Abstract
NMR spectroscopy is a powerful technique for determining structural and functional features of biomolecules in physiological solution as well as for observing their intermolecular interactions in real-time. However, complex steps associated with its practice have made the approach daunting for non-specialists. We introduce an NMR platform that makes biomolecular NMR spectroscopy much more accessible by integrating tools, databases, web services, and video tutorials that can be launched by simple installation of NMRFAM software packages or using a cross-platform virtual machine that can be run on any standard laptop or desktop computer. The software package can be downloaded freely from the NMRFAM software download page (http://pine.nmrfam.wisc.edu/download_packages.html), and detailed instructions are available from the Integrative NMR Video Tutorial page (http://pine.nmrfam.wisc.edu/integrative.html).
Keywords: Automated spectral analysis; Automated structure determination and validation; Chemical shift assignment and validation; Peak identification; Restraint visualization and validation; Visualization of spectra, assignments, and structures
Introduction
NMR spectroscopy is a powerful technique used in many areas of biomolecular research, including structural biology, enzymology, signal transduction, physiology, and drug discovery. NMR enables the collection of atomic-level data under conditions similar to those in cellular systems. Observable NMR parameters such as chemical shifts, peak intensities, scalar and dipolar couplings, line widths, and cross-relaxation provide critical information about target molecules and their interactions. An advantage of NMR, as one of the primary methods for structure determination, is its ability to detect local changes in conformation and dynamics that play functional biological roles.
Despite the growing number of facilities with NMR spectrometers operating at high magnetic fields, the approach has remained largely inaccessible to the larger biological community. In our experience, one reason is the steep learning curve required to become adept at acquiring, processing, and analyzing NMR data. For example, one needs to learn to tailor the experimental approaches and data analysis methods to the aims of the research. In addition, software packages commonly used require different computer operating systems and utilize different standards of atom nomenclature. The fragmentation of protocols presents a high barrier to entry into the field. The Collaborative Computing Project for NMR (CCPN, http://www.ccpn.ac.uk) took steps toward alleviating these problems through its development of CCPNmr Analysis (Vranken et al. 2005). In addition, the WeNMR project offers a number of relevant web-based resources for the process (Wassenaar et al. 2012). Nevertheless, these and other software resources fall short of covering the range of biomolecular experiments in current practice within an integrated package.
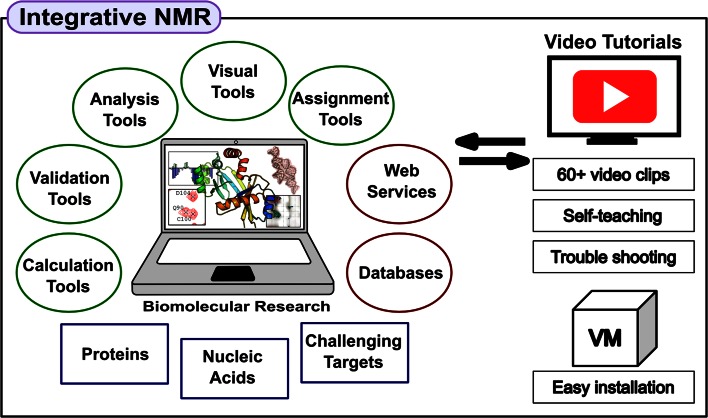
Our approach has been to develop software tools around the popular Sparky software package developed at the University of California, San Francisco (Goddard and Kneller 2008). We refined this platform through a series of nine annual workshops for neophytes held at the National Magnetic Resonance Facility at Madison (NMRFAM). Our objective was to establish a seamless, interactive environment for use by first-time users as well as practiced NMR spectroscopists. Within this platform, tasks are conducted by a series of freely available software packages, including those developed at NMRFAM. This approach has been refined through feedback from workshop students and worldwide users of these tools. The result of this effort is a software platform called Integrative NMR (Fig. 1), which makes biomolecular NMR spectroscopy much more accessible by integrating software tools so that they interact efficiently in ways that support both manual and automated approaches, result validation, and data visualization. Also included are links to web services, databases, and video tutorials. Although the component software packages are available for separate installation, we provide, as an option, all of them pre-installed in a virtual machine that can be run on any standard laptop or desktop computer. The virtual machine avoids the necessity of installing the separate required software programs within different operating systems.
Fig. 1.
The Integrative NMR method for conducting biomolecular research. This integrated set of software packages, which can be installed on a laptop or desktop computer—optionally as part of a virtual machine, cover a wide range of data analysis and visualization steps in the workflows of biomolecular NMR research. The software tools interoperate seamlessly with external servers and databases. Video tutorials cover all operations
Tasks are conducted by enhanced versions of two main software packages, NMRFAM-SPARKY (Lee et al. 2015) and PONDEROSA (Lee et al. 2011, 2014) in which old and new tools are integrated in efficient ways that emphasize visualization. For example, the Dummy Graph tool in NMRFAM-SPARKY depicts regions of the covalent structure of proteins or DNA/RNA molecules along with the status of current chemical shift assignments of their NMR-active atoms. RNA assignments are facilitated by ellipses drawn over spectra to delineate statistical chemical shift assignment regions for atoms in particular bases (Aeschbacher et al. 2013). Experimental data from spectral series, such as pH titrations, molecular interaction studies, or NMR relaxation, can be visualized seamlessly with the NDPPlot (NMR Data Perturbation Plot) tool in NMRFAM-SPARKY. New visual analysis tools in Ponderosa Analyzer simplify many time-consuming tasks to a few screen clicks. An enhanced mode of the PyMOL software package (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.), which supports shortcut commands, enables the visualization of data from the analysis and validation packages of Ponderosa Analyzer. With the virtual machine, a user can launch the calculation of the structure of a protein from NMR data by a few clicks. Without the need to install any individual software packages, the process can make use of APES for peak picking (Shin et al. 2008), PINE for automated assignment (Bahrami et al. 2009), PONDEROSA-C/S for automated structure determination, TALOS-N for shift based torsion angle restraints (Shen and Bax 2013), the PACSY database (Lee et al. 2012), and the CS-Rosetta (Lange et al. 2012) compute server at the Biological Magnetic Resonance data Bank (BMRB). Furthermore, all automated approaches are accompanied by efficient visual verification tools: automated peak picking can be verified by a tool in NMRFAM-SPARKY; errors in automated assignments by PINE can be detected and corrected with PINE-SPARKY (Lee et al. 2009) or ARECA (Dashti et al. 2016); and errors in automated structure calculations can be detected and corrected by the visual tools that are part of Ponderosa Analyzer.
A goal of Integrative NMR is to incorporate multiple approaches to the solution of problems as proposed recently (Dashti et al. 2015). For example, peak assignments can be carried out manually or automatically and NOE peaks can be picked manually or automatically. As its default, PONDEROSA utilizes the Xplor-NIH engine for structure determination, but users with a license for CYANA can use the CYANA engine as an option along with its automated NOE assignment module. The default tools are designed to work with well-folded proteins of small or moderate size. NMRFAM-SPARKY contains additional tools that are useful for larger proteins or intrinsically disordered proteins. The developers of SCAssign (Zhang and Yang 2006) for the assignment of larger proteins and ncIDP-assign (Tamiola and Mulder 2011) for intrinsically disordered proteins have permitted their inclusion in NMRFAM-SPARKY.
All software, including the virtual machine, is freely available from the NMRFAM website (http://pine.nmrfam.wisc.edu/download_packages.html), and video tutorials available from the website cover every step.
Materials and methods
The Integrative NMR platform makes use of several software packages developed at the National Magnetic Resonance Facility at Madison (NMRFAM) and elsewhere. The software packages can be installed separately or can be obtained from NMRFAM installed on a virtual machine that can be used on a variety of computer platforms. This latter approach, which does not entail significantly longer software run times, is particularly useful for non-specialists. The platform provides user-friendly interfaces to freely-available servers in the biomolecular NMR field.
NMRFAM-SPARKY and its tools
The originators of Sparky transferred the development of this popular software package to NMRFAM. We modernized and enlarged many parts of the core engine written in C++ with extensions in Python, added new tools that integrate freely-available tools in the biomolecular NMR field, and released the new version as NMRFAM-SPARKY (Lee et al. 2015). For the benefit of legacy users, we kept changes in user interfaces to a minimum. Continued development described here has focused on the addition of new features and their graphical interfaces and on seamless integration with relevant web services. The tools are menu driven, but Integrative NMR supports many shortcut two-letter commands that more conveniently activate individual tools within NMRFAM-SPARKY (Table 1).
Table 1.
