Abstract
Salmonella enterica serovar 4,[5],12:i:- is a monophasic variant of S. Typhimurium incapable of expressing the second-phase flagellar antigen (fljAB operon), and it is recognized to be one of the most prevalent serovars causing human infections. A clonal lineage characterized by phage type DT193, PulseNet PFGE profile STYMXB.0131 and multidrug resistance to ampicillin, streptomycin, sulphonamides and tetracycline (R-type ASSuT) is commonly circulating in Europe. In this study we determined the deletions affecting the fljAB operon and the resistance region responsible for the R-type ASSuT in a strain of Salmonella enterica serovar 4,5,12:i:- DT193/STYMXB.0131, through an approach based on PCRs and Southern blot hybridization of genomic DNA. Using a set of nine specific PCRs, the prevalence of the resistance region was assessed in a collection of 144 S. enterica serovar 4,[5],12:i:-/ASSuT/STYMXB.0131 strains isolated from Germany, Switzerland and Italy. A 28 kb-region is embedded between the loci STM2759 and iroB, replacing the DNA located in between, including the fljAB operon. It encompasses the genes blaTEM−1, strA-strB, sul2 and tet(B) responsible for the R-type ASSuT together with genes involved in plasmid replication and orfs of unknown function characteristically located on IncH1 plasmids. Its location and internal structure is fairly conserved in S. enterica serovar 4,[5],12:i:-/ASSuT/STYMXB.0131 strains regardless of the isolation source or country. Hence, in the S. enterica serovar 4,[5],12:i:-/ASSuT/STYMXB.0131 clonal lineage widespread in Germany, Switzerland and Italy, a resistance region derived from IncH1 plasmids has replaced the chromosomal region encoding the second flagellar phase and is an example of the stabilization of new plasmid-derived genetic material due to integration into the bacterial chromosome.
Keywords: monophasic Salmonella, antimicrobial resistance, molecular epidemiology, European clone, plasmid origin
Introduction
Multidrug resistant Salmonella enterica subsp. enterica serovar 4,[5],12:i:- (subsequently referred as S. 4,[5],12:i:-), a monophasic variant of S. Typhimurium, is recognized as an emerging public health hazard in Europe (EFSA, 2010, 2014; Hopkins et al., 2010; García et al., 2011, 2014; Gomes-Neves et al., 2014). It is mainly linked to the consumption of contaminated pork, and to a lesser extent, to other sources, such as poultry, cattle and companion animals (EFSA, 2010, 2014; García et al., 2011, 2014; Gomes-Neves et al., 2014). A clonal lineage of S. 4,[5],12:i:- characterized by phage type DT193, PulseNet pulsed field gel electrophoresis (PFGE) profile STYMXB.0131 (XBAI.0027 according to ECDC PFGE nomenclature; van Walle, 2013) and resistance to ampicillin, streptomycin, sulphonamides, and tetracycline (R-type ASSuT) is circulating in Europe (Hauser et al., 2010; Hopkins et al., 2010; Lucarelli et al., 2010; Gallati et al., 2013; Argüello et al., 2014; Barco et al., 2014). In this clonal lineage neither the resistance region nor the deletion affecting the fljAB operon has been determined. Instead in the Italian S. 4,5,12:i:- strain with PulseNet PFGE-XbaI profile STYMXB.0079 (XBAI.0096 according to ECDC) (Lucarelli et al., 2012) R-type ASSuT, the fljAB operon is affected by IS26 insertion and the genes responsible for resistance are located in two genomic regions, named resistance region (RR) 1 and RR2 inserted into two adjacent loci of the bacterial chromosome. A similar region was recently detected in S. 4,5:i:-/ASSuT strains from Belgium (Boland et al., 2015). In contrast, S. 4,[5],12:i:-/ASSuT/STYMXB.0131 strains (mainly phage type DT193) does not harbor equivalent RR1 and RR2 regions.
Therefore, the aim of this study was to determine the deletion occurred at the fljAB locus and the genetic structure and location of the resistance region in a S. 4,5,12:i:-/ASSuT/DT193/STYMXB.0131 strain isolated in Germany. Furthermore, the occurrence of the resistance region was investigated within a collection of strains isolated over the period 2006 to 2012 in Germany, Switzerland and Italy from human, food and animal sources.
