Abstract
Cardiac myosin binding protein-C (cMyBP-C) is a structural and regulatory component of cardiac thick filaments. It is observed in electron micrographs as seven to nine transverse stripes in the central portion of each half of the A band. Its C-terminus binds tightly to the myosin rod and contributes to thick filament structure, while the N-terminus can bind both myosin S2 and actin, influencing their structure and function. Mutations in the MYBPC3 gene (encoding cMyBP-C) are commonly associated with hypertrophic cardiomyopathy (HCM). In cardiac cells there exists a population of myosin heads in the super-relaxed state (SRX), which are bound to the thick filament core with a highly inhibited ATPase activity. This report examines the role cMyBP-C plays in regulating the population of the SRX state of cardiac myosin by using an assay that measures single ATP turnover of myosin. We report a significant decrease in the proportion of myosin heads in the SRX state in homozygous cMyBP-C knockout mice, however heterozygous cMyBP-C knockout mice do not significantly differ from the wild type. A smaller, non-significant decrease is observed when thoracic aortic constriction is used to induce cardiac hypertrophy in mutation negative mice. These results support the proposal that cMyBP-C stabilises the thick filament and that the loss of cMyBP-C results in an untethering of myosin heads. This results in an increased myosin ATP turnover, further consolidating the relationship between thick filament structure and the myosin ATPase.
Keywords: Cardiac SRX, Hypertrophic cardiomyopathy, Myosin binding protein-C (MyBP-C), Myosin II ATPase, Thick filament structure, Cardiac energetics
1. Introduction
Hypertrophic cardiomyopathy (HCM) is a disease of the heart muscle with a prevalence of at least 1 in 500 [1]. It is one of the most commonly inherited cardiac disorders [2], and the most common cause of sudden cardiac death in young, apparently healthy individuals [3]. Clinically, it is usually characterised by asymmetric thickening of the left ventricle (LV) [4] and while systolic function is normally preserved, it is often associated with significant diastolic dysfunction [4]. Mutations in sarcomeric genes are the most common cause of HCM [5] and mutations in the genes encoding myosin heavy chain (MYH7) and cardiac myosin binding protein-C (cMyBP-C encoded by MYBPC3) are the most frequent.
The discovery that mutations in MYBPC3 cause HCM [6, 7] led to a wave of research focused on understanding how these mutations result in the pathology observed in patients with HCM (for a review, see [8]). Homozygous Mybpc3 knockout (cMyBP-C(−/−)) mice do not express cMyBP-C and exhibit significant cardiac hypertrophy compared to both wild type (WT) and heterozygous knockout (cMyBP-C(+/−)) littermates [9] and together they have provided valuable insights into the mechanism that cMyBP-C plays in cardiac muscle regulation and disease. Although the cMyBP-C(−/−) phenotype recapitulates aspects of disease in patients with MYBPC3 mutations (which are most commonly heterozygous), the cMyBP-C(+/−) mice do not display the disease phenotype. It remains unclear whether MYBPC3 mutations in human patients result in a “poison peptide” effect or rather display haploinsufficiency, with reports supporting both possibilities [10-12]. It is possible that both are relevant and that the phenotype depends on the specific mutation of MYBPC3.
The super-relaxed (SRX) state of myosin is a relatively new discovery in the field of muscle biochemistry (for reviews see [13, 14]). It is characterised by a strong inhibition of myosin ATPase activity and has been observed in rabbit, mouse, and tarantula skeletal muscle as well as rabbit cardiac muscle [15-18].
It is believed that myosin heads in the SRX state are aligned along the core of the thick filament, as seen in electron micrographs of isolated thick filaments from a range of striated muscles [19-24]. Three-dimensional reconstructions have revealed multiple specific interactions between neighbouring myosin heads, termed the “interacting heads motif” (for a detailed review, see [25]). Briefly, this motif inhibits the ATPase activity of myosin heads by blocking the ATP-binding site of an adjacent head (free head) and inhibiting the other head from interaction with actin (blocked head). This contrasts to the other state of relaxed myosin, which we refer to as the disordered relaxed (DRX) state. In the DRX state, myosin heads project azimuthally from the thick filament and have an ATPase activity up to ten times faster than those in the SRX state [26].
Our recent review [14] suggested that the SRX state of cardiac myosin may be reduced in mice lacking cMyBP-C because cMyBP-C binds to myosin near the S1/S2 junction and is thus in a position that could influence the interacting heads motif. This hypothesis is supported by published structural data using these mice that shows a disordering of myosin heads in the thick filament [22, 27, 28]. In this report we directly test the hypothesis that cMyBP-C affects the SRX state using cMyBP-C(−/−) and cMyBP-C(+/−) mouse models as well as wild type mice and a mutation negative mouse model of cardiac hypertrophy. Results show that the loss of cMyBP-C reduces the SRX state of cardiac myosin. Recently, it was shown that estradiol deficiency in ovariectomised mice disrupts the SRX state of skeletal muscle [15], however this is the first known report of alterations to the SRX state in a model of cardiac disease.
2. Methods
2.1. Animal Models
Care and handling of all cMyBP-C knockout and littermate control mice was performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of California, Davis, Davis, CA. Heterozygous (cMyBP-C(+/−)) and homozygous (cMyBP-C(−/−)) knockout mice were generated by targeted replacement of exons 3 to 10 encoding for Mybpc3 as previously described [9]. These mice, as well as wild type (WT) littermates were anesthetized using isoflurane and euthanased by cervical dislocation. Hearts were excised and weighed as previously described [29]. LV samples were dissected and frozen immediately in liquid nitrogen and shipped to The University of Sydney on dry ice. LV samples were stored in liquid nitrogen at −196 °C until used.
