Abstract
The Rb-E2F axis is an important pathway involved in cell cycle control that is deregulated in a number of cancers. E2f transcription factors have distinct roles in the control of cell proliferation, cell survival and differentiation in a variety of tissues. We have previously shown that E2fs are important downstream targets of a CSF-1 signaling cascade involved in myeloid development. In cancer, tumor associated macrophages (TAMs) are recruited to the tumor stroma in response to cytokines secreted by tumor cells and are believed to facilitate tumor cell invasion and metastasis. Using the MMTV-Polyoma Middle T antigen (PyMT) mouse model of human ductal carcinoma, we show that the specific ablation of E2f3 in TAMs, but not in tumor epithelial cells, attenuates lung metastasis without affecting primary tumor growth. Histological analysis and gene expression profiling suggests that E2f3 does not impact the proliferation or survival of TAMs, but rather controls a novel gene expression signature associated with cytoskeleton rearrangements, cell migration and adhesion. This E2f3-TAM gene expression signature was sufficient to predict cancer recurrence and overall survival of ER-positive breast cancer patients. Interestingly, we find that E2f3b but not E2f3a levels are elevated in TAMs from PyMT mammary glands relative to controls, suggesting a differential role for these isoforms in metastasis. In summary, these findings identify E2f3 as a key transcription factor in TAMs that influences the tumor microenvironment and tumor cell metastasis.
Keywords: E2fs, metastasis, stroma, macrophages and breast cancer
Introduction
Breast cancer is the second leading cause of cancer mortality in women with approximately 30% patients eventually developing metastasis 34. Metastasis is a complex process during which tumor cells colonize distant organ sites 9, 11. Metastasis contributes to 90% of deaths from solid tumors and displays diverse clinical manifestations 22. The tumor stroma or microenvironment plays a crucial role in the dissemination of tumor cells from the primary cancer. Tumor stroma is composed of extracellular as well as cellular constituents, including fibroblasts, endothelial and immune cells. During tumor progression, the tumor stroma may be altered to influence the neoplastic properties of tumor cells and facilitate their dissemination 12, 22. One of the extreme examples of how stromal cells can drive tumorigenesis was shown by Kim and colleagues where selective deletion of Smad4 in T-cells resulted in formation of epithelial cancer in the gastrointestinal tract of mice 24.
The initial response of immune cells in cancer is to recognize and destroy tumor cells. However, strategies in tumor cells to escape, suppress immune destruction or convert immune cells to have pro-tumorigenic roles often evolve 15. Tumor associated macrophages (TAMs) are recruited to the tumor stroma in response to cytokines secreted by both tumor epithelial cells and other resident stromal cells 7, 12. In addition, TAMs also activate myofibroblasts to secrete cytokines that help in the recruitment of endothelial progenitor cells and promote neovascularization 36. Clinical studies show that infiltration of TAMs correlates with poor prognosis in a variety of cancers 25, 41 and that therapeutic strategies to limit macrophage numbers are effective at decreasing cancer development 2. Similarly, depletion of macrophages using pharmacological agents, such as clodronate reduces tumor angiogenesis and metastasis in various mouse models of cancer 16, 30, 32.
Colony stimulating factor 1 (CSF-1) secreted by tumor cells facilitates the recruitment of TAMs, which in turn promote the invasion and migration of tumor cells by the secretion of EGF 47. Mice lacking the CSF-1 ligand (op/op) have reduced incidence of pulmonary metastasis 27. In human breast cancer overexpression of CSF-1 correlates with poor prognosis and dense leukocyte infiltration 23. The engagement of the CSF-1 receptor by CSF-1 leads to the activation of numerous signaling cascades during normal myeloid development, including the Rb/E2f pathway, which is involved in regulating cell proliferation, survival and differentiation 42.
Genetic alterations of RB have been reported in approximately 20-30% of breast cancers 5, 21. RB exerts its tumor suppressive effects through the regulation of E2F transcription factors 33. Increased E2F3 expression has been reported in prostate, ovarian and lung cancers 13, 14, 18, 35 and amplification of 6p22, which includes the E2F3 locus, has been observed in bladder cancer 35. Introduction of miR-148a and miR-34b/c in tumor cells target E2F3 and result in a reduction of tumor growth and metastasis 28. Collectively, these studies imply that E2F3 is frequently overexpressed or amplified in a number of cancers and that its reduction may deter cancer progression.
We have previously shown that E2fs are important downstream targets in CSF-1 signaling cascade involved in macrophage development 42. However, the role of E2f3 in the tumor microenvironment (stroma) remains to be investigated. Here, we address the potential role of E2f3 in TAMs during mammary tumorigenesis. To this end, we examined the consequence of ablating E2f3 in either TAMs or tumor cells on tumor progression and lung metastasis using the MMTV-Polyoma Middle T antigen (MMTV-PyMT) mouse model of breast cancer. We show that the loss of E2f3 in tumor macrophages, but not in mammary tumor cells, leads to the attenuation of lung metastasis without affecting the growth of the primary tumor.