Materials and methods
Strains and genomic DNA extraction
A collection of 144 epidemiologically unrelated S. 4,[5],12:i:- strains with R-type ASSuT and PFGE-XbaI profile STYMXB.0131 isolated over the period 2006–2012 in Germany, Switzerland and Italy from human, food and animal sources were included in this study (Table S1). For PCR screening, DNA was extracted by thermal cell lysis from a 1 ml aliquot of an overnight culture grown in Luria-Bertani broth at 37°C. The strain 07-2006 (R-type ASSuT, phage type DT193, PFGE profile STYMXB.0131) isolated in 2007 from a pig lymph node in Lower Saxony (Germany) was selected to determine the deletion pattern affecting the fljAB operon and the chromosomal resistance region (Genbank accession no. KR856283). Genomic DNA of strain 07-2006 was purified using the DNeasy Blood & Tissue Kit (Qiagen, Hilden GmbH) according to the manufacturer's instruction.
Characterization of deletions affecting the fljAB region
A set of 19 PCRs were performed to determine the genetic deletion responsible for the loss of the second-flagellar phase expression (fljAB and the surrounding genes) in the strain 07-2006. Usually a PCR contained 0.4 μM of each primer, 200 μM of each dNTP, 1.5 mM MgCl2, 10X PCR buffer, 1 U Platinum Taq polymerase (Invitrogen) and 10 ng of DNA template. PCR conditions were as follows: 95°C for 30 s, 33 cycles of 95°C for 30 s, 55–68°C for 30 s, 72°C for 1–3 min and a final extension at 72°C for 4 min. Primers-used, PCR cycling conditions and sizes of the expected fragments are shown in Table S2.
Localization and genetic structure of the chromosomal resistance region
Long-range PCRs and DNA sequencing
Long-range PCRs were performed using QIAGEN LongRange PCR Kit (Qiagen GmbH, Hilden Germany). All PCRs were performed with Q-Solution. The final concentration of each primer in the reaction was 0.4 μM. As template 5–10 ng genomic DNA of strain 07-2006 was used. PCR conditions were as follows: 93°C for 3 min, 10 cycles of 93°C for 15 s, 62°C for 30 s and 68°C for 20 min followed by 28 cycles with increasing extension time for 20 s per each additional cycle. Primer-sets used in long-range PCR amplifications are shown in Table S3. Long-range PCR products were separated on a 0.8% agarose gel. The PCR fragment of the supposed size (main product) was cut from the agarose gel and purified with the illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Europe GmbH, München, Germany) according to the manufacturer's instruction. All PCR products were sequenced by Qiagen GmbH sequencing service (Hilden, Germany). For sequencing of long-range PCR products a primer walking strategy was applied. Oligonucleotide sequences used for sequencing can be obtained on request.
PCRs for linking long-range PCRs products
Gaps between adjacent long-range PCR products were completed by PCRs and consecutive DNA sequencing of the PCR products. Furthermore, genes STM2759 and iroB were completely sequenced. As indicated before, a PCR usually contained 0.4 μM of each primer, 200 μM of each dNTP, 1.5 mM MgCl2, 10X PCR buffer, 1 U Platinum Taq polymerase (Invitrogen) and 10 ng of DNA template. PCR conditions were as follows: 95°C for 30 s, 33 cycles of 95°C for 30 s, 55–56°C for 30 s, 72°C for 1–3 min and a final extension at 72°C for 4 min. Primers-used for PCRs and sequencing, PCR cycling conditions and size of resulting fragments are shown in Table S4.
Sequence assembling and analysis
Single DNA sequence runs obtained from both long-range and single PCRs were assembled to one contig and analyzed using Lasergene software package (version 8.1; DNASTAR, Madison, WI). Further genomic analysis and annotation was performed using Clone Manager 9 Professional (Scientific & Educational Software, Cary, NC). Sequence comparisons were performed using BLAST search at NCBI (http://blast.ncbi.nlm.nih.gov/).
Southern blot hybridization
The preparation of total DNAs of strain 07-2006 was performed using DNeasy Blood & Tissue Kit (Qiagen, Hilden GmbH). Total DNA (1 μg) was digested with FastDigest EcoRV (Fisher Scientific, Schwerte, Germany) and separated on a 1% agarose gel. As size standard DNA molecular weight marker MIII, DIG-labeled, (Roche Diagnostics GmbH, Mannheim, Germany) was used.