Experimental hypertension was induced by transverse aortic constriction (TAC) in mice under AERC approval (#11/25) at the Victor Chang Cardiac Research Institute. Briefly, LV hypertrophy was induced in 12 week old male C57BL/6J mice by permanent narrowing of the transverse aorta to the diameter of a 27 G needle, as described previously [30], except a 27 G needle was used to create a narrower lumen. Mice displayed significant cardiac hypertrophy after seven days, without decompensation, and were euthanased by cervical dislocation prior to the rapid excision of their hearts, which were frozen for later use. In this report we used six mice each from the WT, cMyBP-C(+/−), and cMyBP-C(−/−) groups, and three mice from the TAC group, with three to five technical repeats per heart.
2.2. SRX solutions
Skinning buffer: NaCl, 100 mM; MgCl2, 8 mM; EGTA, 5 mM; K2HPO4, 5 mM; KH2PO4, 5 mM; NaN3, 3 mM; ATP, 5 mM; DTT, 1 mM; BDM, 20 mM; Triton-X 100, 0.1%, pH 7.0.
Glycerinating solution: K acetate, 120 mM; Mg acetate, 5 mM; K2HPO4, 2.5 mM; KH2PO4, 2.5 mM; MOPS, 50 mM; ATP, 5 mM; BDM, 20 mM; DTT, 2 mM; glycerol, 50% (v/v), pH 6.8.
Rigor buffer: K acetate, 120 mM; Mg acetate, 5 mM; K2HPO4, 2.5 mM; KH2PO4, 2.5; MOPS, 50; DTT, 2 mM (added fresh), pH 6.8.
mATP buffer: Rigor buffer + 250 μM mATP.
ATP chase buffer: K acetate, 120 mM; Mg acetate, 5 mM; K2HPO4, 2.5 mM; KH2PO4, 2.5 mM; ATP, 4 mM; MOPS, 50; DTT, 2 mM (added fresh), pH 6.8.
2.3. Preparation of glycerinated cardiac muscle fibres
Glycerinated muscle fibres were prepared as reported previously, with minor modifications [16]. Cryopreserved LV was fractured under liquid nitrogen in a mortar and pestle. A ~10 mg piece was immersed in skinning solution on ice. This was agitated on ice for 6 hours on an orbital shaker with 3 changes of solution. Skinning solution was then exchanged for glycerinating solution and left overnight (~15 hr) on ice to equilibrate. The glycerinating solution was then refreshed and the glycerinated muscle stored at −20°C for use within 1 week.
2.4. SRX experiments
Small bundles of skinned cardiac muscle fibres (<90 μm in diameter) were dissected in glycerinating solution at 4°C and immobilised on a glass coverslip using two layers of double-sided tape. These coverslips were then placed fibre side down onto a pre-cooled glass slide, creating a flow chamber that allowed rapid flow of buffers around the fibre. Fibres were stored in glycerinating solution on ice and imaged within a few hours. Prior to imaging, fibres were rinsed in rigor buffer to remove ATP, BDM, and glycerol. Rigor buffer was refreshed several times over 5 minutes to ensure complete exchange of rigor buffer. Fibres were then incubated in the fluorescent mATP buffer for 5 minutes prior to imaging. Fibres were imaged at 22°C using a Nikon Ni-E upright epifluorescence microscope with a 20X Plan Apo objective lens (0.75 NA). Fibres were located using bright-field differential interference contrast (DIC) to avoid photobleaching of the mATP probe. Fibres were then excited at 395 nm (DAPI setting) at 10% laser power and 1.5× gain and an exposure time of 20 ms. Images were acquired on a Nikon DS-Qi2 camera at 5 second intervals for 600 seconds for a total exposure time of 2.42 seconds. After 60 seconds, the mATP buffer was flushed with ATP buffer and the decay in fluorescence intensity was recorded. Twenty seconds continuous exposure to the fibre resulted in ~3% decrease in fluorescence intensity, demonstrating that photobleaching of the mATP probe was negligible over the course of the experiment (data not shown).
2.5. SRX analysis
During DIC imaging, two to three rectangular regions of interest (roughly 15 μm × 40 μm) were drawn over randomly selected areas of the muscle fibre as well as over two areas next to the fibre to measure the fluorescence intensity of the fibre and background, respectively. The mean background fluorescence intensity was subtracted from the average of the fibre fluorescence intensity. Each time point was divided by the fluorescence intensity of the final mATP image before washout (t =0). The resulting data were exported to Prism 6.0 (GraphPad Software, Inc.). The decay in fluorescence intensity was then fit using a user-defined two-state exponential:
where: I is fluorescence intensity, P1 and P2 are the initial proportion of fluorescence for the two states, and T1 and T2 are the time constants for the lifetime of these states. P1 and T1 represent the initial rapid decay in fluorescence intensity, which comprises myosin in the DRX state and the release of non-specifically bound mATP. P2 and T2 are representative of the subsequent the slow decrease in fluorescence intensity due to myosin in the SRX state.
2.6. Measurement of phosphorylation
The phosphorylation status of key sarcomeric proteins was measured by Pro-Q Diamond staining of SDS-PAGE gels. Tissue was homogenised and TCA treated in a method similar to [31] to preserve the endogenous phosphorylation levels of the tissue. 30 μg of each sample was then separated on 12% SDS-PAGE gels before fixing and staining with Pro-Q Diamond according to manufacturers instructions. Gels were imaged before overnight staining with total protein stain Sypro Ruby. All imaging was carried out on the Bio-Rad ChemiDoc system with default settings. Pro-Q intensity of proteins of interest were normalised to the Sypro Ruby intensity of the alpha actinin band. The Peppermint Stick molecular weight marker was used to ensure correct staining of Pro-Q and Sypro Ruby.
