Abstract
From 2014 to 2015, three cases of highly pathogenic avian influenza infection occurred in zoo-housed north-east China tigers (Panthera tigris ssp.altaica) and four tigers died of respiratory distress in succession in Yunnan Province, China. We isolated and characterized three highly pathogenic avian influenza A(H5N1) viruses from these tigers. Phylogenetic analysis indicated that A/tiger /Yunnan /tig1404 /2014(H5N1) belongs to the provisional subclade 2.3.4.4e which were novel reassortant influenza A (H5N1) viruses with six internal genes from avian influenza A (H5N2) viruses. The HA gene of the isolated A/tiger /Yunnan /tig1412 /2014(H5N1) virus belongs to the subclade 2.3.2.1b. The isolated A/tiger /Yunnan /tig1508/2015 (H5N1) virus was a novel reassortant influenza A (H5N1) virus with three internal genes (PB2, PB1 and M) from H9N2 virus and belongs to the subclade 2.3.2.1c.
Our results suggested that the reassortant different H5N1 virus sublineages can successfully cross species barriers from avian to mammal and infect north-east China tigers.
Highly pathogenic avian influenza (HPAI) A (H5N1) viruses were first detected in China in 1996 (prototype strain A/goose/Guangdong/1/96 [Gs/GD])1 and have since spread causing outbreaks in poultry and wild birds in more than 70 countries2,3. Recently, HPAI viruses have crossed the species barrier from avian to mammal and caused human and other mammal infections. In 1997, HPAI H5N1 virus caused the first cases of human infection in Hong Kong4,5, and reemerged in 2003. So far, more than 650 cases of human infections HPAI A H5N1 virus (with around 60% fatality) have been reported in 16 countries since 2003 (WHO. WCnochcoaiAHNrt. http://www.who.int/influenza/ human animal interface /EN GIP 20140124 Cumulative Number H5N1cases.pdf.). HPAI A H5N1 viruses infections have also been reported in the globe amongst cats, tigers, leopards and other felids since 20046.
we have reported the novel clade 2.3.4.4 influenza A (H5N1) virus caused a major wave of highly pathogenic avian influenza outbreak in poultry in the Yunnan Province, China from December 2013 to March 20147. Here, we report three cases fatal influenza A (H5N1) virus infection in zoo-housed Tigers in Yunnan Province, China.
From 2014 to 2015, three cases of highly pathogenic avian influenza infection occurred in zoo-housed north-east China tigers (Panthera tigris ssp.altaica) and four tigers died of respiratory distress in Yunnan Province, China. We isolated and characterized three highly pathogenic avian influenza A (H5N1) viruses from tigers. The isolated viruses were named A/tiger /Yunnan /tig1404 /2014(H5N1), A/tiger /Yunnan /tig1412 /2014(H5N1) and A/tiger /Yunnan /tig1508 /2015(H5N1). Full genome influenza sequences and analyses have been performed. Sequence analyses revealed that the three viruses belonged to different clades.
Results
Case Descriptions
On April 8th, 2014, one 12monthold female tiger died in a zoo. The zookeepers reported that there were a total of fifty tigers in the zoo, and four tigers were reared in one cage. The tigers were fed daily with cooked chicken carcasses, pork and beef from the market. Two days prior to its death, the tiger displayed varying degrees of clinical symptoms, including high fever, vomiting with copious amounts of green-yellow liquid and severe respiratory distress. On December 15th, 2014, another 6monthold male tiger died in the zoo with similar clinical symptoms: high fever three days, vomiting and severe respiratory distress. On August 12th, 2015, two more tigers died in the zoo; one 8monthold male tiger died from high fever two days following vomiting and severe respiratory distress, and the other 24monthold female tiger was a sudden death. After the first tiger died, the other tigers were fed daily just with pork and beef, no chicken. But five peacocks died with H5N1 virus infection in the zoo, on June.
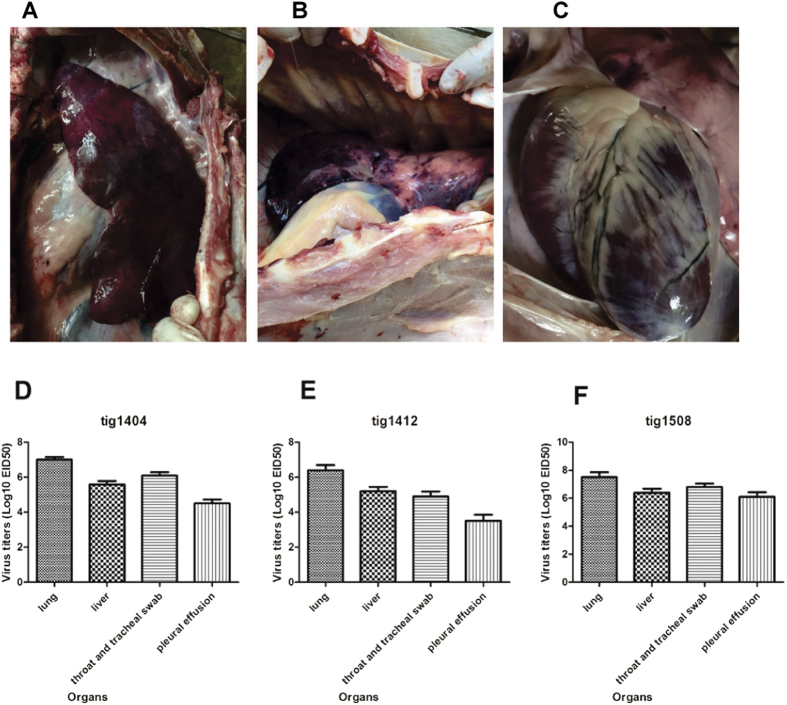
All tigers that died had nasal discharge and neurologic signs of infection. Necropsy was performed at once after their death. The lungs and livers were severely congested with hemorrhaging (Fig. 1A–C, tig1508), the brains had severe congestion; pleural effusion was observed, and serosanguinous exudate was also seen throughout the tracheal and bronchiolar lumen in all deceased tigers. RNA from the lung, liver, pleural effusion, throat and tracheal swab specimens tested positive for the hemagglutinin gene and neuraminidase gene of the H5N1 virus. The homogenates of the RT-PCR positive samples for tigers were centrifuged at lowspeed and either undiluted or 10-fold serially diluted supernatants, and then inoculated into 10-day-old SPF embryonated chicken eggs. The viral titers in the tigers were calculated using the Reed and Muench method and were highest in the lungs (Fig. 1D,E).