Two-letter-codes (case sensitive) used within NMRFAM-SPARKY to activate tools that carryout various operations in Integrative NMR
| Peak identification | |
| ae | APES automated peak picking. Peak positions are identified from local maxima, and peak positions in multiple spectra are compared to flag peaks that are not part of spin systems as noise. Potential noise peaks can be identified and deleted automatically |
| kr | Restricted peak picking. Peaks are identified on the basis of local maxima within search windows specified by peaks in another spectrum |
| LT | Alternate peak list window. Peaks identified from local maxima are sorted by data height; this helps to identify noise peaks which are often have low intensity |
| sp | Strip plot. Once peaks have been identified and noise peaks have been eliminated, the strip plot tool can be used to efficiently delete any remaining false-positive peaks and add missing peaks |
| Automated protein chemical shift assignment | |
| ep | PINE automated assignment. This bring up a window that can be used to specify peak lists from different NMR experiments and launch a submission to the PINE Server to carry out automated protein peak assignments |
| ip | Convert PINE outputs to Sparky. This tool converts probabilistic backbone and sidechain assignment files generated by the PINE Server to a Sparky resonance file that can be read in by two-letter-code rl with the probability set manually |
| rl | Resonance list. This window shows currently assigned resonances with averaged chemical shifts and their deviations. In Integrative NMR, this tool is used to read-in/write-out chemical shifts |
| p2 | PINE2SPARKY converter. PINE2SPARKY generates probable candidates for all peak in the spectra prior to using PINE-SPARKY to verify the PINE output against spectra |
| ab | Assign the Best by PINE. After using PINE2SPARKY to import the probabilistic assignments from PINE to NMRFAM-SPARKY, this tool can be used to set a threshold and to accept all assignments with probabilities that exceed this threshold |
| pp | PINE Graph Assigner. This tool enables graphical examination of all probable assignment candidates on a per-residue and atom-by-atom basis |
| pr | PINE Assigner. This tool enables the examination of all assignment candidates on a peak-by-peak basis |
| Enhanced manual protein chemical shift assignment | |
| ta | Transfer and Simulated Assignments. This versatile tool annotates peaks on a selected spectrum on the basis of assignments from other spectra or predictions. If the assignment is simulated from prediction, the assignment tag contains “_s” to avoid confusion |
| ut | Untag “_s”. This command detaches “_s” from a selected tag for a peak whose assignment has been confirmed |
| cu | Center and Untag “_s”. This command causes a peak identifier to move to the nearest local maximum and detaches the “_s” tag |
| mt | Merge two assignments to a pseudoatom. If two assignments are overlapped after centering and untagging by use of the cu command, the user can merge them as one pseudoatom by typing the mt command |
| Chemical shift validation | |
| lv | Run LACS. This command submits a protein chemical shift file for analysis by LACS (Linear Analysis of Chemical Shifts); the LACS output detects chemical shift outliers and detects chemical shift referencing errors and suggests chemical shift corrections |
| ea | Generate files and export to ARECA. This command opens a window that enables the generation of ARECA input files (peak assignments and NOE peak lists) and opens the ARECA web page to import the files and launch ARECA to validate the assignments |
| ar | ARECA list. This tool enables the user to color peaks and assignments in 3D-NOE spectra according to the assignment probabilities generated by ARECA as a means for their validation |
| Molecular structure visualization | |
| dg | Dummy Graph. This command launches a molecular structure visualization tool that shows the atoms and their assignment status |
| Tools for intrinsically disordered proteins (IDPs) and large proteins | |
| RS | ncIDP Repositioner. Repositions an assigned stretch of protein sequence according to ncIDP chemical shift statistics |
| SG | ncIDP Spin Graph. Spin graph modified for intrinsically disordered proteins (IDPs) |
| sn | SCAssign. Sidechain assignments from 4D-NOESY and CCH-TOCSY data |
| Nucleic acid assignment | |
| ER | Export to RNA-PAIRS. This tool generates RNA-PAIRS inputs and opens the web page of the RNA-PAIRS server |
| SE | RNA statistical ellipses. Draws ellipses on 2D spectra that delineate the ranges of chemical shifts expected for particular RNA bases in the CHESS2FLYA program |
| DG | Dummy Graph for nucleic acids. This tool displays atoms from the covalent structures of DNA/RNA residues and indicates the current status of chemical shift assignments |
| Spectral series | |
| ol | View overlays. Overlays NMR spectra for comparison |
| ct | Color contour levels. This tool enables the user to differentially color the contour plots from overlaid 2D NMR spectra |
| np | Perturbation plot. This tool enables the user to construct plots that compare specified NMR observables from two spectra collected under different conditions |
| ni | Titration plot. This tool traces changes in the chemical shift of a particular resonance in multiple spectra as the function of a variable such as pH or added ligand |
| rh | Peak height analysis. This command enables the plotting of peak heights as a function of assigned residue number or by corresponding resonances in different spectra. The changes in peak height can be saved in tabular form for further analysis. A decaying exponential function is also fit to the data For analysis of T 1/T 2 relaxation data, the peak heights can be fitted to a decaying exponential function. The extracted relaxation constants can then be plotted as a function of residue number |
| eo | Easy overlay dialog. Enables users to easily overlay NMR spectra by a few clicks |
| ec | Easy contour dialog. Enables users to easily adjust contour levels of NMR spectra by a few clicks. |
| ci | Inverse background color. This command changes background color from black to white or from white to black |
| Secondary structure prediction | |
| n6 | PECAN. This command uses assigned chemical shifts as input to PECAN, which carries out probabilistic chemical shift based secondary structure prediction |
| tl | TALOS-N. This command uses assigned chemical shifts as input to TALOS-N , which carries out artificial neural network chemical shift based secondary structure prediction |
| PP | PSIPRED. This command uses amino acid sequence as input to PSIPRED, which carries out Psi-blast sequence based secondary structure prediction |
| Three-dimensional structure prediction | |
| nm | POND-PRED (Ponderosa Prediction Server). This command invokes this server that predicts 3D structure on the basis of amino acid sequence alone. The server uses hydrogen bond constraints from secondary structure predicted by PSIPRED, and distance and angle constraints from the PACSY database to generate structures by simulated annealing from Ponderosa Server |
| ce | CS-Rosetta. This command brings up the BMRB-hosted 3D structure prediction server based on Monte Carlo assembly with chemical shift filtered protein fragments |
| Three-dimensional structure determination | |
| c3 | PONDEROSA-C/S structure calculation. This command carries out automated NOESY peak picking to generate the input for the Ponderosa Server at NMRFAM, which then calculates the 3D structure of the protein |
| cp | Ponderosa Client. This command launches the Ponderosa Client program that enables the specification of additional input for 3D structure calculation, including RDC, SAXS, WAXS, and the use of alternative calculation methods |
| up | Ponderosa Connector. This command establishes a connection between PONDEROSA-C/S and NMRFAM-SPARKY that enables interactive assessment of NOESY peak quality and validation of distance constraints. PONDEROSA-C/S specifies regions of interest to NMRFAM-SPARKY, which displays spectra so that users can decide whether peaks are real and assignments are valid |
| gd | Generate distance constraints. This tool uses the r −3–r −6 approximation to automatically generate distance constraints in PONDEROSA compatible format (DYANA) from assigned NOE cross peaks |
| xf | Manual restraint format. This tool uses a manual binning approach based, as specified, either on peak height or volume to generate distance constraints in PONDEROSA or XPLOR compatible format from assigned NOE cross peaks |
NDPPlot (NMR data perturbation plot)
A feature lacking in the original Sparky software was data visualization from experiments producing spectral series, such as NMR relaxation or titration studies. In order to add a chart plotting tool that works seamless with NMRFAM-SPARKY, we chose Free Pascal and Lazarus IDE (http://www.lazarus-ide.org) for its development because of their convenience in producing statically compiled executable binaries in Windows, Mac, and Linux and because of our prior experience with this IDE (integrated development environment) in developing the Pine2Sparky converter (Lee et al. 2009). The new graphical plotting program is called NDPPlot (NMR data perturbation plot); although it was developed initially for chemical shift tracing, it has proved to be versatile for use in other applications.
Structure calculation
The structure calculation server program, Ponderosa Server, and the NOESY peak picking and data transfer program on the client side, Ponderosa Client, are written in C++ with QT libraries (http://www.qt.io). We developed an interface between the PACSY database and Ponderosa Server to support the AUDANA algorithm (Automated Database-Assisted NOESY Assignment) for automated structure calculation (Lee et al., submitted) and the PACSY-ALIGN algorithm for finding similarities within the protein database (http://pacsy.nmrfam.wisc.edu/pacsyalign). We wrote Xplor-NIH scripts (Schwieters et al. 2003) for structure calculation in Python. We wrote NMRFAM-SPARKYPython extension codes for PONDEROSA-C/S interface that make processes flawless. Furthermore, we built web server for public services with HTML, Apache, CGI, Perl, Python, and MySQL on our Linux cluster system. We prepared 256 CPU cores as structure calculation resources at NMRFAM. We added advanced structural analysis tools (written in Free Pascal with Lazarus IDE) to Ponderosa Analyzer, the program that validates results from structural calculations and assists with iterative calculations. In addition, we created interfaces linking Ponderosa Analyzer, NMRFAM-SPARKY, and PyMOL.
Sample data
NMR data for ubiquitin, SIV, and NANOG were acquired at NMRFAM; data for UbcH5B/CNOT4 was from Dr. A.M.J.J. Bonvin’s web page (http://www.nmr.chem.uu.nl/~abonvin/); and data for OR135 was from the CASD-NMR web page (https://www.wenmr.eu/wenmr/casd-nmr). We used data from ubiquitin (unpublished) and SIV frameshift site RNA (Marcheschi et al. 2007) to develop tools, respectively, for general spectral analysis and assignment of proteins and RNA molecules. We used data from NANOG (unpublished) to develop tools for peak height analysis, UbcH5B/CNOT4 (Dominguez et al. 2004) to develop tools for perturbation/titration analysis, and from OR135 (Rosato et al. 2015) to develop of structure calculation tools (Koga et al. 2012).
Video tutorials
Videos were recorded in OGV format by RecordMyDesktop software (http://recordmydesktop.sourceforge.net), converted to MKV-formatted files, and uploaded onto YouTube (http://www.youtube.com) with added annotations to explain features. Videos can be accessed from (http://pine.nmrfam.wisc.edu/integrative.html); users are encouraged to subscribe to the YouTube channel to receive notifications of uploads of new video tutorials.
Installation of separate modules
We provide simple installers for the software components of Integrative NMR on all supported platforms (Python for Linux and Mac, and Windows Batch for Windows).
Virtual machine
In addition, we make all the software components of Integrative NMR available on a virtual machine. An ISO-formatted 64-bit disk image of Ubuntu MATE 15.04 was downloaded from the Ubuntu MATE web page (http://ubuntu-mate.org) and installed in an ORACLE VM VirtualBox (http://www.virtualbox.org). The software components of Integrative NMR were installed and optimized on this virtual machine. Then, the virtual disk image was exported to Open Virtualization Archive (OVA) format. In addition, we used the 7-zip file compression program (http://www.7-zip.org) to prepare a separated compressed version of the virtual machine for 32-bit operating systems that cannot download files larger than 2 GB from a web browser.
Results and discussion
NMRFAM-SPARKY and its tools
Peak identification
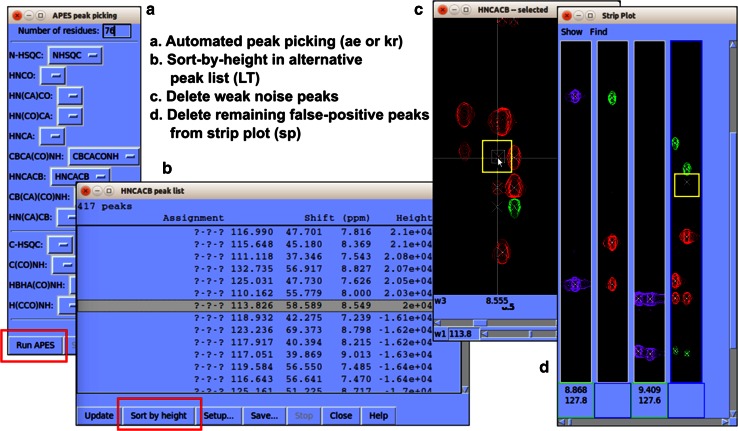
The basic approach to peak identification in 2D biomolecular NMR spectra is to search for local maxima above a chosen contour level. If a graphical tool is used to select the peaks, this algorithm is generally successful; however, when peak picking is automated, too many noise peaks can be included. With spectra of dimension greater than two, visual searching, becomes highly time consuming. Therefore, it is common to use a visual peak picking tool to identify peaks in 2D HSQC spectra first and to use automated peak picking restricted to the chosen frequencies to identify peaks in 3D spectra. As with 2D spectra, the automated approach can include noise and artifacts. To get around this problem NMRFAM-SPARKY employs two advanced automated restricted peak picking tools: APES (Shin et al. 2008) and PONDEROSA. With these tools, one can utilize an alternative peak list window (two-letter-code LT) and strip plot window (two-letter-code sp) to complete the peak picking step as illustrated in Fig. 2.
Fig. 2.
Recommended semi-automated peak identification method in NMRFAM-SPARKY. a Use APES (two-letter-code ae) or restricted peak picking (two-letter-code kr) for automated peak identification. b Use alternative peak list window (two-letter-code LT) to sort peaks by intensities. c Delete weak noise peaks from spectrum view. d Use strip plot (two-letter-code sp) to delete any remaining false-positive peaks and add missing peaks
Automated protein chemical shift assignment
The Integrative NMR suite includes the PINE (Bahrami et al. 2009) assignment engine (two-letter-code ep), which supports probabilistic backbone and sidechain assignments based on available NMR data sets. The ranked assignments proposed by PINE are easily validated and extended through the use of PINE-SPARKY (two-letter-codes ip, p2,ab, pp, and pr), which enables the visualization of proposed assignments against experimental spectral data (Lee et al. 2009).