2.7. Statistical analyses
Individual SRX traces were fit to the above two-state exponential and the resultant outputs were averaged within the respective group. Data for each of these groups are expressed as the mean ± SEM. One-way ANOVA was employed using Prism 6.0, with a Tukey post hoc analysis with significance accepted at p < 0.05.
3. Results
3.1. Hypertrophy in homozygous cMyBP-C knockout mice
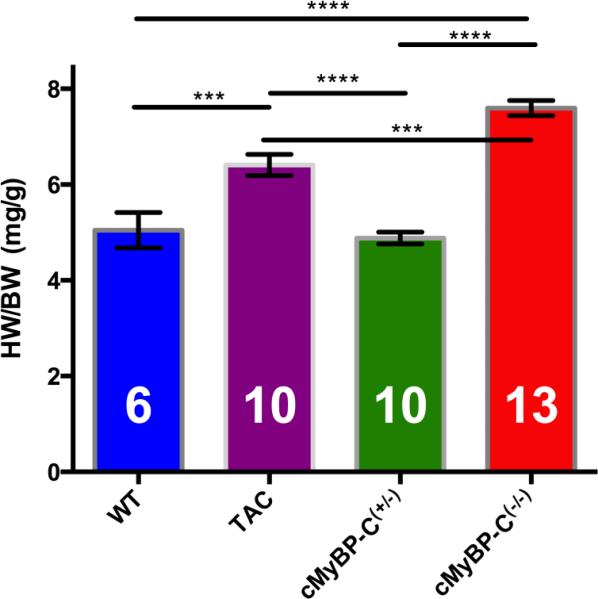
The mean heart-weight to body-weight ratios (HW/BW) of mice from each group is shown in Figure 1. There was a highly significant (p < 0.0001) increase in HW/BW in cMyBP-C(−/−) mice compared to both WT and cMyBP-C(+/−) mice, which is consistent with previously published data [9]. This shows that only homozygous, and not heterozygous deletion of Mybpc3 results in cardiac hypertrophy.
FIGURE 1.
Heart weight to body weight (HW/BW) ratios of WT, TAC, cMyBP-C(−/−) and cMyBP-C(+/−) mouse populations. cMyBP-C(−/−) mice (red) have a significantly increased HW/BW compared to both WT (blue) and cMyBP-C(+/−) mice (green). TAC mice (purple) also exhibited significantly increased HW/BW ratio compared to WT and cMyBP-C(+/−), however this difference didn't appear to be as large as observed in cMyBP-C(−/−) mice. HW/BW ratio was not significantly different between WT and cMyBP-C(+/−). This indicates significant cardiac hypertrophy in cMyBP-C(−/−) and TAC mice. (*** indicates p<0.001, **** indicates p<0.0001, error bars are SEMs). Number of mice used in each measurement inside each column. Note that not all these mice are used in the SRX measurements
TAC mice also exhibited a significantly increased HW/BW ratio compared to WT (p < 0.001) and cMyBP-C(+/−) (p < 0.0001) mice, confirming that this model also results in cardiac hypertrophy. However, the hypertrophy of TAC mice appeared less severe than in cMyBP-C(−/−) mice (p < 0.001).
3.2. Super-relaxed myosin in permeabilised left ventricular muscle fibres from mice
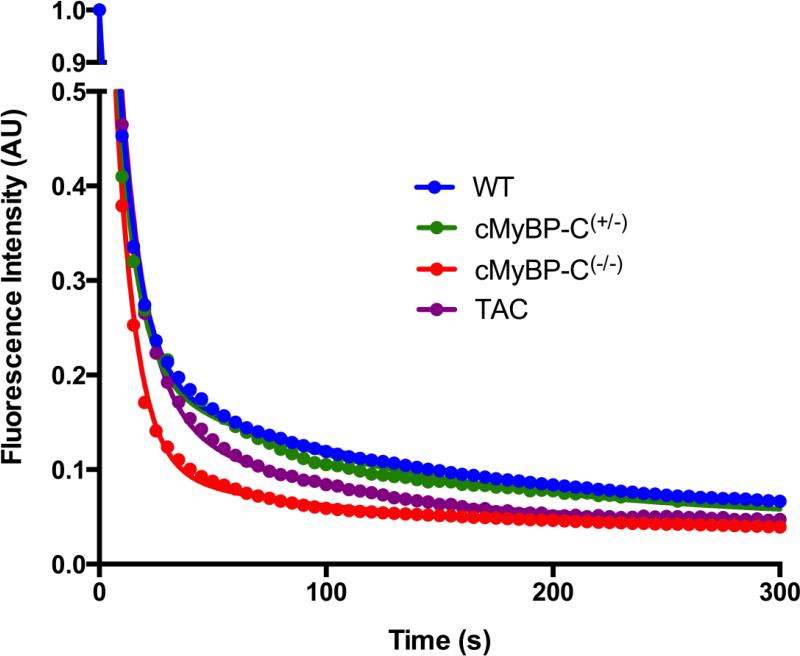
Quantitative epifluorescence microscopy was employed to measure the kinetics of ATP turnover in permeabilised muscle fibres from cryopreserved mouse LV. In these experiments, ATP replaced the bound mATP as the probe was hydrolysed to mADP and Pi and released. The replacement of the fluorescent probe was measured by the decay in fluorescence intensity after ATP chase (Figure 2). The assay was calibrated using rabbit psoas muscle, of which the SRX state has been comprehensively characterised [18], with results that were consistent with published data [18, 26] (see supplementary material). As shown in Figure 2 (blue trace), we found that WT mouse LV cardiomyocytes exhibited a similar decay in fluorescence intensity to that previously described in other models, i.e. a slow turnover of mATP that is associated with the SRX state of myosin. Interestingly, the lifetime of ATP turnover (T2) was similar to the ATP turnover of mouse psoas muscle [15], while the T2 of rabbit cardiac muscle has been reported to be about 1/3rd shorter than rabbit psoas [16, 18]. A strong possibility for this may be differential expression of myosin heavy chain isoforms, and this will be discussed in detail later.
