Abstract
Despite similar components, the heterogeneity of skin characteristics across the human body is enormous. It is classically believed that site-specific fibroblasts in the dermis control postnatal skin identity by modulating the behavior of the surface-overlying keratinocytes in the epidermis. To begin testing this hypothesis, we characterized the gene expression differences between volar (ventral; palmoplantar) and nonvolar (dorsal) human skin. We show that KERATIN 9 (KRT9) is the most uniquely enriched transcript in volar skin, consistent with its etiology in genetic diseases of the palms and soles. In addition, ectopic KRT9 expression is selectively activated by volar fibroblasts. However, KRT9 expression occurs in the absence of all fibroblasts, although not to the maximal levels induced by fibroblasts. Through gain-of-function and loss-of-function experiments, we demonstrate that the mechanism is through overlapping paracrine or autocrine canonical WNT–β-catenin signaling in each respective context. Finally, as an in vivo example of ectopic expression of KRT9 independent of volar fibroblasts, we demonstrate that in the human skin disease lichen simplex chronicus, WNT5a and KRT9 are robustly activated outside of volar sites. These results highlight the complexities of site-specific gene expression and its disruption in skin disease.
Site-specific epidermal differentiation programs define the heterogeneity of skin identity across the human body. It is generally believed that positional skin identity is regulated by epithelial (keratinocytes) and mesenchymal (fibroblasts) interactions.1, 2, 3, 4, 5 Indeed, three-dimensional skin equivalent models and transplantation experiments show that epidermal stratification during the keratinocyte differentiation program is remarkably disrupted in the absence of fibroblasts,2, 4, 5 demonstrating that epidermal development and homeostasis are regulated by extrinsic factors released by fibroblasts. Such interactions have been further clarified through an analysis of the differential localization of epidermal keratins in distinct epidermal layers and body sites.
Volar (palmoplantar) skin is characterized by thick epidermal layers, less pigmentation, and lack of hair. In addition to these features, cytoskeleton KERATIN (KRT) 96 has been thought to be almost exclusively localized to volar keratinocytes,1, 6, 7, 8, 9 indicating that KRT9 can be a unique marker for volar skin. To date, many clinical studies have reported that KRT9 mutations cause epidermolytic palmoplantar keratoderma, characterized by compensatory thickened epidermal layers in the palms and soles.10 Conditional deletion of Krt9 in a murine model demonstrated that Krt9 is responsible for maintaining mechanical integrity and terminal differentiation of volar skin.11 These data highlight not only the functional importance of KRT9 for structural support but also the validity of KRT9 as a marker of positional skin identity. KRT9 can be activated by pressure-activating mitogen-activated protein kinases.12, 13 To further study the mechanism of KRT9 regulation, Moll et al14 reported that volar fibroblasts induce ectopic nonvolar KRT9 expression in keratinocytes, as rarely is the case in vivo. Grafting of human volar fibroblasts together with nonvolar keratinocytes generated thickened KRT9+ human epidermis on severe combined immunodeficiency mice.15 Also, in their in vitro co-culture system, KRT9 was induced in nonvolar keratinocytes after co-culture with volar fibroblasts, but not after co-culture with nonvolar fibroblasts. Therefore, KRT9 is a marker of positional identity, and can be ectopically induced by volar fibroblasts. Herein, we focus on defining the mechanisms behind this interaction.
Recently, the WNT/β-catenin pathway has been shown to be important for differentiation of interfollicular keratinocytes, such as that present in volar skin.16 In mice, deactivation of β-catenin led to a thinner epithelium in the footpad and tongue. Indeed, terminal differentiation of suprabasalar structures, like the filiform papillae of the tongue, did not occur after inhibition of β-catenin. In addition, in humans, decreased WNT/β-catenin signaling from lack of WNT10a leads to a compensatory volar keratoderma and defects in tongue epithelialization.17, 18 Especially given that KRT9 is a suprabasal KERATIN, these findings imply that KRT9 expression might be promoted by β-catenin.
However, the opposite proposal has been made in the literature: because β-catenin is so important for hair follicle development, it has been maintained that β-catenin must be inhibited in hairless volar skin.7, 19 Indeed, although WNT5a has been separately shown to induce KRT920 and KRT1 expression,21 a completely opposite mechanism has been proposed for the mechanism of induction. Rinn et al20 suggested that WNT5a from distal fibroblasts induces KRT9 in keratinocytes in a noncanonical manner by antagonizing β-catenin activity, as also proposed for how Dickkopf-related protein-1 (DKK-1) might promote volar features by Yamaguchi et al.15 In contrast, a recent study showed that autocrine WNT5a signaling stimulates keratinocyte differentiation and expression of KRT1 in a canonical cascade by instead stabilizing β-catenin and augmenting β-catenin–T-cell factor target gene expression.21 However, because volar keratinocytes express both KRT9 and KRT1 coordinately, it is unlikely that WNT5a would induce their expression by diametrically opposite mechanisms. We, therefore, hypothesized that contrary to current proposals,7, 19 the canonical WNT/β-catenin signaling pathway might play a role in KRT9 induction to site-specific gene expression.
Herein, we demonstrate that fibroblasts can inhibit keratinocyte differentiation, but at optimum ratios, KRT9 expression is stimulated by volar fibroblasts. We surprisingly find that keratinocytes can express ectopic KRT9 even independent of all fibroblasts. Using both gain-of-function and loss-of-function approaches, we demonstrated that β-catenin plays a role in keratinocyte KRT9 expression by either an autocrine mechanism in the absence of fibroblasts or a conserved paracrine mechanism in the presence of fibroblasts. Given the potential for volar-fibroblast independent induction of KRT9, we also demonstrate, for the first time, that KRT9 and WNT5a are expressed ectopically in skin diseases, which in some ways recapitulate volar skin. Collectively, our findings provide a new model for how site-specific skin gene expression occurs, and how it is disrupted in disease.
Materials and Methods
Human Samples
Human biopsy specimens were obtained under written informed consent and Johns Hopkins University Institutional Review Board approved protocol (NA_00033375) following Declaration of Helsinki principles. Skin was processed as described,22, 23 and 3-mm skin biopsy specimens were taken from areas of the occipital scalp, palms, soles, and corresponding dorsal positions from the hands and feet. Epidermis and dermis were separated using dispase II protease (Sigma-Aldrich, St. Louis, MO), as described in the literature,22, 24 and subjected for studies as below. Deidentified samples from paraffin-embedded blocks of diagnostic biopsy specimens for normal skins (volar and nonvolar), lichen simplex chronicus (LSC), and psoriasis were used for immunofluorescence analysis, also under Johns Hopkins University Institutional Review Board protocol approval (NA_00072381). Also, specific written consent forms for photography were also completed by subjects whose images are included.
Microarray Analysis
RNAs isolated from volar and nonvolar epidermis were submitted to the Johns Hopkins Medical Institutions Deep Sequencing and Microarray core for Affymetrix Human Exon 1.0 ST microarray chips (Affymetrix, Santa Clara, CA), according to manufacturer's protocols. Raw gene expression signals in the form of Affymetrix CEL files were extracted and normalized with Partek Genomics Suite software version 6.6 using the Robust Multichip Analysis algorithm.25 Significances of gene expression were measured by t-test analysis. These analyses are currently available on Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo; accession numbers GSE39267 and GSE39266).
Primary Skin Cell Isolation and Cultures
Human epidermal keratinocytes were isolated from newborn circumcision foreskin with parents' agreement and from adult abdomen with donor's consent using enzyme digestion method, as described in the literature.22, 24 Briefly, skin samples were cut into small pieces and incubated in 0.4% dispase II solution overnight at 4°C, which allows separating epidermis from dermis. Epidermis was used to isolate keratinocytes using 0.025% trypsin-EDTA (Life Technologies, Grand Island, NY). After removal of epidermis, papillary dermis of volar (palm and sole) and nonvolar (foreskin, scalp, hand, and foot) was placed and incubated for outgrowth of fibroblasts.22, 24 Keratinocytes were cultured in serum-reduced keratinocyte medium with defined growth factors (KGM-GOLD Bullet kit; Lonza, Walkersville, MD) and maintained at preconfluent state to prevent from differentiation until use. For all experiments, keratinocytes were used after two passages to eliminate contaminating cells. Human fibroblasts were maintained in culture Dulbecco's modified Eagle's medium (Corning, Manassas, VA) with 10% fetal bovine serum (Gemini, West Sacramento, CA) and 1× antibiotic-antimycotic cocktail (Life Technologies). To maximize differentiation, keratinocytes were plated by locally dropping (150,000 cells/20 μL of drop; two drops for a 12-well plate) using keratinocyte basal media without growth factors (KBMedia) and KBM, including growth factors (KGMedia). These were described as overconfluent in the text and compared with keratinocytes seeded into wells where all cells were in contact; 150,000 cells but without dropping and into an entire 12-well plate (confluent). Preconfluent keratinocyte group represents undifferentiated keratinocytes and was used to normalize assays and determine keratinocyte differentiation. For co-culture, keratinocytes and fibroblasts were mixed together with different ratios and seeded into a 12-well plate by using the dropping method. Unless otherwise stated, 48 hours after plating, cells were harvested for RNA and protein isolation to study as below.
