Abstract
Purpose
In radiotherapy, PET images can be used to guide the delivery of selectively escalated doses to biologically relevant tumour subvolumes. Validation of PET for such applications requires demonstration of spatial coincidence between PET tracer uptake pattern and the histopathologically confirmed target. This study introduces a novel approach to histopathological validation of PET image segmentation for radiotherapy guidance.
Methods and materials
Sequential tissue sections from surgically excised whole-tumour specimens were used to acquire full 3D-sets of both histopathological images (microscopy) and PET tracer distribution images (autoradiography). After these datasets were accurately registered, a full 3D autoradiographic distribution of PET tracer was reconstructed and used to obtain synthetic PET images (sPET) by simulating the image deterioration induced by processes involved in PET image formation. To illustrate the method, sPET images were used in this study to investigate spatial coincidence between high FDG uptake areas and the distribution of viable tissue in two small animal tumour models.
Results
The reconstructed 3D autoradiographic distribution of the PET tracer was spatially coherent, as indicated by the high average value of the pised pixel-by-pixel correlation of intensities between successive slices (0.84 ± 0.05 and 0.94 ± 0.02). The loss of detail in the sPET images versus the 3D autoradiography was significant as indicated by Dice coefficient values corresponding to the two tumours (0 and 0.1 at 70% threshold). The maximum overlap between the FDG segmented volumes and the extent of the viable tissue as indicated by Dice coefficient values, was 0.8 for one tumour (for the image thresholded at 22% of max intensity) and 0.88 for the other (threshold of 14% of max intensity).
Conclusion
It was demonstrated that the use of synthetic PET images for histopathological validation allows for bypassing a technically challenging and error-prone step of registering non-invasive PET images with histopathology.
Keywords: PET, Histopathological validation, Small-animal imaging, Image-guidance
Positron emission tomography (PET) enables visualisation of tumour microenvironment using radiolabelled biomarker probes. Based on the hypothesis that by segmentation of PET images one can determine the spatial pattern of radioresistant or aggressive subpopulations of tumour cells, these images could be used to guide the delivery of escalated radiation doses with intensity-modulated radiation therapy [1]. The best spatial resolution of PET imaging is about 4–5 mm for clinical whole-body scanners, while for small-animal PET scanners it can approach 1.5 mm [2]. However, the chaotic morphology of cancer tissue results in spatial variations of microenvironment and relevant biological characteristics on a much smaller scale (<250–300 μm) as indicated by histopathology studies [3–7]. When PET imaging is used to guide the delivery of radiotherapy, inherent averaging effects can significantly alter the resulting images [8–10], making it necessary to demonstrate the spatial coincidence between the pattern of tracer uptake as seen in PET images and the pattern of the underlying biological parameter of interest. Therefore, for any PET tracer or segmentation method, validation of PET imaging for image guidance in radiotherapy requires an approach that allows one to test spatial colocalisation of the regions of high tracer uptake, as segmented from PET images, with the spatial distribution of the biological/ morphological features of interest.
One of the standard methods for validating PET image segmentation is histopathological validation. This type of validation can be carried out by registering non-invasive PET images to ex vivo images of histopathological specimens, as to enable the analysis of the spatial association between these images. Unfortunately, the registration of these images is frequently performed in a subjective manner due to lack of common landmarks identifiable on both PET and histopathological images. Furthermore, significant deformation of the tumour specimen following surgical excision can make such registration not feasible [11].
In this study, a new approach to histopathological validation of PET image segmentation, specifically designed for radiotherapy image-guidance applications is presented. Specifically, it is proposed to use excised tumour specimens to acquire both histopathological imaging data, and the distribution of the PET tracer using autoradiography. As demonstrated earlier [12], these datasets can be accurately registered. Autoradiography data can be used to reconstruct 3D distribution of the PET tracer in the tissue and then obtain synthetic PET images (sPET) simulating non-invasive PET images representative of the underlying intratumoural distribution of the PET tracer and the imaging characteristics of the scanner [13].
The use of sPET images for histopathological validation circumvents problems associated with registering in vivo PET images with ex vivo histopathological images. In addition, the natural 3D heterogeneity of the histopathology-based PET creates realistic scenarios for the segmentation algorithms, more so than standard phantom studies. This is an evolution of the two-dimensional analysis of autoradiography versus microscopy studies which is important in determining the specificity of the tracer uptake [14]. Furthermore, the presented approach utilises a large portion of the tumour, eliminating some of the sampling bias that may affect two dimensional studies.
In this study, we demonstrate this approach using a proof-of-principle experiment aimed at validating FDG PET for imaging viable tumour subvolumes [15], by testing the spatial correspondence between regions of high FDG uptake segmented in the sPET image and the viable tissue distribution in a small animal tumour model. However, it should be stressed here that the choice of PET tracer, tumour model, and segmentation algorithm was primarily made with the goal of emphasising the methodology.