Figure 1. Autopsy change and The H5N1 viral titers in different organs of the tigers.
Autopsy change in different organs of tig1508 are shown in (A) (liver), (B) (lung) and (C) (heart); the viral titers in different organs of tigers are shown in (D) (tig1404), (E) (tig1412) and (F) (tig1508).
Virus isolation and genetic identity analysis
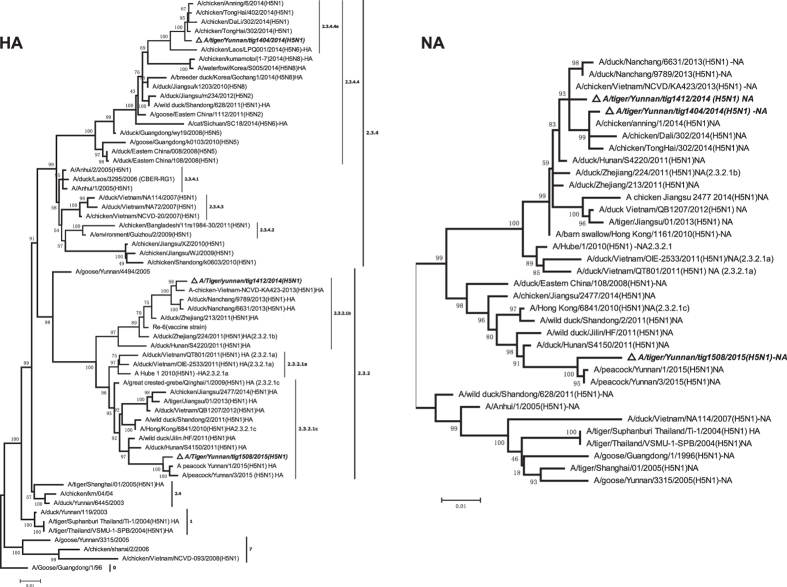
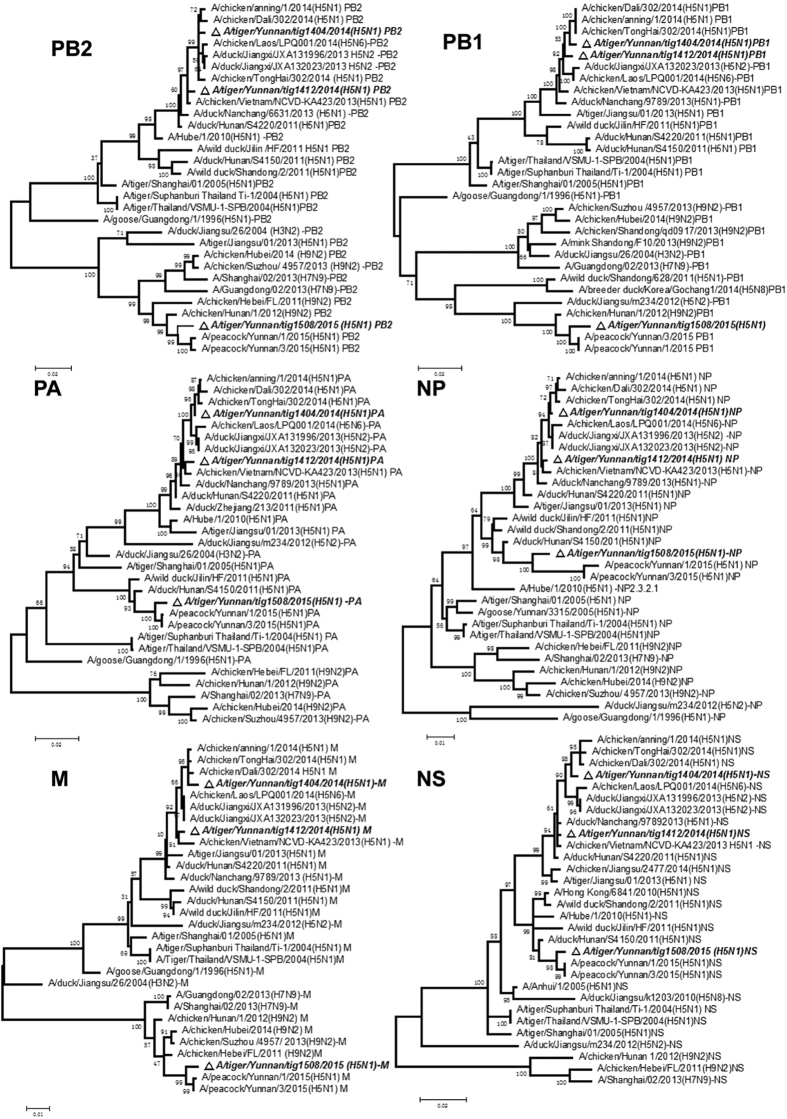
To analyze the correlation of H5N1 virus infection in tigers and peacocks in the zoo, two peacock isolates (A/ peacock/Yunnan /1 /2015(H5N1) and A/peacock /Yunnan /3 /2015(H5N1)) and three tiger originated virus isolates (tig1404, tig1412 and tig1508 isolated from tigers died on April 8th, December 15th, 2014 and August 12th(the 8monthold male tiger), 2015, respectively.) were chosen for full genome sequencing. Sequence analyses revealed that the three viruses belonged to different clades. Phylogenetic analysis indicated that HA, NA and six internal genes of A/tiger /Yunnan /tig1404 /2014(H5N1) shared more 99% homology (Table 1) and clustered with A/chicken /Tonghai/ 302 /2014(H5N1), and belongs to provisional subclade 2.3.4.4e (Figs 2 and 3) which were reassortant influenza A (H5N1) viruses with six internal genes originated from avian influenza A (H5N2) virus: A/duck/Jiangxi/JXA132023/(H5N2) in 2013, and associated with the outbreak of H5N1 occurred in chicken in Yunnan Province from December 2013 to March 20147. The eight gene segments of A/tiger /Yunnan /tig1412 /2014(H5N1) were most closely related to A/chicken /Vietnam /NCVD- KA423 /2013 (H5N1) and A/duck /Nanchang /6631 /2013 (H5N1, isolates from China) from the subclade 2.3.2.1b (Figs 2 and 3) with more 99% nucleotide sequence identity (Table 1).
Table 1. Levels of nucleotide sequence identity of tiger originated AIV H5N1 in Yunnan China, 2014–2015.
| Virus | Gene | Virus with the highest percentage of nucleotide identity | GeneBank accession no. | Identity, % |
|---|---|---|---|---|
| A/Tiger /Yunnan /tig1404 /2014 (H5N1) | PB2 | A/chicken /tonghai/ 302 /2014(H5N1) | KP732544 | 99.3 |
| PB1 | A/chicken /tonghai/ 302 /2014(H5N1) | KP732545 | 99.4 | |