RNA Isolation and RT-qPCR
Total RNA was isolated from skin tissue samples or keratinocytes using RNeasy Mini Kit (Qiagen, Valencia, CA) and treated with DNase I (Qiagen) to eliminate genomic DNA. The purity and concentration of RNAs were analyzed using a NanoDrop 2000c (Thermo Scientific, West Palm Beach, FL). After reverse transcription reactions using high-capacity RNA to cDNA kit (Life Technologies), quantitative RT-PCR (RT-qPCR) was performed to measure target genes using TaqMan probes and Fast reaction master mix reagents (Life Technologies). Relative expression of mRNAs was analyzed by the Ct value of target genes and quantified by normalizing to ribosomal protein large P0 (RPLP0) as a housekeeping gene using the ΔΔCt method.26
siRNA Transfection
β-Catenin and nontargeting control (scramble) siRNAs [β-catenin (SASI_Hs01_00117958); scramble (SIC001); Sigma-Aldrich] were commercially obtained. Keratinocytes (5 × 105 cells per well) in 12-well plates were transfected with 50 nmol/L of siRNAs for 48 hours using Lipofectamine 2000 (Life Technologies), following the manufacturer's directions. At the end of the transfection period, cells were harvested to isolate RNA and protein to perform RT-qPCR and Western blot analysis, respectively.
Western Blot Analysis
Protein samples were extracted from the epidermis layer of skin biopsy specimen and keratinocytes and fibroblasts using M-PER lysis buffer (Thermo Scientific) containing protease inhibitors (Thermo Scientific). Then, protein concentrations were measured by BCA method (Thermo Scientific). Western blot procedures were followed by the manufacturer's protocol (NuPAGE system; Thermo Scientific). Briefly, 5 to 30 μg of protein samples were loaded for electrophoresis and transferred to polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). After blocking for 3 hours in 5% nonfat dry milk, the membrane was incubated with mouse monoclonal anti-human active β-catenin (05-665/clone 8E7; EMD Millipore, Billerica, MA) and rabbit polyclonal anti-human KRT1 (ab93652; Abcam, Cambridge, MA) and KRT9 (ab85283; Abcam) antibody (1:1000 dilution) at 4°C overnight, followed by incubation with anti-mouse and anti-rabbit IgG–horseradish peroxidase (1:1000 dilution) (7074S; Cell Signaling Technology, Danvers, MA) for 1 hour at room temperature. Protein amounts were normalized to rabbit polyclonal anti-human β-actin antibody (1:1000 dilution) (4967L; Cell Signaling Technology). Finally, proteins were visualized using chemiluminescent substrate kit (Thermo Scientific) and saved as image files using ChemiDoc XRS+ (Bio-Rad).
DKK-1 and SB216763 Treatment
Recombinant human (rh) DKK-1 protein (R&D Systems, Minneapolis, MN) and SB216763 (Sigma-Aldrich) were purchased. For rhDKK-1 treatment, 3 × 105 keratinocytes per well or both 3 × 105 keratinocytes per well and 3 × 104 fibroblasts per well plated into 12-well plates were treated with 0 to 200 ng/mL rhDKK-1 in KBMedia for 48 hours. For SB216763 treatment, 3 × 105 keratinocytes per well seeded into 12-well plates were treated with 0 to 5 μmol/L SB216763 in KBMedia for 2 hours, followed by washing cells and additional 48 hours incubation in KBMedia. For cotreatment of rhDKK-1 and SB216763, 3 × 105 keratinocytes per well were plated into 12-well plates and treated with 50 ng/mL rhDKK-1 and 5 μmol/L SB216763 at the same time for 48 hours in KBMedia. RNA and protein samples were analyzed to detect differences of gene expressions.
EGF, Calcium, and SB216763 Treatment
Recombinant human epidermal growth factor (EGF) protein (R&D Systems), calcium chloride (Sigma-Aldrich), and SB216763 (Sigma-Aldrich) were provided commercially. Keratinocytes (3 × 105 cells per well) were plated by locally dropping into 12-well plates and treated with 0 to 1000 ng/mL EGF or 2 mmol/L CaCl2 in KBMedia for 48 hours. For SB216763 treatment, 3 × 105 keratinocytes per well seeded into 12-well plates were treated with 0 to 5 μmol/L SB216763 in KBMedia for 2 hours, followed by washing cells and additional 48 hours incubation in KBMedia. Cells were harvested at the end of incubation and used to analyze differences of gene expressions.
Immunofluorescence
For tissues, paraffin-embedded human skin samples were stained with rabbit anti-human KRT1 (ab93652; Abcam), KRT5 (AF138; Covance, Dedham, MA), and KRT9 (ab85283; Abcam), WNT5a (ab72583; Abcam) antibody (1:200 dilution) overnight at 4°C, followed by incubation with goat anti-rabbit IgG labeled with Alexa Fluor 488 (A11034; Life Technologies) and 594 (A11037; Life Technologies) (1:1000 dilution) for 1 hour at room temperature. For cells, mixture of keratinocytes with sole or foreskin fibroblasts (10:1 ratio) were plated onto coverslip and cultured for 4 days in KBMedia. Then, cells were fixed with 4% paraformaldehyde for 5 minutes and stained with KRT9 antibody (1:100 dilution) overnight at 4°C, followed by incubation with goat anti-rabbit IgG labeled with Alexa Fluor 488 (1:1000) for 1 hour at room temperature. Cell nuclei were determined by mounting medium, including DAPI (Vector Laboratories, Burlingame, CA). Fluorescence images were observed by fluorescence microscopy (Leica Microsystems Inc., Buffalo Grove, IL) and saved as image files.
Immunohistochemistry
Paraffin-embedded human skin tissues were submitted to the Johns Hopkins Medical Institutions Oncology Tissue Services Immunostaining Core Facility and stained with microphthalmia-associated transcription factor to characterize melanocytes in the epidermis. Images were observed under bright field microscopy (Leica Microsystems) and taken with ×100 (low magnification) and ×200 (high magnification). Melanocytes were quantified by counting microphthalmia-associated transcription factor–positive cells and represented with ratios (melanocytes/100 keratinocytes).
Measurement of Cell Metabolic Activity
Keratinocytes were plated into a 96-well plate with a density of 1 × 105 cells in KBMedia. All procedures were followed according to the manual of Cell Counting Kit-8 assay (Dojindo Molecular Technologies, Rockville, MD). The absorbance was measured at 450 nm using a multiplate reader (BioTek, Winooski, VT) at each day. Metabolic activities of keratinocytes were analyzed by comparing OD values on day 1.
Fibroblast-Conditioned Media Experiments
Volar (sole) and nonvolar (foreskin and scalp) fibroblasts were cultured in Dulbecco's modified Eagle's medium with 5% fetal bovine serum. The conditioned medium was then removed and filtered using a 0.22-μm polyvinylidene difluoride syringe filter (Genesee Scientific, San Diego, CA). Keratinocytes were cultured with volar and nonvolar fibroblast-conditioned media for 2 days, and RNA samples were used to analyze KRT9 gene expression.
Mouse Wound Experiments
All animal protocols are approved by the Johns Hopkins University Animal Care and Use Committee. C57BL/6J mice were provided from the Jackson Laboratory (Bar Harbor, ME). Three-week-old male and female mice were used to make a 1-cm2 excisional full-thickness wound on the back skin, as previously described.27, 28, 29 The excised skin was kept for baseline controls of each mouse. At time points after wounding—post wound day 1, 3, and 5—the healing wound was excised with a 1- to 2-mm border of unwounded skin. The baseline and post wound time point samples were then stored in RNAlater solution (Sigma-Aldrich) at −80°C. All samples were used to analyze Krt9 gene expression by using RT-qPCR. Actnb (β-actin) as a housekeeping gene was used for normalization, and differences in gene expression were represented with fold difference.
Genome Annotation
To classify functional gene annotations, 200 genes selected from volar and nonvolar epidermis were uploaded and analyzed by DAVID bioinformatics resource (http://david.abcc.ncifcrf.gov, last accessed July 1, 2015).30 Data were represented with table format, including different annotation categories.
Statistical Analysis
All data were generated by at least three independent experiments and analyzed by t-test (unpaired) to evaluate significant differences. Relative mRNA expression was quantified by normalizing to preconfluent keratinocyte samples. Statistical significance was considered at P < 0.05.