Materials and methods
Experimental setup
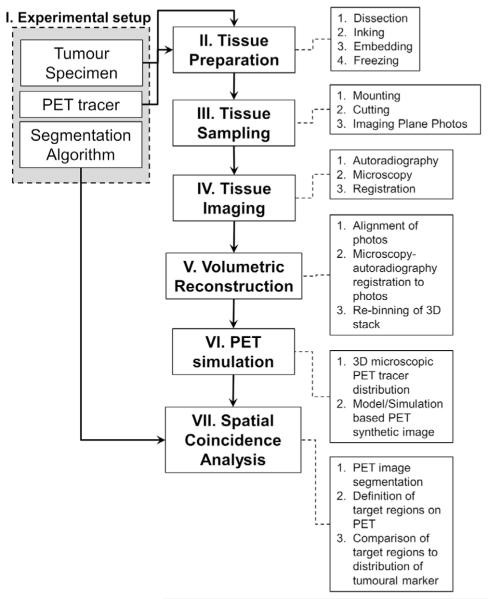
For the proposed approach to histopathological validation of segmentation of PET imaging for defining targets of biological interest, there are three main components defining each validation study: (1) tumour specimen; (2) PET tracer; (3) segmentation algorithm. The methodology presented herein was conceived to adapt to the use of any human or animal tumour resection specimen, any PET tracer that can be used for autoradiography, and any segmentation algorithm. A detailed workflow of the general procedures included in this methodology is presented in Fig. 1.
Fig. 1.
Proposed validation workflow.
Tumour specimen
Two human head and neck tumour xenografts (SQ20B) were used. The xenografts were developed in two different male athymic mice (NCr-nu/nu; National Cancer Institute), from subcutaneous flank inoculation of 106 cells/site. Animals were maintained according to the protocol approved by the Institutional Animal Care and Use Committee at the Virginia Commonwealth University.
PET tracer
14C-labelled 2-Deoxy-2-fluoro-d-glucose (American Radiolabeled Chemicals, Inc.) was used as a tracer under investigation. This radiolabelled compound was synthetised by the manufacturer to be chemically identical to the clinically utilised 18F-FDG.
Segmentation algorithm
Image intensity-based thresholding was utilised in this study as an example of segmentation technique that can be employed to determine high uptake areas of the tumour.
Tissue harvesting and preparation
After the tumours reached 1 cm in the largest diameter as measured by calliper, the animals were anaesthetised with a mixture of oxygen and isoflurane. At 80 min before planned animal euthanasia, a bolus injection containing Hoechst 33342 (Sigma–Aldrich) 0.4 mg/20-g mouse, and 925 kBq of 14C-FDG was administered by tailvein injection. After animal sacrifice, tumours were immediately excised. The entire surface of the tumour specimen was coated with Davidson Marking System black ink (Bradley Products, Inc.). Thereafter, the specimen was embedded in Tissue-Tek OCT (Sakura Finetek).
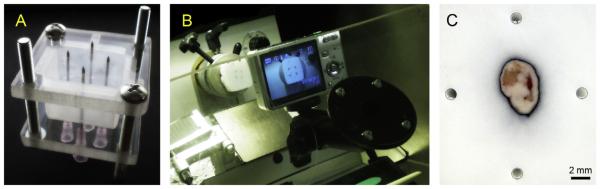
For embedding, a custom-fabricated jig was used (Fig. 2A), which was similar to a previously reported prototype [16]. Specifically, 4 parallel holes were drilled in two rigid plastic plates (6 mm thickness). Their spacing and diameter were chosen such that they would fit 18G needles (BD Biosciences) in a way as to allow a tumour specimen to fit in between the parallel needles. The needles were first inserted into the bottom plastic plate, then through a disposable plastic mould used for specimen freezing. After the excision, the tumour specimen was placed between the needles in the mould containing the OCT. The upper plastic plate was placed on top of the mould, allowing the needles to go through. After the assembly was tightened with bolts, it was placed on dry ice for freezing. When the OCT solidified, the needles were retracted and the tumour blocks were sectioned with a Leica CM1850 UV cryostat (Leica Microsystems) at 8-μm tissue section thickness and mounted on glass microscope slides.
Fig. 2.
(A)Freezing jig for tissue specimen. (B) Cryostat setup with mounted camera on double-jointed adaptor. (C) Example of the tissue specimen digital photo with hollow needle tracks as registration fiducials. The tissue was linked to increase the contrast of the tissue boundary.
Tissue sampling
During cutting, every 120 μm throughout the specimen a stack of 4 consecutive tissue sections was collected. Prior to cutting these 4 sections, a digital image of the cutting plane was acquired utilising a Pentax Optio S7 digital camera with a remote trigger. The camera was mounted on a custom-built plastic frame that fitted the cryostat work-window (Fig. 2B). The double ball-in-socket adjustable locking mount (Ram Mounts Cat# RAM-B-202AU, RAM-B-201U, RAM-B-202U, National Products, Inc.) allowed for an accurate alignment of the camera optical axis with the motion axis of the specimen holder, perpendicular to the cutting plane. Once camera position was selected, the joints of the mount were locked in place, ensuring the reproducibility of the position from which the digital image of the tissue block was taken (Fig. 2C).