PFGE with XbaI enzyme was performed according to the PulseNet Europe protocol (Ribot et al., 2006) and using Lamda PFG Ladder (New England, Biolabs, Ipswich) as molecular weight marker.
Restriction fragments obtained by PFGE and total DNA digestion with EcoRV were transferred onto positively charged nylon membranes (Roche Diagnostics, GmbH, Mannheim, Germany) and hybridized with PCR-generated probes labeled with digoxigenin (DIG) for genes blaTEM−1, sul2, tetA(B), merA, STM2759, iroB, gltS, and methΔ using primers listed in Table S5. Hybridizing bands were detected with the Detection Starter Kit (Roche Diagnostics GmbH).
Prevalence of the identified chromosomal resistance region
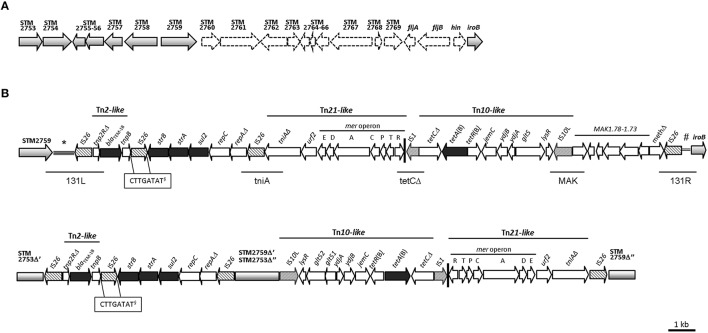
The prevalence of the resistance region was assessed in 144 epidemiologically unrelated S. 4,[5],12:i:-/ASSuT/STYMXB.0131 strains performing specific PCRs for the resistance genes blaTEM−1, strA-strB, sul2, and tet(B) (Lucarelli et al., 2010), for the left (131L) and right (131R) junctions and for the internal region of the resistance region (tniA, tetCΔ, MAK) (Tables S1, S6; Figure 1B).
Figure 1.
(A) Deletion pattern shown by the S. enterica subsp. enterica serovar 4,5,12:i:- strain 07-2006 in the chromosomal region encoding the second flagellar phase. The genetic map is based on S. Typhimurium LT2 (accession no. AE006468). The open reading frames (orfs) and the direction of their transcription are represented by gray (present) and white (absent) arrows. (B) Comparison of the organization, genetic structure and location into the bacterial chromosome of the resistance regions of the strains 07-2006 (accession no. KR856283) and 105/7/03 (accession no. HQ331538). The open reading frames (orfs) and the direction of their transcription are represented by arrows: gray, chromosomal DNA; black, resistance genes; patterned, transposase genes of IS elements; white, other orfs. Vertical black bar indicates the 38 bp terminal imperfect IR of transposon Tn21. (*)indicates the intergenic region between the chromosomal loci STM2759 and STM2760. (#)indicates the intergenic region between the chromosomal genes hin and iroB. (§), DRs of IS26. Left and right junctions, and specific internal regions tested by PCR in a collection of strains are depicted underneath by horizontal lines.
Results
The resistance region (RR3) is a single complex module of 28,204 bp (Figure 1B) that comprises the genes blaTEM−1, strA-strB, sul2 and tet(B) responsible for the R-type ASSuT, genes involved in plasmid replication and open reading frames (orfs) of unknown function. The module is inserted between two chromosomal loci, named STM2759 (encoding a putative phosphotransferase) and iroB (STM2773, a putative glycosyl transferase), causing the deletion of the genomic fragment located in between (15,716 bp). The latter comprises the genes STM2760–STM2769 (encoding different putative proteins), the fljAB operon (encoding a negative regulator of the fliC gene for the first-phase flagellar antigen and the second-phase flagellar antigen, respectively) as well as the hin gene (encoding the DNA invertase responsible for flagellar phase variation). The lacking of these genes in the genome of 07-2006 strain was confirmed by PCR amplification (Figure 1A).