Results
Volar (Palmoplantar) Skin Is Characterized by Exclusive Expression of KRT9
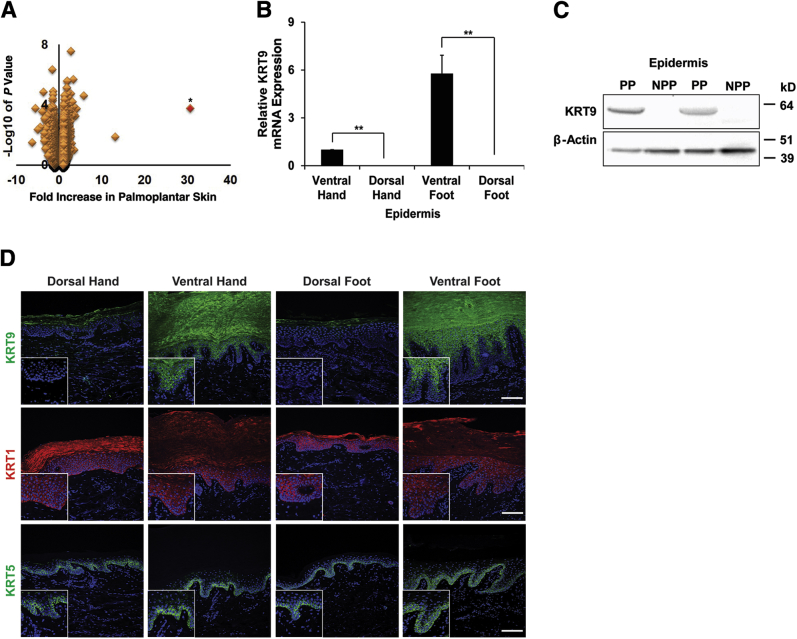
Our first goal was to define the specific shared gene signature for volar skin. Only sparse gene expression profiling has been devoted to learning the basic gene expression differences between native human skin sites. A biopsy specimen was obtained from three human subjects from four distinct sites (palms, soles, and the corresponding dorsal nonvolar positions on the hands and feet), and then isolated RNA from the epidermis for microarrays. Using data from both upper and lower extremity, we ranked individual genes according to their statistical significance and fold change with respect to their selective presence in volar versus nonvolar skin. Genome-wide analysis of overrepresented gene categories identified many significant pathway changes in volar versus nonvolar epidermis, including Wnt pathway genes (Supplemental Figure S1 and Supplemental Table S1). In examining individual genes, we showed that the type I keratin KRT9 mRNA was the most differentially expressed transcript in the volar epidermis, compared with the nonvolar epidermis (Figure 1A). Other significant genes higher in volar skin include HRNR and ITGBL1. We confirmed that KRT9 mRNA (Figure 1B) and protein (Figure 1C) are confined to the volar epidermis. Finally, KRT9 was found to be exclusively localized in suprabasal layers of volar epidermis by immunofluorescence (Figure 1D). The type II keratin KRT1, a possible heterodimer partner for KRT9, was similar between volar and nonvolar epidermis in terms of mRNA (Supplemental Figure S2), but perhaps showed some increased staining in volar skin (Figure 1D). However, another type II keratin, KERATIN 5 (KRT5), is expressed at similar levels in both volar and nonvolar epidermis (Figure 1D). KRT9 is, therefore, a robust marker for volar skin in the samples tested.
Figure 1.
KRT9 is the highest differentially expressed gene in volar keratinocytes. A: Microarray data plotting genes (dots) by P value of differential expression in volar skin (y axis) versus fold increase in volar skin (x axis). Red dot indicates KRT9; nearest neighbor is ITGBL1. B: KRT9 mRNA expression in volar and nonvolar epidermis. C: KRT9 protein expression in volar (PP) and nonvolar epidermis (NPP) from different donors. D: Immunofluorescence staining of KRT1 (red), KRT5 (green), and KRT9 (green) is shown merged with DAPI (blue) nuclear counterstaining. The insets represent enlarged areas from parts of the corresponding images. Relative KRT9 mRNA expression was analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by means of fold difference ± SEM normalized with ventral hand and analyzed by t-test between ventral and dorsal epidermis. n = 3 (A); n = 4 (B). ∗P < 0.05, ∗∗P < 0.01. Scale bar = 100 μm (D, main images). Original maginification, ×200 (D, insets). ITGBL1, integrin beta like 1; KRT, KERATIN; NPP, nonpalmoplantar; PP, palmoplantar.
Keratinocytes without Fibroblasts Can Express KRT9, but More so with Volar Fibroblasts
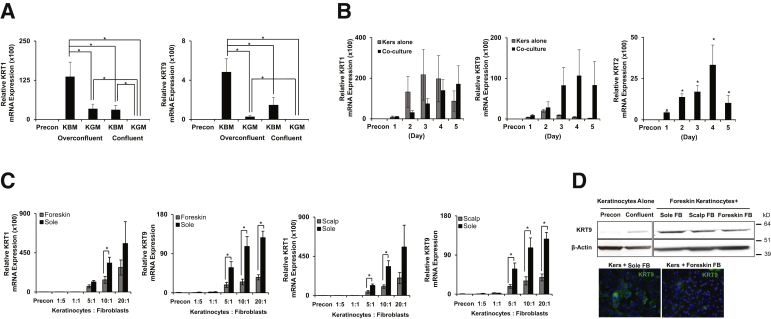
In conducting experiments to measure the capacity for volar fibroblasts to induce KRT9, we were surprised to discover the innate ability for keratinocytes in culture to make the site-specific KRT9. Normally, patients with mutations in KRT9 have no symptoms outside of the hands and feet. We, therefore, tested the innate capacity of foreskin keratinocytes to induce KRT9. Keratinocytes can be induced to differentiate in vitro by increasing the cell density and reducing the concentration of growth factors,31 although calcium concentration changes do the same (Supplemental Figure S3). Given that KRT9 is a suprabasal keratin, we initially analyzed the level of both KRT1 and KRT9 in keratinocytes cultured alone at different cell densities in various culture media. To identify the effect of growth factors, keratinocytes were cultured in basal media in the presence (KGMedia) or absence (KBMedia) of growth factors for 48 hours. Although KRT9 is not detected in foreskin keratinocytes from preconfluent cultures, KRT1 and KRT9 are significantly induced in the setting of KBMedia at overconfluency and in the absence of all fibroblasts (Figure 2A), or after calcium switch (Supplemental Figure S3).
Figure 2.
KRT9 expression is induced by volar fibroblasts, but also independent of them at lower levels. A: KRT1 and KRT9 mRNA expression in keratinocytes alone increases with higher cell density and culture media conditions promoting differentiation. B: KRT1, KRT9, and then KRT2 mRNA expression increase with differentiation in time-course experiments, with greater expression of KRT9 in co-cultures with volar fibroblasts compared with keratinocytes alone. C: Sole fibroblasts preferentially induce KRT1 and KRT9 mRNA expression in keratinocytes cultured with foreskin or sole, and scalp or sole fibroblasts. D: KRT9 protein expression is highest in co-cultures with sole fibroblast, but also detectable in the absence of all fibroblasts. Co-cultures of foreskin keratinocytes with none, sole, scalp, or foreskin fibroblasts were analyzed by Western blot stained for KRT9 and β-actin expression. KRT9 protein (green) in the co-culture system was confirmed by immunocytochemistry and was shown merged with DAPI (blue) nuclear counterstaining. Relative KRT mRNA expression was analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by means of fold-difference ± SEM normalized with preconfluent keratinocytes only. n = 6 (A); n = 3 (B); n = 4 (C). ∗P < 0.05. Original magnification, ×400 (D). FB, fibroblast; KBM, keratinocyte basal media; Kers, keratinocytes cultured alone; KGM, KBMedia plus growth factors; KRT, KERATIN; Precon, preconfluent keratinocytes only.
To determine the time point at which KRT9 expression reached its peak in vitro, and to compare solo cultures of keratinocytes to co-cultures with fibroblasts, cells were cultured in KBMedia at high density, and the level of KRT1 and KRT9 mRNA was analyzed on each day for 5 days. KRT1 mRNA expression gradually increased until day 3 and decreased on days 4 and 5 in keratinocyte solo cultures, but gradually increased in co-cultures (Figure 2B). On the other hand, KRT9 mRNA expression peaked on day 2 and then declined (Figure 2B) in co-cultures, but peaked at a higher level on day 4 in co-cultures. We also found that KRT1 transcription is more robust and stable than that of KRT9 (Figure 2B), suggesting that the KRT1 promoter is more responsive than the KRT9 promoter to confluence conditions. As in vivo, KRT2 expression peaked last. Because of growth factor depletion, cells were less metabolically active with time, and were not cultured beyond 5 days (Supplemental Figure S4).