| PA | A/chicken /tonghai/ 302 /2014(H5N1) | KP732546 | 99.7 | |
| HA | A/chicken /tonghai/ 302 /2014(H5N1) | KP732547 | 99.4 | |
| NP | A/chicken /tonghai/ 302 /2014(H5N1) | KP732548 | 99.6 | |
| NA | A/chicken /tonghai/ 302 /2014(H5N1) | KP732549 | 99.7 | |
| M | A/chicken /tonghai/ 302 /2014(H5N1) | KP732550 | 99.5 | |
| NS | A/chicken /tonghai/ 302 /2014(H5N1) | KP732551 | 99.0 | |
| A/Tiger /Yunnan /tig1412 /2014 (H5N1) | PB2 | A/chicken/Vietnam/NCVD-KA423/2013(H5N1) | KP097840 | 99.7 |
| A/duck /Nanchang/6631/2013(H5N1) | KP288313 | 99.0 | ||
| PB1 | A/chicken/Vietnam/NCVD-KA423/2013(H5N1) | KP288314 | 99.2 | |
| A/duck /Nanchang/6631/2013(H5N1) | KP097861 | 99.0 | ||
| PA | A/chicken/Vietnam/NCVD-KA423/2013(H5N1) | KP097882 | 99.4 | |
| A/duck /Nanchang/6631/2013 (H5N1) | KP288315 | 99.1 | ||
| HA | A/chicken/Vietnam/NCVD-KA423/2013(H5N1) | KP097915 | 99.2 | |
| A/duck /Nanchang/6631/2013 (H5N1) | KP288316 | 98.7 | ||
| NP | A/chicken/Vietnam/NCVD-KA423/2013(H5N1) | KP097938 | 99.6 | |
| A/duck /Nanchang/6631/2013 (H5N1) | KP288317 | 99.4 | ||
| NA | A/chicken/Vietnam/NCVD-KA423/2013(H5N1) | KP097971 | 99.6 | |
| A/duck /Nanchang/6631/2013 (H5N1) | KP288318 | 99.3 | ||
| M | A/chicken/Vietnam/NCVD-KA423/2013(H5N1) | KP097994 | 99.2 | |
| A/duck /Nanchang/6631/2013 (H5N1) | KP288319 | 99.0 | ||
| NS | A/chicken/Vietnam/NCVD-KA423/2013(H5N1) | KP098015 | 99.8 | |
| A/duck /Nanchang/6631/2013 (H5N1) | KP288320 | 99.6 | ||
| A/Tiger /Yunnan /tig1508/2015 (H5N1) | PB2 | A/chicken/Hunan/1/2012(H9N2) | KF714772 | 98.1 |
| PB1 | A/chicken/Hunan/1/2012(H9N2) | KF714773 | 98.2 | |
| PA | A/duck/Hunan/S4150/2011(H5N1) | CY146691 | 98.2 | |
| HA | A/duck/Hunan/S4150/2011(H5N1) | CY146692 | 97.6 | |
| NP | A/duck/Hunan/S4150/2011(H5N1) | CY146693 | 98.2 | |
| NA | A/duck/Hunan/S4150/2011(H5N1) | CY146694 | 97.1 | |
| M | A/chicken/Hebei/FL/2011(H9N2) | KC821206 | 98.4 | |
| A/chicken/Hunan/1/2012(H9N2) | KF714778 | 98.1 | ||
| NS | A/duck/Hunan/S4150/2011(H5N1) | CY146696 | 98.8 |
*PB2, basic polymerase 2; PB1, basic polymerase 1; PA, acidic polymerase; HA, hemagglutinin; NP, nucleoprotein; NA, neuraminidase; M, matrix; NS, nonstructural protein.