Tissue imaging
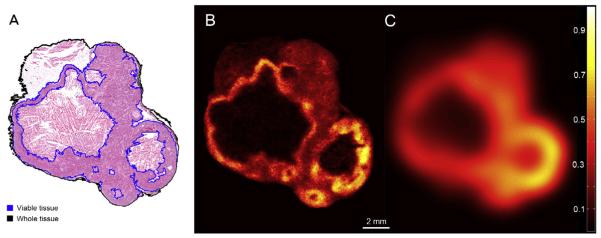
One section from each stack of consecutive tissue sections obtained at every sampling location was used for autoradiography. These sections were covered with a thin plastic wrap and placed in contact with the phosphor plates in light-tight cassettes for 5 days at room temperature. Thereafter, the imaging plates were read with a Typhoon 9410 Variable Mode Imager (GE Healthcare) to obtain images of the FDG intratumoural distribution (B). The reader output was digitised in grayscale TIFF images with 25 μm pixel size. The grayscale intensities were converted to activity concentrations based on the images of 14C standards that were co-exposed with the tissue sections on the same imaging plates (Fig. 3B).
Fig. 3.
(A) Microscopy image of haematoxylin and eosin staining. Image was acquired at 20× using Olympus BX61 microscope equipped with a motorised stage. (B) Autoradiography image. (C) Simulated PET image slice at the same location. Both B and C images were pised to the maximum activity value in the whole tumour. All images are shown on the same scale. All images were obtained at the same tumour location. Microscopy image was obtained from tissue section adjacent to the one used for autoradiography.
Tissue sections adjacent to the ones used for autoradiography were utilised to image the other relevant aspects of tumour morphology and/or microenvironment. For this study Meyer’s haematoxylin and eosin (H&E) staining was performed. Tissue characterised by intact cellular structure as seen in H&E images was considered viable. The H&E microscopy images were deformably registered to the corresponding autoradiography images using previously described methods, in this case utilising Hoechst images of blood flow [12]. The registered H&E images were utilised to segment the extent of the tumour tissue (whole tissue mask) and the subset corresponding to the viable tumour tissue (viable tissue mask) (Fig. 3A). The results of the viable tissue segmentation in selected images were reviewed by a pathologist to confirm validity. Any pathologist-validated method of delineating the viable tissue and/or tissue outline can be used.
3D reconstruction
In order to reconstruct the 3D spatial distribution of the intratumoural FDG uptake along with the 3D distribution of the viable tissue, a two-step process was utilised. In the first step, the digital photo images of the cutting plane containing the inked tissue specimen (Fig. 2C) were rigidly registered using the visible needle tracks as corresponding reference points. Specifically, the top tissue section was considered the reference image and the rest of the section images were rigidly registered to it by aligning the needle tracks. In the second step, the pairs of already registered autoradiography images and H&E-based tissue masks from each sampling location were registered to their corresponding aligned digital photos (obtained from the same location in the tumour) using tissue boundaries visible on both photographic and H&E images.
Both registration steps were implemented in MATLAB R2010a (MathWorks) using the rigid and non-rigid applications of the coherent point drift algorithm available for research use. The coherent point drift algorithm is a probabilistic point set registration algorithm, and it has been shown to produce accurate transformations for 2D and 3D problems, even in the case of missing data, poor initialisation, and in the presence of noise. No specific correspondences need to be defined between the point sets. The algorithm aligns two sets of points by creating a coherent transformation of the secondary point set to the primary point set. For rigid transformations, coherence is established by reparametrising the location of each point of the secondary set using rigid parameters and constraining the scale ratios. In the non-rigid case, the obtained deformable vector field is regularised with an elastic model [17].
A contour-based deformable registration was used for the second step of the registration process. Therein, the utilised point sets used in the coherent point drift algorithm corresponded to the contours obtained from the inked tissue boundaries, which remained intact during tissue section transfer onto the glass slide. Using intensity-based image segmentation, the primary contours (obtained from the digital photographs) were obtained by taking advantage of the high-contrast between the black ink and the white OCT background. The secondary contours were obtained from the H&E-based whole tissue mask by choosing the perimeter pixels of the mask. A deformable transformation was obtained by registering the secondary contours (H&E) to the primary contours (photographs). This transformation was then applied to the autoradiography and viable tissue mask, already registered to the H&E image.
At the end of the registration process, all images were stacked into three-dimensional matrices, and re-binned at 50 × 50 × 120 μm per voxel utilising the mean values, in order to eliminate any residual misalignments. The resulting 3D autoradiography data set was analysed for three-dimensional consistency.
Consistency was defined in terms of smoothness of the signal spatial distribution between consecutive slices [18]. Specifically, a pised correlation coefficient was calculated between each pair of consecutive slices, and the mean and standard deviation of the correlation coefficients was reported as a measure of the 3D consistency of the reconstructed volumes.