Results
Loss of one copy of E2f3 reduces lung metastasis
To investigate the role of E2f3 during mammary gland tumorigenesis in mice, we used the MMTV-Polyoma Middle T antigen (PyMT) model of breast cancer. The advantage of this model is that there is rapid tumor progression accompanied by immune infiltration and lung metastasis. We generated PyMT;E2f3+l+ (n=36) and PyMT;E2f3+/− (n=19) female mice to determine whether the loss of one E2f3 allele would affect tumor growth and metastasis. Mice were palpated twice a week and time to tumor was recorded once the tumor volume reached 5mm3. Kaplan Meier plots show that loss of one copy of E2f3 had no effect on the timing of tumor onset (Figure 1A) or tumor burden (Figure 1B), suggesting that loss of a single E2f3 allele does not affect tumor development by the PyMT oncogene. Moreover, histopathological analysis of tumors isolated from PyMT;E2f3+/+ and PyMT;E2f3+/− mice revealed no gross pathological differences between the two groups of mice (Figure 1C). However, examination of lungs at end-stage showed a significant reduction of metastasis in PyMT;E2f3+/− mice (Figures 1D and 1E). In addition, to the lungs we also examined the bones, spleens and livers from PyMT;E2f3+/-and found no signs of metastasis at these sites.
Figure 1. Loss of one E2f3 allele results in reduction of lung metastasis.
(A) Kaplan–Meier tumor-free curves (time from birth to tumor onset) of MMTV-PyMT; E2f3+/+ (black line) and MMTV-PyMT; E2f3+/− (red line) mice. The number of mice in each cohort is indicated by n.
(B) Bar graphs showing tumor burden of mice described above. The mammary tumors were harvested from mice at termination of the study at 110 days. Percent tumor burden was calculated as described in methods. No significant difference was observed between MMTV-PyMT; E2f3+/+ (black bar) and MMTV-PyMT; E2f3+/− (red bar). Values represent mean ± SD.
(C) Paraffin sections of mammary tumors from 110-day old mice MMTV-PyMT;E2f3+/+ (left) and MMTV-PyMT;E2f3+/− (right) mice were stained with H&E (top panel), Ki67 (middle panel) or F4/80 (bottom panel). The Ki67 or F4/80 positive cells were quantified from section as described above.
(D) Analysis of the metastatic area in lungs isolated from MMTV-PyMT; E2f3+/+ (black diamonds) and MMTV-PyMT; E2f3+/− (red diamonds). Percentage metastatic area was calculated from the H&E section from all five lobes of lungs. The data represents the mean of each genotype indicated by horizontal line. Significant difference was observed between the two groups (p<0.003).
(E) Representative picture of H&E stained lung showing metastatic lesion in mice from MMTV-PyMT; E2f3+/+ (left) and MMTV-PyMT; E2f3+/− (right).
Since E2f3 null mice are not viable, we used a conditional E2f3 allele and a bitransgenic system where the expression of Cre is under the control of the mouse mammary tumor virus promoter (MMTV-rtTa;teto-Cre) to specifically ablate E2f3 in the mammary ductal epithelium. Two cohorts of female mice were generated (PyMT;E2f3loxp/loxp;MMTV-rtTa and PyMT;E2f3loxp/loxp;MMTV-rtTa;teto-Cre) and fed with food containing doxycycline (1mg/kg). Surprisingly, ablation of E2f3 in PyMT;E2f3loxp/loxp;MMTV-rtTa; teto-Cre mice resulted in a slight but significant delay in tumor initiation (p<0.003), but had little impact on tumor burden and metastasis to the lungs (Supplementary Figure S1A-S1C). Examination of mammary glands revealed no change in tumor cell proliferation or macrophage infiltration between the two cohorts of mice (Supplementary Figure S1D). Together, these results suggest that the reduction of lung metastasis initially observed in PyMT; E2f3+/− mice may be mediated through a cell non-autonomous mechanism.
E2f3 ablation in TAMs leads to the reduction of lung metastasis
Macrophages are an important part of tumor stroma and have been shown to be important in tumor cell dissemination during metastasis 12, 22. Given that global deletion of one allele of E2f3 led to a decrease in lung metastasis, we entertained the hypothesis that E2f3 in macrophages may contribute to metastasis in mice. To test this hypothesis we used the Lysozyme Cre (LysCre) transgene to conditionally inactivate E2f3 in macrophages and compare tumor growth and metastasis in PyMT;LysCre;E2f3loxp/loxp, PyMT;LysCre; E2f3+/loxp and PyMT;E2f3loxp/loxp mice. PCR genotyping of macrophages isolated from tumors and lungs showed that Cre expression resulted in deletion of E2f3, albeit not in one hundred percent of cells (Figure 2A).
Figure 2. Deletion of E2f3 in macrophages leads to attenuation of lung metastasis.
(A) E2f3 genotyping on genomic DNA isolated from TAMs of MMTV-PyMT; LysCre; E2f3loxp/loxp (lanes 1, 3, 4 and 6) and MMTV-PyMT; LysCre; E2f3+/loxp (lanes 2 and 5) mice.
(B) Kaplan–Meier tumor-free curves (time from birth to tumor onset) of MMTV-PyMT; E2f3loxp/loxp (black line), MMTV-PyMT;LysCre;E2f3+/loxp (pink line) and MMTV-PyMT; LysCre;E2f3loxp/loxp (red line) cohorts. The number of mice in each cohort is indicated by n.
(C) Bar graphs showing tumor burden of mice. The tumors were harvested from mice at termination of the study at 110 days. Percent tumor burden was calculated as described in methods. MMTV-PyMT; E2f3loxp/loxp (black bar), MMTV-PyMT; LysCre; E2f3+/loxp (pink bar) and MMTV-PyMT; LysCre; E2f3loxp/loxp (red bar). Values represent mean ± SD.