In particular, the acquired DNA adjacent to STM2759 comprises a truncated Tn2 transposon flanked by two copies of IS26 in opposite orientation. This Tn2-like transposon consists of a partial resolvase gene (tnp2RΔ), the gene encoding a β-lactamase of type TEM-1B (blaTEM−1) and a presumptive transposase gene (tnpB). Downstream, the strB and strA phosphotransferase genes for streptomycin resistance, and sul2 encoding a dihydropteroate synthase enzyme responsible for resistance to sulphonamides were found, followed by repC and repAΔ, plasmid replication genes, and a third copy of an IS26 element.
Downstream a partial Tn21 transposon included tniAΔ and urf2 genes, encoding a putative transposase and a hypothetical protein, respectively, and the mercury resistance operon was found. After a defective Tn10 transposon included an IS1, tetC(B)Δ gene, tetR(B), and tetA(B) genes encoding the tetracycline repressor and resistance proteins, respectively, jemC, ydjB, ydjA, gltS, and lysR genes, with hypothetical functions and a truncated IS10-left transposase. The Tn10 transposon was followed by six orfs of unknown function together with a putative DNA modification methylase gene (methΔ) truncated by another IS26 element, which represent the right-hand end of the region.
In order to confirm the structure of the resistance region RR3, Southern blot hybridizations of genomic DNA were performed with relevant probes. According with the predicted structure of the nucleotide sequence (Genbank accession no. KR856283) two hybridization signals were obtained in the XbaI-pattern (Figure S1): a fragment of about 500 kb using probes for the genes iroB, gltS and methΔ, and a fragment of estimated 30 kb for genes blaTEM−1, sul2, tetA(B), merA, and STM2759. The occurrence of a XbaI restriction site within tetR(B) (nt position 20,248) explains the separation of the observed hybridization signals. Likewise, a 13.6 and 4.9 kb fragments were observed in EcoRV-pattern (Figure S2) when using probes for strA-strB and sul2, or blaTEM−1 and STM2759, respectively. Altogether, these results confirmed the expected number of gene copies and position.
A PCR screening of 144 S. 4,[5],12:i:-/ASSuT/ STYMXB.0131 strains (Table S1) revealed that the resistance region is highly conserved in this clonal lineage along recent years (2006–2012) regardless of source (human, food, or animal) or country of isolation (Germany, Switzerland, or Italy; Table 1). In fact, 84% of strains (n = 121) were positive for all five PCRs, which include three internal targets and the left and right junctions of the resistance module. Thus, only 16% were negative for at least one of the five expected PCR products, being the more variable loci 131L (left junction) and tniA (internal junction between repAΔ and tniAΔ; Table S1).
Table 1.
Results of the PCRs for the presence of RR3 in 144 epidemiologically unrelated S. 4,[5],12:i:-/ASSuT/STYMXB.0131 strains.
| PCR profile | Number of strains | |||||||
|---|---|---|---|---|---|---|---|---|
| 131L | tniA | tetCΔ | MAK | 131R | Total (%) | Germany | Switzerland | Italy |
| + | + | + | + | + | 121 (84) | 46 | 36 | 39 |
| − | + | + | + | + | 4 (2.8) | 0 | 0 | 4* |
| − | − | + | + | + | 2 (1.4) | 1 | 0 | 1 |
| − | + | − | - | + | 1 (0.7) | 1 | 0 | 0 |
| − | + | + | + | − | 2 (1.4) | 1 | 0 | 1 |
| − | − | − | + | − | 1 (0.7) | 0 | 0 | 1 |
| + | − | + | + | + | 8 (5.6) | 3 | 4 | 1 |
| + | + | + | − | + | 1 (0.7) | 1 | 0 | 0 |
| + | + | + | + | − | 4 (2.8) | 1 | 2 | 1 |
| TOTAL | 144 (100) | 54 | 42 | 48 | ||||
PCR-profile is based on the amplicons represented in Figure 1. Individual features of strains are compiled in Table S1.
indicates a fragment with aberrant molecular weight.