PET simulation
In this study, the simulated PET (sPET) image was obtained by convolving 3D autoradiographic data sets with a kernel representing 3D centre field-of-view point spread function of the Siemens Inveon small-animal PET scanner (Siemens Medical Solutions, Inc.) using MATLAB R2010a (MathWorks). The point spread function was obtained utilising a custom-built cubic water-equivalent phantom containing a line source. The line source was a mouse tail-vein catheter (Braintree Scientific, Inc.), with an inner diameter of 0.17 mm, filled with 26.6 MBq/ml of 18F solution. The PET scan setup was replicated according to NEMA recommendations for spatial resolution measurements [19]. The point spread function in each dimension was derived from the Gaussian fits of the line profiles [20]. The results obtained in-house were consistent with previously published data [2,21].
Spatial coincidence analysis
As an illustration of the proposed analysis methodology, the spatial coincidence between the areas of high FDG uptake in the sPET images and the spatial distribution of the viable tissue, as defined based on H&E segmentation (Fig. 3A), was analysed utilising overlap analysis described in a previous work [8]. In summary, for each unique activity value found inside the volume defined by the whole tumour mask, the sPET image was thresholded obtaining a distinct segmentation 3D volume. These 3D volumes were compared against the defined region of biological interest: the H&E-based viable tissue delineation (ground truth), and different intersection sets were obtained: true positive, true negative, false positive and true negative sets [22]. Based on these, Dice coefficient values were calculated to determine the spatial overlap between segmented volumes from sPET images and the viable tumour tissue at each threshold value. Receiver-operating-characteristic (ROC) curves were utilised to evaluate the overall performance of segmentation of FDG sPET image at classifying tissue voxels as viable or non-viable. The area under the curve was used as a measure of how well thresholding of FDG PET can potentially determine the spatial extent of the tumour viable tissue.
Results
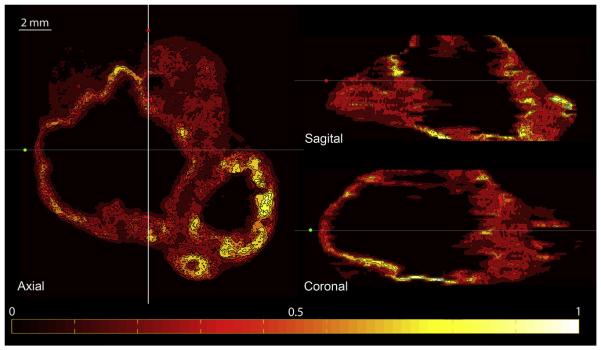
Fig. 4 depicts orthogonal slices though the reconstructed 3D FDG autoradiography image set, confirming its three-dimensional coherence. It was also confirmed by an analytic measure of 3D consistency of the analysed sets: there was high correlation of the pised intensity values between pairs of successive slices in the reconstructed autoradiography volume. The mean and standard deviation for the two tumours were 0.84 ± 0.05 (range: 0.73– 0.91) and 0.94 ± 0.02 (0.86–0.97).
Fig. 4.
Orthogonal slices through the reconstructed 3D FDG autoradiography image set for one of the tumours. All images are pised to the maximum tumour activity registered in the aligned autoradiography stack. Colour bar is linear and it indicates [0, 1] range. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The 3D Gaussian kernel representative of the point spread function as measured at the centre of the scanner’s field of view has the following dimensions: 1.56 mm tangential FWHM, 1.62 mm radial FWHM, and 2.12 mm axial FWHM. Fig. 3C presents a slice through the resulting sPET image set at the location corresponding to the autoradiography image shown in Fig. 3B. The heterogeneity of the tracer uptake as seen in sPET image is significantly different from that seen in autoradiography, as indicated by the overlap between the volumes obtained by thresholding of sPET data set and the actual activity map reconstructed from autoradiography: 0.0 and 0.1 (70% threshold) for tumour A and B, respectively, 0.02 and 0.34 (50% threshold), 0.28 and 0.47 (30% threshold), 0.87 and 0.79 (10% threshold). The difference in volumetric heterogeneity between sPET and 3D autoradiography is further illustrated in Fig. 5, which shows 3D rendering of thresholded volumes on the different data sets.
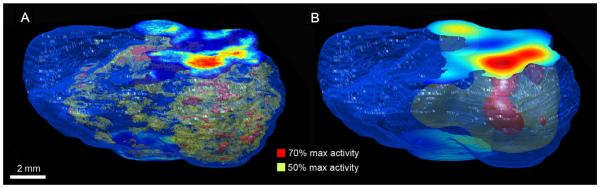
Fig. 5.
Rendered tissue outline with volumes segmented from the FDG images: (A) 3D reconstructed autoradiography; (B) sPET. Threshold values represent % of maximum intensity.
Only to illustrate the proposed validation method, we present the results of the experiment testing thresholding of FDG PET images as a way of viable tumour tissue delineation. Fig. 5B depicts the distinct tumour subvolumes resulting from sPET image segmentation performed using different threshold values. To evaluate the classification performance of the thresholding operation performed on FDG sPET images as a method of viable tissue delineation, the area under the ROC curve (AUC) was computed. The results show that FDG segmentation is a good classifier for voxels containing viable tissue (0.74, 0.81). These high AUC values confirm that the higher FDG uptake values are spatially co-localised with regions of viable tissue within the tumour.