(D) Analysis of metastasis in lungs isolated from MMTV-PyMT; E2f3loxp/loxp (black diamonds), MMTV-PyMT; LysCre; E2f3+/loxp (pink diamonds) and MMTV-PyMT; LysCre; E2f3loxp/loxp (red diamonds). Percentage metastatic area was calculated from the H&E section from all five lobes of lungs. The data represents the mean of each genotype indicated by horizontal line.
(E) Representative picture of H&E stained lung showing metastatic lesion in mice from MMTV-PyMT; E2f3loxp/loxp, MMTV-PyMT; LysCre; E2f3+/loxp and MMTV-PyMT; LysCre; E2f3loxp/loxp.
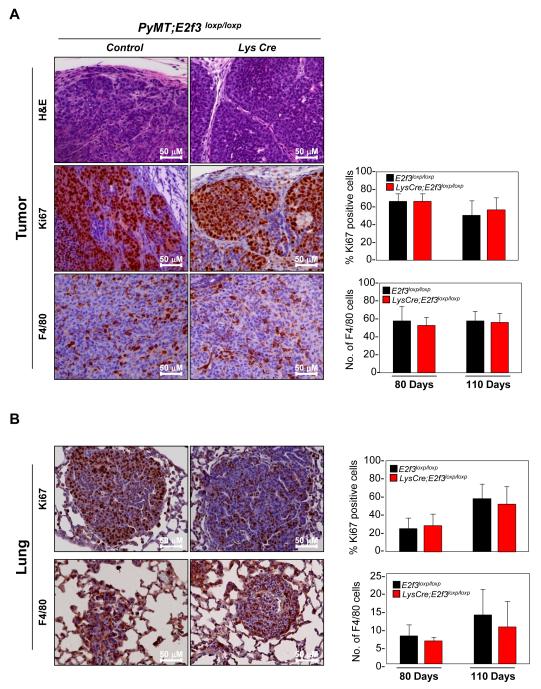
While the tumor burden in the three cohorts of mice was similar (Figure 2B and 2C), lung metastasis in PyMT;LysCre; E2f3loxp/loxp and PyMT;LysCre; E2f3+/loxp were significantly reduced (p<0.01) when compared to PyMT;E2f3loxp/loxp (control) mice (Figure 2D and 2E). We measured macrophage numbers by immuno-histochemistry (IHC) staining using F4/80-antibody on early adenomas (80-old day tumor mice) and late stage carcinomas (110- day old tumor mice). This analysis revealed no significant changes in macrophage numbers in tumors at either early or late stages of progression or in lung metastasis when E2f3 was deleted in macrophages (Figures 3A-3B and S2-S3), suggesting that E2f3 is not required for the proliferation of TAMs. Consistent with the absence of an effect on tumor burden, E2f3 deletion in macrophages had no effect on the proliferation of tumor cells in the primary tumor or metastatic sites (Figures 3A-3B and S2). Analysis of inguinal mammary glands from virgin, pregnant and involuting LysCre; E2f3loxp/loxp and E2f3loxp/loxp mice revealed normal mammary gland development 44 (Supplementary Figure S4). Together, these results suggest that E2f3 ablation in TAMs results in a significant reduction of lung metastasis without affecting mammary development or tumor initiation and growth.
Figure 3. E2f3 is not required for macrophages proliferation.
H&E and immuno-histochemistry of tumors isolated from 110- or 80- day old MMTV-PyMT;E2f3loxp/loxp (control) and MMTV-PyMT; LysCre;E2f3loxp/loxp mice. (A) Mammary tumors and (B) Lungs. The paraffin sections were stained using Ki67- and F4/80- antibodies. Quantification of Ki67- and F4/80- positive cells (right panels).
E2f3 in lung and tumor macrophages promotes metastasis
Macrophages have been shown to contribute to tumor cell dissemination from the primary tumor or in the establishment of a metastatic niche at distant organ sites. To explore the role of E2f3 in TAMs we first orthotopically injected highly metastatic syngeneic MVT-1 cells into the fat pad of E2f3loxp/loxp and LysCre;E2f3loxp/loxp mice and evaluated lung metastasis 4 weeks later. While tumors in both groups reached the same size, there were significantly fewer metastatic lesions in lungs of LysCre;E2f3loxp/loxp mice compared to controls (Figure 4A). F4/80 and Ki67 antibody staining of primary tumors and metastases showed no differences in either macrophage infiltration or in the proliferation of tumor cells (4B).
Figure 4. E2f3 deletion leads to reduction in lung metastasis after orthotopic injection of tumor cells into fat pad.
(A) H&E stained section of lung showing metastatic lesions, collected 28 days after injection of MVT-1 cells into mammary fat pad of LysCre;E2f3loxp/loxp and E2f3loxp/loxp mice. (top panel). A significant reduction in lung metastasis between the two cohorts can be observed (p<0.008) (top right panel). Lung section of the indicated genotype stained with Ki67 (middle panel) or macrophage specific F4/80 antibody (bottom panel).
(B) Mammary tumor isolated from the mice described above stained with Ki67 (top panel) and F4/80 (bottom panel). Quantification is depicted in the right panels.