Figure 2. Phylogenetic tree of influenza A (H5N1) hemagglutinin (HA) gene and neuraminidase(NA) gene sequences from the tiger isolates in Yunnan and related reference viruses retrieved from the GenBank database.
The phylogenetic tree was generated in MEGA version 6 (www.megasoftware. net), using maximum likelihood (ML) analysis with 1,000 bootstrap replicates. The HA tree was rooted to prototype strain A/goose/Guangdong/1/96 [Gs/GD]. Scale bar indicates nucleotide substitutions per site. The viruses reported in this paper were highlighted using black triangle and bold italic. Note*:some 2.3.4.4 sub-clades such as 2.3.4.4e have just been proposed but not yet been confirmed by the WHO/OIE/FAO H5N1 nomenclature working group.
Figure 3. Phylogenetic analyses of six internal genes of the tiger isolates in Yunnan and related reference viruses.
The viruses reported in this paper were highlighted using black using black triangle and bold italic. PB2, basic polymerase 2; PB1, basic polymerase 1; PA, acidic polymerase; NP, nucleoprotein; M, matrix protein and NS, nonstructural protein.
Homological analysis showed that A/tiger /Yunnan /tig1508 /2015(H5N1) and peacock isolates (including A/peacock/Yunnan /1 /2015(H5N1) and A/peacock /Yunnan /3 /2015(H5N1)) shared 99.0%, 98.8%, 98.3%, 98.6%, 99.1%, 97.5%, 99.2% and 99.3% nucleotide identities with HA, NA, PB2, PB1, PA, NP, M and NS genes, respectively. The highest homology of the tig1508 isolate virus genome were as follows (in Table 1): 97.6%, 97.1%, 98.2%, 98.2% and 98.8% homology with the HA, NA, PA, NP and NS genes of A/duck /Hunan /S4150 /2011(H5N1) belonging to Clade 2.3.2.1c, 98.1% and 98.2% with the PB2 and PB1 genes of A/chicken/ Hunan/1/2012(H9N2) and 98.4% with the M gene of A/chicken/ Hebei /FL/ 2011(H9N2). Evolutionary analysis showed that A/tiger /Yunnan /tig1508 /2015(H5N1) including peacock isolates is a novel reassortant virus originating from H5N1 and H9N2 subtypes influenza A virus. The HA, NA and three internal genes (PA, NP, NS) of the novel H5N1 virus originated from A/duck /Hunan /S4150 /2011(H5N1) belonging to subclade 2.3.2.1c and the other three internal genes originated from avian influenza A (H9N2) virus: PB2 and PB1 segments originated from A/chicken/ Hunan/1/2012(H9N2) and M segment originated from A/chicken/ Hebei /FL/ 2011(H9N2) (Figs 2 and 3).
Molecular features associated with AIV virulence, transmissibility, and antiviral resistance
We analyzed the molecular features of tiger originated viruses associated with H5N1 virus virulence, transmissibility, and antiviral resistance. The three isolates displayed some molecular markers associated with increased virulence and transmission in mammals according to the H5N1 Genetic Changes Inventory (http://www.cdc.gov/ flu/ pdf/ avianflu/ h5n1-inventory.pdf). The isolated hemagglutinin cleavage sites possess a multibasic amino acid cleavage site (Table 2). Although the three isolates possessed a conserved amino acid motif Q-R-G (Gln Ser Gly equivalent to clades 2.3.4 and 2.3.2) at residues 222–224 (H3 numbering 226–228) of the hemagglutinin protein indicating no substantial changes in avian-like receptor binding preferences8,9, the HA of the viruses isolate from the tigers harbored some mutations (such as Asp94Asn in tig1404 and tig1508, Ser133Ala in tig1404, tig1412 and tig1508, Thr156Ala in tig1412, Thr188Ile in tig1412 and tig1508, and Lys189Arg in tig1412), which several studies have suggested increased the binding of the H5N1 virus to the sialic acid (SA) α-2, 6-Gal (α2-6) receptor10,11,12. The neuraminidase stalk deletion (49–68 deletion) was detected in the three isolates, which could play a role in enhancing H5N1 virulence in mice13. The mutations/substitutions involved in drug resistance and transmission to mammals, were observed in the NA (His254Tyr) of the three isolates and PB2 (Asp701Asn) proteins of tig1508 isolate (Table 2)14,15. The three isolates possessed Asn30Asp and Thr215Ala mutation in M1 protein, which are associated with increased virulence in mice16. The Ser31Asn mutation was exhibited in the M2 protein of the tig1508 virus, which confers resistance to the antiviral drug adamantane and rimantadine17,18. The mutations of NS1 protein plays an important role in enhancing the virulence of H5N1 viruses in mice, such as 80–84 deletion, Leu98Phe, Ile101Met and PDZ ligand motif ESEV in 222–225 sites19,20, were observed in the three isolates from the tigers and Asp87Glu in the tig1404 and tg1412 isoslates.