Enhanced manual protein chemical shift assignment
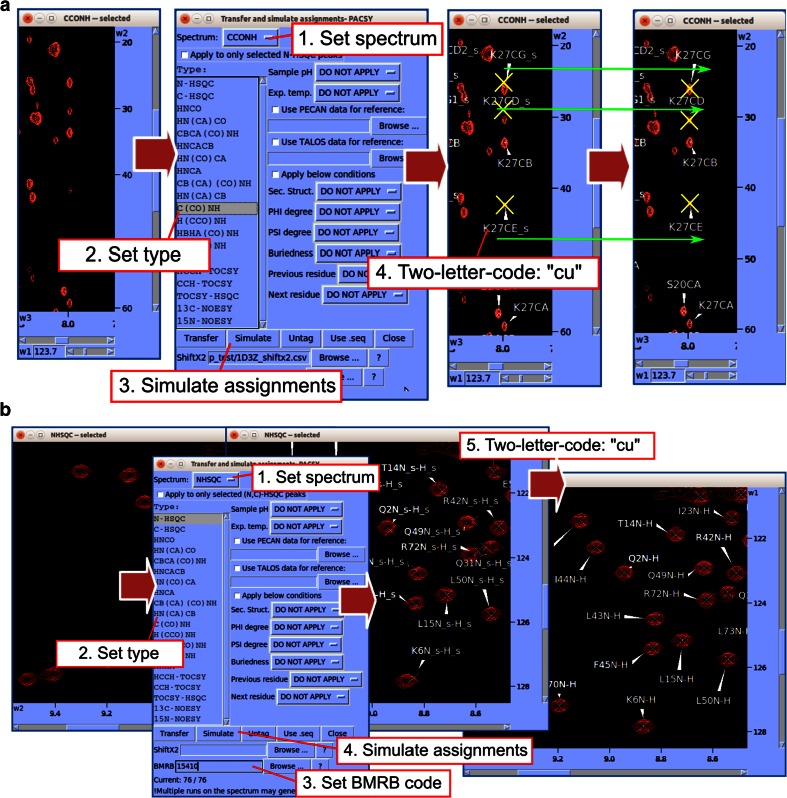
Transfer and Simulated Assignments (two-letter-code ta) is a versatile assignment tool recently developed under NMRFAM-SPARKY that uses the PACSY database (Lee et al. 2012) to enable a new assignment method, predict-and-confirm. This approach greatly accelerates assignments by eliminating the redundant procedures and potential user errors associated with the traditional pick-and-assign method. Transfer and Simulated Assignments was originally devised for fast side chain assignment from spectra such as C(CO)NH, H(CCO)NH, and HBHA(CO)NH (Fig. 3a); however, as shown in Fig. 3b, if a corresponding BMRB entry exists, the approach can be used for one-shot assignments based entirely on 2D HSQC spectra.
Fig. 3.
Predict-and-confirm method for fast semi-automated chemical shift assignment. a Efficient method for assigning a C(CO)NH spectrum. In the Transfer and simulate assignment window (two-letter-code: ta), set Spectrum and Type; then click the Simulate button to annotate predicted assignments on the experimental spectrum (yellow ‘X’s). Drag each yellow X onto the nearest peak in the spectrum, and type “cu” to confirm the assignment. The yellow ‘X’s are now centered on the position of the experimental peak. b Illustration of how Predict-and-confirm can be used to import assignments from a BMRB entry. The BMRB assignments are displayed over the spectrum, and the user can then adjust and confirm them
Chemical shift validation
Linear Analysis of Chemical Shifts (LACS) is supported by NMRFAM-SPARKY (two-letter-code lv); LACS detects and corrects errors in chemical shift referencing (Wang et al. 2005). ARECA (Assessment of the REliability of Chemical shift Assignments) is a tool for validating protein chemical shift assignments on the basis of NOE data (Dashti et al. 2016). The input can be prepared by either NMRFAM-SPARKY (two-letter-code ea) or Ponderosa Client from 15N- and/or 13C-filtered NOE experiments (two-letter-code pc). The NMRFAM-SPARKY extension for ARECA (two-letter-code ar) handles data analysis. Chemical shifts can be validated in advance of a structure determination to minimize subsequent refinement steps. Validated assignments are also important for other types of experiments, such as ligand binding or dynamics studies.
Molecular structure visualization
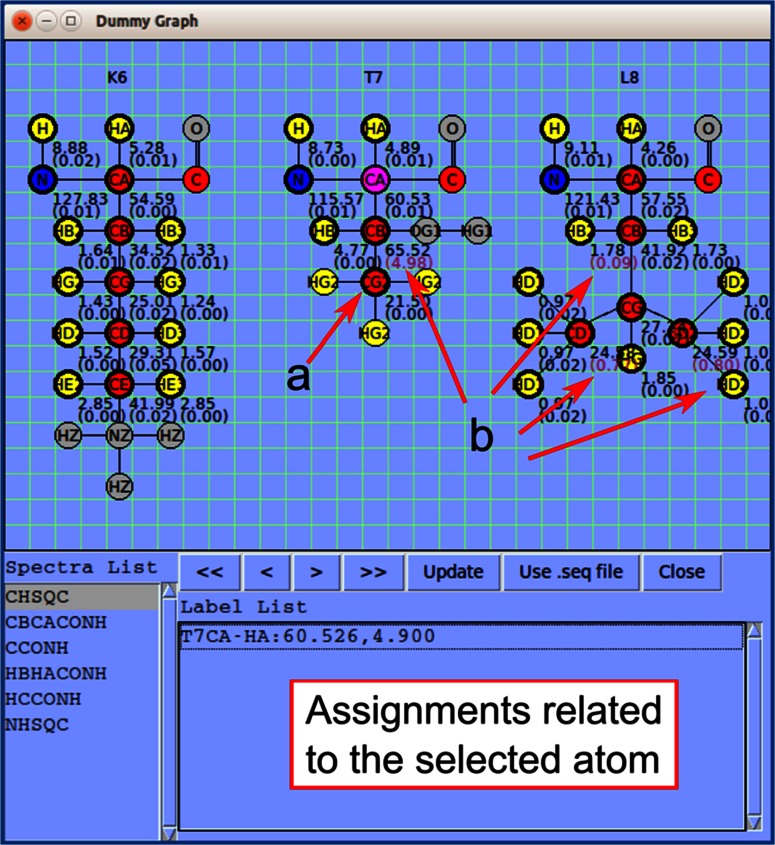
Pine Graph Assigner, the visual tool for molecular structure visualization in the original PINE-SPARKY (Lee et al. 2009), has been simplified and generalized for universal use as Dummy Graph (Fig. 4, two-letter-code dg). Dummy Graph shows atoms to be assigned along with average and standard deviation of assigned chemical shifts; it also shows the assignment labels for a selected atom and enables the user to visualize the place in a given spectrum where the assigned peak is located. Missing assignments (Fig. 4a) and erroneous assignments (Fig. 4b) can be recognized by direct visualization.
Fig. 4.
Dummy Graph is molecular structure visualization tool that shows the atoms in a three-residue stretch and indicates which atoms have been assigned. Also shown is the average chemical shift and standard deviation (in ppm) from the positions of all peaks in the different spectra supporting a given assignment. An atom can be selected by clicking; this turns its background pink as shown for T7CA. The panel at the bottom shows assignments (in the Label List) related to the selected atom from the spectrum selected in the Spectra List. a A thick line around an atom and chemical shift indicates that the atom is already assigned. Otherwise, a thin line around an atom indicates that it is not yet assigned. b Large chemical shift deviations are written in purple to alert the user to the possibility of an erroneous assignment
Tools for intrinsically disordered proteins (IDPs) and large proteins
NMRFAM-SPARKY supports the assignment of challenging targets such as IDPs and large proteins. For IDP assignment, NMRFAM-SPARKY includes the set of tools developed by Mulder group including their IDP chemical shift statistics (Tamiola and Mulder 2011). The ncIDP-assign package, which consists of ncIDP Repositioner (two-letter-code RS) and ncIDP Spin Graph (two-letter-code SG), is pre-installed. For large proteins, the SCAssign package (two-letter-code sn) supports assignments based on 4D 13C-,15N-edited NOESY and 3D CCH-TOCSY spectra (Zhang and Yang 2006). See http://yangdw.science.nus.edu.sg/SCAssign for an online tutorial from the Yang group. These approaches become more powerful within NMRFAM-SPARKY because they can take advantage of the predict-and-confirm and Dummy Graph methods described above.
Tracking of manual assignments
NMRFAM-SPARKY supports the annotation module of CONNJUR R (Fenwick et al. 2015), which records information about peaks that have been reassigned manually. This functionality can be used to improve the reproducibility of NMR structure determinations. This feature is available on the virtual machine, Linux, and Mac versions of Integrative NMR.
Nucleic acid assignment
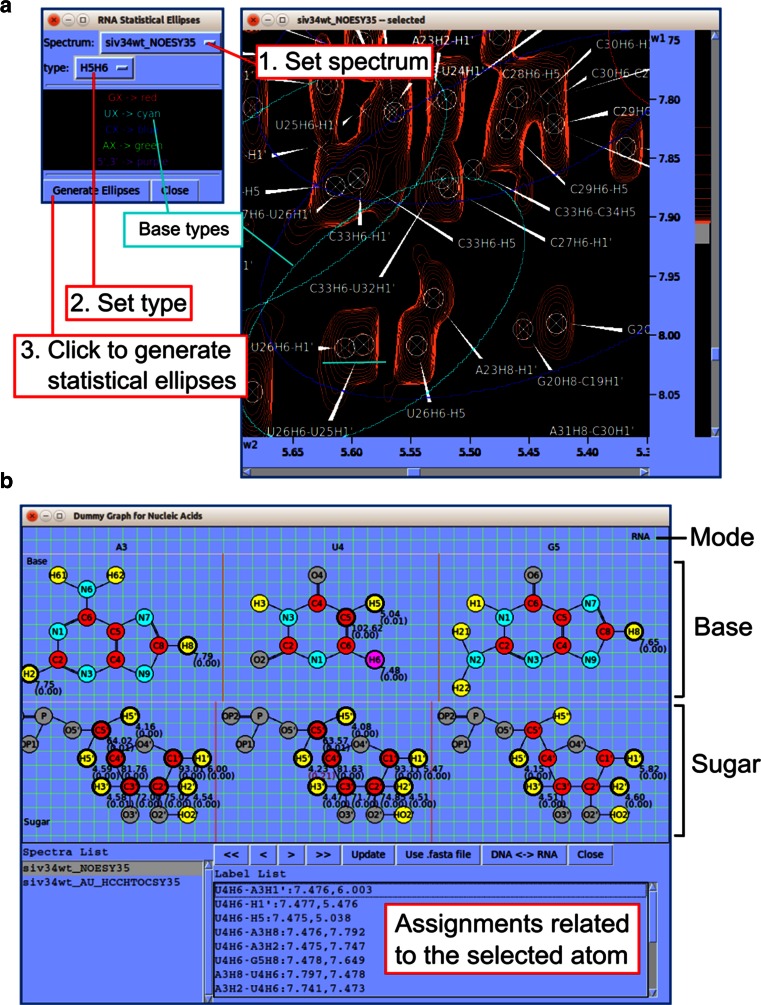
RNA-PAIRS is an algorithm for automated RNA imino resonance assignment (Bahrami et al. 2012). NMRFAM-SPARKY contains a link (two-letter-code ER) that generates RNA-PAIRS inputs and redirects the user’s web browser to the RNA-PAIRS web server page at NMRFAM. RNA chemical shift statistics calculated by the Schubert group (Aeschbacher et al. 2013) suggested covariance statistics for 1H and 13C chemical shifts. The RNA Statistical Ellipses window in NMRFAM-SPARKY (Fig. 5a) displays the statistical ellipses overlaid on RNA spectra to assist chemical shift assignment. A nucleic acid version of Dummy Graph (Fig. 5b) displays the atomic structure of the DNA or RNA molecule being assigned.
Fig. 5.