Next, we injected MVT-1 cells into the tail vein of LysCre; E2f3loxp/loxp and E2f3loxp/loxp mice and examined the seeding and establishment of tumor metastases in lungs. Compared to controls, deletion of E2f3 in macrophages resulted in a significant reduction (p<0.01) of the metastatic lung area (Figure 5A). Macrophage infiltration and tumor cell proliferation at the metastatic site was similar in both groups of mice (Figure 5B). Together, these results suggest that E2f3 in macrophages promotes the metastasis of mammary tumor cells to the lung.
Figure 5. E2f3 deletion leads to reduction in lung metastasis after tail vein injection of tumor cells.
(A) H&E stained section of lung showing metastatic lesions, collected 14 days after the injection of MVT-1 cells into tail vein of LysCre;E2f3loxp/loxp and E2f3loxp/loxp mice. A significant reduction in lung metastasis between the two cohorts can be observed (p<0.01). The data represents the mean of each genotype and is indicated by horizontal line. Quantification of lung metastasis from H&E stained lung sections (top right panel).
(B) Lung sections stained with Ki67 antibody (top panel) or F4/80 antibody (bottom panel). Quantification of Ki67 and F4/80 positive cells. (right panel).
Loss of E2f3 leads to disruption of cell adhesion and cytoskeleton genes
To explore the underlying molecular mechanism by which E2f3 in macrophages promote lung metastasis, we analyzed global gene expression in control and E2f3 deleted TAMs using an Affymetrix platform. To this end, RNA from F4/80+ TAMs collected from PyMT; E2f3loxp/loxp (control) and PyMT; LysCre; E2f3loxp/loxp mice was processed and used to query mouse Affymetrix 430 2.0 oligo-arrays. An unbiased method similar to Gene Set Enrichment Analysis 29 was used to identify genes that were differentially expressed between the two groups. Surprisingly, the vast majority of the 104 differentially expressed genes were upregulated in the E2f3 deleted TAMs compared to controls (>1.5 fold, p<0.001; Figure 6A). Quantitative real time (RT-PCR) expression analysis confirmed the increased levels of a subset of these mRNAs in E2f3 depleted TAMs (Figure 6B). Gene ontology of differentially expressed genes showed an enrichment of gene functions related to cell migration, adhesion, cytoskeleton and signaling networks and a conspicuous absence of cell cycle related functions (Figure 6C and Supplementary Table 1).
Figure 6. Loss of E2f3 in TAMs leads to disruption of genes involved in cell adhesion.
(A) Heatmap showing hierarchical clustering analysis of genes differentially expressed between MMTV-PyMT;E2f3loxp/loxp (control) and MMTV-PyMT; LysCre;E2f3loxp/loxp mice (p<0.001). TAMs isolated from mice of the indicated genotype were used for microarray analysis. Four independent samples were analyzed from each genetic group.
(B) Quantitative real-time PCR to validate the expression of some differentially expressed genes. The value on the y-axis represents fold induction.
(C) Gene ontology performed on the differentially expressed genes as determined by the Ingenuity Pathway Analysis (p<0.001).
(D) Representative image showing the rate of migration of MVT-1 cells in response to conditioned media obtained from TAMs of indicated genotypes (left panel). Quantification of number of migrated MVT-1 cells (right panel). Values represent mean ± SD
(E) The figure depicts percentage of E2F sites conserved between the human and mouse promoter.
(F) ChIP assay showing E2F3 recruitment on promoters of genes in wild type TAMs. No E2F3 loading is observed on promoters devoid of E2f binding site ‘no site (NS)’.
The matricellular proteins (Sparcl1, Jam2 and Fbln5) participate in the assembly and stabilization of extracellular matrix structures and are involved in cell adhesion and migration. Their expression has been found to be down regulated in a number of cancers 20, 38, 48. Our microarray and RT-PCR data showed increased expression of these genes in E2f3 deleted TAMs, suggesting that their increased expression may lead to reduced migration of tumor cells. Based on these findings, we evaluated whether deletion of E2f3 in TAMs might impair tumor cell migration. Migration assays were performed using MVT-1 cells and conditioned media collected from TAMs of PyMT;E2f3loxp/loxp (control) and PyMT;LysCre;E2f3loxp/loxp mice. These assays showed a significant reduction in tumor cell migration (p<0.0001) when conditioned media from E2f3 ablated TAMs was used (Figure 6D and Supplementary Figure S5A).
Sequence analysis of gene promoters revealed that 51 out of the 104 differentially expressed genes contained consensus E2F binding sites within 1kb from the transcriptional start (TS) site, and 14 of the 51 putative target promoters had E2F consensus sites that were conserved between mouse and human (Figure 6E). Analysis of a subset of these using Chromatin immunoprecipitation (ChIP) assays and F4/80+ sorted TAMs from tumors showed E2f3 directly bound to these conserved gene promoters (Figure 6F and Supplementary Figure S5B).
The E2f3 locus encodes two isoforms, E2f3a and E2f3b, whose expression is driven by distinct promoters 26. Based on their expression pattern during the cell cycle and their ability to physically interact with Rb, it was suggested that E2f3a may function as a transcriptional activator and E2f3b as a transcriptional repressor 1. Interestingly, RT-PCR gene expression analysis on FACS sorted F4/80+ macrophages showed that E2f3b levels were significantly upregulated in TAMs relative to normal macrophages, whereas E2f3a expression was unchanged (Supplementary Figure S6). From these experiments we conclude that E2f3b expression in macrophages is responsive to tumor cells. While the individual roles of E2f3a and E2f3b remain to be determined, the fact that both isoforms are deleted in PyMT;LysCre;E2f3loxp/loxp mice, may suggest that E2f3b may have a specific function in the repression of TAM related gene expression.