Table 2. Analysis of molecular features associated with AIV virulence, transmissibility, and antiviral resistance in H5N1 AIV isolated from tigers in Yunnan China, 2014–2015.
| Protein | Molecular feature or amino acid substitution | Phenotypic effect | tig1404 | ig1412 | tig1508 |
|---|---|---|---|---|---|
| HA | Asp94Asn | Increased virus binding to α2-6; enhanced virus fusion | N(Asn) | S(Ser) | N(Asn) |
| Ser133Ala | Increased psuedovirus binding to α2-6 | A(Ala) | A(Ala) | A(Ala) | |
| Ser155Asn | Increased virus binding to α2-6 | D(Asp) | D(Asp) | D(Asp) | |
| Thr156Ala | Increased virus binding to α2-6 and increased transmission in guinea pigs | T(Thr) | A(Ala) | T(Thr) | |
| Asp183Gly | Increased virus binding to α2-6 | N(Asn) | D(Asp) | D(Asp) | |
| Thr188Ile | Increased psuedovirus binding to α2-6 | T(Thr) | I(Ile) | I(Ile) | |
| Lys189Arg | Increased virus binding to α2-6 | N(Asn) | R(Arg) | T(Thr) | |
| Gln192Arg, Gln192His | Increased virus binding to α2-6 | K(Lys) | K(Lys) | Q(Gln) | |
| Lys218Glu | Altered pathogenicity and tissue tropism in mice, emerged in the course of virus replication in a patient | Q(Gln) | R(Arg) | K(Lys) | |
| Gln222Leu | Increased virus binding to α2-6 | Q(Gln) | Q(Gln) | Q(Gln) | |
| Ser223Asn | Increased virus binding to α2-6, emerged in the course of virus replication in a patient (fatal case) | R(Arg) | R(Arg) | S(Ser) | |
| Gly224Ser | Increased virus binding to α2-6 | G(Gly) | G(Gly) | G(Gly) | |
| 323 to 330 (R-X-R/K-R) | Polybasic cleavage motif sequence required for high pathogenicity of H5N1 avian influenza viruses | RERRR*KR | IERRRRKR | RERRR*KR | |
| NA | 49–68 deletion | Enhanced virulence in mice | deletion | deletion | deletion |
| Gln116Leu/Lys/Arg (136 in N2) | Reduced susceptibility to zanamivir and oseltamivir | Q(Gln) | Q(Gln) | H(His) | |
| His254Tyr/Arg (274 in N2) | Reduced susceptibility to oseltamivir and peramivir | Y(Tyr) | Y(Tyr) | Y(Tyr) | |
| PB2 | Glu627Lys | Increased virulence in mice | V(Val) | E(Glu) | E(Glu) |
| Asp701Asn | Mammalian host adaptation Enhanced replication efficiency increased virulence and transmission in guinea pigs | D(Asp) | D(Asp) | N(Asn) | |
| M1 | Asn30Asp | Increased virulence in mice | D(Asp) | D(Asp) | D(Asp) |
| Thr215Ala | Increased virulence in mice | A(Ala) | A(Ala) | A(Ala) | |
| M2 | Val27Ala | Reduced susceptibility to amantadine and rimantadine | V(Val) | V(Val) | G(Gly) |
| Ser31Asn/Gly | Reduced susceptibility to amantadine and rimantadine | S(Ser) | S(Ser) | N(Asn) | |
| NS1 | Pro42Ser | Increased virulence in mice | S (Ser) | S (Ser) | S (Ser) |
| 80–84 deletion | Increased virulence in mice | deletion | deletion | deletion | |
| Asp87Glu | Increased virulence in mice | E(Glu) | E(Glu) | D(Asp) | |
| Leu98Phe | Increased virulence in mice | F(Phe) | F(Phe) | F(Phe) | |
| Ile101Met | Increased virulence in mice | M(Met) | M(Met) | M(Met) | |
| 222–225 (PDZ ligand domain) | Increased virulence in mice | ESEV | ESEV | ESEV |
PB2, basic polymerase 2; PB1, basic polymerase 1; PA, acidic polymerase; HA, hemagglutinin; NP, nucleoprotein; NA, neuraminidase; M, matrix; NS, nonstructural protein.
The N-Glycosylation sites were predicted to examine the sequence context of Asn-Xaa-Ser/Thr sequins by the NetNglyc server 1.0. In addition to seven possible N-glycosylation sites observed for other serotype H5N1 in hemagglutinin proteins9, the predicted results of the N-Glycosylation sites indicated that the HA segments of each isolate harbored new N-glycosylation sites: Asn84 and Asn154 in the tig1404 isolate, Asn273 in the tig1412 isolate, and Asn154 in the tig1508 isolate, respectively.
In summary, all of mutations/substitutions of the gene segments of the tiger originated viruses could contribute to the enhancement of virulence or the increase of the H5N1 virus binding to the α2-6 receptor.