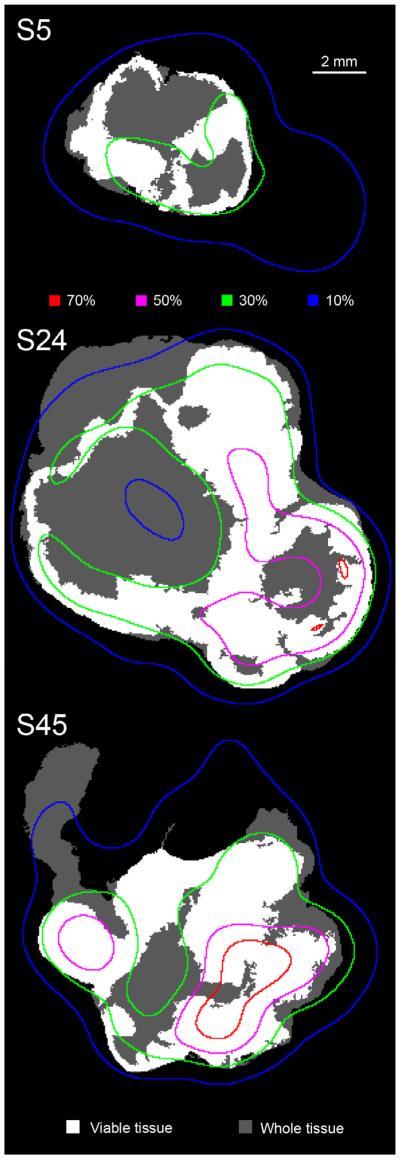
The maximum overlap between the FDG segmented volumes and the extent of the viable tissue as indicated by Dice coefficient values, was 0.8 for one tumour (at 22% of max intensity) and 0.88 for the other (at 14% of max intensity). FDG segmentation was highly sensitive at determining the extent of the viable tissue at these thresholds (true positive fraction: 0.94, 0.99), but it was not very specific (true negative fraction: 0.17, 0.36). Fig. 6 presents a sequence of viable tissue masks from one of the tumours, with overlaid isointensity lines obtained from the corresponding sPET image set.
Fig. 6.
Three sections through tumour B at different levels (tissue section index is labelled), presenting viable tumour tissue (white) as a subset of the whole observable tissue (grey). The coloured lines represent iso-intensity levels from the sPET image sequence. Thresholds are given as a percent of the maximum intensity. All images are shown on the same scale. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Discussion
The purpose of this study is to introduce a new approach to histopathological validation of PET imaging for image-guidance in radiotherapy. The presented method of histopathological validation of PET imaging for image-guidance in radiotherapy does not require registration of in vivo PET data with histopathological images, a step prone to error and subjectivity. Instead, the whole tumour specimen is used to obtain both histopathological data and the intratumoural distribution of the tracer, which is subsequently used to generate a synthetic PET image. In this manner, many confounding factors associated with the registration of PET images to histopathological images (tissue deformation post-excision) are bypassed, since they affect the entire specimen before the sPET image is created. The only registration errors that can potentially affect the spatial colocalisation analysis are the residual misalignments between autoradiography and microscopy images obtained at each tumour sampling location. Using previously reported methods, these misalignments induced by tissue distortion due to freezing/fixation, and also by cutting and mounting on glass slides, can be reduced to under 45 μm [12]. While larger tissue specimens inherently create more complicated logistics when it comes to obtaining images of the entire tumour, solutions exist to mediate these situations [23].
By studying the problem from the bottom-up, the experimental inaccuracies, such as the uncertainty associated with registration of non-invasive PET images with histopathological images, that undermines the accuracy of the data are minimised/eliminated. By combining a series of available techniques, we envisioned creating an infrastructure which can facilitate various types of studies investigating spatial colocalisation of intratumoural tracer uptake pattern as seen in PET images with any biological features of interest that can be identified in histopathological images. Here, the specific choices of tracer, tumour line, and segmentation method were made with the sole purpose of demonstrating the implementation of the method. Furthermore, the specific methods for obtaining volumetric microscopy and autoradiography images, generation of simulated PET images, and data analysis can be substituted for other similar techniques, as long as they fit the general workflow presented herein and can be implemented with similar/better accuracy. Potentially, this method can be used to benchmark any PET segmentation algorithm against the ground truth of histopathological data.
A factor that could affect the accuracy of the reconstruction of PET tracer distribution and the corresponding formation of the sPET image set is the accuracy of the digital photo image registration. In a separate experiment, the accuracy of the rigid registration process used to align the consecutive digital photos of the cutting plane was proven to be better than 10 microns. However, the innovative approach presented in this validation methodology allows for certain flexibility in the accuracy of the 3D reconstruction. As long as the in plane registration errors are minimised (between the autoradiography data and the microscopy data), the 3D reconstruction errors, even if present, affect both image sets in a similar manner.