E2f3 TAM gene signature predicts good prognosis in breast cancer patients
To determine the relevance of these findings to human breast cancer, we used the E2F3-related TAM gene expression signature identified in Figure 6A to query previously published global expression profiles derived from laser-captured tumor stroma (49 samples) and adjacent normal stroma (52 samples) in breast cancer patients 17. The 104-gene signature derived from mouse E2f3 deficient TAMs was able to perfectly segregate normal from tumor stroma (Figure 7A). Many of the genes in this signature were expressed at higher levels in normal stroma when compared to tumor-stroma (Supplementary Table 2), consistent with lower levels of E2F3 expression in the normal stroma compartment (p<0.015; data not shown).
Figure 7. E2f3 TAM gene signature predicts survival in human breast cancer patient datasets.
(A) Heat map displaying the human orthologs corresponding to differentially expressed genes from mouse E2f3 TAM list using Finak dataset. The dendogram obtained from a hierarchical clustering with Euclidean distances is shown on top of the heat map. Red and green regions in the heat map indicate genes that are up-regulated or down-regulated with respect to the median gene expression over the patients.
(B) Kaplan-Meier plots predicting overall survival using Finak, Wang, NKI and Stockholm breast cancer datasets.
To determine whether the E2F3 TAM gene signature correlated with clinical outcome, we used unsupervised clustering of expression data obtained from four independent breast cancer patient datasets. The comparison of mouse E2f3 TAM gene signature with human breast cancer patient datasets (Finak, NKI, Stockholm and Wang) predicted a significant better overall survival (p<0.05) (Figure 7B). Because the NKI and Wang datasets have a large cohort of ER-positive and ER-negative breast cancer patients, we queried these two patient subsets independently. This analysis shows that the E2f3 mouse gene signature predicted good prognosis in ER-positive, but not in ER-negative, breast cancer patients (Supplementary Figure S7). From these results we conclude that the E2f3 TAM gene signature identified in E2f3 deleted macrophages is represented within normal stroma and is associated with favorable clinical outcome in breast cancer patients.
Discussion
The Rb-E2F axis represents an important pathway involved in cell cycle control that is disrupted in cancer 10. Alterations in RB or components that regulate the RB pathway occur in the majority of cancers, including breast cancer 5, 21. Recent studies have demonstrated the involvement of E2Fs in driving Her2/Neu- and Myc-induced mammary tumorigenesis in mice 46. However, the role of E2f3 in tumor stroma during cancer progression and metastasis remains to be investigated. Using a PyMT mammary cancer model we show here that specific ablation of E2f3 in TAMs attenuates lung metastasis without having a major effect on the growth of the primary tumor.
Increased E2F3 expression has been observed in human prostate, ovarian and lung cancers 13, 18, 35. In a mouse model of thyroid cancer, a role for E2f3 has been implicated in the metastasis of tumor cells to the liver and lung 49, however, whether cell autonomous or non-autonomous mechanisms were at play was not determined. Using conditional knockout approaches we show that ablation of E2f3 in TAMs results in decreased pulmonary metastasis without affecting the growth of the primary tumor, suggesting that E2f3 in the tumor stroma has an important cell non-autonomous role in promoting the metastatic potential of tumor cells.
The E2F family of transcription factors is conserved from nematodes to mammals, with three genes encoding one activator and two repressors in C. elegans and eight genes encoding three activators and five repressors in humans 8. E2Fs were identified as factors that control the cell cycle, apoptosis, differentiation and stress responses, thus E2Fs have been bestowed with a role as master regulators of cell proliferation 33. Studies in flies, worms and mice support a role for E2Fs beyond cell cycle control. For example, loss-of-function studies identified roles for lin-35 (RB1 orthologue) and efl-1 (E2F1-3 orthologue) in epidermal growth factor mediated cell fate determination in C. elegans during vulval development 8. Additional roles for mammalian E2Fs in angiogenesis, adipogenesis and cell migration have also been described 3, 8, 31.
The E2f3 locus encodes two isoforms, E2f3a and E2f3b, whose expression is driven by distinct promoters 26. Previous studies from the Tlsty lab show that inactivation of Rb pathway and elevated E2f3 expression are characteristic features of basal-like cancer 19. A recent study from our laboratory has shown increased E2f3a expression in tumor cells overexpressing the Erb2/Her2 oncogene 46. Consistent with these findings, we show here that deletion of E2f3 in the mammary epithelium of PyMT mice also delays tumor onset, suggesting that the E2f3a isoform may have oncogenic functions, likely through promoting cell proliferation and cell survival, in a number of mammary tumor models. Whereas E2f3a is highly expressed in tumor cells, we observe that E2f3b is highly expressed in TAMs. We find that E2f3 in TAMs directly binds to regulatory sequences of a number of genes that negatively regulate tumor cell metastasis. The expression of these genes in TAMs is increased upon the loss of the E2f3 allele. Thus, we suggest that E2f3b is the likely isoform encoded by the E2f3 locus that functions as a transcriptional repressor in TAMs to promote metastasis. Because the Cre mediated ablation of the E2f3 floxed allele used here and in previous studies results in deletion of both E2f3 isoforms, the relative contributions of each isoform during tumorigenesis remains to be rigorously determined. In the present study, we show that E2F3 controls a gene expression signature associated with cytoskeletal rearrangements and cell adhesion. Surprisingly, E2f3 ablation in TAMs had no effect on their proliferation nor in the expression of cell cycle regulated genes. It is well recognized that the ECM is not simply a scaffold for tumor cells but instead provides critical signals that affect tumor cell growth, survival and migration 4. For instance, one of the proteins that promote ECM deposition is secreted protein acidic and rich in cysteine (Sparc). The absence of Sparc is associated with decreased collagen deposition and Sparc−/− mice have enhanced tumorigenesis 4, 6.