Tools developed for assisting chemical shift assignment of nucleic acids. a NMRFAM-SPARKY supports covariance ellipses for RNA 1H and 13C chemical shifts first introduced in the CHESS2FLYA program. Statistical ellipses is a tool in NMRFAM-SPARKY (two-letter-code SE) that overlays color-coded ellipses on the spectra to outline the range of chemical shifts expected for different RNA bases. The peaks underlined in cyan are near the center of the cyan ellipse, which corresponds to Uracil. b Dummy Graph for Nucleic Acids (two-letter-code DG) is a tool for visualizing atoms of DNA or RNA residues; base atoms of are shown at the top and those of the sugar and phosphate are shown at the bottom. The panel below shows assignments related to the selected atom shown in pink (U4H6) from the selected spectrum
Spectral series
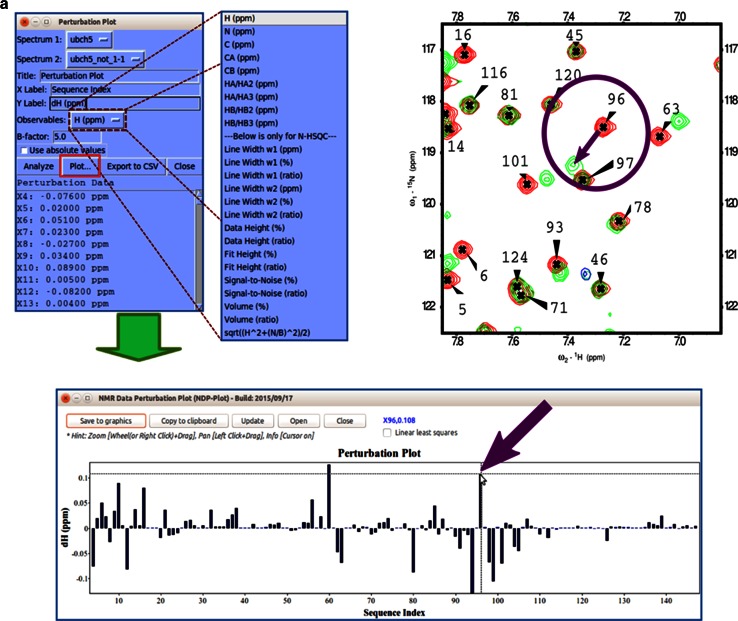
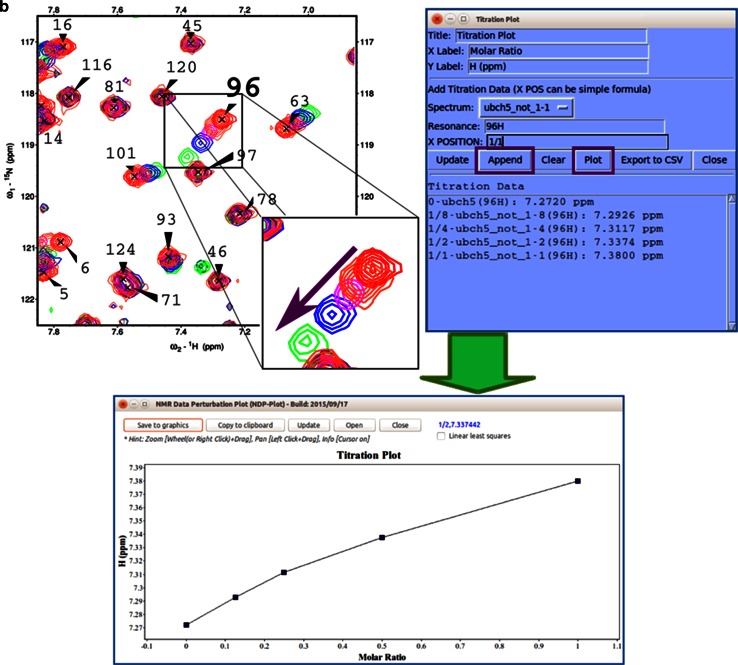
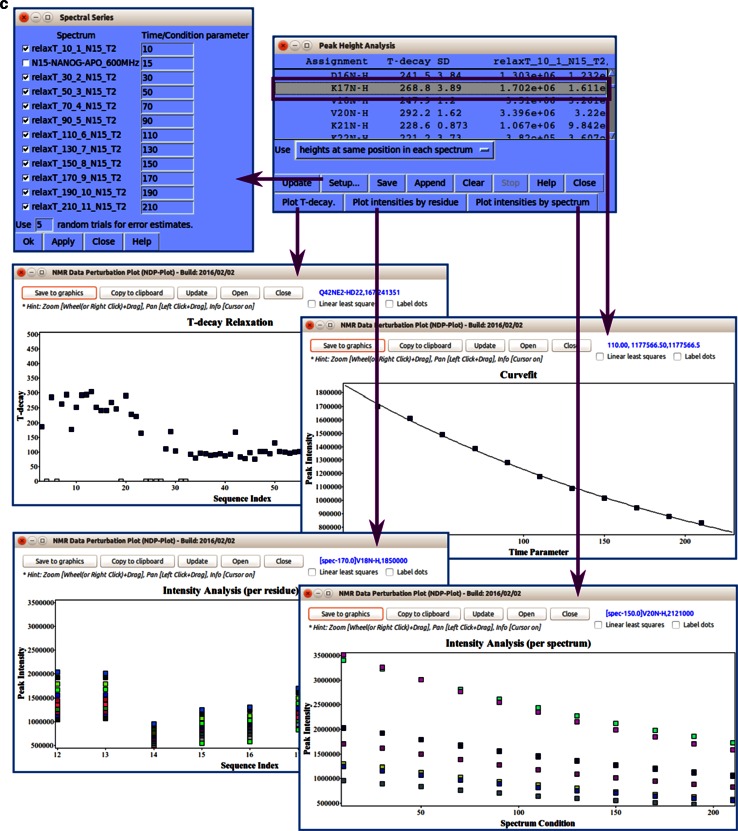
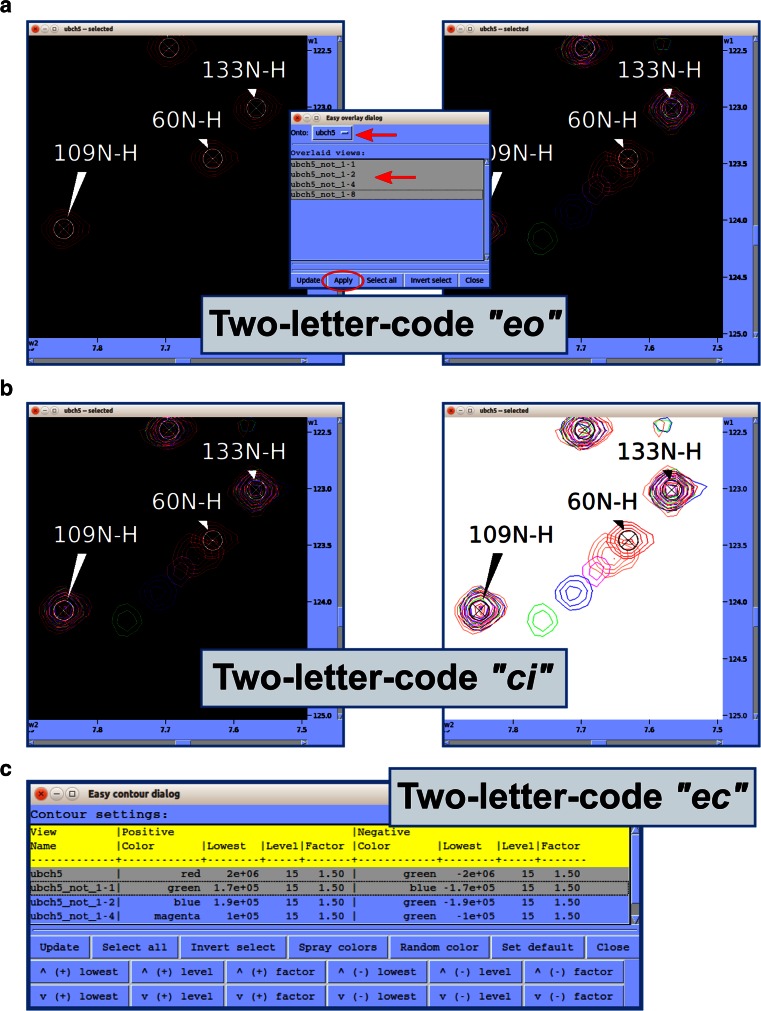
The graphical chart tool, NMR Data Perturbation Plot (NDPPlot), which was originally an internal chart module of Ponderosa Analyzer, has been isolated from the program to be an independent program and also integrated into NMRFAM-SPARKY. NDPPlot supports seamless visualization of a series of NMR spectra, such as time series or titrations. Perturbation Plot (two-letter-code np, Fig. 6a) displays global spectral changes resulting from a change in solution conditions or composition. Titration Plot (two-letter-code ni, Fig. 6b) traces changes in the chemical shift of a particular resonance as the function of a variable such as pH or added ligand. Peak Height Analysis (two-letter-code rh, Fig. 6c) is used in the analysis of data for relaxation measurements. With a few clicks (Save to graphics button), NDPPlot is capable of generating figures and plots in the popular scalable vector graphics format (SVG). The NDPPlot program accepts INI (ititialization) format as input and saves graphics files. It includes useful mouse functions, such as entity identification, zoom in, zoom out and pan. This program is designed for visualizing and analyzing spectral series data; however, we started providing NDPPlot compatible files from our PINE and PECAN web servers because we found that the zooming capability of NDPPlot improved the visualization of data from larger proteins. Because the traditional overlay dialog (two-letter-code ol) is limited to overlaying one spectral view at a time, we added Easy overlay dialog (two-letter-code eo), which lets users select multiple spectral views for overlay onto a specified view (Fig. 7a). A white background, which is better for visualizing differently colored data from multiple spectra, can be selected (two-letter-code ci, Fig. 7b). The Easy contour dialog (two-letter-code ec, Fig. 7c) box enables the adjustment of contour threshold, levels, and colors for multiple spectra.
Fig. 6.
NDPPlot is a tool fully integrated into NMRFAM-SPARKY for visualizing spectral changes. a Perturbation plot (two-letter-code np). The arrows point to the change in the chemical shift of residue 96. The contour plot overlays (two-letter-code ol or eo) two selected 1H–15N HSQC spectra: one recorded with (green contour) and one without (red contour) added substrate. The displacement of the signal assigned to residue 96 is highlighted within the circle. NDPPlot generates the bar chart shown to plot the chemical shift differences between two spectra along the sequence; this is achieved by choosing the two spectra and the observable to be compared (in this case, the chemical shift of amide protons) in the Perturbation plot window and by clicking Plot. b Titration Plot (two-letter-code ni) visualizes chemical shift changes from a titration experiment. Results are plotted by the NDPPlot program. This example shows how the 1H NMR chemical shift of residue 96 from the 1H-15N HSQC spectra changes upon the addition of a substrate. The spectrum shows the overlap (two-letter-code ol) of spectra without and with four increasing levels of added substrate. Contour colors are set by the two-letter-code ct (with 0 for red, 1/8 for tomato, 1/4 for magenta, 1/2 for blue, 1 for green). In the Titration Plot window, by choosing the spectra and the observable to be compared (in this case, the 1H chemical shift of H96) and clicking the Plot button, NDPPlot graphs 1H NMR chemical shift of H96 as a function of the molar ratio. c The Peak Height Analysis tool (two-letter-code rh) can be used to analyze results from relaxation experiments or any other peak intensity related experiments. After choosing a series of assigned spectra with different time/condition parameters, clicking the Plot T-decay button can be used to visualize the relaxation time constant as a function of residue number or by choosing a single residue, the decay in its peak intensity over time/condition. Alternatively, per residue and per spectrum intensity analysis options are available for observing overall differences between residues or spectra
Fig. 7.