Pathogenicity of the tigers originated H5N1 viruses in mice
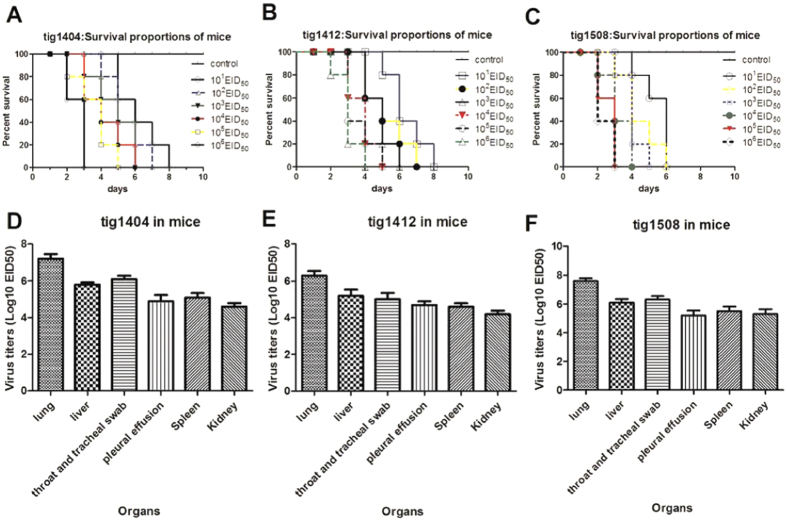
To further characterize the virulence of these novel tigers-originated H5N1 viruses in mammals, we infected mice with these viruses and observed for 10 days for morbidity. The signs of illness, anorexia and dyspnea were observed in the mice inoculated with 101-106 ELD50 and the mice inoculated with 106.0 EID50 began to die at 2 days post infection (dpi); by 8 dpi, all mice in the experimental groups had died(Fig. 4A–C). Among the mice infected with tig1404, tig1412 and tig1508, viruses were positive with RT-PCR and were successfully re-isolated from lung, liver, pleural effusion, throat and tracheal swab, Kidney and Spleen tissues. Viral titers in eggs were calculated and shown in the Fig. 4 (Fig. 4D–F). Virus replication was not detected in any of the direct contact mice in the tig1404 and tig1412 groups. But in the Tig1508 group, virus replication was detected in one direct contact naïve mouse. These results indicated that these tigers-originated H5N1 viruses caused infection and death in mice and the tig1508 virus maybe have the ability for transmission and infection in mice through direct contact.
Figure 4. Virulence of tigers-originated- H5N1 viruses in mice.
The survival rate of infected mice are shown in (A) (tig1404), (B) (tig1412) and (C) (tig1508); the tigers-originated- H5N1 viruses were detected in lung, liver, pleural effusion, throat and tracheal swab, Kidney and Spleen of infected mice and the viral titers in each organ of mice after challenge with influenza are shown in (D) (tig1404), (E) (tig1412) and (F) (tig1508).
The hemagglutination inhibition (HI) test
The HI titers of Re-5 antiserum against subtype tig1404, tig1412 and tig1508 isolate was 4.825 ± 1.083, 4.289 ± 1.160 and 4.232 ± 1.212 log2, respectively. Re-6 antiserum specimens (n = 38) collected from healthy chickens vaccinated twice with the Re-6 vaccine strain (belonging to clade 2.3.2.1b) (Jan 2014). The HI titers of Re-6 antiserum against subtype tig1404, tig1412 and tig1508 isolate was 4.475 ± 1.753, 6.75 ± 0.840 and 3.675 ± 1.163 log2, respectively(Table 3). If HI titers of the tested serum were higher than 5 log2, they were determined to be positive. As suggested by the HA1 protein sequence differences, clade 2.3.2.1b antiserum (from the RE-6 vaccine strain) inhibited hemagglutination of a representative clade 2.3.2.1b virus including tig1412 isolate, but did not inhibit hemagglutination of clade 2.3.2.1c such as tig1508 isolate, a representative clade 2.3.4 RE-5 (vaccine strain) and tig1404 isolate.
Table 3. Results of HI assays using Re-5 antiserum and Re-6 antiserum for avian influenza A(H5N1) isolated from tigers in Yunnan China, 2014–2015*.
| Isolate | Isolation date | HI titer ± SD, log2 | |
|---|---|---|---|
| Re-5 antiserum | Re-6 antiserum | ||
| A/tiger/Yunnan/Tig1404/2014 | 2014 Apr | 4.825 ± 1.083 | 4.475 ± 1.753 |
| A/tiger/Yunnan/Tig1412/2014 | 2014 Dec | 4.289 ± 1.160 | 6.75 ± 0.840 |
| A/tiger/Yunnan/Tig1508/2015 | 2015 Aug | 4.232 ± 1.212 | 3.675 ± 1.163 |
| A/peacock/Yunnan/1/2015 | 2015 Jun | 4.116 ± 1.141 | 3.525 ± 1.339 |
| Re-6 diagnostic antigen† | NA | 4.245 ± 1.791 | 8.875 ± 1.090 |
| Re-5 diagnostic antigen† | NA | 7.250 ± 0.909 | 5. 375 ± 1.330 |
*Re-5 (n = 38) and Re-6 (n = 40) antiserum were generated by vaccinating specific-pathogen free chickens with the commercial Re-5 and Re-6 vaccine (Harbin Weike biologic Technology Development Company, Harbin, China). HI titers against the homologous antigen/virus are shown in boldface. HI, hemagglutination inhibition; NA, not applicable. The titre differences were statistically significant by One Way ANOVA (P < 0.05).