Importantly, it was shown that Sparc produced by host leukocytes, rather than tumor cells, promotes the assembly and function of tumor-associated stroma through the organization of collagen type IV 39. Similarly, Jam2, Fbln5 and Sparcl1 are matricellular proteins that participate in the assembly and stabilization of extracellular matrix structures involved in cell adhesion and migration. The expression of these genes is downregulated in a number of human malignancies 20, 48. The role of these proteins in inhibiting the migration of cancer cells is well documented 4, 40. However, the cells that produce these factors and the mechanisms involved in their gene regulation remain poorly understood. We find that ablation of E2f3 in TAMs leads to increased expression of Jam2, Fbln5, Col18A1, Sparcl1 and other secreted factors with roles in cell migration and adhesion, which are known to impede migration of tumor cells. Consistent with the upregulation of this secretory cell adhesion signature in E2f3 deficient TAMs, analysis of the Finak stroma-specific dataset 17 revealed that expression of this signature was downregulated in tumor stroma relative to normal stroma. This gene signature also predicted better survival outcomes in the Finak, NKI, Wang and Stockholm datasets 17, 37, 43, 45. We would thus suggest that elevation of E2f3b expression and repression of this secretory cell adhesion signature in TAMs may play a role during tumor progression and metastasis in breast cancer patients.
Materials and Methods
Mammary Tumor Models
All animals used in the study were 5th generation FVB/N background. Mice carrying PyMT oncogene under control of MMTV promoter were used in this study. Macrophage specific deletion of E2f3f/f was achieved by breeding them with the mice carrying Cre recombinase driven by the lysozyme promoter (LysCre). To delete E2f3loxp/loxp in mammary epithelial cells, mice were bred with MMTV–rtTA;teto-Cre mice. Induction of the MMTV-rtTA/teto-Cre system was achieved by administration of doxycycline in food (1mg/kg) at 4 weeks of age and continued until animals were sacrificed. Mice were palpated twice a week for tumors, they were considered tumor free until the tumors were 5mm. Mice were sacrificed when they were 110- days old or reached early removal criteria (ERC). Genotyping was performed on genomic DNA isolated from tail clips of the mice.
Orthotropic Injections
Briefly 2×105 MVT-1 cells were injected either through the tail vein or subcutaneously into the fat pad of 8-10 week old E2f3loxp/loxp and LysCre; E2f3loxp/loxp mice. Mice were sacrificed 2-4 weeks after the injection.
Histology and Immunohistochemistry
For whole mount staining of mammary glands, inguinal gland #4 or #9 were removed, fixed in Carnoys fixative at 4°C overnight, re-hydrated and stained with Carmine Red. Tumors and lungs removed from the mice were fixed in 10% buffered formalin and embedded in paraffin. To quantify metastatic foci in the lungs, each lobe was sectioned and stained with hemotoxylin and eosin (H&E). Image J software was used to quantify the metastatic area in each lobe of the lung. For H&E and IHC staining, 5um sections were cut, deparaffinized in xylene and rehydrated with graded ethanol series. The tumor and lung sections were then stained with either α-F4/80 (Invitrogen MF-48004) or α-Ki67 (Pharmingen 550609) antibodies as described below. All primary and secondary antibodies were diluted in DAKO diluent (DAKO). Sections were blocked with M.O.M. blocking reagent (Vector Labs) for 30min and incubated with primary antibody for 30min. The sections were rinsed and incubated with a biotinylated secondary antibody for 15min. Following the biotinylated secondary antibody, the streptavidin-Alexa dye conjugate was applied for 15min. Sections were washed for 5min with Phosphate-buffered saline (PBS) before incubating with DAB for 2min. Sections were washed in water and counter stained using Meyers hematoxylin followed by a brief rinse in DI water and mounted using Gel/Mount (Biomed, Foster City, CA).
Image Analysis
Images of histological and IHC sections were taken with Axio digital camera (Zeiss) mounted on the Axioskop microscope (Zeiss). Whole mount images were taken with a Coolpix 5700 digital camera (Nikon). Image files were processed using AxioVision 4.3 software (Zeiss).
Flow Cytometry
Tumors harvested from the mice were minced and digested with collagenase. Single cell suspensions were prepared after passing through a 40 micron mesh and stained with fluorochrome conjugated F4/80 antibody (Invitrogen, MF48020). After staining the cells were sorted using FACS Aria (Becton Dickinson, BD).
Microarray analysis
F4/80+ TAMs were isolated from tumor samples by FACS as described above. Four independent samples from each genetic group were used for analysis by Affymetrix microarray. RNA was isolated using TRIzol reagent. RNA was then subjected to purification and processed for hybridization to Affymetrix Mouse Genome 430 2.0 Arrays. Expression values were normalized and log transformed using RMAExpress, normalized data was then analyzed using BRB-ArrayTools 3.7.0. Differentially expressed genes that increased or decreased at p<0.001 in PyMT;LysCre;E2f3loxp/loxp samples relative to control samples were used to generate the heatmaps. The microarray data were deposited with Gene Expression Omnibus (GEO) and can be viewed at http://www.ncbi.nlm.nih.gov/geo/ GSE67081.