The tissue processing workfiow is highly adaptable and inexpensive to implement for a wide range of cryostat microtomes. Different tissue mounting jigs can be built for different tumour sizes using readily available materials. While there are institutions with highly specialised infrastructure for 3D tissue reconstruction [24], the method presented in this study meets the demands of the validation goal: to obtain a 3D intratumoural distribution of a PET tracer that can be used to generate sPET images precisely registered to histopathological data. The 3D distribution of FDG uptake, as obtained in this study, demonstrated good spatial correlation between neighbouring slices, even though a non-negligible interslice distance was utilised. Assuming that 100–150 μm range can be considered as a typical size of the microenvironmental feature, e.g. change in oxygenation or proliferation status of the tissue, the chosen distance of 120 μm between sampling locations ensured that all major spatial features of different aspects of the tumour microenvironment were sampled, allowing for a coherent representation of 3D spatial distributions of tracer activity and tumour morphology, as well as any other tumour microenvironmental parameter (e.g. hypoxia, cell proliferation, etc.), while allowing for a feasible tissue collection time. For the two tumours presented here, the total number of collected 4-section stacks were 41 and 48. The recorded time stamps on the digital photos indicated that for an experienced user, one 4-section stack could be consistently obtained every two minutes (120 μm inter-stack distance). Theoretically, it is possible to collect tissue sections at smaller intervals (down to continuous sampling), capturing biological features in more detail. However, it will result in increased demands on existing resources and decreased practicality of the method. The information gain from more frequent sampling has not been investigated as a part of this study. Furthermore, by increasing the time between tumour resection and autoradiography, the residual tissue activity is reduced (in the case of 18F imaging) resulting in the decreased signal-to-noise ratio of autoradiography imaging.
The FWHM size of the convolution kernel adopted for the formation of the sPET images was considered appropriate since the reconstructed tumours were generally small (≤ cm in largest diameter) and the spatial resolution was shown to be constant between the centre of the field of view and approximately 15 mm radial offset [2,21] for the scanner in question. However, in case of larger resection specimens (>1 cm), and situations where the specimen is positioned away from the centre of the field of view, using a variable convolution kernel (spatially dependant on position inside the scanner bore) could be required in order to provide a reasonable approximation of the PET image. An even more accurate approach to obtaining sPET images would be to use readily available Monte Carlo models for clinical and research PET scanners [25–27] that simulate the physical processes involved in PET image formation in a specific PET scanner. Starting with a given heterogeneous source of activity, such as a 3D autoradiography microscopic tracer uptake map, these models can be utilised to generate the raw data that can be used to reconstruct corresponding realistic PET images, taking into account the variable spatial resolution of the acquisition system.
The obvious effect of the convolution of the autoradiography-based activity maps is the dramatic change in spatial heterogeneity of the tracer uptake (Fig. 5). As indicated by the overlap analysis between sPET and 3D autoradiography, there is little to no overlap between the segmented volumes obtained using thresholds of higher activity values. This result is similar to previous studies illustrating the averaging effects of PET imaging with FDG [10]. Currently, clinical whole p PET imaging resolution (using 18F and an 80 cm bore scanner) is about 4–5 mm. It is theoretically limited at about 2 mm FWHM because of positron travel and annihilation photon non-colinearity [28]. The inherent blurring introduced only by these physical properties of PET image acquisition has a profound effect on the spatial concordance between tracer uptake as seen in PET images and the actual microscopic pattern of the tracer uptake. This aspect emphasises the need for the type of validation experiment presented herein, if clinical PET images are to be used for treatment guidance. For relatively large tumours, heterogeneity in tracer uptake is consistently observed [29]. Therefore, there is a need for validating that the observed heterogeneity captures correctly the underlying pattern of the biological feature of interest. All steps in the validation methodology presented can be directly applied to human specimen studies as well with only minimal adaptations required.
Currently, a number of 18F-labelled PET tracers targeting different aspects of tumour biology are being developed [1] and new PET-guided dose escalation clinical trials [30,31] are proposed and conducted. The method of PET tracer validation for image-guided radiotherapy presented here has the potential to streamline the validation of the novel tracers and provide the needed experimental basis for the clinical implementation of PET-guided biologically adaptive radiotherapy. It is possible to assemble a database of histopathological human tumour specimens with corresponding 3D sPET images registered to spatial distributions of biological features/subvolumes of interest. Using these databases, different segmentation techniques could be tested to ascertain which one is the best at reproducing the location and extent of the biological features to be targeted. By studying the probability of correctly delineating a biological target within a sPET segmented volume, the validation databases can provide needed information for the definition of boost volumes. For example, in the provided illustration involving the analysis of the colocalisation between high uptake regions of FDG and the spatial distribution of viable tissue, it was observed that if the FDG PET image were segmented with a 30% of maximum uptake threshold, more than 80% of the viable tissue would be included in the delineated subvolumes for both tumours (illustrated in Fig. 6). By extending this type of analysis over a population of patients with similar tumours (size and phenotype), the delineation uncertainty associated with clinical segmentation protocols can be directly quantified. Analysis of the validation databases would also provide an indication of the efficacy with which biologically targeted radiotherapy could be implemented for a given segmentation protocol.