Fibroblasts delay keratinocyte differentiation (Figure 2B),2 but specify differentiation programs. We, therefore, next cultured keratinocytes with different ratios of fibroblasts from volar and nonvolar dermis in KBMedia at high density for 48 hours, the earliest time point for inductivity. KRT1 mRNA expression showed mild, but statistically insignificant, increases in volar fibroblast-keratinocyte co-culture compared with nonvolar fibroblast-keratinocyte co-culture (Figure 2C). However, the level of KRT9 mRNA expression induced by volar fibroblasts was more robust: 4.5-fold greater than the level induced by nonvolar fibroblasts at the intermediate (10:1) ratio of keratinocytes/fibroblasts (Figure 2C). Western blot analysis and immunofluorescence confirmed these RT-qPCR results (Figure 2D). These results demonstrate that, although fibroblasts modulate KRT9 expression by paracrine signaling, it also can be expressed as a default keratin through autocrine signaling, despite its limited expression in human skin.
β-Catenin Promotes KRT9 Expression
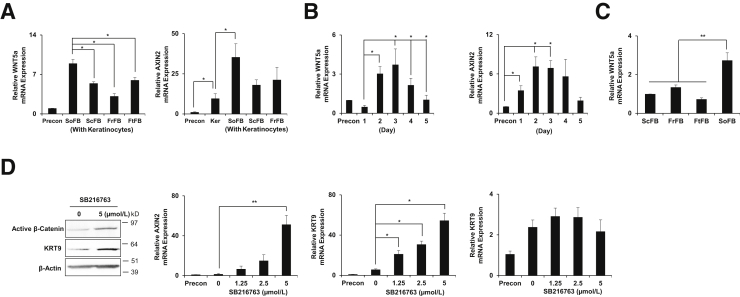
If keratinocytes alone could produce KRT9, but normally do not in vivo, volar fibroblasts might mimic the same signaling pathway that endows the keratinocytes with an innate ability to produce KRT9. Thus, the same signaling molecules might be present in both the context of autocrine signaling in keratinocytes alone and paracrine signaling between volar fibroblasts and keratinocytes. Previous studies provided evidence that differentiated keratinocytes secrete WNT5a and induce KRT1 in a β-catenin–dependent promoting canonical cascade.22 Also, separate studies have shown that WNT5a from volar fibroblasts induce ectopic KRT9 expression in nonvolar keratinocytes,20, 21 although it was further proposed that WNT5a acts as a proximal-distal signal and induces KRT9 in a noncanonical cascade by inhibiting β-catenin activity.7, 21 Because WNT5a expression gradually increases during keratinocyte differentiation (Figure 3B), we speculated instead that the canonical WNT signaling pathway might promote KRT9 expression in both autocrine and paracrine contexts.
Figure 3.
Keratinocytes induce KRT9 through canonical activation of β-catenin. A: WNT5a and AXIN2 mRNA are elevated in co-culture with foreskin keratinocytes and plantar (sole) fibroblasts analyzed by quantitative RT-PCR. B: WNT5a and AXIN2 mRNA are induced in keratinocytes alone at higher cell densities and culture media conditions promoting differentiation in time-course experiments. C: mRNA of WNT5a is elevated in volar fibroblasts comparing with nonvolar fibroblasts. D: Active β-catenin protein, AXIN2 mRNA, and KRT9 mRNA and protein are induced with 5 μmol/L of SB216763, but not KRT6a mRNA. ∗P < 0.05, ∗∗P < 0.01. n = 3 (A); n = 3 to 6 (B); n = 3 to 4 (C); n = 3 to 5 (D). FrFB, foreskin; FtFB, dorsal foot; KRT, KERATIN; Precon, preconfluent keratinocytes only; ScFB, scalp; SoFB, sole fibroblast.
From a second cohort of three human subjects, we obtained biopsy specimens from paired volar and nonvolar skin of the upper and lower extremities, as we had performed before (Figure 1A). Herein, we expanded fibroblasts as used in the above co-cultures, but instead subjected them to gene expression analysis. Again, we analyzed the transcripts for a conserved signature between individuals and body sites for volar skin fibroblasts. We determined that WNT5a was the WNT ligand most up-regulated in volar fibroblasts, and the 168th highest fold expressed gene of approximately 47,000 transcripts. Therefore, we focused on WNT5a as a likely volar-specific fibroblast WNT.
We first determined the level of WNT5a mRNA in co-cultures using RT-qPCR, and found that the level was higher in co-cultures containing keratinocytes with volar fibroblasts than co-cultures containing keratinocyte with scalp, foreskin, or foot fibroblasts (Figure 3A). We next examined the level of WNT5a mRNA in keratinocytes and fibroblasts separately. The level of WNT5a mRNA was highest in keratinocytes cultured in KBMedia at high density, the same conditions where we saw maximal KRT9 induction (Figure 3B). Also, the time course of WNT5a mRNA expression showed the highest level around days 2 and 3 (Figure 3B), which coincided with the highest level of KRT1 and KRT9 mRNA in the same experiment using keratinocytes alone (Figure 2B). Consistent with these observations, the level of WNT5a mRNA is higher in volar epidermis (Supplemental Figure S5), and higher in volar fibroblasts than in nonvolar fibroblasts (Figure 3C), confirming our initial microarray results. Finally, we measured AXIN2, a reporter gene for canonical β-catenin activity, and found a similar pattern of expression to WNT5a. These results demonstrate a correlation between WNT ligand expression and β-catenin activation in conditions where KRT9 is activated in both autocrine and paracrine contexts.
WNT5a increases KRT9 levels; suggested to be through the inhibition of β-catenin,7 we next tested this hypothesis. To test whether KRT9 expression is regulated by the canonical WNT signaling pathway, we first increased the level of β-catenin by treating keratinocytes with a specific glycogen synthase kinase-3 inhibitor (SB216763) that allows for β-catenin translocation to the nucleus and activation of the WNT pathway. Keratinocytes were treated with SB216763 at indicated concentrations (0 to 5 μmol/L) for 2 hours and further incubated for 48 hours. Western blot analysis results showed that active β-catenin is successfully induced by SB216763 (Figure 3D). Supporting the stabilization of β-catenin, expression of AXIN2, a direct target of a canonical WNT signaling, was analyzed to monitor transcriptional activities of β-catenin in the nucleus. The AXIN2 level increased significantly in response to SB216763 (Figure 3D). With activated β-catenin signaling, KRT9 also increased substantially after SB216763 treatment (Figure 3D), indicating that β-catenin is sufficient to induce keratinocyte differentiation. SB216763 had similar effects on adult abdominal keratinocytes (Supplemental Figure S6). However, KERATIN 6a (KRT6a), type II, also significantly elevated in volar skin, also suprabasal, and also induced by differentiation (Supplemental Figure S7), was expressed independent of β-catenin signaling (Figure 3D). Nevertheless, SB216763 could induce involucrin and loricrin (Supplemental Figure S8). To check for the specificity of the SB216763 compound, we added the canonical β-catenin pathway antagonist, DKK-1,32 which partially reversed the effects (Figure 4C). These results demonstrate the selective sufficiency of β-catenin to enhance KRT9, but not KRT6a, expression.
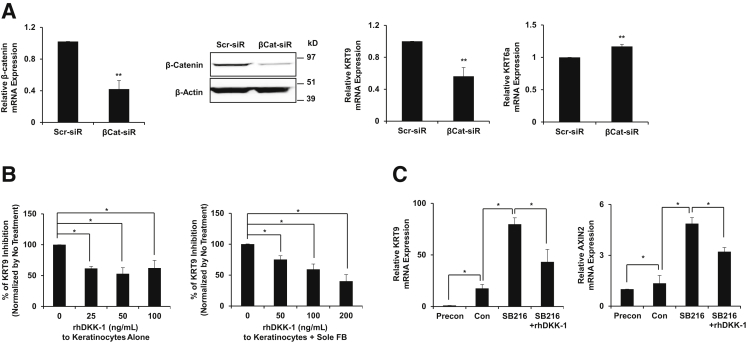
Figure 4.
β-Catenin is partially necessary for KRT9 expression in keratinocytes. A: β-Catenin knockdown (βCat-siR) in keratinocytes inhibits β-catenin protein and KRT9, but not KRT6a, mRNA compared with Scr-siR. B: rhDKK-1 inhibits KRT9 expression in either solo keratinocyte cultures or co-cultures with sole FBs. C: Effects of SB216763 to increase KRT9 and AXIN2 are reversible by rhDKK-1 in keratinocytes. ∗P < 0.05, ∗∗P < 0.01. n = 3 to 4 (A); n = 4 (B); n = 3 (C). Con, keratinocytes treated with vehicle; FB, fibroblast; Precon, preconfluent keratinocytes only; rh, recombinant human; Scr-siR; scramble siRNA.