FIGURE 2.
Representative single traces from WT, cMyBP-C (+/−), cMyBP-C(−/−), and TAC mouse models. The fluorescence intensity decreases much faster in cMyBP-C(−/−) than in WT and cMyBP-C(+/−), which are almost coincident. The TAC also exhibits a faster decrease in fluorescence than WT and cMyBP-C(+/−), but to a lesser extent than cMyBP-C(−/−) muscle fibres. This indicates a severe disruption of the SRX state in the LV of cMyBP-C(−/−) mice. For individual fits of these traces, see supplementary data.
3.3. Comparison of SRX in wild type and cMyBP-C knock out, and TAC mouse LV cardiomyocytes
Figure 2 shows representative traces of ATP chases in permeabilised muscle fibres from WT, cMyBP-C(+/−), cMyBP-C(−/−) and TAC mice. The fluorescence intensity of the cMyBP-C(−/−) cardiomyocytes clearly decays faster than measured in the WT and cMyBP-C(+/−) muscle fibres. The fluorescence intensity in the TAC cardiomyocytes, a model of cardiac hypertrophy independent of cMyBP-C, decays slower than the cMyBP-C(−/−) trace, however the decay is faster than the WT and cMyBP-C(+/−) traces, which are almost coincident.
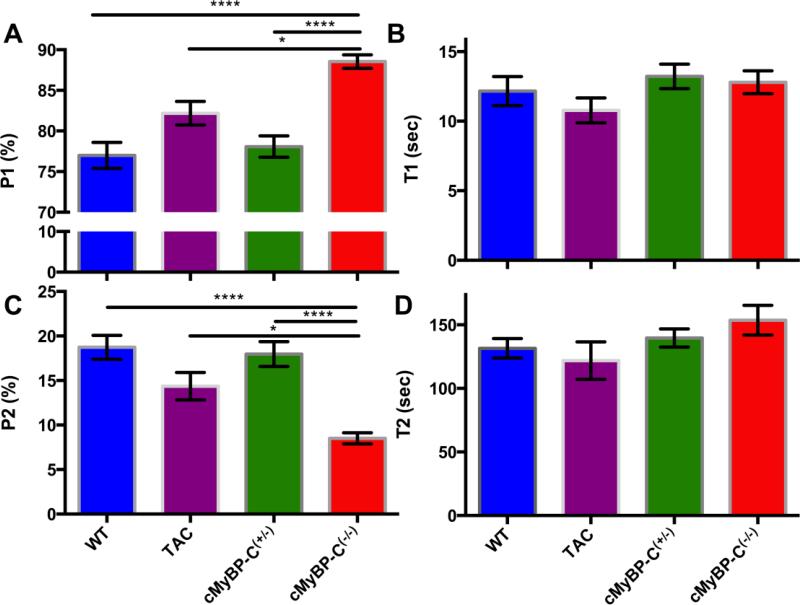
The averages of all experimental fits are summarised in Figure 3, and Table 1. There is a significant increase in the P1 of cMyBP-C(−/−) mice compared to wild type and cMyBP-C(+/−), p < 0.0001. This difference is coupled to a highly significant decrease in the proportion of myosin heads in the SRX state (P2) of cMyBP-C(−/−) muscle fibres compared to WT and cMyBP-C(+/−) mice (p<0.0001).
FIGURE 3.
A summary of the parameters derived from the two-state exponential (P1, T1 and P2, T2) in the studied mouse models. The fluorescence decay of the fast component (P1) is increased in cMyBP-C(−/−) mice compared to other models (Panel A). This is coupled to a decrease in the P2 state (Panel C) indicating a shift of myosin heads from the SRX state to the DRX state in the cMyBP-C(−/−) mice. The lifetimes of ATP turnover in both these states remain unchanged (Panels B and D). Data are expressed as mean ± SEM (n=6 for WT, cMyBP(+/−), and cMyBP-C(−/−) and n=3 for TAC; **** indicates p<0.0001, * indicates p < 0.05).
Table 1.
Comparison of parameters of the SRX state in WT, TAC, cMyBP-C(+/−), cMyBP-C(−/−) mice. n represents the number of mice used in this study, whereas the number in the parentheses is the total number of technical repeats. P2 is interpreted as the proportion of myosin heads in the SRX state, while T2 is the lifetime of nucleotide turnover by these srx myosin. Data expressed as mean ± SEM.
| n | P1% | T1 (sec) | P2% | T2 (sec) | |
|---|---|---|---|---|---|
| WT | 6 (21) | 77 ± 2# | 12 ± 1 | 19 ± 1# | 132 ± 8 |
| TAC | 3 (12) | 82 ± 1* | 10.8 ± 0.9 | 14 ± 2* | 122 ± 15 |
| cMyBP-C(+/−) | 6 (17) | 78 ± 1# | 13.2 ± 0.9 | 18 ± 1# | 140 ± 7 |
| cMyBP-C(−/−) | 6 (17) | 89 ± 1 | 12.8 ± 0.8 | 8.5 ± 0.6 | 154 ± 12 |
indicates p<0.0001 compared to Homozygous cMyBP-C knockout
indicates p<0.05 compared to Homozygous cMyBP-C knockout
We also examined the SRX state in a mutation negative model of cardiac hypertrophy generated by transverse aortic constriction (TAC). Interestingly, the SRX state of cMyBP-C(−/−) mice was also reduced compared to TAC mice. There was a modest increase in the P1 of cMyBP-C(−/−) mice compared to the TAC mice (89 ± 1% vs. 82 ± 1%, p < 0.05). Again this was coupled with a reduction of the P2 of cMyBPC(−/−) mice (8.5 ± 0.6% vs. 14 ± 2%, p < 0.05). There was also an apparent reduction in the P2 of TAC mice compared to WT and cMyBP-C(+/−) mice, although this difference was not significant. The lifetimes of ATP turnover (T2) were unchanged between groups.