Discussion
The resistance region (RR3) is a single complex module that comprises the genes responsible for the R-type ASSuT, genes involved in plasmid replication and open reading frames (orfs) of unknown function. The module is inserted between two chromosomal loci, named STM2759 and iroB (STM2773), causing the deletion of the genomic fragment located in between. Regarding the flanking chromosomal DNA of RR3, it shows high-level sequence identity to the corresponding regions of the biphasic S. Typhimurium LT2 genome (GenBank accession no. AE006468). Indeed, 99% sequence identity was observed for the segment located upstream of the resistance module, spanning from the 5′ end of STM2758 to the intergenic region between STM2759 and STM2760, and 100% for the downstream segment, that includes the iroB gene and the intergenic non-coding sequence next to hin. Previous studies have described different deletion patterns affecting the fljAB operon and surrounding genes as a consequence of a single IS26 insertion (Soyer et al., 2009; EFSA, 2010; Laorden et al., 2010; Lucarelli et al., 2012; García et al., 2013; Boland et al., 2015). In contrast, a large resistance region bounded by direct copies of IS26 along with a 15-kb deletion was found at this location in the S. 4,[5],12:i:-/ASSuT/STYMXB.0131 strain, being responsible for the monophasic phenotype. However, the IS26 located at the fljAB operon is inserted at the same nt position (2,916,036 of GenBank accession no AE006468) of the intergenic region between the hin and iroB genes as in the 4 strains described by Lucarelli et al. (2012) and in the chromosomal resistance regions identified in S. 4,5:i:- strains from Belgium (GenBank accession no. KJ999732; Boland et al., 2015). According to previous observations (Lucarelli et al., 2012; García et al., 2013; Boland et al., 2015), our results support the hypothesis that the region encoding the second-phase flagellar antigen, having a lower average of GC content in comparison with the Salmonella core genome (45 vs. 52.2%), could be an integration hotspot for foreign DNA (Bäumler and Heffron, 1998). Therefore, it could be hypothesized that one copy of IS26 located at the fljAB region could be acted as a target or recognition site for the incorporation of additional antibiotic resistance gene(s) by another IS26 element, and so on (Reid et al., 2015). These IS26-mediated events may occur sequentially explaining the acquisition of multiple tandem copies of IS26 (four in total) and leading the assembly of the final resistance region identified in the chromosome of the S. 4,[5],12:i:-/ASSuT/STYMXB.0131 strain.
The region including the three copies of IS26 elements (Figure 1) shows 99% sequence identity with RR1, also named as Tn6029E in Reid et al. (2015), located in the chromosome of the Italian S. 4,5,12:i:- ASSuT/STYMXB.0079 strain (GenBank accession no. HQ331538) and 100% sequence identity with the Belgium S. 4,5:i:-/ASSuT strain (GenBank accession no. KJ999732). As reported by others (Hall and Cain, 2012), this element could correspond to an intermediate structure in the derivation of transposon Tn6029 which differs only in the reversed orientation of the segment strA-strB-sul2-repC-repAΔ (Cain and Hall, 2012), or be related with part of the transposon Tn6026 after the inversion of the same genetic segment (Reid et al., 2015). In fact, a BLAST search (NCBI, February 2016) revealed that the region identified in this study shares high-level identity with these composite transposons identified in other bacterial chromosomes such as E. coli O104:H4 (GenBank accession no. CP003297 and CP003301; Ahmed et al., 2012; Chowdhurry et al., 2015) or S. Typhi (SGI1, GenBank accession no. KM023773; Chiou et al., 2014); as well as carried by a number of IncH1 plasmids, for example pO111_1 (E. coli, GenBank accession no. AP010961; Ogura et al., 2009), pSRC27-H (S. Typhimurium, GenBank accession no. HQ840942; Cain and Hall, 2012) and p109/9 (S. Typhimurium, GenBank accession no. KP899805).