Because activation of β-catenin promoted KRT9 expression, we next tested the effects of loss of function. We reduced β-catenin expression using β-catenin siRNA and examined the levels of KRT6a and KRT9 mRNA in β-catenin knockdown keratinocytes. Transfection and knockdown efficiency of β-catenin siRNA (50 nmol/L) was evaluated by RT-qPCR and Western blot analysis. β-Catenin knockdown keratinocytes (approximately 60% reduction versus Scr-siR) expressed a lower level of KRT9 mRNA than nontargeting scramble siRNA (Scr-siR)–transfected keratinocytes, indicating that β-catenin is necessary to induce KRT9 during keratinocyte differentiation. However, the level of KRT6a mRNA was slightly increased in β-catenin knockdown keratinocytes (Figure 4A), suggesting that the regulatory mechanism of KRT6a is different from that of KRT9. KRT1 showed intermediate results (Supplemental Figure S2). These results demonstrate the selective necessity of β-catenin to support KRT9, but not KRT6a, expression.
Given that the adhesion functions of β-catenin could be affected in siRNA knockdown studies, we next tested inhibition of the canonical WNT cell signaling pathway. When we treated keratinocytes with rhDKK-1, the level of KRT9 mRNA decreased by approximately half (Figure 4B). Underlining the shared mechanisms of KRT9 induction, addition of rhDKK-1 also inhibited the ability of volar fibroblasts to induce KRT9 in co-cultures (Figure 4B). Collectively, these data show that β-catenin–dependent canonical WNT signals play an important role in KRT9 induction in keratinocytes via a shared autocrine and paracrine mechanism.
Human Skin Diseases Ectopically Express KRT9 in Nonvolar Epidermis
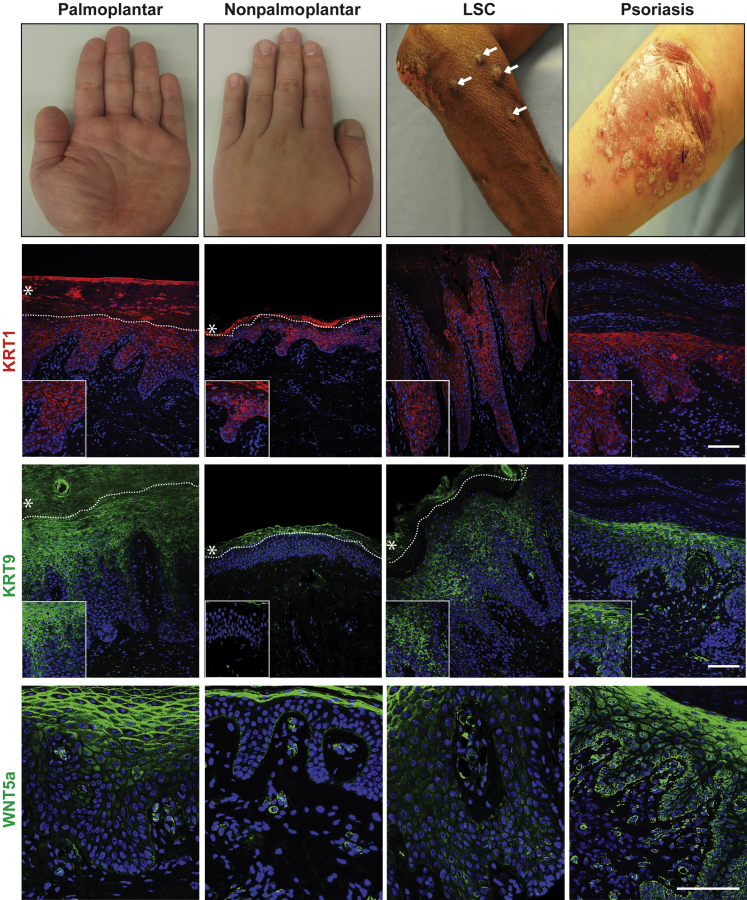
Because nonvolar keratinocytes have an intrinsic capability to express KRT9 in vitro, we hypothesized that KRT9 could be induced ectopically in nonvolar skin in certain disease conditions. We first tested lichen simplex chronicus (LSC), a human skin disease that results from chronic rubbing or scratching. LSC demonstrates a thickened and hyperproliferative epidermis and occasional decreased activity of melanocytes (Figure 5 and Supplemental Figure S9) (ie, clinical and histopathological features of volar skin). We also tested psoriasis as a second condition known to have a hyperproliferative epidermis (Figure 5). We verified that volar skin and some areas of LSC lesions have fewer melanocytes (Supplemental Figure S9). We examined KRT9 expression using immunofluorescence. Normal volar and nonvolar skin tissues were used as a positive and negative control, respectively. To quantify the level of KRT9 expression, percentage of KRT9+ cells was calculated of total DAPI-stained keratinocytes in the epidermis. Consistent with Figure 1D, KRT1 was constitutively expressed in all samples, with perhaps some increase in volar skin. We also found that KRT9 was expressed in nonvolar skin of LSC (71.3%) similar to volar skin (67.7%), whereas psoriasis samples expressed a lower level of KRT9 (29.7%) (Figure 5 and Table 1). The percentage of KRT9-positive cells in LSC was as significantly different to nonvolar fibroblasts as volar fibroblasts were to nonvolar fibroblasts (P < 0.01; n = 3), and appeared somewhat higher in nodular areas. In staining these four conditions, we also found that epidermal WNT5a expression matched KRT9 expression. Collectively, these results confirmed that WNT5a and KRT9 can be ectopically expressed independent of volar fibroblasts in nonvolar keratinocytes in human subjects and represent a partial conversion of skin identity in LSC.
Figure 5.
Ectopic overlapping expression of KRT9 and WNT5a in the absence of volar fibroblasts in lesions of LSC and psoriasis. Immunofluorescence studies demonstrating KRT1, KRT9, and WNT5a expression in volar (palmoplantar) skin and nonvolar skin from human sole and scalp skin, respectively, as well as from disease states with features of volar skin. KRT1 is shown by red, KRT9, and WNT5a in green, and nuclei are stained blue. Dotted lines reveal inferior boundary of stratum corneum, where non-specific staining can be seen (asterisks). Images from normal palmoplantar and nonpalmoplantar skin and patients with LSC and psoriasis. Arrows in LSC skin indicate prurigo nodules with skin thickening and loss of pigmentation. Insets represent enlarged areas of the corresponding images. Scale bar = 100 μm. Original magnification: ×200 (KRT1 and KRT9); ×400 (insets and WNT5a). KRT, KERATIN; LSC, lichen simplex chronicus.
Table 1.
KRT9+ Cells
| Skin tissue | Palmoplantar | Nonpalmoplantar | LSC | Psoriasis |
|---|---|---|---|---|
| % of KRT9 | 67.7 (±1.3) | 0∗∗ | 71.1 (±6.9)†‡ | 29.7 (±11.7)∗ |
Percentage of keratinocytes with positive staining for KRT9 of DAPI-stained keratinocytes in each respective skin sample. KRT9 and DAPI stained keratinocytes were counted and the number of KRT9+ cells were divided by total number of DAPI stained keratinocytes to calculate percentage of KRT9+ cells. Results were represented by average percentage of KRT9+ cells ± SEM (n = 3).
∗P < 0.05, ∗∗P < 0.01 vs volar.
†P < 0.05 vs nonvolar.
‡P < 0.05 vs psoriasis.
KRT, KERATIN; LSC, lichen simplex chronicus.
Discussion
Despite constant cellular turnover, skin identity is remarkably fixed. Even adjacent areas of skin, such as at the transition from volar to nonvolar type in the hand or foot, maintain their distinct identity throughout a human's lifespan. A deeper understanding of how skin identity is specified and maintained will enable us to reprogram skin identity for therapeutic purposes. For example, our group is interested in reprogramming the identity of stump skin in an amputee to the volar type of skin found in the palms and soles. This will enhance the use of prosthetics, and is only one example of possible clinical applications of modifying human skin identity. To promote the success of such therapies, a deeper understanding of how tissue identity is initiated and maintained is necessary.
Herein, we focused on the establishment of volar skin identity. KRT9 is thought to be responsible for the structural resiliency of volar skin. The molecular mechanisms that regulate KRT9 expression remain largely unknown, although pressure and mitogen-activated protein kinase signaling contribute.12, 13 Until now, it was assumed that KRT9 expression was mostly under the control of site-specific volar fibroblasts. Indeed, despite a mixed primary literature,33, 34, 35, 36, 37 it has been commonly hypothesized that primary signals from fibroblasts exclusively dictate epidermal skin identity through epithelial-mesenchymal interactions. In this article, we have discovered that epithelium has intrinsic differentiation programs, which include volar KRT9 expression, and this is even manifest in human skin disease. Specifically, we discovered ectopic expression of KRT9 in epidermal layers of nonvolar skin lesions from psoriasis and LSC tissues. The number of KRT9+ cells in LSC is similar to that in volar skin (Figure 5 and Table 1). Clinically, hypopigmentation is a frequent feature of LSC in those with darker skin types (Figure 5 and Supplemental Figure S9), suggesting that chronic scratching results in thickened and less pigmented epidermis, underlining a partial phenocopy of volar skin. Therefore, our studies reflect a more nuanced model of epithelial-mesenchymal interactions.