Useful tools for spectral series analysis. a Easy overlay dialog. We developed much simpler tool for spectral overlay than traditional “ol” tool particularly for many spectra. It is activated by two-letter-code “eo”. Simply, user can choose a spectrum “onto” and select spectra to “overlay” by mouse dragging and clicking with Ctrl and Shift keys. When most of all spectra in the project need to be overlaid, user can simply click Select all button and exclude spectra by Ctrl key and mouse clicks. b Change inverse background. We understood the visual limitation of black background color for spectral series analysis if user needs to color spectra differently. Thus, we developed two-letter-code “ci” to change background color from black to white and from white to black. The background color can be set differently and if ornament color is black and white, they are automatically changed to the other color, and so markers such as crosshair are. c Easy contour dialog. This tool is activated by two-letter-code “ec”. It can be used to adjust contour color, level and threshold of multiple spectra all at once
Secondary structure prediction
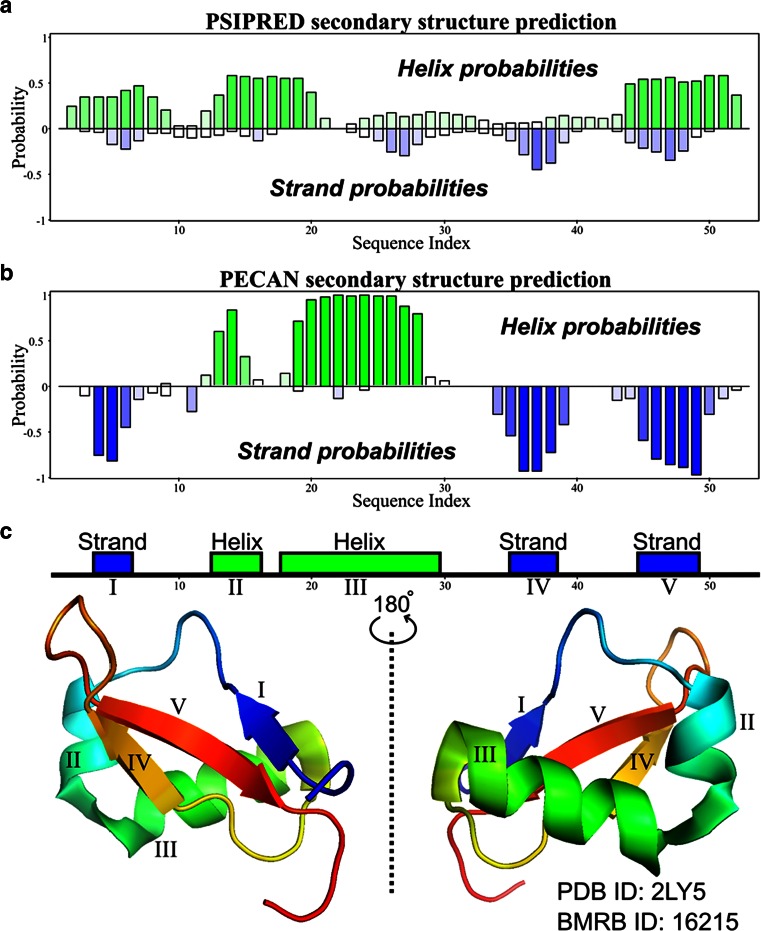
NMRFAM-SPARKY supports both sequence-only (PSIPRED) (Jones et al. 1999) and chemical shift-based methods (PECAN) (Eghbalnia et al. 2005) or (TALOS-N) (Shen and Bax 2013) for secondary structure prediction. Generally, sequence-only methods yield 70–80 % accuracy, and the accuracy can be improved by using chemical-shift-based methods (Fig. 8). For example, we determined that PECAN surpassed PSIPRED in predicting the secondary structure of the small protein brazzein (PDB ID: 2LY5, BMRB ID: 16215) (Cornilescu et al. 2013). Ponderosa Server is a part of the PONDEROSA-C/S package (Lee et al. 2014) that automatically runs TALOS-N and applies optimized torsion angle constraints for the structure calculation. Ponderosa Analyzer, another component of PONDEROSA-C/S, offers tools for refining torsion angle constraints.
Fig. 8.
Sequence-based and chemical shift-based predictions from NMRFAM-SPARKY of the secondary structure of the protein brazzein (PDB ID: 2LY5, BMRB ID: 16215). The illustration was made by NDPPlot. a Sequence-based prediction by PSIPRED (two-letter-code PP) could not predict secondary structures correctly and the probabilities are not confident. b Chemical shift-based prediction by PECAN (two-letter-code n6). c Secondary structures from PDB deposited structure. The PECAN prediction matched perfectly with the secondary structural elements determined from the structure
Three-dimensional structure prediction of proteins
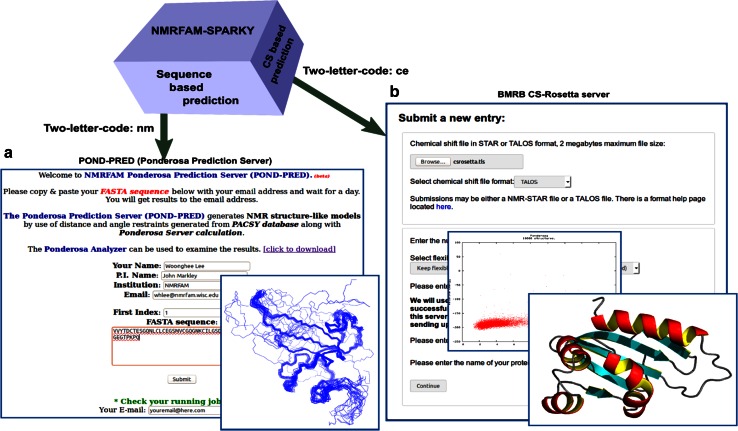
Integrative NMR supports predictions of protein 3D structure either on the basis of amino acid sequence alone and on the basis of assigned NMR chemical shifts. Jobs to be carried out on external servers are launched from NMRFAM-SPARKY. The sequence-only method, POND-PRED (Ponderosa Prediction Server), which is carried out on an NMRFAM server (http://ponderosa.nmrfam.wisc.edu/model.html), predicts hydrogen bond constraints from PSIPRED results and analyzes the PACSY database to generate distance and angle constraints. This method generates structures by simulated annealing as in typical NMR structure calculations (Fig. 9a). The chemical-shift-based method utilizes CS-Rosetta calculations (Shen et al. 2008) carried out on a server at BMRB (https://csrosetta.bmrb.wisc.edu/csrosetta) that employs the Condor (Thain et al. 2003) grid computing system (Fig. 9b).
Fig. 9.
Three-dimensional structures can be predicted by sequence-based method and chemical shift-based methods in NMRFAM-SPARKY. a POND-PRED (two-letter-code nm) is a webserver offered by NMRFAM for predicting protein 3D structures from amino acid sequences. b CS-Rosetta is a chemical shift-based 3D structure prediction program; it is accessibly from NMRFAM-SPARKY (two-letter-code ce) on a web server hosted by BMRB
PONDEROSA-C/S
Three-dimensional structure determination
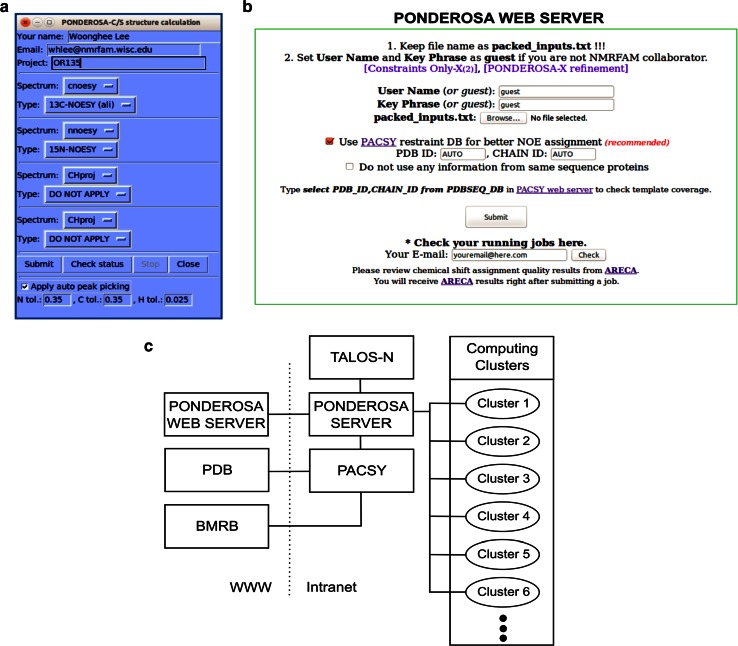
Integrative NMR supports a complete environment for structure calculation. The initial version of PONDEROSA demonstrated its potential by generating accurate structures from raw NOESY spectra in the second round of the CASD-NMR competition (Rosato et al. 2015). The newer version, PONDEROSA-C/S, that is part of Integrative NMR isolates the computation module on a server allowing the user to focus on the input and output data. The integration of NMRFAM-SPARKY with PONDEROSA-C/S makes it possible to calculate and verify structures with a few clicks. For example, a new structure calculation (two-letter-code c3) requires only clicking to specify the assignment file and raw NOESY spectra and entering the user’s e-mail address (Fig. 10a). Then, after clicking the ‘Submit’ button, NOE cross peaks from the spectra are picked and evaluated, and a pre-packed Ponderosa Server input file is sent to the Ponderosa Web Server for structure determination (Fig. 10b). Details are provided below.
Fig. 10.
NMR-based 3D protein structure calculation. a Calculation 3D structure by PONDEROSA-C/S (two-letter-code c3) offers direct job submission from NMRFAM-SPARKY to the Ponderosa Web Server. It supports fully automated mode with and without automated NOESY peak picking and semi-automated mode with partially or fully assigned NOESY data. b Ponderosa Web Server is a freely available web resource that transmits the structure calculation command to the Ponderosa Server running at NMRFAM. c Diagram showing the integrated architecture of Ponderosa Web Server and Ponderosa Server at NMRFAM
Ponderosa web server
The Ponderosa web server is a free computational resource for structure calculation (Fig. 10b, http://ponderosa.nmrfam.wisc.edu/ponderosaweb.html) maintained by NMRFAM. The server benefits from monthly updates of the PACSY DB and offers the most recent version of the Ponderosa Server software (Fig. 10c). As a default, the structure calculation utilizes Xplor-NIH and includes the AUDANA algorithm and water refinement. The final stage of the structure determination calculates 100 structures with constraints obtained from AUDANA by setting the option to Constraints only for final step in the Ponderosa Client program (Lee et al. submitted).
Ponderosa client
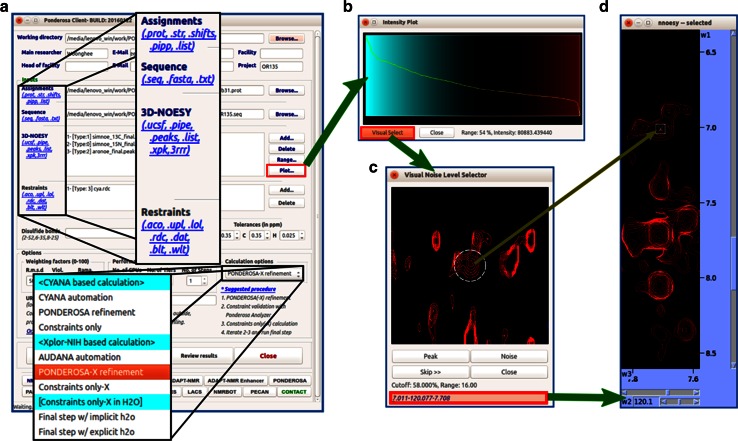
The Ponderosa client accepts a wide range of inputs in addition to NOESY spectra. Also supported are: residual dipolar coupling (RDC), small angle X-ray scattering (SAXS), and wide angle X-ray scattering (WAXS) data (Fig. 11a). Manual constraints can be added and combined with automated NOE assignments. Intensity Plot automatically analyzes the intensities from long range peaks and uses an r−6 approximation to predict the 5.5 Å intensity threshold (Fig. 11b); signals beyond this threshold are considered to be noise.