Conclusion
This study presents a novel method of histopathological validation of PET for image-guidance applications in radiotherapy. This method is specifically aimed at probing the spatial coincidence of the areas of high PET tracer uptake, as seen in PET images, and the biological features of interest. The method allows for experimental validation of PET imaging for image guidance in radiotherapy while circumventing the difficulties associated with registration of in vivo PET data to ex vivo histopathological images.
The method is illustrated by carrying out the analysis of the spatial coincidence between the intratumoural pattern of FDG up-take and the spatial distribution of viable tissue. The method can be expanded to surgically-excised human tumour specimens. Furthermore, it can be utilised to evaluate performance of different PET image segmentation algorithms proposed for automated target definition in PET-guided selective dose escalation radiation therapy.
Acknowledgements
We would like to acknowledge the expertise Dr. Ema Dragoescu, MD, who assisted with the interpretation of the H&E images, and that of Mark Hile, who built the freezing jigs. This study was supported by funds from the Department of Radiation Oncology and Massey Cancer Centre support grant 2P30CA016059-28.
Footnotes
Conflict of interest statement
No conflicts of interest.
References
- [1].Bussink J, Kaanders JHAM, van der Graaf WTA, Oyen WJG. PET-CT for radiotherapy treatment planning and response monitoring in solid tumors. Nat Rev Clin Oncol. 2011;8:233–42. doi: 10.1038/nrclinonc.2010.218. [DOI] [PubMed] [Google Scholar]
- [2].Constantinescu C, Mukherjee J. Performance evaluation of an Inveon PET preclinical scanner. Phys Med Biol. 2009;54:2885–99. doi: 10.1088/0031-9155/54/9/020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Konerding MA, Malkusch W, Klapthor B, et al. Evidence for characteristic vascular patterns in solid tumours: quantitative studies using corrosion casts. Br J Cancer. 1999;80:724–32. doi: 10.1038/sj.bjc.6690416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ljungkvist ASE, Bussink J, Rijken PFJW, Kaanders JHAM, van der Kogel A, Denekamp J. Vascular architecture, hypoxia, and proliferation in first-generation xenografts of human head-and-neck squamous cell carcinomas. Int J Radiat Oncol Biol Phys. 2002;54:215–28. doi: 10.1016/s0360-3016(02)02938-3. [DOI] [PubMed] [Google Scholar]
- [5].Rijken PF, Bernsen HJ, Peters JP, Hodgkiss RJ, Raleigh JA, van der Kogel AJ. Spatial relationship between hypoxia and the (perfused) vascular network in a human glioma xenograft: a quantitative multi-parameter analysis. Int J Radiat Oncol Biol Phys. 2000;48:571–82. doi: 10.1016/s0360-3016(00)00686-6. [DOI] [PubMed] [Google Scholar]
- [6].Rijken PFJW, Peters JPW, Van der Kogel AJ. Quantitative analysis of varying profiles of hypoxia in relation to functional vessels in different human glioma xenograft lines. Radiat Res. 2002;157:626–32. doi: 10.1667/0033-7587(2002)157[0626:qaovpo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [7].van Laarhoven HWM, Kaanders JHAM, Lok J, et al. Hypoxia in relation to vasculature and proliferation in liver metastases in patients with colorectal cancer. Int J Radiat Oncol Biol Phys. 2006;64:473–82. doi: 10.1016/j.ijrobp.2005.07.982. [DOI] [PubMed] [Google Scholar]
- [8].Axente M, He J, Bass CP, et al. Tumour microenvironment heterogeneity affects the perceived spatial concordance between the intratumoural patterns of cell proliferation and 18F-fluorothymidine uptake. Radiother Oncol. 2012;105:49–56. doi: 10.1016/j.radonc.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Busk M, Horsman M, Overgaard J. Resolution in PET hypoxia imaging: voxel size matters. Acta Oncol. 2008;47:1201–10. doi: 10.1080/02841860802307716. [DOI] [PubMed] [Google Scholar]
- [10].Christian N, Lee J, Bol A, De Bast M, Jordan B, Gregoire V. The limitation of PET imaging for biological adaptive-IMRT assessed in animal models. Radiother Oncol. 2009;91:101–6. doi: 10.1016/j.radonc.2008.11.014. [DOI] [PubMed] [Google Scholar]
- [11].Schormann T, Dabringhaus A, Zilles K. Statistics of deformations in histology and application to improved alignment with MRI. IEEE Trans Med Imaging. 1995;14:25–35. doi: 10.1109/42.370399. [DOI] [PubMed] [Google Scholar]
- [12].Axente M, He J, Bass C, et al. Comprehensive approach to coregistration of autoradiography and microscopy images acquired from a set of sequential tissuesections. J Nucl Med. 2011;52:1621–9. doi: 10.2967/jnumed.111.091595. [DOI] [PubMed] [Google Scholar]