3.4. Phosphorylation of sarcomeric proteins
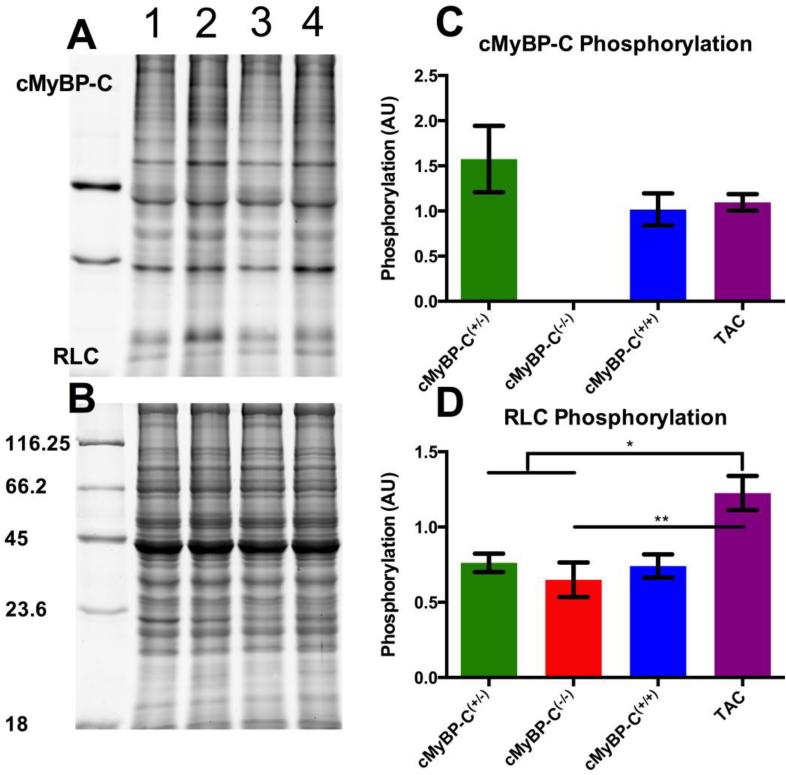
We measured the phosphorylation of cMyBP-C, troponin-T (TnT), troponin-I (TnI) and the regulatory light chain (RLC) by Pro-Q Diamond staining (Figure 4A). The density of each of these bands was normalised to the alpha-actinin band stained by Sypro Ruby (Figure 4B). cMyBP-C phosphorylation was stable between WT, cMyBP-C(+/−) and TAC mice (Figure 4C). TnT and TnI also remained unchanged between groups (data not shown), with a slight qualitative increase in the TnI of TAC and cMyBP-C(−/−) mice. There were no differences in the RLC phosphorylation between WT, cMyBP-C(+/−) and cMyBP-C(−/−) mice, however this was found to be increased in TAC mice compared to all other groups (Figure 4D).
Figure 4.
Phosphorylation of sarcomeric protein by Pro-Q Diamond staining. A) Pro-Q diamond stained gel showing cMyBP-C(+/−) (1), cMyBP-C(−/−) (2), cMyBP-C(+/+) (3) and TAC (4) mice. B) The corresponding Sypro Ruby stained gel showing nearly equal loading of protein for each lane. The numbers on the side represent the molecular weights (kDa) for each band in the molecular weight marker. cMyBP-C phosphorylation was unchanged amongst those samples expressing the protein (panel C), while phosphorylation RLC was significantly increased in the TAC model compared to all other groups (panel D). Data is represented as mean ± SEM (n=2 for each group, 2 technical repeats, * indicates p < 0.05, ** < 0.01).
4. Discussion
4.1. Super-relaxed myosin in mouse LV cardiomyocytes
Our experiments are the first to demonstrate the presence of super-relaxed myosin in mouse cardiomyocytes. Our results indicate that 19 ±1% (P2) of myosin is in the SRX state and turnover one ATP molecule every 132 ± 8 seconds (T2) in WT mice. Surprisingly, the SRX states of mouse cardiomyocytes and psoas fibres (P2 = 17 ± 2%, T2 = 102 ± 7 sec) are similar [15]. This is in contrast to previously published data, which shows a reduced T2 in rabbit cardiac muscle compared to rabbit psoas (144 ± 10 seconds vs. 230 ± 24 seconds) [16, 18]. This is likely due to the fact that the mouse psoas and LV muscle both express fast myosin heavy chains, while the rabbit psoas and LV muscle express fast and slow myosin heavy chains, respectively. These two isoforms have different rates of ATP hydrolysis [32], and thus may be important in the regulation of the lifetime (T2) of the SRX state of myosin. Further supporting this argument is the similarity in the T2 of rabbit soleus (156 ± 20 seconds) and LV (144 ± 10 seconds) muscle fibres, which both express slow myosin heavy chains [16, 18].