The composite structure of incompletes Tn21 and Tn10 transposons shows a 99% of nucleotide identity with the RR2 region of the S. 4,5,12:i:- ASSuT/STYMXB.0079 Italian strain but with some differences: (i) the segment stands inverted with Tn21-like at the left side; (ii) the gltS gene encodes a single hypothetical protein lacking the frameshift detected by Lucarelli et al. (2012); and (iii) IS10L is not contiguous with S. Typhimurium chromosomal DNA. Indeed, immediately downstream of the Tn10-like transposon and adjacent to iroB a segment of DNA, not present in RR2, was identified. It comprises six orfs of unknown function together with a putative DNA modification methylase gene (methΔ) truncated by another IS26 element. BLAST analysis (NCBI, February 2016) of these additional orfs (4709 bp; nt 25,595–30,303) revealed they are commonly part of the conserved backbone of IncH1 plasmids showing 99% sequence identity with nine of them: pO111_1 (E. coli, GenBank accession no. AP010961; Ogura et al., 2009), pSRC27-H (S. Typhimurium, GenBank accession no. HQ840942; Cain and Hall, 2012), p109/9 (S. Typhimurium, GenBank accession no. KP899805), pB71 (S. Typhimurium, GenBank accession no. KP899806), pF8475 (S. Typhimurium, GenBank accession no. KP899804), pMAK1 (GenBank S. Choleraesuis, accession no. AB366440), pHCM1 (S. Typhi, GenBank accession no. AL513383; Parkhill et al., 2001), as well as pEQ1 and pEQ2, two sequenced plasmids recovered from equine E. coli strains (GenBank accession no. KF362121 and KF362122; Dolejska et al., 2014). It is remarkable that both Tn21 and Tn10, either truncated or entire, and contiguous or not to Tn6029, have been detected among IncH1 plasmids. Therefore, it could be hypothesized that in strain S. 4,5,12:i:-/STYMXB.0079 and in strain S. 4,5,12:i:-/STYMXB.0131 two independent and different acquisitions of the same regions from IncH1 plasmids occurred, with exception of the six orfs present only in the latter strain. Similar events of DNA acquisition have been proposed for the S. 4,5:i:- Belgium strain which harbors part of the Tn21-Tn10-like structure and the mentioned IncH1 region identified in S. 4,5,12:i:-/STYMXB.0131 (Boland et al., 2015).
Despite the presence of numerous transposable elements (four copies of IS26, one copy of IS1 and one copy of IS10), a PCR screening of 144 S. 4,[5],12:i:-/ASSuT/ STYMXB.0131 strains revealed that the resistance region is highly conserved in 84% of strains regardless of source or country of isolation. The observed variability in 16% of strains could be explained, at least in part, by the repeated presence of IS26 which could recombine yielding different genetic variants. In addition, the presence of the IS26 in the same position of the fljAB operon also in other strains (Lucarelli et al., 2012; Boland et al., 2015) suggest the existence of a common ancestor harboring an IS26 in this position, where occurred IS26-mediated homologous recombination events, responsible for (i) acquisition of the new region present in the chromosome of S. 4,[5],12:i:-/ASSuT/ STYMXB.0131 07-2006, and (ii) deletion of the second phase flagellar antigen genes.
In conclusion, this study describes a resistance region identified on the chromosome of a clonal lineage of monophasic S. 4,[5],12:i:- widely distributed in at least Germany, Switzerland, and Italy. The region has replaced genes responsible for the expression of the second phase flagellar antigen in biphasic S. Typhimurium strains and represents an example of the stabilization of new plasmid-derived material into the bacterial chromosome.
Further studies are needed in order to establish if mercury operon is expressed and if this resistance island could have any effect on the bacterial physiology, as well as metabolic pathways, growth, and virulence.
Author contributions
PG, BM, and MR were responsible for sequencing the structure of the genomic resistance region. BM, RS, HH, CL provided further strains and characterized them. PG, BM, BG, and CL drafted the manuscript. PG, BM, HH, and CL designed the study and edited the manuscript. All authors read, commented on, and approved the final manuscript.