Real-Time RT-PCR
RNA was isolated using Trizol reagent from F4/80+ TAM. Reverse transcription of total RNA was performed using Superscript III reverse transcriptase (Invitrogen) and RNAse Inhibitor (Roche) according to the manufacturer’s protocol. Real-time PCR was performed using a BioRad iCycler and reactions were performed in triplicate and relative amounts of cDNA were normalized to RPL4.
Migration Assay
TAMs were isolated from mammary tumors of PyMT;E2f3loxp/loxp (control) and PyMT;LysCre;E2f3loxp/loxp mice as described above. After FACS sorting TAMs were plated in 6 well plates in serum free media overnight following which the conditioned media was collected and used for the migration assay. Transwell inserts containing 1×105 MTV-1 cells were placed in 24-well plates containing conditioned media obtained from TAMs. The migration of MVT-1 cells was allowed to proceed overnight. The inserts were stained using the standard protocol following manufactures instructions. Images were acquired using Nikon microscope and number of cells were quantified using image J software.
ChIP Assays
For ChIP assays, F4/80+ TAMs were crosslinked and chromatin was sonicated to an average size of 200-1000 bp. Lysates were subsequently pre-cleared with Salmon Sperm DNA/Protein G agarose slurry. Antibodies specific to α-E2F3 (2ug, SC-878) were then added to each sample and incubated overnight at 4°C. Antibody-protein-DNA complexes were recovered by addition of Salmon Sperm DNA/Protein G agarose slurry after incubation for 1h at 4°C. Following extensive washing, the complexes were eluted and decrosslinked at 65°C for 4h. Finally, samples were treated with Proteinase K (Roche), RNase A (Roche) and purified through Qiaquick columns (Qiagen). Real-time PCR quantification of immunoprecipitated DNA was performed using the Biorad iCycler with primers specific for the indicated promoter regions. Sequences for the primers used are available upon request.
Human Stroma Heatmap
Analysis of the mouse E2f3 TAM microarray data led to the identification of 104 differentially expressed unique genes. These genes were queried against a published human breast stroma microarray dataset (GSE4823, henceforth referred as Finak dataset), which is publicly available and accessible through the NCBI GEO website. A heat map was generated using these genes on the Finak human stroma dataset (52 normal stroma and 49 tumor stroma samples) using hierarchical clustering with Euclidean distance measures.
Survival analyses of breast cancer patient cohorts
A Cox proportional hazards model is applied to predict an outcome for each patient from the gene expression of the human orthologs that correspond to differentially regulated genes from mouse E2f3 TAMs. The patients are then split into two groups (high-risk and low-risk patients) and displayed in a Kaplan-Meier plot. A log-rank test was applied to evaluate statistical significance between the two groups. Details are provided in the supplementary methods.
Statistical Analyses
Data are expressed as mean ± SD. Statistical significance was determined using ANOVA and Wilcoxon Rank Sum tests.
Supplementary Material
Acknowledgement
We thank Bryan Mcelwain for technical assistance with flow cytometry. This work was funded by NIH grants R01CA121275, R01CA098956 and P01CA097189 to GL. P.T and N.S are recipients of the T32 fellowship (CA 106196-06).
Footnotes
Conflict of Interest: “The authors disclose no potential conflicts of interest.”
References
- 1.Adams M, Sears R, Nuckolls F, Leone G, Nevins J. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol Cell Biol. 2000;20:3633–3639. doi: 10.1128/mcb.20.10.3633-3639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit Rev Oncol Hematol. 2008;66:1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clapé C, Chavey C, et al. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol. 2011;13:1146–1152. doi: 10.1038/ncb2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol. 2002;14:608–616. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 5.Bosco EE, Knudsen ES. RB in breast cancer: at the crossroads of tumorigenesis and treatment. Cell Cycle. 2007;6:667–671. doi: 10.4161/cc.6.6.3988. [DOI] [PubMed] [Google Scholar]
- 6.Brekken RA, Puolakkainen P, Graves DC, Workman G, Lubkin SR, Sage EH. Enhanced growth of tumors in SPARC null mice is associated with changes in the ECM. J Clin Invest. 2003;111:487–495. doi: 10.1172/JCI16804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol. 2012;14:159–167. doi: 10.1038/ncb2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceol CJ, Horvitz HR. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol Cell. 2001;7:461–473. doi: 10.1016/s1097-2765(01)00194-0. [DOI] [PubMed] [Google Scholar]
- 9.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Tsai S, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang A, Massagué J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condeelis J, Pollard J. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Cooper C, Nicholson A, Foster C, Dodson A, Edwards S, Fletcher A, et al. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer. 2006;54:155–162. doi: 10.1016/j.lungcan.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 14.De Meyer T, Bijsmans IT, Van de Vijver KK, Bekaert S, Oosting J, Van Criekinge W, et al. E2Fs mediate a fundamental cell-cycle deregulation in high-grade serous ovarian carcinomas. J Pathol. 2009;217:14–20. doi: 10.1002/path.2452. [DOI] [PubMed] [Google Scholar]
- 15.DeNardo DG, Johansson M, Coussens LM. Immune cells as mediators of solid tumor metastasis. Cancer Metastasis Rev. 2008;27:11–18. doi: 10.1007/s10555-007-9100-0. [DOI] [PubMed] [Google Scholar]