4.2. Ablation of cMyBP-C disrupts the SRX state in cardiomyocytes
The major finding of this study is that cMyBP-C appears to modulate the SRX state of myosin in mouse LV muscle fibres. The P2 (Figure 3C and Table 1) of cMyBP-C(−/−) LV cardiomyocytes was reduced by 55% compared to WT mice (8.5 ± 0.6% vs. 19 ± 1%, respectively). This was matched to a nearly identical increase in P1 (Figure 3A, Table 1) in cMyBP-C(−/−) mice compared to WT (89 ± 1% and 77 ± 2%, respectively). Together, these data strongly suggest that myosin heads are shifted from the SRX state into the DRX state in the absence of cMyBP-C. Phosphorylation of the regulatory light chain of myosin has previously been shown to diminish the SRX state in rabbit psoas [18], and is thus a possibility for the disruption seen in these mice. Given that the phosphorylation status of the RLC remains unchanged between WT, cMyBP-C(+/−) and cMyBP-C(−/−) models (Figure 4D) [33, 34], it is likely that the reduction of SRX myosin results from structural deficiencies of the thick filament rather than post-translational modifications of contractile proteins in the cMyBP-C(−/−) mice.
It is important to note that the actual proportion of myosin heads in the SRX state is larger than the reported P2, due to non-specific binding of the mATP within the fibre. The level of non-specific binding is measured by comparing the fluorescence intensity of the fibre when incubated in 250 μM mATP and 250 μM mATP plus 4 mM ATP. This found that roughly 50% of the fluorescence intensity arises from non-specific binding (see supplementary data). Thus the proportion of SRX myosin heads is about twice the P2. Thus the shift of myosin heads in the SRX state from WT to cMyBP-C(−/−) mice is actually closer to 21%. This is similar to the incidence of myosin heads in contact with cMyBP-C (assuming the C-zone comprises approximately 50% of the thick filament and cMyBP-C is present in every third crown of heads). This is consistent with the notion that loss of cMyBP-C is responsible for the disruption of the SRX state.
An alternate explanation for the loss of SRX myosin heads is the significant remodelling seen in HCM [5]. In order to determine whether the reduction of myosin in the SRX state is due to the loss of cMyBP-C or hypertrophic remodelling, we studied another mouse model of cardiac hypertrophy induced by transverse aortic constriction (TAC). Compared to the TAC mice, cMyBP-C(−/−) LV cardiomyocytes exhibited a 39% relative reduction (or 5.5% total change) in the proportion (P2 Fig. 3B, Table 1) of myosin heads in the SRX state (14 ± 2% vs. to 8.5 ± 0.6%, p<0.05). This was coupled with a 7% absolute increase in P1 of cMyBP-C(−/−) mice (82 ± 1% vs. to 89 ± 1%, p<0.05). Interestingly, there was a small decrease in the average P2 values (Fig. 3A, Table 1) for the TAC mice compared to WT mice (19 ± 1% vs. to 14 ± 2%), however this difference was not statistically significant. This reduction suggests that although absence of cMyBP-C results in a far more dramatic loss of SRX myosin, a level of SRX disruption may be common to all HCM. If so, it is an intriguing possibility that disruption of the SRX state contributes to energetic imbalances that lead to heart failure.
As the TAC mice in our study did not exhibit the same extent of remodelling as the cMyBP-C(−/−) mice, we cannot eliminate the possibility that reduction of the SRX state may be related to the severity of hypertrophy (measured by HW/BW) or due to other post translational changes. However, we found that the phosphorylation of the RLC of TAC mice was increased compared to the other groups (Figure 4D). This is a possible explanation for the slight reduction of the SRX state in the TAC mice. Therefore we suggest that the dramatic reduction of the SRX state in cMyBP-C results from the loss of cMyBP-C rather than a higher severity of hypertrophy.
It has previously been documented that LV from cMyBP-C(−/−) hearts display a partial isoform switch from α- to β-myosin heavy chain (MHC) [9], and this switch has also been demonstrated in TAC mice [35]. It is possible that these changes may affect the SRX state in these mice. However, given that the T2 of the cMyBP-C(+/−) mice (no shift in MHC isoform) is intermediate to the TAC and cMyBP-C(−/−) mice, we do not believe this affects the lifetime of the SRX state. Additionally, there were no reported differences in the P2 between fast and slow skeletal and between skeletal and cardiac muscles [16, 18]. Thus the reduction in P2 reported in this study is unlikely due to isoform switching.
Importantly, the current data agree with structural studies from MyBP-C(−/−) mice where myosin heads were more disordered compared to WT mice in electron micrographs of isolated thick filaments [28]. Three-dimensional reconstructions of these thick filaments revealed myosin heads that were less well resolved and fewer of them displayed the interacting heads motif in these cMyBP-C(−/−) mice [22]. This suggests a shift of myosin heads away from the SRX state into the DRX state. X-ray diffraction data also reveal that myosin heads in cMyBP-C(−/−) mice move closer to the actin filament and azimuthally away from the thick filament surface, suggesting a loss of the SRX state [27]. Furthermore, addition of MyBP-C to myosin filaments produces more ordered synthetic thick filaments than those lacking MyBP-C, indicating the importance of MyBP-C in thick filament stability [36].
4.3. Functional consequences of loss of thick filament structural order
The disruption of the SRX state is consistent with the contractile changes reported in cMyBP-C(−/−) mice. Ablation of cMyBP-C increases rates of force redevelopment and loaded shortening at submaximal calcium concentrations [37, 38]. Additionally the power output measured in cMyBP-C(−/−) mice was increased by 26%, which is consistent with our findings (21% decrease in proportion of myosin heads in the SRX state) [37]. Stretch activated responses of the cMyBP-C(−/−) mice were also accelerated compared to WT [39]. This is consistent with the notion that the loss of MyBP-C promotes an azimuthal shift of myosin heads away from the thick filament closer to the thin filaments where activation time will be reduced.