Fig. 11.
Ponderosa Client supports several formats and settings for structure calculation. a Ponderosa Client accepts several types of input including NOE (raw spectra: .ucsf, .pipe; peak list: .peaks, .list, .xpk, 3rrr), RDC (.rdc), and SAXS (.dat) for Xplor-NIH based structural calculations. b The optimized noise threshold for NOESY peak picking is determined automatically by Intensity Plot, which ranks the intensities of NOE peaks and uses an r −6 (r: distance between two protons) approach to estimate the intensity corresponding to the 5.5 Å cutoff (blue robust range, black mixture of real peaks and noise, red noise range). c Alternatively, the user can employ the Visual Select tool to determine the noise level. This tool randomly selects a position where the chemical shift assignments suggest that a peak should be found, and the user decides whether the signal is a real peak or noise. d The Visual Select tool is integrated with NMRFAM-SPARKY for better decision making. Clicking the peak position in the Visual Select tool enables the user to navigate (two-letter-code up) to the position of the peak in a spectrum displayed by NMRFAM-SPARKY
The Visual Select tool in Intensity Plot (NMRFAM-SPARKY two-letter-code up; Fig. 11c) supports more refined manual noise threshold adjustment. It visualizes positions at which real peaks are predicted to appear at a certain threshold and allows user to decide whether the data support a real peak. The user is guided to find the optimal noise threshold level by a few clicks. This feature also can be used to determine positions of peaks in overlapped regions of strip plots (Fig. 11d).
Ponderosa server
Structure determination jobs are submitted to the Ponderosa Server. Once the calculations are completed, the user is sent an email containing the URL from which the results can be downloaded. We keep upgrading the program and installing in NMRFAM servers. Thus, a user using our server always uses the latest version at the time without any other installation.
Ponderosa analyzer
Ponderosa Analyzer, which integrates an enhanced version of PyMOL and NMRFAM-SPARKY (Fig. 12), is designed to analyze not only coordinates but also essential characteristics of the protein. Enhanced PyMOL is activated by launching regular PyMOL from Ponderosa Analyzer, which includes several tools described below that can be used to refine the input used for structure determinations.
Fig. 12.
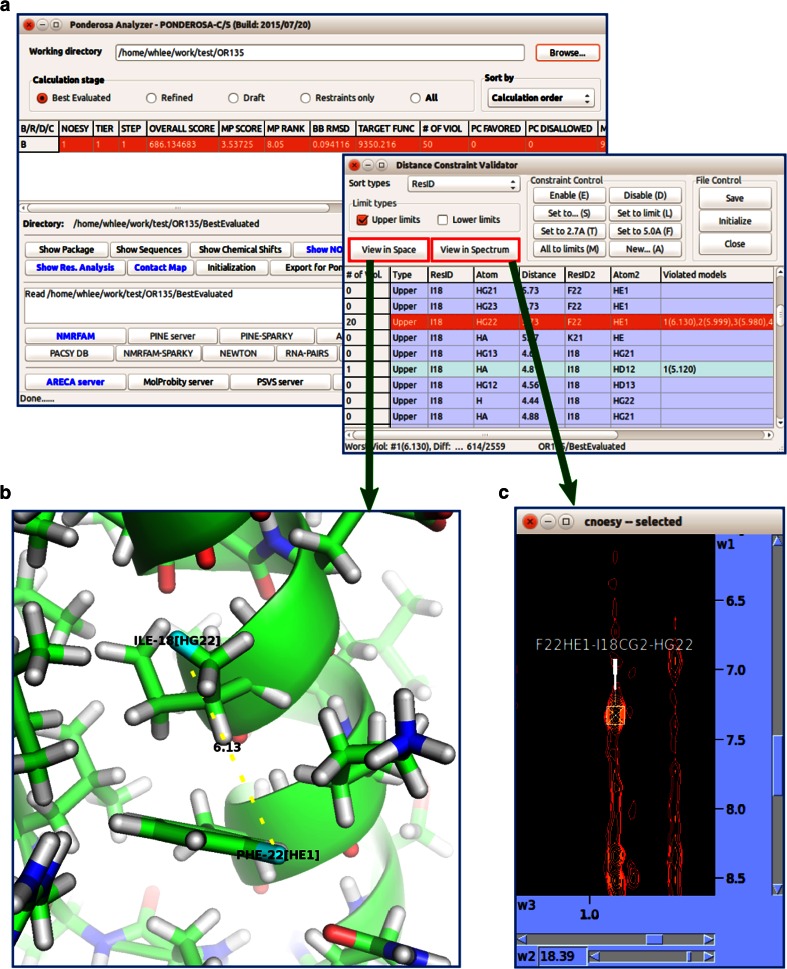
Ponderosa Analyzer provides tools for validating assignments, constraints and structures that are integrated with Enhanced PyMOL and NMRFAM-SPARKY. a Distance Constraint Validator is a tool for analyzing distance information extracted from NOESY data. b 3D illustration of the constraint selected (Enhanced PyMOL command @p). c NOESY spectrum highlighting the experimental evidence for the selected constraint (NMRFAM-SPARKY two-letter-code up). The user can manually adjust or exclude the examined constraint for the next run by means of the Constraint Control buttons in the Distance Constraint Validator
Constraint validation
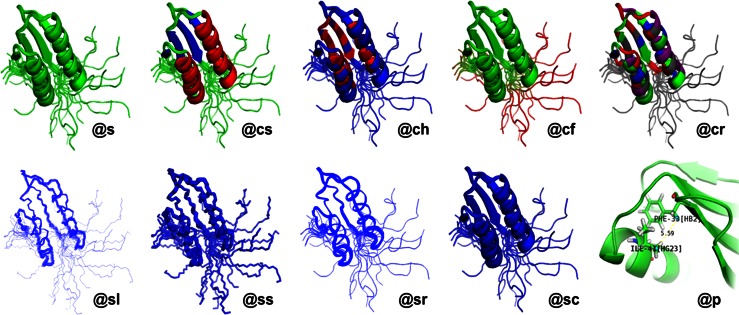
Distance constraint validator (Fig. 12a) is a validation tool for distance information extracted from NOESY data that integrates Enhanced PyMOL (command @p, Fig. 12b) and NMRFAM-SPARKY (two-letter-code up, Fig. 12c). Distance Constraint Validator enables the user to exclude or adjust erroneously extracted inter-proton constraints by simply clicking buttons in the control panel of the program. Ponderosa Violation Investigator(Ponderosa VI) is a simplified validator that runs independently from Ponderosa Analyzer and supports quick violation lookup.
Hydrogen bond constraints
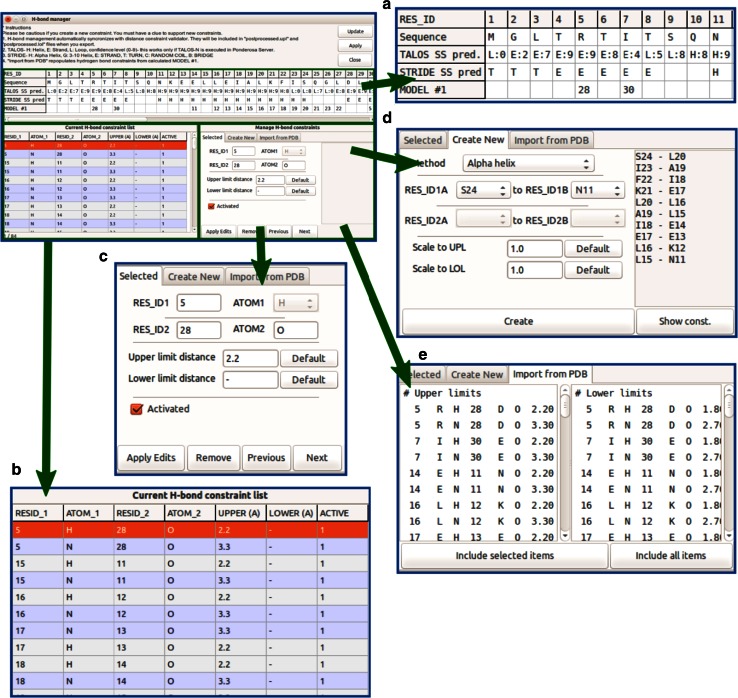
With H-bond Manager (Fig. 13), the user can add or remove hydrogen bond information on the basis of experimental H/D exchange data, characteristic NOE patterns, patterns of secondary chemical shifts, trans H-bond couplings, or results from previous calculations.
Fig. 13.
H-bond manager is a Ponderosa Analyzer tool that provides an easy way to add or remove hydrogen bond constraints. a The H-bond manager panel shows secondary structure information from TALOS-N prediction and from close distances detected structural models from a previous calculation. b Current hydrogen bond constraints are listed in the lower-left panel. During the first structure calculation, they are generated automatically from NOE cross peak patterns. The updates in the lower-right panel change the content in this panel. c An H-bond constraint selected in the lower-right panel can be modified or removed. d New constraints from characteristic secondary structures can be easily added. e Close atom distances from the most recent structure determination can be reviewed as possible H-bonds and can be added as constraints for the next structure determination
Management of constraint types
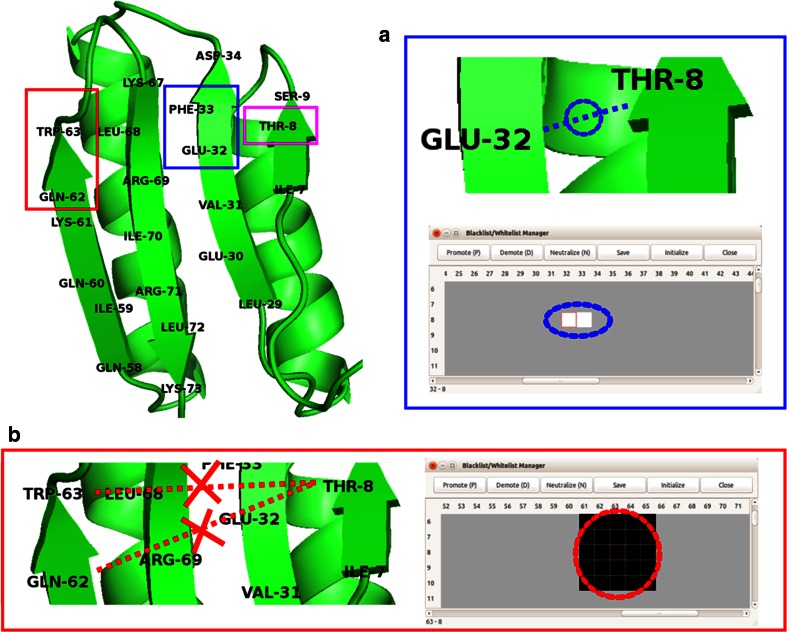
The Blacklist/Whitelist Manager (Fig. 14) provides a graphical user interface that enables the user to modify the weighting factors of inter-residue contacts. For example, if the user determines that two protons are close enough to have an NOE cross peak, the weighting can be promoted. Alternatively, if an NOE connectivity is determined to be erroneous, its weighting can be demoted. Revised constraint files are automatically generated when the user selects ‘Export to the Ponderosa Client’ in the main window of Ponderosa Analyzer.