†The commercial Re-5 and Re-6 diagnostic antigen (including positive and negative control serum) are from Harbin Weike biologic Technology Development Company, Harbin, China.
Discussion
In this study, we provide the evidence of fatal H5N1 AIV infection in zoo-housed Tigers in Yunnan Province. In comparisons of previously published nucleotide sequences with those of other avian influenza viruses from public databases, the isolated three tiger-originated-viruses (specially the tig1404 and tig1508 isolate) had a high level of homology with the recently identified HPAI H5N1 viruses, which circulates mainly in chickens and other avians in Yunnan province at the same time7 and the southern provinces of China or Southeast Asia, but low levels of homology with isolates from tigers in Shanghai, Jiangsu and Thailand6,21,22. Thanawongnuwech et al. previously reported that H5N1 influenza virus transmission occurred between tigers in Thailand in 200423. In the case On August 12th, 2015, RNAs from the two died tigers tested positive for the HA and NA gene of the H5N1 virus. The virulence in mice indicated that the tig1508 virus maybe have the ability for transmission and infection in mice through direct contact.
Our results suggested that the reassortant different H5N1 virus subclades can successfully cross species barriers from avian to mammal and infect north-east China tigers which might be with the contribution that mutations/substitutions of the gene segments in the tiger originated viruses could enhance virulence or increase the H5N1 virus binding to the α2-6 receptor.
Evolutionary analysis showed that A/tiger /Yunnan /tig1508 /2015(H5N1) including A/peacock/ Yunnan /1 /2015(H5N1) virus and A/peacock/ Yunnan /3 /2015(H5N1) virus which circulates in peacocks and other poultry is a novel reassortant virus originating from H5N1 and H9N2 subtypes influenza A virus. The HI assay demonstrated antiserum from the RE-6 vaccine strain did not inhibit hemagglutination of clade 2.3.2.1c such as tig1508 and peacock isolates, and this change must be considered when evaluating and selecting prepandemic candidate vaccine viruses for the region.
Materials and Methods
Tissue samples of all deceased tigers, including throat and tracheal swab, lung, liver, spleen, kidney, cardiac with aquae pericardii, and cerebrospinal fluid, were collected to determine the cause of death. Testing for detection of influenza A virus was performed by a reverse transcription PCR method after RNA extraction using a viral RNA kit (Invitrogen, USA)24. RNA from the lung, liver, cardiac with aquae pericardii, throat and tracheal swab specimens tested positive for the hemagglutinin gene and neuraminidase gene of the H5N1 virus.
Virus isolation and gene sequencing
To isolate and characterize the H5N1 viruses, isolates from H5N1 virus RNA positive dead tiger’s lung samples and peacocks’s cloacal swab specimens were injected into 10-day-old specific pathogen free embryonated chicken eggs in a Biosafety Level-3 laboratory. Two peacock isolates (A/peacock/Yunnan /1 /2015(H5N1) and A/peacock /Yunnan /3 /2015(H5N1)) and three tiger originated virus isolates (tig1404, tig1412 and tig1508 isolated from tigers died on April 8th, December 15th, 2014 and August 12th, 2015, respectively.) were chosen for full genome sequencing. The specific RT-PCRs were performed as described previously25. The homogenates of the RT-PCR positive samples for tigers, including the lung, liver, cardiac with aquae pericardii, throat and tracheal swab specimens, were centrifuged at lowspeed (6,000 × g) for 10 min at 4 °C, treated with 100,000 U/ml penicillin and 100 μg/ml streptomycin, and either undiluted or 10-fold serially diluted supernatants, and then inoculated into 10-day-old SPF embryonated chicken eggs. Viral titers were then calculated using the Reed and Muench method.
Genetic and phylogenetic analysis
The nucleotide sequences were analyzed using DNAman (version 6.0) and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic analyses were performed using the maximum likelihood (ML) method (MEGA, version 6.0)26. The N-Glycosylation sites were predicted to examine the sequence context of Asn-Xaa-Ser/Thr sequins by the NetNglyc server 1.0. The full genome sequences of all isolates have been sumbitted to the GenBank database and GenBank accection numbers are KU057261-KU057300.
Pathogenicity of the tigers originated H5N1 viruses in mice
To further characterize the virulence of these novel H5N1 viruses in mice, groups of five mice under light CO2 anesthesia were inoculated intranasally with 101−106 50% egg lethal dose (ELD50) of tested virus in a volume of 50 μl. Meanwhile, a group of five mice were inoculated with an equal volume of PBS as negative control. All the mice were monitored for mortality daily for 10 days. For virus infectivity and replication testing, groups of three mice were lightly anesthetized with CO2 and inoculated intranasally with 106 ELD50 of tested virus in a volume of 50 μl, every three inoculated mice were euthanized at 2–5 days post-inoculation, and their organs were collected for virus re-isolation and RT-PCR test. To investigate the virus transmission ability, after 24 hours post-inoculation, three naïve mice were placed in direct contact with the inoculated mice. Virus re-isolation was performed in 10-day-old SPF embryonated chicken eggs.
The hemagglutination inhibition (HI) test
To test the antigenic relationship of three tiger viruses, we examined serologic cross-reactivity between the tiger originated isolates and the diagnostic antigen of the widely used inactivated reassortant vaccine Re-5 and vaccine Re-6 in Yunnan Province (Table 3). A hemagglutination-inhibition test was conducted to test for Re-5 and Re-6 antiserum against tiger’s viruses27. Forty serum specimens were collected from healthy chickens were vaccinated twice with RE-5 vaccine strain (collection date = 2012).