Funding
This work was supported by the Federal Institute for Risk Assessment (BfR: project no.s 46-001 and 46-003, Germany), the “Fondo de Investigación Sanitaria” of the “Instituto de Salud Carlos III” (FIS PI11-00808, Spain), co-funded by European Regional Development Fund of the European Union: a way to making Europe. PG was the recipient of a grant from the “Fundación para el Fomento en Asturias de la Investigación Científica Aplicada y la Tecnología” (FICYT, Ref. BP08-031). She performed a short stay at the Department of Biological Safety of the Federal Institute for Risk Assessment (BfR), Berlin, Germany, supported by the same grant. This work was partly supported by a grant from the Italian Ministry of Health Centro Nazionale per la Prevenzione ed il Controllo delle Malattie (CCM; project “Sorveglianza delle malattie trasmesse da alimenti e acqua (EnterNet): adeguamento del sistema italiano al quadro normativo europeo”) and by the Swiss Federal Office of Public Health, Division Communicable Diseases.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ernst Junker and Laura Busi for technical assistance. The European Centre for Disease Prevention and Control has no responsibility for the views expressed in this article.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00680
References
- Ahmed S. A., Awosika J., Baldwin C., Bishop-Lilly K. A., Biswas B., Broomall S., et al. (2012). Genomic Comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2. PLoS ONE 7:e48228. 10.1371/journal.pone.0048228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello H., Sørensen G., Carvajal A., Baggesen D. L., Rubio P., Pedersen K. (2014). Characterization of the emerging Salmonella 4,[5],12:i:- in Danish animal production. Foodborne Pathog. Dis. 11, 366–372. 10.1089/fpd.2013.1672 [DOI] [PubMed] [Google Scholar]
- Barco L., Ramon E., Cortini E., Longo A., Dalla Pozza M. C., Lettini A. A., et al. (2014). Molecular characterization of Salmonella enterica serovar 4,[5],12:i:- DT193 ASSuT strains from two outbreaks in Italy. Foodborne Pathog. Dis. 11, 138–144. 10.1089/fpd.2013.1626 [DOI] [PubMed] [Google Scholar]
- Bäumler A. J., Heffron F. (1998). Mosaic structure of the smpB-nrdE intergenic region of Salmonella enterica. J. Bacteriol. 180, 2220–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland C., Bertrand S., Mattheus W., Dierick K., Jasson V., Rosseel T., et al. (2015). Extensive genetic variability linked to IS26 insertions in the fljB promoter region of atypical monophasic variants of Salmonella Typhimurium. Appl. Environ. Microbiol. 81, 3169–3175. 10.1128/AEM.00270-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain A. K., Hall R. M. (2012). Evolution of a multiple antibiotic resistance region in IncH1 plasmids: reshaping resistance region in situ. J. Antimicrob. Chemother. 67, 2848–2853. 10.1093/jac/dks317 [DOI] [PubMed] [Google Scholar]
- Chiou C. S., Alam M., Kuo J. C., Liu Y. Y., Wang P. J. (2014). Chromosome-mediated multidrug resistance in Salmonella enterica serovar Typhi. Antimicrob. Agents Chemother. 59, 721–723. 10.1128/AAC.04081-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhurry P. R., Charles I. G., Djordjevic S. P. (2015). A role for Tn6029 in the evolution of the complex antibiotic resistance gene loci in genomic island 3 in enteroaggregative hemorrhagic Escherichia coli O104:H4. PLoS ONE 10:e0115781. 10.1371/journal.pone.0115781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejska M., Villa L., Minoia M., Guardabassi L., Carattoli A. (2014). Complete sequences of IncHI1 plasmids carrying blaCTX-M-1 and qnrS1 in equine Escherichia coli provide new insights into plasmid evolution. J. Antimicrob. Chemother. 69, 2388–2393. 10.1093/jac/dku172 [DOI] [PubMed] [Google Scholar]
- EFSA (2010). Scientific Opinion on monitoring and assessment of the public health risk of “Salmonella Typhimurium-like” strainS. EFSA J 8:1826 10.2903/j.efsa.2010.1826 [DOI] [Google Scholar]
- EFSA (2014). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2012. EFSA J. 12:3547 10.2903/j.efsa.2014.3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallati C., Stephan R., Hächler H., Malorny B., Schroeter A., Nüesch-Inderbinen M. (2013). Characterization of Salmonella enterica subsp. enterica serovar 4,[5],12:i:- clones isolated from human and other sources in Switzerland between 2007 and 2011. Foodborne Pathog. Dis. 10, 549–554. 10.1089/fpd.2012.1407 [DOI] [PubMed] [Google Scholar]