Fig. 14.
Blacklist and Whitelist are constraint types supported exclusively by PONDEROSA-C/S for resolving ambiguity in NOESY data by applying different weighting factors to the inter-residue contacts. The Blacklist/Whitelist Manager provides a graphical user interface to change the weighting of individual residue–residue contacts. a As an example, if the user is certain that T8-E32 and T8-E33 are close enough to have NOE cross peaks, the corresponding grids can be promoted and colored white. b If the user is certain that T8-W63, T8-Q62 and the surrounding residues are too distant to produce NOE cross peaks, the corresponding grids can be blacked out to avoid errors in automated NOE assignment and structure calculation by the Ponderosa Server
Analysis of contacts
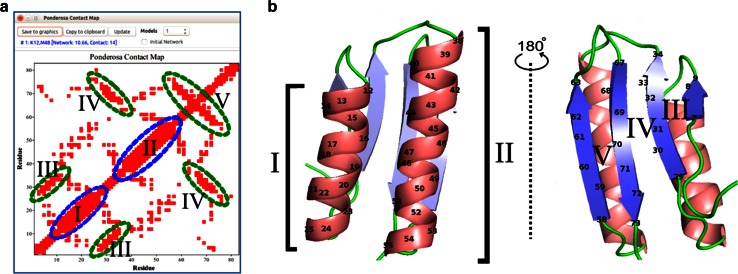
Contact Map illustrates residue–residue contacts from a three-dimensional structural model as a simple two-dimensional plot that reveals secondary structural features (Fig. 15). Contact Map can be used to identify inter proton distances that are shorter than 5.5 Å and are predicted to give rise to NOE cross peaks.
Fig. 15.
Contact Map is a Ponderosa Analyzer tool that assists with structural and spectral analysis of the protein. Because it displays inter-proton distances shorter than 5.5 Å, unlike maps that simply show Cα–Cα contacts, it can be used to predict cross peaks in NOESY spectra. a Contact Map displays patterns that identify secondary structure. b Cartoon representation of the 3D structure of the protein with secondary structural elements colored (Enhanced PyMOL commands @sc and @cs). Good agreement is seen between the secondary structural elements shown in a and b (I and II, alpha helices; III, parallel sheet; IV, anti-parallel sheet consisting of two strands far apart in the sequence; V, anti-parallel sheet consisting of two strands close in the sequence connected by a short turn)
Analysis of backbone dihedral angles
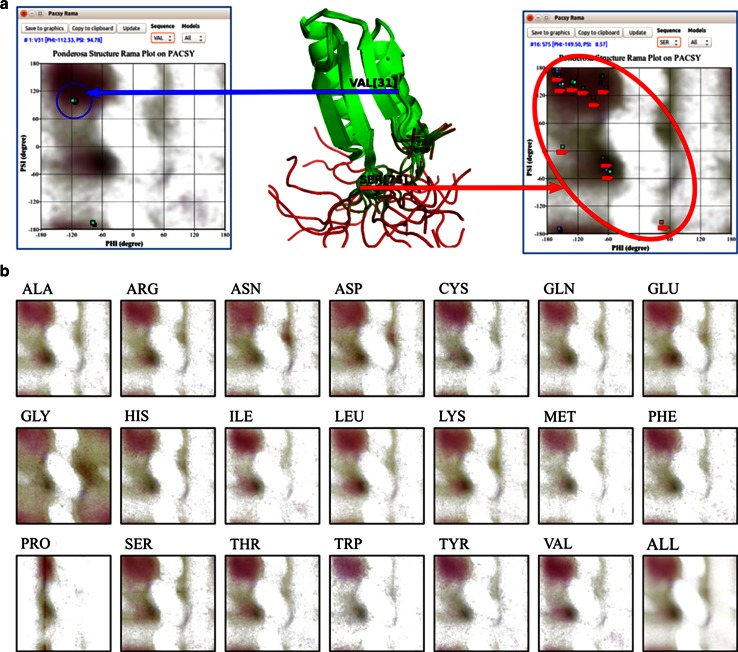
Plotting backbone φ/ψ dihedral angles onto a Ramachandran plot provides a useful way of assessing structural quality. A ‘good’ structure is expected to have backbone dihedral angles clustered in the statistically favorable areas, and outliers may be indicative of errors or the presence of forces that perturb the structure to a higher energy state. In addition, large deviations provide evidence for structural flexibility, such as that for S24 in the example shown in Fig. 16a. Pacsy Rama is a tool that enables the user to display the φ/ψ values for residues in a set of structural models against a consensus Ramachandran plot or a Ramachandran plot for the specific residue type (Fig. 16b). Specialized Ramachandran plots were derived from the PACSY database by counting the occurrences of dihedral angles within 4° × 4° φ/ψ voxels restricted by secondary structure type or amino acid type. From these, image files were created for each of the 20 standard amino acid residues and also for the consensus. The images, which are in PNG (Portable Network Graphics) format, are downloadable from (http://pine.nmrfam.wisc.edu/download_packages.html).
Fig. 16.
Pacsy Rama provides a set of images, derived from quantitative analysis of φ and ψ angles of proteins in the PACSY database, that provide a visual representation of favorable dihedral regions according to amino acid type or for all amino acid types. The images are useful for assessing the structural quality and dynamic characteristics of NMR solution structure of a protein. a In this example, the φ/ψ angles for V31 in the 20 models representing the structure, are closely clustered in an energetically favorable region, whereas the φ/ψ angles for S75 from the various models are highly dispersed consistent with the residue being present in an ill-defined region of the protein. b Pacsy Rama images for the 20 common amino acids and all combined. These images are downloadable from (http://pine.nmrfam.wisc.edu/download_packages.html)
Analysis of distance constraints
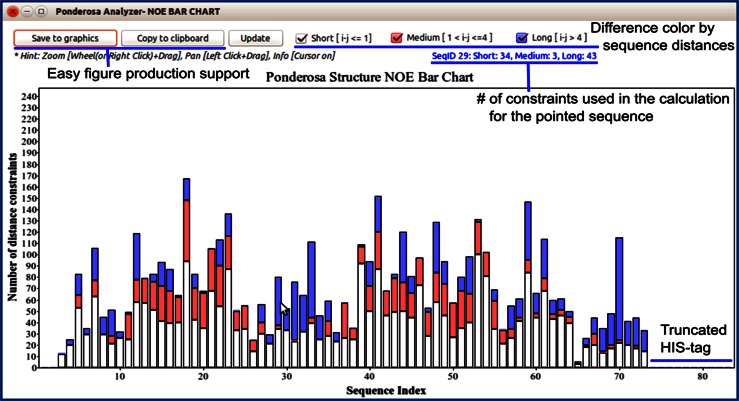
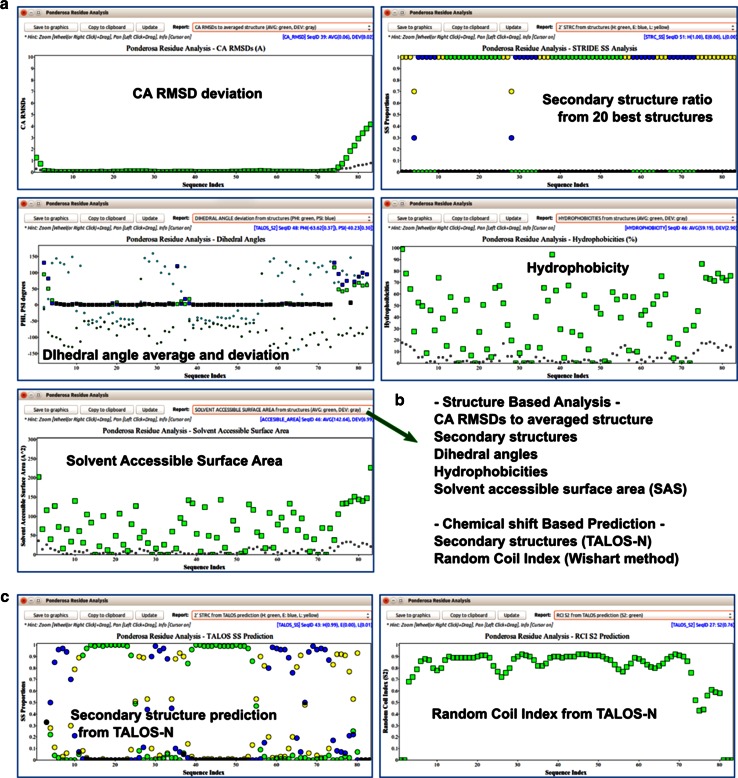
NOE Bar Chart is a tool that shows the number of distance constraints for each residue used in the structure calculation (Fig. 17). These numbers provide an indication of the quality of the structure and identify regions that are ill-defined. The results may indicate that more effort needs to be expended in identifying additional distance constraints. The Cα RMSD Chart and Random Coil Index Prediction Chart (Fig. 18a) can be used to identify disordered regions. In addition, the Color by Flexibility command (@cf) for Enhanced PyMol can be used to distinguish well-defined from ill-defined regions of the protein (Fig. 19; Table 2). Additional information about internal mobility may be available from cross-relaxation and relaxation results. If heteronuclear NOE data are available, they can be visualized in NMRFAM-SPARKY by means of Perturbation Plot (two-letter-code np, Fig. 7a), and if T1/T2 relaxation data have been collected, they can be visualized using Peak Height Analysis (two-letter-code rh, Fig. 6c).
Fig. 17.
The NOE Bar Chart tool in Pacsy Analyzer represents the number and type (white short range; red medium range; blue long range) of distance constraints for each residue used in a structure calculation
Fig. 18.
The Residue Analysis tool in Ponderosa Analyzer provides visual representation of the properties of amino acid residues plotted by sequence number. a Examples of structure-based properties derived from the 20 best models from the structure determination. b List of the features supported by structure-based analysis and chemical shift-based prediction. c Examples of chemical shift-based predictions. Secondary structure and random coil index (S2) values from TALOS-N based on backbone chemical shifts. The user can compare properties predicted from the backbone chemical shifts with those from the calculated structures
Fig. 19.
Ponderosa Analyzer offers these 10 preset drawing modes in Enhanced PyMOL as assigned by the shortcut codes shown. See Table 2 for descriptions
Table 2.
Shortcut commands used by Enhanced PyMOL for custom visualization
| Command | Description |
|---|---|
| @s | Split models in the ensemble |
| @cs | Color by secondary structure (red: helix, blue: strand, green: loop) |
| @ch | Color by hydrophobicity (red: <10 %, blue: ≥30 %, gradient from red to blue between 10 and 30 %) |
| @cf | Color by flexibility (red: ≥2.0 Å for average Cα RMSD, gradient from green to red between 0 and 1.5 Å) |
| @cr | Color by magnitude of RDC violation (red: ≥4.5 Hz, purple: 3.0–4.5 Hz, blue: 1.5–3.0 Hz, green: <1.5 Hz) |
| @sl | Show backbone as lines |
| @ss | Show backbone as sticks |
| @sr | Show backbone as ribbon |
| @sc | Show backbone as cartoon |
| @p | Display selected distance/angle constraints from Ponderosa Analyzer |
Secondary structure analysis
The Residue Analysis tool is a visual chart tool for easy recognition of structural properties on a residue basis. Residue Analysis supports structure-based analysis (Fig. 18a) and chemical-shift-based prediction (Fig. 18c). For the best 20 models from the structure calculation, the structure-based analysis provides a visualization of Cα atom RMSDs to the average structure, secondary structure, dihedral angles (φ and ψ), hydrophobicities, and solvent accessible surface area (SAS). Chemical-shift-based prediction provides a visualization of secondary structure and random coil index derived order parameters (S2) predicted from TALOS-N (Fig. 18b). Comparison of the results can yield insights about the quality of the structure determination and the particular characteristics of the protein.