- 16.Diel I, Solomayer E, Bastert G. Bisphosphonates and the prevention of metastasis: first evidences from preclinical and clinical studies. Cancer. 2000;88:3080–3088. doi: 10.1002/1097-0142(20000615)88:12+<3080::aid-cncr27>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 17.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 18.Foster C, Falconer A, Dodson A, Norman A, Dennis N, Fletcher A, et al. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23:5871–5879. doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 19.Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurley PJ, Marchionni L, Simons BW, Ross AE, Peskoe SB, Miller RM, et al. Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1) is down regulated in aggressive prostate cancers and is prognostic for poor clinical outcome. Proc Natl Acad Sci U S A. 2012;109:14977–14982. doi: 10.1073/pnas.1203525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Z, Deng T, Jones R, Li H, Herschkowitz JI, Liu JC, et al. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120:3296–3309. doi: 10.1172/JCI41490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joyce J, Pollard J. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kacinski B. CSF-1 and its receptor in ovarian, endometrial and breast cancer. Ann Med. 1995;27:79–85. doi: 10.3109/07853899509031941. [DOI] [PubMed] [Google Scholar]
- 24.Kim BG, Li C, Qiao W, Mamura M, Kasprzak B, Kasperczak B, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441:1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 25.Leek R, Lewis C, Whitehouse R, Greenall M, Clarke J, Harris A. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56:4625–4629. [PubMed] [Google Scholar]
- 26.Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, et al. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lujambio A, Calin G, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mootha V, Lindgren C, Eriksson K, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 30.Mundy G, Yoneda T. Bisphosphonates as anticancer drugs. N Engl J Med. 1998;339:398–400. doi: 10.1056/NEJM199808063390609. [DOI] [PubMed] [Google Scholar]
- 31.Myers TR, Greenwald I. lin-35 Rb acts in the major hypodermis to oppose ras-mediated vulval induction in C. elegans. Dev Cell. 2005;8:117–123. doi: 10.1016/j.devcel.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Nasser MW, Qamri Z, Deol YS, Ravi J, Powell CA, Trikha P, et al. S100A7 enhances mammary tumorigenesis through upregulation of inflammatory pathways. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 34.O'Shaughnessy J. Extending survival with chemotherapy in metastatic breast cancer. Oncologist. 2005;10(Suppl 3):20–29. doi: 10.1634/theoncologist.10-90003-20. [DOI] [PubMed] [Google Scholar]
- 35.Olsson A, Feber A, Edwards S, Te Poele R, Giddings I, Merson S, et al. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene. 2007;26:1028–1037. doi: 10.1038/sj.onc.1209854. [DOI] [PubMed] [Google Scholar]
- 36.Orimo A, Gupta P, Sgroi D, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 37.Pawitan Y, Bjöhle J, Amler L, Borg AL, Egyhazi S, Hall P, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds LE, Watson AR, Baker M, Jones TA, D'Amico G, Robinson SD, et al. Tumour angiogenesis is reduced in the Tc1 mouse model of Down's syndrome. Nature. 2010;465:813–817. doi: 10.1038/nature09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sangaletti S, Stoppacciaro A, Guiducci C, Torrisi MR, Colombo MP. Leukocyte, rather than tumor-produced SPARC, determines stroma and collagen type IV deposition in mammary carcinoma. J Exp Med. 2003;198:1475–1485. doi: 10.1084/jem.20030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stetler-Stevenson WG, Aznavoorian S, Liotta LA. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu Rev Cell Biol. 1993;9:541–573. doi: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- 41.Talmadge J, Donkor M, Scholar E. Inflammatory cell infiltration of tumors: Jekyll or Hyde. Cancer Metastasis Rev. 2007;26:373–400. doi: 10.1007/s10555-007-9072-0. [DOI] [PubMed] [Google Scholar]
- 42.Trikha P, Sharma N, Opavsky R, Reyes A, Pena C, Ostrowski MC, et al. E2f1-3 are critical for myeloid development. J Biol Chem. 2011;286:4783–4795. doi: 10.1074/jbc.M110.182733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 44.Van Nguyen A, Pollard JW. Colony stimulating factor-1 is required to recruit macrophages into the mammary gland to facilitate mammary ductal outgrowth. Dev Biol. 2002;247:11–25. doi: 10.1006/dbio.2002.0669. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 46.Wu L, de Bruin A, Wang H, Simmons T, Cleghorn W, Goldenberg LE, et al. Selective roles of E2Fs for ErbB2- and Myc-mediated mammary tumorigenesis. Oncogene. 2013 doi: 10.1038/onc.2013.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyckoff J, Wang W, Lin E, Wang Y, Pixley F, Stanley E, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 48.Yue W, Sun Q, Landreneau R, Wu C, Siegfried JM, Yu J, et al. Fibulin-5 suppresses lung cancer invasion by inhibiting matrix metalloproteinase-7 expression. Cancer Res. 2009;69:6339–6346. doi: 10.1158/0008-5472.CAN-09-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziebold U, Lee EY, Bronson RT, Lees JA. E2F3 loss has opposing effects on different pRB-deficient tumors, resulting in suppression of pituitary tumors but metastasis of medullary thyroid carcinomas. Mol Cell Biol. 2003;23:6542–6552. doi: 10.1128/MCB.23.18.6542-6552.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.