Statistical analysis
The statistical significance of differences between groups was determined using the Student’s t-test (Graphpad prism 5). A P value < 0.05 was considered statistically significant.
Ethics Statement
The animal experiment was conducted in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People’s Republic of China. All procedures using animals were approved by the Animal Care and Use Committees of Centre for Disease Control and Prevention, Chengdu Military Region.
Additional Information
How to cite this article: Hu, T. et al. Fatal influenza A (H5N1) virus Infection in zoo-housed Tigers in Yunnan Province, China. Sci. Rep. 6, 25845; doi: 10.1038/srep25845 (2016).
Acknowledgments
This work was financially sponsored by The Science and Technology Program from Yunnan Province (2012CH002), the Key Programs (no. AWS11L009) and the Science and Technology Programs of Military (CWS12J075, 13BJYZ37, A12005 and BWS14J025), and the Youth Fund Program (no. 81101618) from the National Natural Science Foundation of China.
Footnotes
Author Contributions T.H., Q.F. and F.Z. are co-corresponding authors, who conceived and designed the research. T.H., Y.Z. and Q.C. wrote and revised the manuscript. T.H., H.Z., Q.K. and Z.Z. performed the experiments. Y.Z., W.Z., W.Q. and B.D. performed sample collections and prepared figures. All authors read and approved the final manuscript.
References
- Duan L. et al. Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: implications for the origin of highly pathogenic H5N1 viruses. J Virol 81, 7529–7539 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmayanti N. L. et al. Genetic Characterization of Clade 2.3.2.1 Avian Influenza A(H5N1) Viruses, Indonesia, 2012. Emerg Infect Dis 20, 677–680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M. et al. Novel variants of clade 2.3.4 highly pathogenic avian influenza A(H5N1) viruses, China. Emerg Infect Dis 19, 2021–2024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K. Y. et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet (London, England) 351, 467–471 (1998). [DOI] [PubMed] [Google Scholar]
- Hui D. S. Review of clinical symptoms and spectrum in humans with influenza A/H5N1 infection. Respirology (Carlton, Vic 13 Suppl 1, S10–13 (2008). [DOI] [PubMed] [Google Scholar]
- Keawcharoen J. et al. Avian influenza H5N1 in tigers and leopards. Emerg Infect Dis 10, 2189–2191 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T. et al. Emergence of novel clade 2.3.4 influenza A (H5N1) virus subgroups in Yunnan Province, China. Infect Genet Evol 33, 95–100 (2015). [DOI] [PubMed] [Google Scholar]
- Gambaryan A. S. et al. Differences between influenza virus receptors on target cells of duck and chicken and receptor specificity of the 1997 H5N1 chicken and human influenza viruses from Hong Kong. Avian Dis 47, 1154–1160 (2003). [DOI] [PubMed] [Google Scholar]
- Stevens J. et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410 (2006). [DOI] [PubMed] [Google Scholar]
- Yang Z. Y. et al. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317, 825–828 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Yang H. Y., Zhang B. J., Jia H. L. & Tien P. Analysis of a point mutation in H5N1 avian influenza virus hemagglutinin in relation to virus entry into live mammalian cells. Arch Virol 153, 2253–2261 (2008). [DOI] [PubMed] [Google Scholar]
- Suguitan A. L. Jr. et al. The multibasic cleavage site of the hemagglutinin of highly pathogenic A/Vietnam/1203/2004 (H5N1) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J Virol 86, 2706–2714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. et al. The special neuraminidase stalk-motif responsible for increased virulence and pathogenesis of H5N1 influenza A virus. PloS one 4, e6277 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Q. M. et al. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437, 1108 (2005). [DOI] [PubMed] [Google Scholar]
- Bogs J. et al. Reversion of PB2-627E to -627K during replication of an H5N1 Clade 2.2 virus in mammalian hosts depends on the origin of the nucleoprotein. J Virol 85, 10691–10698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S. et al. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology 384, 28–32 (2009). [DOI] [PubMed] [Google Scholar]
- Pielak R. M., Schnell J. R. & Chou J. J. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc Natl Acad Sci USA 106, 7379–7384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc Natl Acad Sci USA 110, 1315–1320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S. H., Hoffmann E. & Webster R. G. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nature medicine 8, 950–954 (2002). [DOI] [PubMed] [Google Scholar]
- Jackson D., Hossain M. J., Hickman D., Perez D. R. & Lamb R. A. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci USA 105, 4381–4386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq M. H. et al. Complete genome analysis of a highly pathogenic H5N1 influenza A virus isolated from a tiger in China. Arch Virol 153, 1569–1574 (2008). [DOI] [PubMed] [Google Scholar]
- He S. et al. Lethal infection by a novel reassortant H5N1 avian influenza A virus in a zoo-housed tiger. Microbes and infection/Institut Pasteur 17, 54–61 (2015). [DOI] [PubMed] [Google Scholar]
- Thanawongnuwech R. et al. Probable tiger-to-tiger transmission of avian influenza H5N1. Emerg Infect Dis 11, 699–701 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R. A. et al. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol 38, 4096–4101 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E., Stech J., Guan Y., Webster R. G. & Perez D. R. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146, 2275–2289 (2001). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayali G. et al. Testing human sera for antibodies against avian influenza viruses: horse RBC hemagglutination inhibition vs. microneutralization assays. J Clin Virol 43, 73–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]