We also investigated the conflicts in the literature about how WNT signals might regulate KRT9. Despite previous models where inhibition of β-catenin might explain both the lack of hair follicles and expression of KRT9,7, 19 we show that canonical β-catenin activation actually stimulates KRT9 via shared autocrine or paracrine mechanisms, depending on the context. We suggest that site-specific epidermal differentiation is modulated by both these intrinsic and extrinsic signaling mechanisms.
These results raise a larger question for the field of developmental biology in the skin: how can the same WNT/β-catenin signaling pathway promote such different forms of differentiation, ranging from hair follicle to volar-specific genes? Clearly, other signaling molecules modulate gene expression. However, we were unable to optimize conditional media approaches because of possible close-proximity paracrine signaling (Supplemental Figure S10); we showed that EGF inhibits KRT9 and WNT5a expression (Supplemental Figure S11). In addition, we found that KRT9 expression in nonvolar keratinocytes is not induced during the wound healing process by analyzing wounded mouse back skins (Supplemental Figure S12). Although context dependence is well appreciated, defining the exact epigenetic and signaling components that complement β-catenin (eg, EGF signaling) to establish specific forms of epithelial differentiation is an exciting future path for research.
In conclusion, the present study refines a more nuanced model that site-specific epidermal differentiation is regulated both by keratinocytes intrinsically and by fibroblasts extrinsically. Our data provide a framework for understanding the mechanism of site-specific epidermal differentiation in skin regeneration and have translational potential for therapeutic reprogramming of skin identity (eg, at stump sites of amputees).
Acknowledgments
We thank Conover Talbot, Jr. (Johns Hopkins University Microarray Core) for assistance with microarray analysis and Dr. Pierre Coulombe and his laboratory for critical discussions.
D.K. and L.G. conceived the hypothesis, designed experiments, interpreted data, and assembled the manuscript; D.K. performed experiments with assistance from M.Z.H., A.N., L.G., N.B.Y., S.M.O., A.P., S.H., T.S.R., and J.Q; S.K. provided assistance in hypothesis generation; and J.M.T. assisted in human tissue acquisition and interpretation.
Footnotes
Supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, part of the NIH, under award R01AR064297, by the Department of Defense, Defense Advanced Research Projects Agency (N66001-10-1-4081) and Armed Forces Institute of Regenerative Medicine, Extremities Regeneration (AFIRM2-ER11), Northrop Grumman Electronic Systems (Veteran/Amputee Skin Regeneration Program Initiative), the Thomas Provost, MD, Young Faculty Development Fund of Johns Hopkins Dermatology (L.A.G.), and Howard Hughes Medical Institute Medical Research Fellowship (A.N.).
Disclosures: None declared.
Current address of M.Z.H., Department of Pharmacy, BRAC University, Dhaka, Bangladesh.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2015.12.030.
Supplemental Data
Functional annotation of gene family differences between volar and nonvolar epidermis. Listed are overrepresented classes of genes present in the top 250 genes present in volar skin (A) versus the top 250 genes present in nonvolar skin (B). Lists are ranked according to number of genes in each category and generated using DAVID.
In vivo KRT1 expression in volar and nonvolar skin and in vitro regulation by β-catenin. A: Relative KRT1 mRNA expression in volar and nonvolar epidermis. B: KRT1 protein and gene expression induced by 0 to 5 μmol/L SB216763. C: β-Catenin knockdown inhibits KRT1 mRNA expression in keratinocytes. Knockdown efficiency of β-catenin was analyzed by Western blot analysis and quantitative RT-PCR (RT-qPCR) compared with nontargeting Scr-siR. Relative KRT1 mRNA expression was analyzed by RT-qPCR and normalized with RPLP0 housekeeping gene. Results were represented by means of fold-difference ± SEM normalized with ventral hand, preconfluent keratinocytes, and Scr-siR to analyze significances between ventral and dorsal epidermis and control and treatment using t-test. n = 4 (A and B); n = 3 (C). ∗P < 0.05. βCat-siR, β-catenin siRNA; KRT, KERATIN; Precon, preconfluent keratinocytes only; Scr-siR, Scramble siRNA.
KRT9 expression is induced in calcium-mediated keratinocyte differentiation. Relative mRNA expression of KRT1 (A), KRT9 (B), and KRT10 (C) increases in calcium-treated keratinocytes. Relative gene expression levels were analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by fold-difference from two replicate experiments and compared with preconfluent keratinocytes. Error bar represents SD from two replicate experiments. n = 2 (A–C). Ca, calcium; KRT, KERATIN; Precon, preconfluent keratinocytes only.
Supplemental Figure S4.
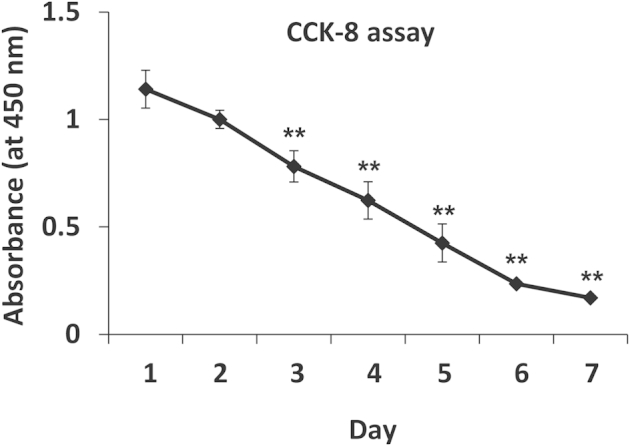
Decreasing metabolic activity of keratinocytes cultured in KBMedia without growth factors. Metabolic activity of keratinocytes cultured in KBMedia was analyzed by Cell Counting Kit-8 (CCK-8) assay in time course. Metabolic activity is compared with day 1 and gradually decreases. ∗∗P < 0.01. n = 4.
WNT5a mRNA is higher in volar epidermis. Relative WNT5a mRNA expression in volar and nonvolar epidermis. Relative WNT5a mRNA expression was analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by means of fold-difference ± SEM normalized with ventral hand and analyzed by t-test between ventral and dorsal epidermis. ∗P < 0.05. n = 3.
Similar KRT9 induction in adult abdominal versus neonatal foreskin keratinocytes. A: KRT9 mRNA expression increases with differentiation in time-course experiments using abdominal adult keratinocytes. AXIN2 (B) and KRT9 (C) mRNA significantly increase in response to 0 to 5 μmol/L SB216763 using abdominal adult keratinocytes. Relative AXIN2 and KRT9 mRNA expression levels were analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by fold-difference from a representative experiment. KRT, KERATIN; Precon, preconfluent keratinocytes only.
KRT6a mRNA is up-regulated in volar skin and with keratinocyte confluence. A: Relative KRT6a mRNA expression in volar and nonvolar epidermis. B: KRT6a mRNA expression in keratinocytes sole culture. Relative KRT6a mRNA expression was analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by means of fold-difference ± SEM normalized with ventral hand (A) and sole epidermis (B) to measure significances using t-test. n = 3 (A); n = 3 to 4 (B). ∗P < 0.05, ∗∗P < 0.01. Confluent, keratinocytes cultured by dropping method for 2 days; KRT, KERATIN; Precon, preconfluent keratinocytes only; Sole, sole epidermis.
IVL and LOR gene expression is induced by β-catenin–mediated signaling in keratinocytes. IVL (A) and LOR (B) mRNA expression significantly increase in response to 0 to 5 μmol/L SB216763. Relative IVL and LOR mRNA expression levels were analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by fold-difference from a representative experiment. ∗P < 0.05. n = 3. IVL, involucrin; LOR, loricrin; Precon, preconfluent keratinocytes only.
LSC subject with localized decreased pigmentation and decreased melanocyte density, similar to volar skin. A: Top panels (from left to right): MITF+ cells in volar skin, nonvolar skin, psoriasis with low magnification (LM) and high magnification (HM). Middle panels (from left to right): MITF+ cells in hyperpigmented areas of LSC with low magnification and high magnification. MITF+ cells in hypopigmented areas of LSC with LM and HM. Bottom panels (from left to right): KRT9 staining in hyperpigmented areas of LSC with LM and HM, KRT9 staining in hypopigmented areas of LSC with LM and HM. B: Ratios of melanocytes in normal skin and skin diseases. C: Ratios of melanocytes in hyperpigmented (HMel) and hypopigmented (LMel) areas of LSC skin. ∗∗P < 0.01, ∗∗∗P < 0.001. n = 3 (B and C). Original magnification: ×100 (A, volar skin, nonvolar skin, and psoriasis with LM); ×200 (A, psoriasis with HM). HMel, high melanocytes; KRT, KERATIN; LMel, low melanocytes; LSC, lichen simplex chronicus; MITF, microphthalmia associated transcription factor.