- [13].Beare R, Richards K, Murphy S, Petrou S, Reutens D. An assessment of methods for aligning two-dimensional microscope sections to create image volumes. J Neurosci Methods. 2008;170:332–44. doi: 10.1016/j.jneumeth.2008.01.012. [DOI] [PubMed] [Google Scholar]
- [14].Pugachev A, Ruan S, Carlin S, et al. Dependence of FDG uptake on tumor microenvironment. Int J Radiat Oncol Biol Phys. 2005;62:545–53. doi: 10.1016/j.ijrobp.2005.02.009. [DOI] [PubMed] [Google Scholar]
- [15].Purandare NC, Kulkarni AV, Kulkarni SS, et al. 18F-FDG PET/CT-directed biopsy: does it offer incremental benefit? Nucl Med Commun. 2013;34:203–10. doi: 10.1097/MNM.0b013e32835c5a57. [DOI] [PubMed] [Google Scholar]
- [16].Simonetti A, Elezi V, Farion R, et al. A low temperature embedding and section registration strategy for 3D image reconstruction of the rat brain from autoradiographic sections. J Neurosci Methods. 2006;158:242–50. doi: 10.1016/j.jneumeth.2006.06.004. [DOI] [PubMed] [Google Scholar]
- [17].Myronenko A, Song X. Point set registration: coherent point drift. IEEE Trans Pattern Anal Mach Intell. 2010;32:2262–75. doi: 10.1109/TPAMI.2010.46. [DOI] [PubMed] [Google Scholar]
- [18].Palm C, Vieten A, Salber D, Pietrzyk U. Evaluation of registration strategies for multi-modality images of rat brain slices. Phys Med Biol. 2009;54:3269–89. doi: 10.1088/0031-9155/54/10/021. [DOI] [PubMed] [Google Scholar]
- [19].National Electrical Manufacturers A NEMA standards publication NU 4-2008: performance measurements of small animal positron emission tomographs. 2008.
- [20].Marchand EW. Derivation of the point spread function from the line spread function. J Opt Soc Am. 1964;54:915–9. [Google Scholar]
- [21].Kemp B, Hruska C, McFarland A, Lenox M, Lowe V. NEMA NU 2-2007 performance measurements of the Siemens Inveon preclinical small animal PET system. Phys Med Biol. 2009;54:2359–76. doi: 10.1088/0031-9155/54/8/007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shattuck D, Prasad G, Mirza M, Narr K, Toga A. Online resource for validation of brain segmentation methods. NeuroImage. 2009;45:431–9. doi: 10.1016/j.neuroimage.2008.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Capek M, Bruza P, Jancek J, Karen P, Kubnov L, Vagnerov R. Volume reconstruction of large tissue specimens from serial physical sections using confocal microscopy and correction of cutting deformations by elastic registration. Microsc Res Tech. 2009;72:110. doi: 10.1002/jemt.20652. [DOI] [PubMed] [Google Scholar]
- [24].Sieren J, Weydert J, Namati E, et al. A process model for direct correlation between computed tomography and histopathology application in lung cancer. Acad Radiol. 2010;17:169–80. doi: 10.1016/j.acra.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schmidtlein CR, Kirov A, Nehmeh S, et al. Validation of GATE Monte Carlo simulations of the GE advance/discovery LS PET scanners. Med Phys. 2006;33:198–208. doi: 10.1118/1.2089447. [DOI] [PubMed] [Google Scholar]
- [26].Merheb C, Petegnief Y, Talbot JN. Full modelling of the MOSAIC animal PET system based on the GATE Monte Carlo simulation code. Phys Med Biol. 2007;52:563–76. doi: 10.1088/0031-9155/52/3/002. [DOI] [PubMed] [Google Scholar]
- [27].Canadas M, Arce P, Rato Mendes P. Validation of a small-animal PET simulation using GAMOS: a GEANT4-based framework. Phys Med Biol. 2011;56:273–88. doi: 10.1088/0031-9155/56/1/016. [DOI] [PubMed] [Google Scholar]
- [28].Levin CS, Hoffman EJ. Calculation of positron range and its effect on the fundamental limit of positron emission tomography system spatial resolution. Phys Med Biol. 1999;44:781–99. doi: 10.1088/0031-9155/44/3/019. [DOI] [PubMed] [Google Scholar]
- [29].Bradshaw TJ, Bowen SR, Jallow N, Forrest LJ, Jeraj R. Heterogeneity in intratumor correlations of 18F-FDG, 18F-FLT, and 61Cu-ATSM PET in canine sinonasal tumors. J Nucl Med. 2013;54:1931–7. doi: 10.2967/jnumed.113.121921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Duprez F, De Neve W, De Gersem W, Coghe M, Madani I. Adaptive dose painting by numbers for head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2011;80:1045–55. doi: 10.1016/j.ijrobp.2010.03.028. [DOI] [PubMed] [Google Scholar]
- [31].Madani I, Duthoy W, Derie C, et al. Positron emission tomography-guided, focal-dose escalation using intensity-modulated radiotherapy for headand neck cancer. Int J Radiat Oncol Biol Phys. 2007;68:126–35. doi: 10.1016/j.ijrobp.2006.12.070. [DOI] [PubMed] [Google Scholar]