An alteration to cardiac energetics is another possible consequence of a reduced SRX state in the cMyBP-C(−/−) mice. The shift of myosin heads from the energetically economical SRX state into the DRX state would generate a large increase in myocardial ATP consumption (see [16] for a quantitative discussion). Although this effect is smaller than that from active heads, in the long term this may result in myocardial energy imbalances, which have been hypothesised to play a role in the pathogenesis of cardiac hypertrophy [40].
We expect a similar, but less severe loss of the SRX state when cMyBP-C is phosphorylated. X-ray diffraction studies comparing untreated and PKA-treated cardiac fibres reported a shift of mass away from the thick filament, towards the thin filament, indicative of thick filament disorder [33]. This represents a shift of myosin out of the SRX state into the DRX state.
5. Summary and Conclusions
This report investigated the role of cMyBP-C in regulating the SRX state of myosin in murine LV cardiomyocytes. We compared a cMyBP-C(−/−) mouse model of hypertrophic cardiomyopathy to WT and cMyBP-C(+/−) littermate mice that exhibit no cardiac hypertrophy. Results showed a significant decrease in the proportion of myosin heads in the SRX state in permeabilised cardiomyocytes from cMyBP-C(−/−) mice, consistent with previous reports of disordered myosin heads in the absence of cMyBP-C [22, 27, 28]. We also investigated the SRX state of a mouse model of induced hypertrophy (TAC) without genetic mutation and found a slight but non-significant reduction in the SRX state. These results may suggest that while the HCM phenotype of cardiac hypertrophy may contribute slightly to the disruption of myosin in the SRX state, it is the absence of cMyBP-C that results in a more dramatic reduction of the SRX state in cMyBP-C(−/−) mice. This is consistent with the hypothesis that cMyBP-C has an important role in regulating the structure of the thick filament, and also that this thick filament structure is closely related to myosin ATPase regulation.
In the future, we will determine whether this reduction of myosin in the SRX state also occurs in cardiomyocytes from human HCM patients with MYBPC3 mutations. Given the differences between human HCM patients with MYBPC3 mutations and the murine model studied here, without additional work we cannot draw definitive conclusions about how the SRX state is altered in human disease. Further studies will also aim to determine what fragment, if any, of cMyBP-C is sufficient to restore the SRX state of cMyBP-C(−/−) mice to that seen in WT mice.
Supplementary Material
Highlights.
The inhibited super-relaxed state of myosin was measured in permeabilised mouse left ventricles.
Severe hypertrophy induced by cMyBP-C ablation was accompanied by a loss of SRX regulation.
An intermediate, non-significant decrease in the SRX was noted in trans-aortic constricted mice.
Acknowledgements
We would like to thank Dr. Ying Ying Su (Australian Centre for Microscopy and Microanalysis) and Dr. Louise Cole (Bosch Advanced Microscopy Facility) for their technical expertise. SL, AL and JM are supported by the NG Macintosh Memorial Fund. SL and AL are supported by the National Heart Foundation. This work was supported in part by NIH R01 HL080367 (SPH). SvD is supported in part by a postdoctoral fellowship from the American Heart Association. RC would like to acknowledge that he is on the scientific advisory board of Myokardia, in South San Francisco.
Abbreviations
- SRX
super-relaxed state
- DRX
disordered-relaxed state
- cMyBP-C
cardiac myosin binding protein-C
- MYBPC3/Mybpc3
cardiac myosin binding protein-C gene in human/mouse
- HCM
hypertrophic cardiomyopathy
- MOPS
3-(N-morpholino) propanesulfonic acid
- DTT
dithiothreitol
- BDM
2,3-butanedione monoxime
- mATP
2’-(or-3’)-O-(N-methylanthraniloyl) adenosine 5-triphosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–9. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 2.Elliott P, McKenna WJ. Hypertrophic cardiomyopathy. Lancet. 2004;363:1881–91. doi: 10.1016/S0140-6736(04)16358-7. [DOI] [PubMed] [Google Scholar]
- 3.McKenna WJ, Behr ER. Hypertrophic cardiomyopathy: management, risk stratification, and prevention of sudden death. Heart. 2002;87:169–76. doi: 10.1136/heart.87.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Towbin JA. Hypertrophic Cardiomyopathy. Pacing Clin Electrophysiol. 2009;32:S23–S31. doi: 10.1111/j.1540-8159.2009.02381.x. [DOI] [PubMed] [Google Scholar]
- 5.Houston BA, Stevens GR. Hypertrophic cardiomyopathy: a review. Clin Med Insights Cardiol. 2014;8:53–65. doi: 10.4137/CMC.S15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:438–40. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- 7.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–7. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 8.Schlossarek S, Mearini G, Carrier L. Cardiac myosin-binding protein C in hypertrophic cardiomyopathy: mechanisms and therapeutic opportunities. J Mol Cell Cardiol. 2011;50:613–20. doi: 10.1016/j.yjmcc.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 10.Helms AS, Davis FM, Coleman D, Bartolone SN, Glazier AA, Pagani F, et al. Sarcomere mutation-specific expression patterns in human hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2014;7:434–43. doi: 10.1161/CIRCGENETICS.113.000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marston S, Copeland O, Gehmlich K, Schlossarek S, Carrier L. How do MYBPC3 mutations cause hypertrophic cardiomyopathy? J Muscle Res Cell Motil. 2012;33:75–80. doi: 10.1007/s10974-011-9268-3. [DOI] [PubMed] [Google Scholar]