- García P., Guerra B., Bances M., Mendoza M. C., Rodicio M. R. (2011). IncA/C plasmids mediate antimicrobial resistance linked to virulence genes in the Spanish clone of the emerging Salmonella enterica serotype 4,[5],12:i-. J. Antimicrob. Chemother. 66, 543–549. 10.1093/jac/dkq481 [DOI] [PubMed] [Google Scholar]
- García P., Hopkins K. L., García V., Beutlich J., Mendoza M. C., Threlfall J., et al. (2014). Diversity of plasmids encoding virulence and resistance functions in Salmonella enterica subsp. enterica serovar Typhimurium monophasic variant 4,[5],12:i:- strains circulating in Europe. PLoS ONE 9:e89635. 10.1371/journal.pone.0089635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García P., Malorny B., Hauser E., Mendoza M. C., Rodicio M. R. (2013). Genetic types, gene repertoire, and evolution of isolates of the Salmonella enterica serovar 4,5,12:i:- Spanish clone assigned to different phage typeS. J. Clin. Microbiol. 51, 973–978. 10.1128/JCM.02777-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Neves E., Antunes P., Manageiro V., Gärtner F., Caniça M., da Costa J. M., et al. (2014). Clinically relevant multidrug resistant Salmonella enterica in swine and meat handlers at the abattoir. Vet. Microbiol. 168, 229–233. 10.1016/j.vetmic.2013.10.017 [DOI] [PubMed] [Google Scholar]
- Hall R. M., Cain A. K. (2012). Comment on: Nucleotide sequence of the chromosomal region conferring multidrug resistance (R-type ASSuT) in Salmonella Typhimurium and monophasic Salmonella Typhimurium strains. J. Antimicrob. Chemother. 67, 785. 10.1093/jac/dkr525 [DOI] [PubMed] [Google Scholar]
- Hauser E., Tietze E., Helmuth R., Junker E., Blank K., Prager R., et al. (2010). Pork contaminated Salmonella enterica serovar 4,[5],12:i:, an emerging health risk for humans. Appl. Environ. Microbiol. 76, 4601–4610. 10.1128/AEM.02991-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins K. L., Kirchner M., Guerra B., Granier S. A., Lucarelli C., Porrero M. C., et al. (2010). Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Euro. Surveill. 15:19580. Available online at: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19580 [PubMed] [Google Scholar]
- Laorden L., Herrera-Leon S., Martinez I., Sanchez A., Kromidas L., Bikandi J., et al. (2010). Genetic evolution of the Spanish multidrug-resistant Salmonella enterica 4,5,12:i:- monophasic variant. J. Clin. Microbiol. 48, 4563–4566. 10.1128/JCM.00337-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucarelli C., Dionisi A. M., Filetici E., Owczarek S., Luzzi I., Villa L. (2012). Nucleotide sequence of the chromosomal region conferring multidrug resistance (R-type ASSuT) in Salmonella Typhimurium and monophasic Salmonella Typhimurium strains. J. Antimicrob. Chemother. 67, 111–114. 10.1093/jac/dkr391 [DOI] [PubMed] [Google Scholar]
- Lucarelli C., Dionisi A. M., Torpdahl M., Villa L., Graziani C., Hopkins K., et al. (2010). Evidence for a second genomic island conferring multidrug resistance in a clonal group of strains of Salmonella enterica serovar Typhimurium and its monophasic variant circulating in Italy, Denmark, and the United Kingdom. J. Antimicrob. Chemother. 48, 2103–2109. 10.1128/jcm.01371-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y., Ooka T., Iguchi A., Toh H., Asadulghani M., Oshima K., et al. (2009). Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 17939–17944. 10.1073/pnas.0903585106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Dougan G., James K. D., Thomson N. R., Pickard D., Wain J., et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852. 10.1038/35101607 [DOI] [PubMed] [Google Scholar]
- Reid C. J., Chowdhury P. R., Djordjevic S. P. (2015). Tn6026 and Tn6029 are found in complex resistance regions mobilized by diverse plasmids and chromosomal islands in multiple antibiotic resistant Enterobacteriaceae. Plasmid 80, 127–137. 10.1016/j.plasmid.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Ribot E. M., Fair M. A., Gautom R., Cameron D. N., Hunter S. B., Swaminathan B., et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3, 59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- Soyer Y., Moreno Switt A., Davis M. A., Maurer J., McDonough P. L., Schoonmaker-Bopp D. J., et al. (2009). Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 47, 3546–3556. 10.1128/JCM.00546-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Walle I. (2013). ECDC starts pilot phase for collection of molecular typing data. Euro. Surveill. 18: pii: 20357 Available online at: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20357 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.