew (Sorex araneus, Insectivora, mammalia). Cytogenet. Cell Genet. 1998;83:199–206. doi: 10.1159/000015178. [DOI] [PubMed] [Google Scholar]
- Zedek F, Bureš P. Evidence for centromere drive in the holocentric chromosomes of Caenorhabditis. PLoS One. 2012;7:e30496. doi: 10.1371/journal.pone.0030496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.