RDC analysis
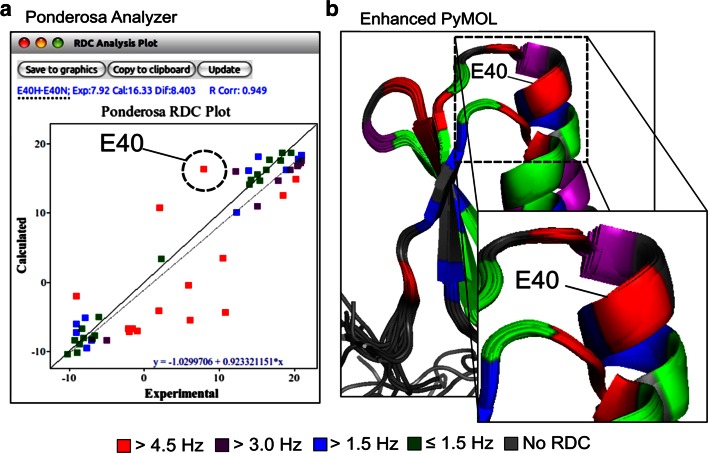
RDC data provide global information about the orientations of individual bonds or entire secondary structure elements and can be used to validate or refine structures determined from NOESY data. This is particularly useful for all α-helical proteins or large proteins. RDC data can be included in the input to the Ponderosa Server and used to enhance automated NOE assignments. The RDC Analysis tool from Ponderosa Analyzer can be used to create a plot of experimental RDC data versus RDCs calculated from the structure (Fig. 20a). The linear least squares fitted line (gray dashed line) indicates the agreement between the experimental RDCs and the RDCs calculated from the structure generated by Ponderosa Server. Enhanced PyMOL (command @cr, Fig. 19) can be used to visualize the correlation between experimental and calculated RDCs and to depict potential errors in the 3D structure. In the illustration shown, residue E40 (colored in red) in the calculated structure does not agree with the input RDC data; thus, E40 is flagged both by the RDC Analysis tool in Ponderosa Analyzer (Fig. 20a) and by Enhanced PyMOL (Fig. 20b).
Fig. 20.
RDC Analysis Plot tool in Ponderosa Analyzer. a The RDC Analysis tool plots experimental RDC data versus RDC data calculated from the structure generated by Ponderosa Server. The linear least squares fitted line (gray dashed line) indicates the agreement between the experimental and calculated RDCs. b The shortcut command @cr (color by RDC violations) in Enhanced PyMOL uses the RMSD report generated by Ponderosa Server to visualize the correlation between experimental and calculated RDCs as a means for identifying potential errors in the 3D structure (or the in the RDC data)
Data visualization with enhanced PyMOL
As part of Integrative NMR, Enhanced PyMOL offers a range of data visualization options activated two- or three-letter shortcuts typed into the PyMOL command-line (Table 2; Fig. 19).
Data output
Chemical shift assignments and peak lists generated by Integrative NMR can be outputted in NMR-STAR format (Markley et al. 2003) for direct deposition to the BMRB or wwPDB.
Video tutorials
Structural analysis method described above for biomolecules are produced as video clips for any non-NMR expert to easily use NMR data in their research. They are freely available from the NMRFAM-SPARKY web page: (http://www.nmrfam.wisc.edu/nmrfam-sparky-distribution.htm), from the PONDEROSA-C/S web page: (http://ponderosa.nmrfam.wisc.edu/videos.html), or from the combined video playlist page: (http://pine.nmrfam.wisc.edu/integrative.html).
Software availability and installation
All NMRFAM software is freely available from the NMRFAM Software Download page (http://pine.nmrfam.wisc.edu/download_packages.html). The Integrative NMR method requires the installation of NMRFAM-SPARKY, Ponderosa Analyzer, Ponderosa Client and PyMOL. The website provides instructions, installation scripts and video tutorials for their installation. The NMRFAM Virtual Machine is recommended for non-specialists, because it contains pre-installed versions of scientific software packages developed by NMRFAM and elsewhere, including NMRFAM-SPARKY, Ponderosa Client, Ponderosa Analyzer, Ponderosa VI, and PyMOL along with its Adaptive Poisson-Boltzmann Solver (APBS) plugin (Baker et al. 2001). A set of examples based on target OR135 from the second round of CASD-NMR is also included. The virtual machine (VM) can be run under a number of different virtualization software programs (VirtualBox and VMware among others) that support the Open Virtualization Format (.ovf, .ova). These virtualization programs are available for a wide variety of different popular host computers and operating systems (Windows, Mac OSX, Linux). A VM emulates a complete computer system. For example, the base operating system of the Integrative NMR VM is Ubuntu Mate 15.04 (64 bit Linux) (https://ubuntu-mate.org); the virtualization software allows this Linux VM to run natively on any host computer.
Acknowledgments
This work was supported by a Grant (P41GM103399) from the Biomedical Technology Research Resources (BTRR) Program of the National Institute of General Medical Sciences (NIGMS), National Institutes of Health (NIH). We are grateful to Dr. Daiwen Yang from National University of Singapore, Dr. Alexandre Bonvin from Utrecht University, and Dr. Frans Mulder from Aarhus University for permitting their contributions to be included in NMRFAM-SPARKY. We also thank the WeNMR Project (European FP7 e-Infrastructure Grant, Contract No. 261572, www.wenmr.eu), supported by the European Grid Initiative (EGI) through the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, South Africa, Malaysia, Taiwan, the Latin America GRID infrastructure via the Gisela Project, the International Desktop Grid Federation (IDGF) with its volunteers and the US Open Science Grid (OSG) for the sufficient example sets for the development of tools.
Contributor Information
Woonghee Lee, Phone: +1-608-263-1722, Email: whlee@nmrfam.wisc.edu.
John L. Markley, Phone: +1-608-263-9349, Email: markley@nmrfam.wisc.edu
References
- Aeschbacher T, Schmidt E, Blatter M, Maris C, Duss O, Allain FH, Guntert P, Schubert M. Automated and assisted RNA resonance assignment using NMR chemical shift statistics. Nucleic Acids Res. 2013;41:e172. doi: 10.1093/nar/gkt665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput Biol. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami A, Clos LJ, II, Markley JL, Butcher SE, Eghbalnia HR. RNA-PAIRS: RNA probabilistic assignment of imino resonance shifts. J Biomol NMR. 2012;52:289–302. doi: 10.1007/s10858-012-9603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu CC, Cornilescu G, Rao H, Porter SF, Tonelli M, Derider ML, Markley JL, Assadi-Porter FM. Temperature-dependent conformational change affecting Tyr11 and sweetness loops of brazzein. Proteins. 2013;81:919–925. doi: 10.1002/prot.24259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti H, Lee W, Tonelli M, Cornilescu CC, Cornilescu G, Assadi-Porter FM, Westler WM, Eghbalnia HR, Markley JL. NMRFAM-SDF: a protein structure determination framework. J Biomol NMR. 2015;62:481–495. doi: 10.1007/s10858-015-9933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti H, Tonelli M, Lee W, Westler WM, Cornilescu G, Ulrich EL, Markley JL. Probabilistic validation of protein NMR chemical shift assignments. J Biomol NMR. 2016;64:17–25. doi: 10.1007/s10858-015-0007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Eghbalnia HR, Wang L, Bahrami A, Assadi A, Markley JL. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J Biomol NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- Fenwick M, Hoch JC, Ulrich E, Gryk MR. CONNJUR R: an annotation strategy for fostering reproducibility in bio-NMR-protein spectral assignment. J Biomol NMR. 2015;63:141–150. doi: 10.1007/s10858-015-9964-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG (2008) SPARKY 3. (San Francisco, University of California, San Francisco), p. Sparky version (3.115). www.cgl.ucsf.edu/home/sparky/manual/index.html
- Jones DT, Tress M, Bryson K, Hadley C. Successful recognition of protein folds using threading methods biased by sequence similarity and predicted secondary structure. Proteins Struct Func Genet Suppl. 1999;3:104–111. doi: 10.1002/(SICI)1097-0134(1999)37:3+<104::AID-PROT14>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci. USA. 2012;109:10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Westler WM, Bahrami A, Eghbalnia HR, Markley JL. PINE-SPARKY: graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics. 2009;25:2085–2087. doi: 10.1093/bioinformatics/btp345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Kim JH, Westler WM, Markley JL. PONDEROSA, an automated 3D-NOESY peak picking program, enables automated protein structure determination. Bioinformatics. 2011;27:1727–1728. doi: 10.1093/bioinformatics/btr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Yu W, Kim S, Chang I, Lee W, Markley JL. PACSY, a relational database management system for protein structure and chemical shift analysis. J Biomol NMR. 2012;54:169–179. doi: 10.1007/s10858-012-9660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Stark JL, Markley JL. PONDEROSA-C/S: client–server based software package for automated protein 3D structure determination. J Biomol NMR. 2014;60:73–75. doi: 10.1007/s10858-014-9855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Petit CM, Cornilescu G, Stark JL, Markley JL (submitted) The AUDANA algorithm for automated protein 3D structure determination from NMR NOE data [DOI] [PMC free article] [PubMed]
- Marcheschi RJ, Staple DW, Butcher SE. Programmed ribosomal frame shifting in SIV is induced by a highly structured RNA stem-loop. J Mol Biol. 2007;373:652–663. doi: 10.1016/j.jmb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley JL, Ulrich EL, Westler WM, Volkman BF. Macromolecular structure determination by NMR spectroscopy. Methods Biochem Anal. 2003;44:89–113. [PubMed] [Google Scholar]
- Rosato A, Vranken W, Fogh RH, Ragan TJ, Tejero R, Pederson K, Lee HW, Prestegard JH, Yee A, Wu B. The second round of Critical Assessment of Automated Structure Determination of Proteins by NMR: CASD-NMR-2013. J Biomol NMR. 2015;62:413–424. doi: 10.1007/s10858-015-9953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Lee W, Lee W. Structural proteomics by NMR spectroscopy. Expert Rev Proteomics. 2008;5:589–601. doi: 10.1586/14789450.5.4.589. [DOI] [PubMed] [Google Scholar]
- Tamiola K, Mulder FA. ncIDP-assign: a SPARKY extension for the effective NMR assignment of intrinsically disordered proteins. Bioinformatics. 2011;27:1039–1040. doi: 10.1093/bioinformatics/btr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain D, Tannenbaum T, Livny M, Berman F, Hey AJG, Fox G (2003) Condor and the grid. In: Berman F, Hey AJG, Fox GC (Eds) Grid computing: making the global infrastructure a reality, Wiley, New York
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wang L, Eghbalnia HR, Bahrami A, Markley JL. Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J Biomol NMR. 2005;32:13–22. doi: 10.1007/s10858-005-1717-0. [DOI] [PubMed] [Google Scholar]
- Wassenaar TA, van Dijk M, Loureiro-Ferreira N, van der Schot G, de Vries SJ, Schmitz C, van der Zwan J, Boelens R, Giachetti A, Ferella L, et al. WeNMR: Structural Biology on the Grid. J Grid Comput. 2012;10:743–767. doi: 10.1007/s10723-012-9246-z. [DOI] [Google Scholar]
- Zhang L, Yang D. SCAssign: a sparky extension for the NMR resonance assignment of aliphatic side-chains of uniformly 13C,15N-labeled large proteins. Bioinformatics. 2006;22:2833–2834. doi: 10.1093/bioinformatics/btl477. [DOI] [PubMed] [Google Scholar]