KRT9 mRNA expression of keratinocytes cultured in FB–conditioned media. KRT9 mRNA was measured in keratinocytes cultured with volar (sole) and nonvolar (foreskin and scalp) fibroblast-conditioned media (not significant). Relative mRNA of KRT9 was analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. n = 4. KRT, KERATIN; FB, fibroblast.
rhEGF inhibits KERATIN and WNT5a expression. Relative mRNA expression of KRT1 (A), KRT9 (B), KRT10 (C), and WNT5a (D) decreases in keratinocytes in the presence of 0 to 1000 ng/mL rhEGF. Relative gene expression levels were analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by fold-difference from replicate experiments. ∗P < 0.05. n = 3 to 7. Precon, preconfluent keratinocytes only. rhEGF, recombinant human epidermal growth factor.
Krt9 expression during mouse wound healing. Krt9 gene expression was measured from mouse back skins at different time points after wound. During wound healing, Krt9 expression was detected at extremely low levels (Ct values: 33 to 37; 12,000-fold less than in volar skin) and not induced after wounding. Relative mRNA of Krt9 was analyzed by quantitative RT-PCR and normalized with Actnb housekeeping gene. n = 6. pwd, post wound day; w/o, without. Krt, keratin (mouse).
References
- 1.Compton C.C., Nadire K.B., Regauer S., Warland G., Landry D.B., Simon M., O'Connor N.E., Gallico G.G. Cultured human sole-derived keratinocyte grafts re-express site-specific differentiation after transplantation. Differentiation. 1998;64:45–53. doi: 10.1046/j.1432-0436.1998.6410045.x. [DOI] [PubMed] [Google Scholar]
- 2.El-Ghalbzouri A., Gibbs S., Lamme E., Van Blitterswijk C.A., Ponec M. Effect of fibroblasts on epidermal regeneration. Br J Dermatol. 2002;147:230–243. doi: 10.1046/j.1365-2133.2002.04871.x. [DOI] [PubMed] [Google Scholar]
- 3.Ehama R., Ishimatsu-Tsuji Y., Iriyama S., Ideta R., Soma T., Yano K., Kawasaki C., Suzuki S., Shirakata Y., Hashimoto K., Kishimoto J. Hair follicle regeneration using grafted rodent and human cells. J Invest Dermatol. 2007;127:2106–2115. doi: 10.1038/sj.jid.5700823. [DOI] [PubMed] [Google Scholar]
- 4.Driskell R.R., Lichtenberger B.M., Hoste E., Kretzschmar K., Simons B.D., Charalambous M., Ferron S.R., Herault Y., Pavlovic G., Ferguson-Smith A.C., Watt F.M. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuan T.L., Keller L.C., Sun D., Nimni M.E., Cheung D. Dermal fibroblasts activate keratinocyte outgrowth on collagen gels. J Cell Sci. 1994;107(Pt 8):2285–2289. doi: 10.1242/jcs.107.8.2285. [DOI] [PubMed] [Google Scholar]
- 6.Langbein L., Heid H.W., Moll I., Franke W.W. Molecular characterization of the body site-specific human epidermal cytokeratin 9: cDNA cloning, amino acid sequence, and tissue specificity of gene expression. Differentiation. 1993;55:57–71. doi: 10.1111/j.1432-0436.1993.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi Y., Passeron T., Hoashi T., Watabe H., Rouzaud F., Yasumoto K., Hara T., Tohyama C., Katayama I., Miki T., Hearing V.J. Dickkopf 1 (DKK1) regulates skin pigmentation and thickness by affecting Wnt/β-catenin signaling in keratinocytes. FASEB J. 2008;22:1009–1020. doi: 10.1096/fj.07-9475com. [DOI] [PubMed] [Google Scholar]
- 8.Swensson O., Langbein L., McMillan J.R., Stevens H.P., Leigh I.M., McLean W.H., Lane E.B., Eady R.A. Specialized keratin expression pattern in human ridged skin as an adaptation to high physical stress. Br J Dermatol. 1998;139:767–775. doi: 10.1046/j.1365-2133.1998.02499.x. [DOI] [PubMed] [Google Scholar]
- 9.Rinn J.L., Wang J.K., Liu H., Montgomery K., van de Rijn M., Chang H.Y. A systems biology approach to anatomic diversity of skin. J Invest Dermatol. 2008;128:776–782. doi: 10.1038/sj.jid.5700986. [DOI] [PubMed] [Google Scholar]
- 10.Reis A., Hennies H.C., Langbein L., Digweed M., Mischke D., Drechsler M., Schrock E., Royer-Pokora B., Franke W.W., Sperling K., Kuster W. Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK) Nat Genet. 1994;6:174–179. doi: 10.1038/ng0294-174. [DOI] [PubMed] [Google Scholar]
- 11.Fu D.J., Thomson C., Lunny D.P., Dopping-Hepenstal P.J., McGrath J.A., Smith F.J.D., Irwin McLean W.H., Pedrioli D.M. Keratin 9 is required for the structural integrity and terminal differentiation of the palmoplantar epidermis. J Invest Dermatol. 2014;134:754–763. doi: 10.1038/jid.2013.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gormar F.E., Bernd A., Bereiter-Hahn J., Holzmann H. A new model of epidermal differentiation: induction by mechanical stimulation. Arch Dermatol Res. 1990;282:22–32. doi: 10.1007/BF00505641. [DOI] [PubMed] [Google Scholar]
- 13.Kippenberger S., Bernd A., Loitsch S., Guschel M., Muller J., Bereiter-Hahn J., Kaufmann R. Signaling of mechanical stretch in human keratinocytes via MAP kinases. J Invest Dermatol. 2000;114:408–412. doi: 10.1046/j.1523-1747.2000.00915.x. [DOI] [PubMed] [Google Scholar]
- 14.Moll I., Heid H., Franke W.W., Moll R. Distribution of a special subset of keratinocytes characterized by the expression of cytokeratin 9 in adult and fetal human epidermis of various body sites. Differentiation. 1987;33:254–265. doi: 10.1111/j.1432-0436.1987.tb01565.x. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi Y., Itami S., Tarutani M., Hosokawa K., Miura H., Yoshikawa K. Regulation of keratin 9 in nonpalmoplantar keratinocytes by palmoplantar fibroblasts through epithelial-mesenchymal interactions. J Invest Dermatol. 1999;112:483–488. doi: 10.1046/j.1523-1747.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- 16.Choi Y.S., Zhang Y., Xu M., Yang Y., Ito M., Peng T., Cui Z., Nagy A., Hadjantonakis A.K., Lang R.A., Cotsarelis G., Andl T., Morrisey E.E., Millar S.E. Distinct functions for Wnt/beta-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell Stem Cell. 2013;13:720–733. doi: 10.1016/j.stem.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adaimy L., Chouery E., Megarbane H., Mroueh S., Delague V., Nicolas E., Belguith H., de Mazancourt P., Megarbane A. Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. Am J Hum Genet. 2007;81:821–828. doi: 10.1086/520064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrof G., Fong K., Lai-Cheong J.E., Cockayne S.E., McGrath J.A. Schopf-Schulz-Passarge syndrome resulting from a homozygous nonsense mutation, p.Cys107X, in WNT10A. Australas J Dermatol. 2011;52:224–226. doi: 10.1111/j.1440-0960.2011.00788.x. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi Y., Itami S., Watabe H., Yasumoto K., Abdel-Malek Z.A., Kubo T., Rouzaud F., Tanemura A., Yoshikawa K., Hearing V.J. Mesenchymal-epithelial interactions in the skin: increased expression of dickkopf1 by palmoplantar fibroblasts inhibits melanocyte growth and differentiation. J Cell Biol. 2004;165:275–285. doi: 10.1083/jcb.200311122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinn J.L., Wang J.K., Allen N., Brugmann S.A., Mikels A.J., Liu H., Ridky T.W., Stadler H.S., Nusse R., Helms J.A., Chang H.Y. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22:303–307. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popp T., Steinritz D., Breit A., Deppe J., Egea V., Schmidt A., Gudermann T., Weber C., Ries C. Wnt5a/[beta]-catenin signaling drives calcium-induced differentiation of human primary keratinocytes. J Invest Dermatol. 2014;134:2183–2191. doi: 10.1038/jid.2014.149. [DOI] [PubMed] [Google Scholar]
- 22.Garza L.A., Yang C.C., Zhao T., Blatt H.B., Lee M., He H., Stanton D.C., Carrasco L., Spiegel J.H., Tobias J.W., Cotsarelis G. Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. J Clin Invest. 2011;121:613–622. doi: 10.1172/JCI44478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Normand J., Karasek M.A. A method for the isolation and serial propagation of keratinocytes, endothelial cells, and fibroblasts from a single punch biopsy of human skin. In Vitro Cell Dev Biol Anim. 1995;31:447–455. doi: 10.1007/BF02634257. [DOI] [PubMed] [Google Scholar]
- 24.Aasen T., Belmonte J.C. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat Protoc. 2010;5:371–382. doi: 10.1038/nprot.2009.241. [DOI] [PubMed] [Google Scholar]