- 12.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JMJ, Winegrad S, et al. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–83. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 13.Cooke R. The role of the myosin ATPase activity in adaptive thermogenesis by skeletal muscle. Biophys Rev. 2011;3:33–45. doi: 10.1007/s12551-011-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNamara JW, Li A, dos Remedios CG, Cooke R. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys Rev. 2015;7:5–14. doi: 10.1007/s12551-014-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colson BA, Petersen KJ, Collins BC, Lowe DA, Thomas DD. The myosin super- relaxed state is disrupted by estradiol deficiency. Biochem Biophys Res Commun. 2014 doi: 10.1016/j.bbrc.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooijman P, Stewart MA, Cooke R. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys J. 2011;100:1969–76. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naber N, Cooke R, Pate E. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J Mol Biol. 2011;411:943–50. doi: 10.1016/j.jmb.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci U S A. 2010;107:430–5. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AL-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. Atomic model of the human cardiac muscle myosin filament. Proc Natl Acad Sci U S A. 2013;110:318–23. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.AL-Khayat HA, Morris EP, Kensler RW, Squire JM. Myosin filament 3D structure in mammalian cardiac muscle. J Struct Biol. 2008;163:117–26. doi: 10.1016/j.jsb.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodhead JL, Zhao F-Q, Craig R, Egelman EH, Alamo L, Padron R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–9. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 22.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci U S A. 2008;105:2386–90. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto A, Sanchez F, Alamo L, Padron R. The myosin interacting-heads motif is present in the relaxed thick filament of the striated muscle of scorpion. J Struct Biol. 2012;180:469–78. doi: 10.1016/j.jsb.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Sola M, Al-Khayat HA, Behra M, Kensler RW. Zebrafish cardiac muscle thick filaments: isolation technique and three-dimensional structure. Biophys J. 2014;106:1671–80. doi: 10.1016/j.bpj.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–12. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Wilson C, Naber N, Pate E, Cooke R. The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle. Biophys J. 2014;107:1637–46. doi: 10.1016/j.bpj.2014.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colson BA, Bekyarova T, Fitzsimons DP, Irving TC, Moss RL. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J Mol Biol. 2007;367:36–41. doi: 10.1016/j.jmb.2006.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kensler RW, Harris SP. The structure of isolated cardiac Myosin thick filaments from cardiac Myosin binding protein-C knockout mice. Biophys J. 2008;94:1707–18. doi: 10.1529/biophysj.107.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dijk SJ, Witt CC, Harris SP. Normal cardiac contraction in mice lacking the proline-alanine rich region and C1 domain of cardiac myosin binding protein C. J Mol Cell Cardiol. 2015;88:124–32. doi: 10.1016/j.yjmcc.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du XJ, Fang L, Gao XM, Kiriazis H, Feng X, Hotchkin E, et al. Genetic enhancement of ventricular contractility protects against pressure-overload- induced cardiac dysfunction. J Mol Cell Cardiol. 2004;37:979–87. doi: 10.1016/j.yjmcc.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Zaremba R, Merkus D, Hamdani N, Lamers JMJ, Paulus WJ, dos Remedios C, et al. Quantitative analysis of myofilament protein phosphorylation in small cardiac biopsies. Proteomics Clin Appl. 2007;1:1285–90. doi: 10.1002/prca.200600891. [DOI] [PubMed] [Google Scholar]
- 32.Pope B, Hoh J, Weeds A. The ATPase activities of rat cardiac myosin isoenzymes. FEBS Lett. 1980;118:205–8. doi: 10.1016/0014-5793(80)80219-5. [DOI] [PubMed] [Google Scholar]
- 33.Colson BA, Bekyarova T, Locher MR, Fitzsimons DP, Irving TC, Moss RL. Protein kinase A–mediated phosphorylation of cMyBP-C increases proximity of myosin heads to actin in resting myocardium. Circ Res. 2008;103:244–51. doi: 10.1161/CIRCRESAHA.108.178996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mamidi R, Gresham KS, Li A, dos Remedios CG, Stelzer JE. Molecular effects of the myosin activator omecamtiv mecarbil on contractile properties of skinned myocardium lacking cardiac myosin binding protein-C. J Mol Cell Cardiol. 2015;85:262–72. doi: 10.1016/j.yjmcc.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorn GW, Robbins J, Ball N, Walsh RA. Myosin heavy chain regulation and myocyte contractile depression after LV hypertrophy in aortic-banded mice. Am J Physiol Heart Circ Physiol. 1994;267:H400–H5. doi: 10.1152/ajpheart.1994.267.1.H400. [DOI] [PubMed] [Google Scholar]
- 36.Koretz JF. Effects of C-protein on synthetic myosin filament structure. Biophys J. 1979;27:433–46. doi: 10.1016/S0006-3495(79)85227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korte FS, McDonald KS, Harris SP, Moss RL. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ Res. 2003;93:752–8. doi: 10.1161/01.RES.0000096363.85588.9A. [DOI] [PubMed] [Google Scholar]
- 38.Stelzer JE, Fitzsimons DP, Moss RL. Ablation of myosin-binding protein-C accelerates force development in mouse myocardium. Biophys J. 2006;90:4119–27. doi: 10.1529/biophysj.105.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stelzer JE, Dunning SB, Moss RL. Ablation of cardiac myosin-binding protein- C accelerates stretch activation in murine skinned myocardium. Circ Res. 2006;98:1212–8. doi: 10.1161/01.RES.0000219863.94390.ce. [DOI] [PubMed] [Google Scholar]
- 40.Ashrafian H, Redwood C, Blair E, Watkins H. Hypertrophic cardiomyopathy:a paradigm for myocardial energy depletion. Trends Genet. 2003;19:263–8. doi: 10.1016/S0168-9525(03)00081-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.