- 25.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Ito M., Yang Z., Andl T., Cui C., Kim N., Millar S.E., Cotsarelis G. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 28.Nelson A.M., Loy D.E., Lawson J.A., Katseff A.S., FitzGerald G.A., Garza L.A. Prostaglandin D(2) inhibits wound-induced hair follicle neogenesis through the receptor, Gpr44. J Invest Dermatol. 2013;133:881–889. doi: 10.1038/jid.2012.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson Amanda M., Reddy Sashank K., Ratliff Tabetha S., Hossain M.Z., Katseff Adiya S., Zhu Amadeus S., Chang E., Resnik Sydney R., Page C., Kim D., Whittam Alexander J., Miller Lloyd S., Garza Luis A. dsRNA released by tissue damage activates TLR3 to drive skin regeneration. Cell Stem Cell. 2015;17:139–151. doi: 10.1016/j.stem.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Poumay Y., Pittelkow M.R. Cell density and culture factors regulate keratinocyte commitment to differentiation and expression of suprabasal K1/K10 keratins. J Invest Dermatol. 1995;104:271–276. doi: 10.1111/1523-1747.ep12612810. [DOI] [PubMed] [Google Scholar]
- 32.Mao B., Wu W., Davidson G., Marhold J., Li M., Mechler B.M., Delius H., Hoppe D., Stannek P., Walter C., Glinka A., Niehrs C. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 33.Billingham R.E., Silvers W.K. Studies on the conservation of epidermal specificies of skin and certain mucosas in adult mammals. J Exp Med. 1967;125:429–446. doi: 10.1084/jem.125.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doran T.I., Vidrich A., Sun T.T. Intrinsic and extrinsic regulation of the differentiation of skin, corneal and esophageal epithelial cells. Cell. 1980;22:17–25. doi: 10.1016/0092-8674(80)90150-6. [DOI] [PubMed] [Google Scholar]
- 35.Mackenzie I.C., Hill M.W. Connective tissue influences on patterns of epithelial architecture and keratinization in skin and oral mucosa of the adult mouse. Cell Tissue Res. 1984;235:551–559. doi: 10.1007/BF00226952. [DOI] [PubMed] [Google Scholar]
- 36.Boukamp P., Breitkreutz D., Stark H.J., Fusenig N.E. Mesenchyme-mediated and endogenous regulation of growth and differentiation of human skin keratinocytes derived from different body sites. Differentiation. 1990;44:150–161. doi: 10.1111/j.1432-0436.1990.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 37.Konstantinova N.V., Lemak N.A., Duong D.M., Chuang A.Z., Urso R., Duvic M. Artificial skin equivalent differentiation depends on fibroblast donor site: use of eyelid fibroblasts. Plast Reconstr Surg. 1998;101:385–391. doi: 10.1097/00006534-199802000-00019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional annotation of gene family differences between volar and nonvolar epidermis. Listed are overrepresented classes of genes present in the top 250 genes present in volar skin (A) versus the top 250 genes present in nonvolar skin (B). Lists are ranked according to number of genes in each category and generated using DAVID.
In vivo KRT1 expression in volar and nonvolar skin and in vitro regulation by β-catenin. A: Relative KRT1 mRNA expression in volar and nonvolar epidermis. B: KRT1 protein and gene expression induced by 0 to 5 μmol/L SB216763. C: β-Catenin knockdown inhibits KRT1 mRNA expression in keratinocytes. Knockdown efficiency of β-catenin was analyzed by Western blot analysis and quantitative RT-PCR (RT-qPCR) compared with nontargeting Scr-siR. Relative KRT1 mRNA expression was analyzed by RT-qPCR and normalized with RPLP0 housekeeping gene. Results were represented by means of fold-difference ± SEM normalized with ventral hand, preconfluent keratinocytes, and Scr-siR to analyze significances between ventral and dorsal epidermis and control and treatment using t-test. n = 4 (A and B); n = 3 (C). ∗P < 0.05. βCat-siR, β-catenin siRNA; KRT, KERATIN; Precon, preconfluent keratinocytes only; Scr-siR, Scramble siRNA.
KRT9 expression is induced in calcium-mediated keratinocyte differentiation. Relative mRNA expression of KRT1 (A), KRT9 (B), and KRT10 (C) increases in calcium-treated keratinocytes. Relative gene expression levels were analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by fold-difference from two replicate experiments and compared with preconfluent keratinocytes. Error bar represents SD from two replicate experiments. n = 2 (A–C). Ca, calcium; KRT, KERATIN; Precon, preconfluent keratinocytes only.
WNT5a mRNA is higher in volar epidermis. Relative WNT5a mRNA expression in volar and nonvolar epidermis. Relative WNT5a mRNA expression was analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by means of fold-difference ± SEM normalized with ventral hand and analyzed by t-test between ventral and dorsal epidermis. ∗P < 0.05. n = 3.
Similar KRT9 induction in adult abdominal versus neonatal foreskin keratinocytes. A: KRT9 mRNA expression increases with differentiation in time-course experiments using abdominal adult keratinocytes. AXIN2 (B) and KRT9 (C) mRNA significantly increase in response to 0 to 5 μmol/L SB216763 using abdominal adult keratinocytes. Relative AXIN2 and KRT9 mRNA expression levels were analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by fold-difference from a representative experiment. KRT, KERATIN; Precon, preconfluent keratinocytes only.
KRT6a mRNA is up-regulated in volar skin and with keratinocyte confluence. A: Relative KRT6a mRNA expression in volar and nonvolar epidermis. B: KRT6a mRNA expression in keratinocytes sole culture. Relative KRT6a mRNA expression was analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by means of fold-difference ± SEM normalized with ventral hand (A) and sole epidermis (B) to measure significances using t-test. n = 3 (A); n = 3 to 4 (B). ∗P < 0.05, ∗∗P < 0.01. Confluent, keratinocytes cultured by dropping method for 2 days; KRT, KERATIN; Precon, preconfluent keratinocytes only; Sole, sole epidermis.
IVL and LOR gene expression is induced by β-catenin–mediated signaling in keratinocytes. IVL (A) and LOR (B) mRNA expression significantly increase in response to 0 to 5 μmol/L SB216763. Relative IVL and LOR mRNA expression levels were analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by fold-difference from a representative experiment. ∗P < 0.05. n = 3. IVL, involucrin; LOR, loricrin; Precon, preconfluent keratinocytes only.
LSC subject with localized decreased pigmentation and decreased melanocyte density, similar to volar skin. A: Top panels (from left to right): MITF+ cells in volar skin, nonvolar skin, psoriasis with low magnification (LM) and high magnification (HM). Middle panels (from left to right): MITF+ cells in hyperpigmented areas of LSC with low magnification and high magnification. MITF+ cells in hypopigmented areas of LSC with LM and HM. Bottom panels (from left to right): KRT9 staining in hyperpigmented areas of LSC with LM and HM, KRT9 staining in hypopigmented areas of LSC with LM and HM. B: Ratios of melanocytes in normal skin and skin diseases. C: Ratios of melanocytes in hyperpigmented (HMel) and hypopigmented (LMel) areas of LSC skin. ∗∗P < 0.01, ∗∗∗P < 0.001. n = 3 (B and C). Original magnification: ×100 (A, volar skin, nonvolar skin, and psoriasis with LM); ×200 (A, psoriasis with HM). HMel, high melanocytes; KRT, KERATIN; LMel, low melanocytes; LSC, lichen simplex chronicus; MITF, microphthalmia associated transcription factor.
KRT9 mRNA expression of keratinocytes cultured in FB–conditioned media. KRT9 mRNA was measured in keratinocytes cultured with volar (sole) and nonvolar (foreskin and scalp) fibroblast-conditioned media (not significant). Relative mRNA of KRT9 was analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. n = 4. KRT, KERATIN; FB, fibroblast.
rhEGF inhibits KERATIN and WNT5a expression. Relative mRNA expression of KRT1 (A), KRT9 (B), KRT10 (C), and WNT5a (D) decreases in keratinocytes in the presence of 0 to 1000 ng/mL rhEGF. Relative gene expression levels were analyzed by quantitative RT-PCR and normalized with RPLP0 housekeeping gene. Results were represented by fold-difference from replicate experiments. ∗P < 0.05. n = 3 to 7. Precon, preconfluent keratinocytes only. rhEGF, recombinant human epidermal growth factor.
Krt9 expression during mouse wound healing. Krt9 gene expression was measured from mouse back skins at different time points after wound. During wound healing, Krt9 expression was detected at extremely low levels (Ct values: 33 to 37; 12,000-fold less than in volar skin) and not induced after wounding. Relative mRNA of Krt9 was analyzed by quantitative RT-PCR and normalized with Actnb housekeeping gene. n = 6. pwd, post wound day; w/o, without. Krt, keratin (mouse).